Significance
Natural transformation is a major mechanism of horizontal gene transfer (HGT) by which bacteria take up exogenous DNA directly in their environment and integrate it in their genome. Acquiring new genetic information may confer an adaptive advantage but an uncontrolled uptake of foreign DNA may be harmful. We document a previously unsuspected means to control HGT by natural transformation in the human pathogen Legionella pneumophila. We found that the DNA uptake system required for natural transformation is subjected to silencing. A member of the widespread ProQ/FinO domain-containing protein family acts as an RNA chaperone and allows the targeting of the mRNAs of the genes coding the DNA uptake system by a newly identified trans-acting small RNA.
Keywords: natural transformation, RNA chaperone, non-coding RNA, Legionella pneumophila, ProQ/FinO
Abstract
A highly conserved DNA uptake system allows many bacteria to actively import and integrate exogenous DNA. This process, called natural transformation, represents a major mechanism of horizontal gene transfer (HGT) involved in the acquisition of virulence and antibiotic resistance determinants. Despite evidence of HGT and the high level of conservation of the genes coding the DNA uptake system, most bacterial species appear non-transformable under laboratory conditions. In naturally transformable species, the DNA uptake system is only expressed when bacteria enter a physiological state called competence, which develops under specific conditions. Here, we investigated the mechanism that controls expression of the DNA uptake system in the human pathogen Legionella pneumophila. We found that a repressor of this system displays a conserved ProQ/FinO domain and interacts with a newly characterized trans-acting sRNA, RocR. Together, they target mRNAs of the genes coding the DNA uptake system to control natural transformation. This RNA-based silencing represents a previously unknown regulatory means to control this major mechanism of HGT. Importantly, these findings also show that chromosome-encoded ProQ/FinO domain-containing proteins can assist trans-acting sRNAs and that this class of RNA chaperones could play key roles in post-transcriptional gene regulation throughout bacterial species.
Natural transformation is a common mode of horizontal gene transfer in bacteria. It results from the intrinsic capacity of bacteria to import exogenous DNA and integrate it by recombination in their chromosome (1). Active acquisition of random parts of the genetic material released by phylogenetically close organisms produces genetic polymorphism and functions as “localized sex” in reference to the function of sex in eukaryotic organisms (2). In addition, evidence suggests that genetic transformation can also occur with distantly related and even damaged DNA (3, 4). Depending on the bacteria’s biotope, the constant and random acquisition of genetic material could be as harmful as it is beneficial, and in most transformable species, natural transformation is a strictly regulated process (1, 5). Natural transformation only occurs when bacteria enter a specialized physiological state, called competence, during which a DNA uptake system is set up (6). This system generally involves a type IV pilus exposed at the cell surface and a transporter associated to the cytoplasmic membrane (1, 6). The type IV pilus is thought to initially interact with DNA and convey it to the small ComEA protein that binds double-strand DNA (dsDNA) (7). The captured dsDNA is then converted into single-strand DNA (ssDNA) and transported across the cytoplasmic membrane through a transmembrane channel formed by ComEC (8). The ssDNA entering the cytoplasm is rapidly loaded with ssDNA binding proteins SsbB and DrpA and can recombine with the chromosome (9, 10).
The concerted expression of the DNA uptake system during competence invariably relies on transcriptional activation (1). In Gram-positive bacteria, it results from the action of a transcriptional activator (e.g., ComK in Bacillus subtilis) or sigma factors (e.g., σX in Streptococcus pneumoniae, σH in Staphylococcus aureus). In the Gram-negative Haemophilus influenzae and Vibrio cholerae, it involves the transcription activator TfoX/Sxy. However, a number of species that are naturally transformable lack these known competence activators. One such species is the Gram-negative pathogen Legionella pneumophila, which was found to develop competence under microaerophilic growth, exposure to DNA-damaging agents, or suboptimal growth temperature (11–13). Possible competence regulatory elements in L. pneumophila were revealed by a genetic screen that identified a gene annotated “proQ, activator of osmoprotectant ProP” as a repressor of natural transformation (14). The proQ gene was initially identified in Escherichia coli as an activator of the proline transporter ProP (15), but recent evidence suggests that this effect is indirect (16). More significantly, ProQ bears the conserved PFAM domain 04352 (named the ProQ/FinO domain), which has been functionally and structurally studied in the F plasmid-encoded FinO protein (17, 18). FinO is an RNA chaperone that down-regulates the conjugative transfer of IncF plasmids between Enterobacteriaceae by controlling the function of an antisense RNA, FinP (19). FinO facilitates the interaction between FinP and the complementary 5′ untranslated region (UTR) of the mRNA of traJ (20, 21). Sense–antisense pairing prevents translation of the TraJ transcriptional activator of the plasmid tra operon and thereby inhibits plasmid conjugation. Although finO is restricted to IncF plasmids, genome-encoded proteins with a ProQ/FinO domain have been identified, but their function remains elusive (19). As the L. pneumophila ProQ represses natural transformability (14) and to avoid any unfounded inference with proline metabolism, we will refer to it by its locus tag name in the Paris strain, Lpp0148.
We characterize here Lpp0148 and describe its biological function in interaction with the first trans-acting sRNA-negative regulator of competence. We document an entirely novel mechanism of competence regulation by post-transcriptional silencing, diverging from the current notion that competence strictly depends on transcriptional activation. Our study expands the gene regulatory functions of ProQ/FinO domain-containing proteins by demonstrating that this class of RNA chaperones can act on regulatory trans-acting sRNAs.
Results
The ProQ/FinO Domain-Containing Protein Lpp0148 Specifically Controls a Competence Regulon.
To determine if Lpp0148 is a specific repressor of competence, we analyzed the global transcriptional activity in a lpp0148 mutant created by the introduction of a premature stop codon (denoted lpp0148TAA) (22). The lpp0148TAA mutant, which is highly transformable (Fig. S1), was grown to exponential growth phase (OD600 of 0.8) and subjected to RNA-seq transcriptional profiling (Table S1). The regulon controlled by Lpp0148 (fold change > 2, P < 0.01) consists of 11 genes up-regulated in the mutant strain and arranged in seven potential transcriptional units (Table S1). Among those genes, six are homologous to either genes encoding elements of the DNA uptake system (comEA, comEC, and comF) or genes previously found induced in competent bacteria (comM, radC, and mreB) (23–25). lpp1976 is the last gene of an operon with lpp1977 and lpp1978, both of which are moderately induced (fold change, 1.8 and 1.3, respectively; P < 0.01). This operon structure and the domains of these proteins suggest that they are pseudopilins involved in the biogenesis of a type IV pilus, a required appendage of transformable bacteria. The remaining four genes are of unknown function (lpp0851) or belong to the transcription units of the induced genes radC and comM (lpp2554, lpp2555/lepB, and lpp0639). Most likely due to the premature stop codon present in the lpp0148TAA mutant, the operon formed by lpp0148 and lpp0149 appears down-regulated (fold change < –2, P < 0.01). The function of lpp0149 is unknown, but it is not needed to repress competence (Fig. S1). The only other down-regulated gene (lpp0712) is expressed at a very low level, specific to the Paris strain, and of unknown function. The data show that the constitutive natural transformability of the lpp0148TAA mutant (Fig. S1) is not part of a pleiotropic effect. Rather, Lpp0148 is a specific repressor of a small competence regulon.
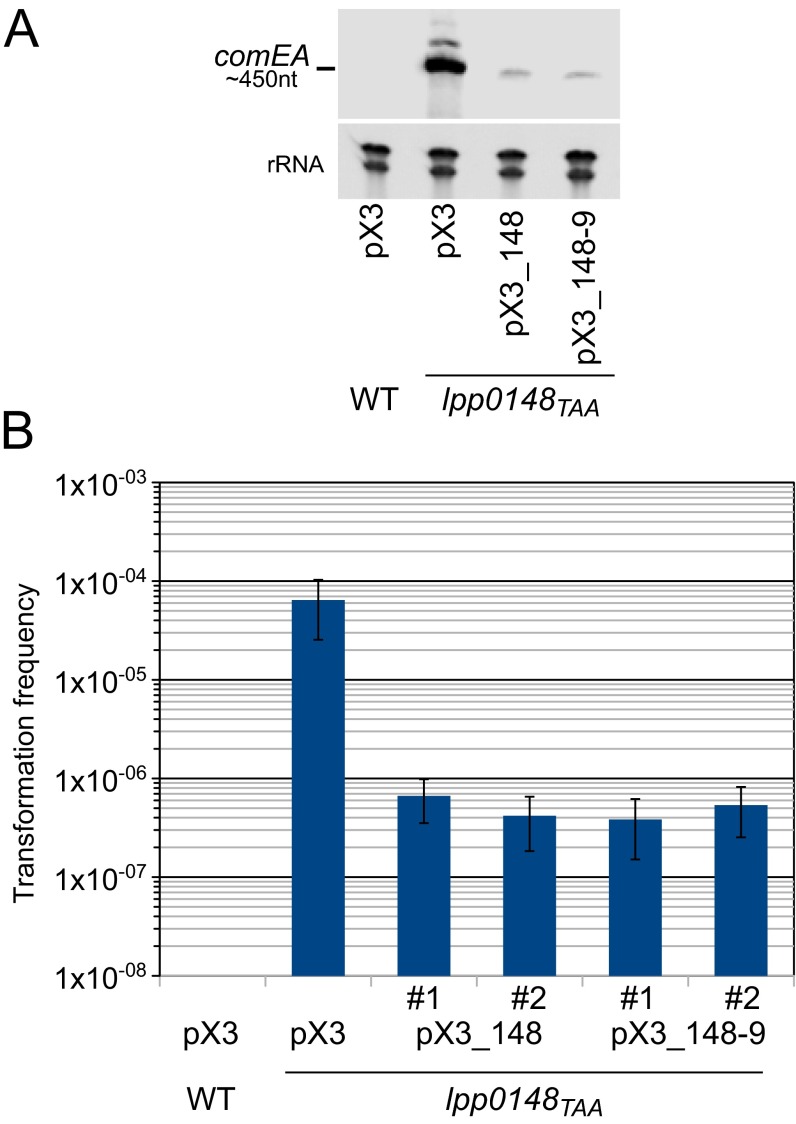
Fig. S1.
Complementation of L. pneumophila Paris lpp0148TAA mutant. (A) Northern blot analysis of comEA expression in the Paris wild-type strain and lpp0148TAA mutant transformed with the empty plasmid pX3, the pX3 plasmid carrying the lpp0148 gene alone (pX3-0148), or the putative lpp0148-0149 operon (pX3-0148-9). (B) Determination of the transformation frequency of the same strains. Two clones for each of the complementing plasmids were tested (#1 and #2). Bacteria were exposed to transforming DNA and then plated on selective and nonselective media. The transformation frequency is the ratio of the number of cfus counted on selective medium divided by the number of cfus counted on nonselective medium. Transformation frequency for the Paris strain was below the detection limit (<1.10−9). Error bars represent SE of experimental determination of cfu counts.
Table S1.
Comparison between the L. pneumophila Paris wild-type strain and its lpp0148TAA mutant derivative by RNA-seq analysis
| RNA-seq lpp0148TAA versus WT (fold change > 2; Padj < 0.01) | |||
| Feature | Fold change | Padj | Name and/or function |
| lpp2554 | 10.54 | 3.6E-147 | Hypothetical |
| lpp0872 | 7.99 | 2.0E-168 | comEA |
| lpp0640 | 7.13 | 1.9E-208 | comM |
| lpp2553 | 3.82 | 2.4E-50 | radC |
| lpp2280 | 3.14 | 1.4E-41 | comF |
| lpp2555 | 2.97 | 8.5E-51 | lepB |
| lpp0639 | 2.86 | 1.8E-32 | Hypothetical |
| lpp0873 | 2.51 | 1.6E-34 | mreB |
| lpp0680 | 2.18 | 3.3E-13 | comEC |
| lpp0851 | 2.08 | 8.8E-14 | Hypothetical; transposase IS200 like |
| lpp1976 | 2.06 | 1.5E-14 | Hypothetical; pilin homolog |
| lpp0712 | −2.03 | 8.6E-11 | Hypothetical |
| lpp0148 | −3.96 | 2.1E-53 | lpp0148 (ProQ/FinO domain) |
| lpp0149 | −6.65 | 1.2E-85 | Hypothetical |
Total RNA from bacterial cultures grown to an OD600 of 0.8 at 37 °C was extracted, purified, and sequenced (Illumina). This was done three times for each strain. The number of reads was normalized, and the statistical significance (Padj) of the obtained enrichment was calculated as described in SI Materials and Methods.
Lpp0148 Binds a Highly Conserved Intergenic sRNA Repressor of Competence.
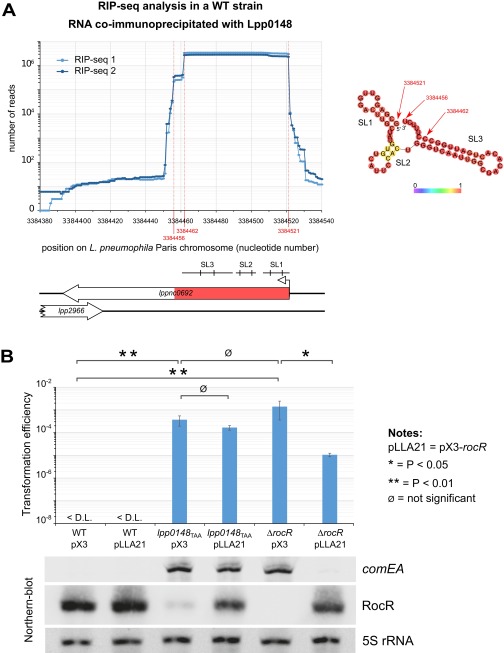
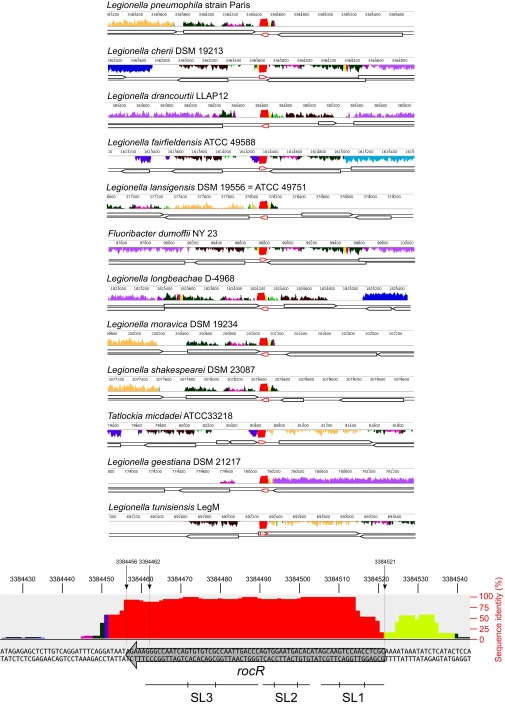
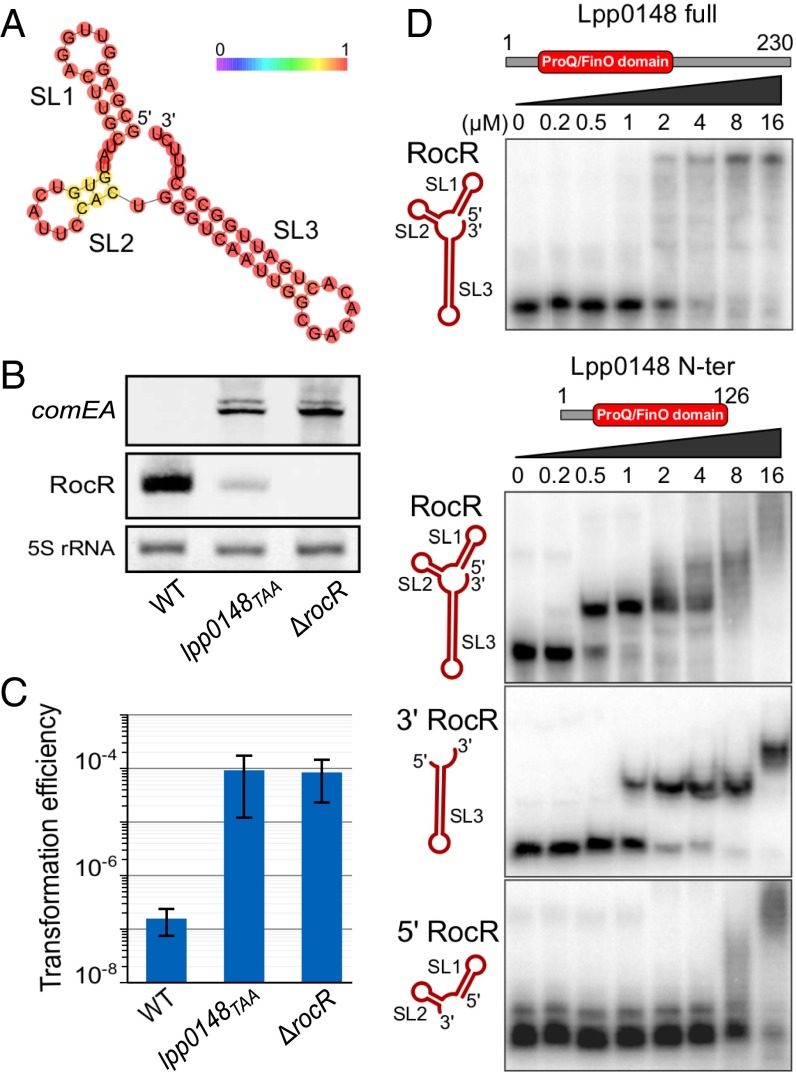
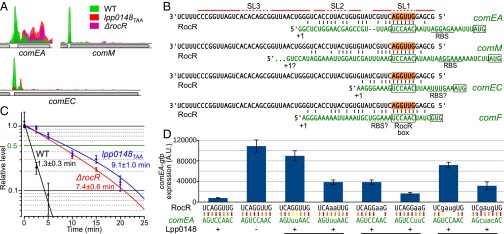
Because Lpp0148 carries an RNA-binding domain, we used an RNA immunoprecipitation technique coupled to deep sequencing (RIP-seq) to identify Lpp0148-bound RNAs. Following a reverse transcription protocol that preserves their 5′- and 3′-end sequences, the Lpp0148-bound RNAs were sequenced on an Illumina platform. Illumina reads mapping on five annotated features, including four sRNA features (lppnc0692, lppnc0344, lppnc0319, and lppnc0187), were found significantly enriched in the eluted RNAs from the wild-type strain compared with the lpp0148TAA mutant (Table S2). The lppnc0692 feature showed the highest enrichment (>200-fold), and most importantly, the normalized count of reads mapping on lppnc0692 represents 99.98% of all combined normalized read counts (Table S2). The reads mapping on lppnc0692 correspond to a 66-nt sequence beginning at the previously mapped transcription start site (26) and terminating at a predicted Rho-independent transcription terminator (Fig. S2A). The RIP-seq data suggest that Lpp0148 nearly exclusively binds in vivo a 66-nt sRNA expressed from the lppnc0692 feature (Fig. S2A). Interestingly, this enriched 66-nt sequence is present in all sequenced Legionella species with a sequence identity of over 95%, indicating a functional constraint (Fig. S3). Indeed, RNA fold predicts that this putative sRNA adopts a stable secondary structure with two strong stem-loop structures (SL1 and SL3) and a weak one (SL2) (Fig. 1A). Northern blot analysis confirmed the expression of this 66-nt sRNA in the wild-type strain (Fig. 1B). Importantly, deletion of the coding region for this sRNA resulted in an increase in comEA expression similar to that observed in the lpp0148TAA mutant (Fig. 1B) as well as a dramatically enhanced transformability (Fig. 1C). Expression of this 66-nt sRNA from a multicopy plasmid reduces transformability and comEA mRNA levels in the sRNA deletion mutant but not in the lpp0148TAA strain (Fig. S2B). Altogether the data demonstrate that this sRNA is a competence repressor whose function requires the Lpp0148 protein. These results prompted us to name it RocR, for “Repressor of competence, RNA.”
Table S2.
Comparison between RNAs coimmunoprecipitated using an anti-Lpp0148 antibody in the L. pneumophila Paris wild-type strain and its lpp0148TAA mutant derivative by RIP-seq analysis
| RIP-seq WT versus lpp0148TAA; enriched RNA with Padj < 0.01 | |||||
| Feature | Start position | Normalized counts | Fold enrichment | Padj | |
| Average in WT | Average in lpp0148TAA | ||||
| lppnc0692 (rocR) | 3384521 | 718,134.1 | 3,356.3 | 214 | 2.3E-12 |
| lppnc0344 | 1593477 | 24.8 | 0.4 | 69 | 1.3E-4 |
| lpp2366 | 2713176 | 85.7 | 2.2 | 40 | 6.2E-3 |
| lppnc0319 | 1498044 | 8.4 | 0.4 | 23 | 4.0E-4 |
| lppnc0187 | 863098 | 8.7 | 0.6 | 15 | 7.0E-3 |
At an OD600 of 0.8 exponentially growing bacterial cultures were fixed with 1% formaldehyde, and Lpp0148 and its partners were immunoprecipitated using a specific antibody. RNA was extracted, purified, and sequenced (Illumina). This was repeated three times in the WT strain and two times in the lpp0148TAA strain. The number of reads was normalized, and the statistical significance (Padj) of the obtained enrichment was calculated as described in SI Materials and Methods.
Fig. S2.
RocR is the cognate sRNA of Lpp0148 and represses competence (related to Fig. 1). (A) Determination of the principal RNA partner of Lpp0148 by the RIP-seq method in L. pneumophila Paris. Exponentially growing bacterial culture was fixed with 1% formaldehyde for 30 min. Lpp0148 was immunoprecipitated using specific affinity-purified antibodies, and bound RNAs were extracted and purified. cDNA libraries were prepared following 3′-end polyadenylation and 5′-end RNA adapter ligation and sequenced. The graph shows the number of reads per nucleotide obtained in two biological replicates at the lppnc0692 locus. The locus is schematized under the graph, with the enriched 66 nt highlighted in red and the position of the stem-loop structures predicted by RNA-fold (SL1, SL2, and SL3). (Right) The RNA fold structure prediction of this enriched 66-nt sRNA. (B) Complementation of the ΔrocR mutant in the L. pneumophila Paris strain. The wild-type, lpp0148TAA, and ΔrocR strains were transformed with the empty plasmid pX3 or the pX3 plasmid carrying the rocR gene (pLLA21). The transformability of the resulting strains was determined and the quantity of comEA and RocR was analyzed by Northern blot at an OD600 of 0.8 at 37 °C. For transformation experiments, bacteria were exposed to transforming DNA and then plated on selective and nonselective media. The transformation frequency is the ratio of the number of cfus counted on selective medium divided by the number of cfus counted on nonselective medium. Transformation frequency for the Paris strain was below the detection limit (<1.10−9). Error bars represent SD from the mean of three independent experiments. Differences in transformation frequencies were considered significant when P values of Welch’s t tests on log-transformed data were below 0.05.
Fig. S3.
Conservation of RocR in all sequenced species of Legionellales (related to Fig. 1). The 10 sequenced species of Legionellales were aligned at the rocR locus with the MAUVE software (http://darlinglab.org/mauve/mauve.html). Bottom shows a zoom-in on the rocR sequence from the L. pneumophila Paris strain with the percentage of sequence identity as well as the position of the three stem loops of RocR.
Fig. 1.
The intergenic sRNA RocR directly interacts with the ProQ/FinO domain of Lpp0148 and represses competence. (A) RNA fold predicted secondary structure of the intergenic 66-nt-long sRNA RocR. Bases are colored according to base-pairing probabilities (see color scale). See also Fig. S2. (B) Northern blot analyses of comEA and RocR expression in L. pneumophilla Paris wild-type, lpp0148TAA, and ΔrocR strains at an OD600 of 0.8 at 37 °C. (C) Natural transformability of the same strains as in B. Error bars represent SD from the mean of three independent experiments. (D) EMSAs of RocR–Lpp0148 complexes. RocR full size, 5′ (SL1 + 2), or 3′ (SL3) parts were incubated with increasing concentrations of Lpp0148 full size or its ProQ/FinO domain in the presence of unlabeled tRNA in excess and run in a native acrylamide gel.
The ProQ/FinO Domain of Lpp0148 Interacts with the SL3 of RocR.
To test the hypothesis that Lpp0148 directly binds RocR, we tested their interaction in vitro by electrophoretic mobility shift assay (EMSA) (Fig. 1D). The results indicate that full-length Lpp0148, as well as a truncated protein containing only its ProQ/FinO domain, can bind RocR in the presence of an excess of unlabeled competitor RNA (Fig. 1D), suggesting a specific interaction. Moreover, the ProQ/FinO domain of Lpp0148 binds RocR with a higher apparent affinity than full-length Lpp0148 and yields a well-defined shifted band. This suggests a more kinetically stable and homogeneous complex and is reminiscent of FinO, whose proteolytically stable ProQ/FinO domain binds RNA significantly more tightly than the intact protein (20). Our results also indicate that the ProQ/FinO domain of Lpp0148 binds tightly to the 3′ region of RocR (SL3) but only nonspecifically to the 5′ region (SL1 and SL2) (Fig. 1D). Taken together, these results indicate that, similarly to FinO (27), Lpp0148 uses its conserved ProQ/FinO domain to directly bind the 3′ rho-independent terminator hairpin and polyU tail of RocR.
The FinO/ProQ Domain of Lpp0148 Protects RocR from Degradation.
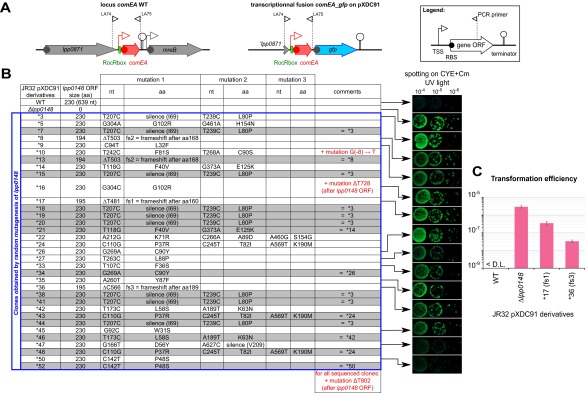
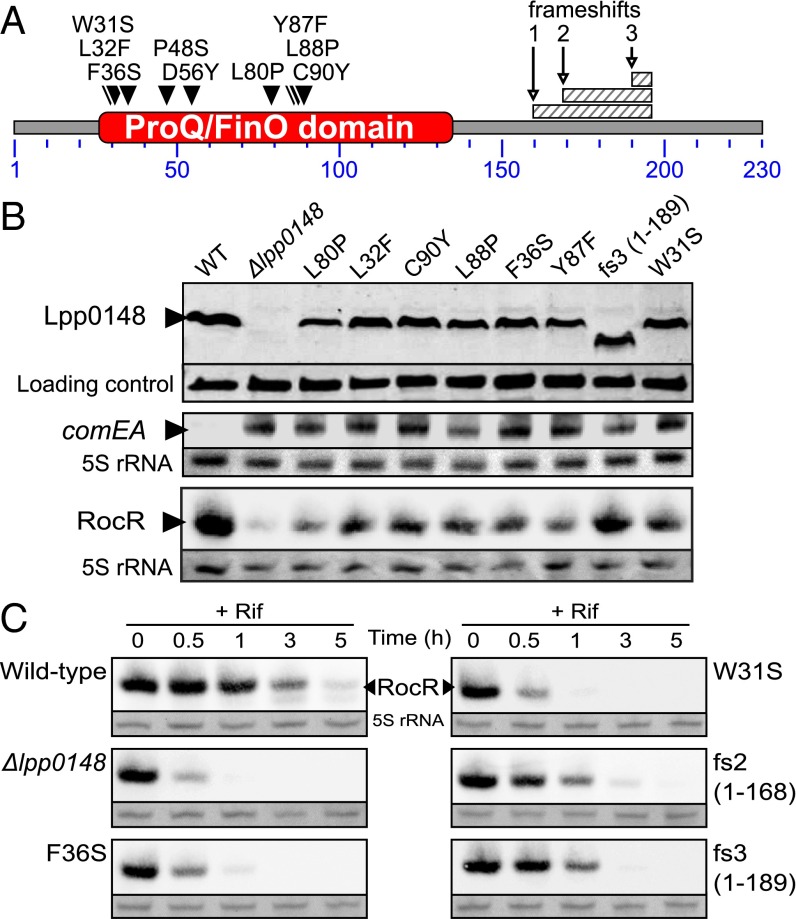
As a consequence of the hypercompetent phenotype, a plasmid-borne comEA-gfp transcriptional fusion is strongly induced in a lpp0148 deletion mutant and results in bright green colonies (12) (Fig. S4). We used this phenotype to perform a loss-of-function genetic screen. We subjected lpp0148 to random mutagenesis and isolated 34 lpp0148 alleles that could not repress competence (Fig. 2A and Fig. S4). Among those, we obtained three frameshift mutations leading to premature stop codon and C-terminal truncated proteins (Fig. 2 A and B and Fig. S4). All other mutants had acquired at least one nonsynonymous mutation in the ProQ/FinO domain, including 10 single mutations that did not alter protein expression (Fig. 2 A and B and Fig. S4). In these mutants, Northern blot analysis confirmed induction of the chromosomal comEA gene to levels similar to those observed in a Δlpp0148 mutant (Fig. 2B). Northern blot analysis also revealed that RocR was less expressed in the lpp0148TAA mutant (Fig. 1B) as well as in the Δlpp0148 mutant and in the strains expressing mutated Lpp0148 proteins than in the wild-type strain (Fig. 2B). RNA decay experiments showed that RocR is highly stable with a half-life (∼2 h) that exceeds the L. pneumophila doubling time in a wild-type strain, but it is relatively unstable in the Δlpp0148 mutant (Fig. 2C). Nonsynonymous mutations in the ProQ/FinO domain that result in the loss of competence repression also resulted in decreased stability of RocR (Fig. 2C). Thus, a function of Lpp0148 is to maintain the steady-state level of RocR. However, this may not be sufficient to repress competence, as overexpression of RocR cannot restore competence repression in the lpp0148TAA mutant (Fig. S2B). Deletions of the C-terminal domain (through frameshift mutations) did not impact RocR expression and stability to the same extent as the mutations in the ProQ/FinO domain; however, these mutations significantly abrogated repression of competence (Fig. 2 B and C and Fig. S4). This result is consistent with the finding that the C-terminal domain is not required to bind RocR (Fig. 1D) and suggests that this domain could instead be involved in RocR remodeling to promote duplex formation of RocR with its mRNA targets.
Fig. S4.
Hypercompetent mutants obtained by random mutagenesis of lpp0148 in the L. pneumophila JR32, pXDC91 strain (related to Fig. 2). (A) The two diagrams above the table show the comEA JR32 wild-type locus and the transcriptional fusion comEA-gfp borne by the pXDC91 (CmR). The Inset at the top right corner explains the symbols used. (B) The table shows the different mutants obtained. For each single mutant, 10-fold dilution of a cell suspension at an OD600 of 1 was spotted on CYE + Cm and incubated for 3 d at 37 °C. The colonies under UV exposure are shown on the Right. (C) Effect of different lpp0148 mutations on the transformation efficiency of the JR32 pXDC91 strain. Bacteria were exposed to transforming DNA and then plated on selective and nonselective media. The transformation frequency is the ratio of the number of cfus counted on selective medium divided by the number of cfus counted on nonselective medium. Transformation frequency for the Paris strain was below the detection limit (<1.10−9). Error bars represent SD from the mean of three independent experiments.
Fig. 2.
Mutations of the ProQ/FinO domain of Lpp0148 impair its ability to repress competence and to stabilize RocR. (A) Diagram of the L. pneumophila Lpp0148 protein. The ProQ/FinO PFAM domain (PF04352) is shown in red. Loss-of-function mutations are indicated as downward-facing black triangles or hatched box for mutations resulting in a frameshift. See also Fig. S4. (B) Western blot analysis of Lpp0148 and Northern blot analysis of the competence-induced comEA gene in the L. pneumophila JR32 wild-type strain and its mutant derivatives (Δlpp0148 and mutated lpp0148 alleles). A cross-reacting band and the 5S rRNA were used as loading controls for Western blot and Northern blot, respectively. (C) Northern blot analysis of the decay of RocR following transcription inhibition with rifampicin (100 µg/mL) at an OD600 of 0.8 in the L. pneumophila JR32 wild-type strain and its mutant derivatives.
Lpp0148 and RocR Target the 5′ UTR of mRNAs Encoding the DNA Uptake System.
Lpp0148 interaction with RocR suggests that this ribonucleoprotein complex may directly interact with the mRNAs of the Lpp0148-repressed genes. To identify a possible interaction site, we reexamined the results of the RIP-seq experiment. While specifically looking at Lpp0148-repressed genes (comEA, comEC, and comM) we noticed a sharp peak of coverage at their 5′ UTR (Fig. 3A). These sequences were absent or much less abundant in the RNA samples immunoprecipitated from the lpp0148TAA and ΔrocR mutants, suggesting that they were specifically interacting with Lpp0148 and RocR (Fig. 3A). Indeed, RNAfold predicted a potential duplex between the 5′ end region of RocR and the 5′ UTR of Lpp0148-repressed genes (Fig. 3B). All predicted duplexes contain a stretch of 7–10 pairing nucleotides near the putative ribosome binding site (RBS) with a shared 6-nt sequence, which we named RocR box, complementary to the first exposed loop of RocR (in SL1). The predicted duplexes are consistent with the prototypical mechanism of sRNA-mediated silencing: binding of the sRNA masks the RBS, thereby preventing translation and exposing the target mRNAs to ribonucleases (28). In agreement with this model, the 1.5-min half-life of the comEA mRNA in the wild-type strain is increased to 8–9 min in either the lpp0148TAA or ΔrocR mutants (Fig. 3C). Substitution of the two consecutive GC pairs of the predicted RocR–comEA duplex by two weaker GU wobble pairs resulted in almost complete loss of comEA repression (Fig. 3D). A double UA pair restored repression, albeit to a lesser extent than the original and stronger GC pairs (Fig. 3D). Similarly, changing the following two UA pairs into non-pairing AA affected repression, which was restored by AU pairs. Repression was also affected if the original AGGU sequence of RocR was scrambled into GAUG, but it was restored when the complementary bases were introduced in the comEA 5′ UTR (Fig. 3D). We conclude that RocR acts by a base-pairing mechanism using its first stem loop as a seed sequence to form a duplex with the 5′ UTR of targeted mRNAs and negatively impacts their steady-state levels.
Fig. 3.
Lpp0148 and RocR destabilize the comEA mRNA by base pairing. (A) Strand-specific read coverage of three competence loci obtained by RIP-seq with anti-Lpp0148 antibodies in L. pneumophila Paris wild-type (green), lpp0148TAA (red), and ΔrocR (pink) strains. (B) Predicted duplex formation between RocR and the mRNA of Lpp0148-repressed genes. (C) comEA mRNA half-life determination by RT-qPCR. Decay of comEA was followed after transcription was stopped with rifampicin at an OD600 of 0.8. Data, expressed as the relative amount of mRNA before the addition of rifampicin (t = 0), were fit to a first-order exponential decay and half-lives were calculated from three quantifications. (D) Cultures of different JR32 strains of L. pneumophila harboring the plasmid carrying the comEA-gfp fusion with wild-type (pXDC91) or mutated (pLLA27-28-29) RocR box were analyzed by flow cytometry. GFP levels were measured in 5.105 cells per sample; error bars represent the SD. The strains are JR32 pXDC91 (RocR box WT), JR32 Δlpp0148 pXDC91, JR32 pLLA27 (RocR box m3), JR32 rocRm3 pLLA27, JR32 rocRm4 pXDC91, JR32 rocRm4 pLLA28 (RocR box m4), JR32 rocRm5 pXDC91, and JR32 rocRm5 pLLA29 (RocR box m5).
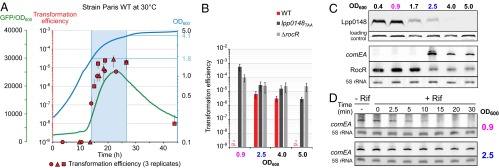
Lpp0148 and RocR Control the Development of Natural Transformability.
The L. pneumophila Paris strain naturally and transiently develops competence for natural transformation when grown at 30 °C (22). Analyses of the comEA-gfp fusion expression (Fig. 4A) and chromosomal comEA expression by Northern blot (Fig. 4C) show that competence (i.e., expression of the DNA uptake system) begins at the midlog phase (OD600 > 1.8) and ends before entering the stationary phase (OD600 > 4). Expression levels of comEA correlate with natural transformability, which is below detection level (<1 × 10−9) in the exponential phase but goes up to a frequency of 2 × 10−5 when comEA is expressed, before going back to basal level in the stationary phase (Fig. 4 A and B). The observed reduced transformability in the stationary phase is consistent with a previous report that quorum sensing by the Lqs (Legionella quorum sensing) system represses competence in the stationary phase (29). In contrast to the wild-type strain, both the lpp0148TAA and ΔrocR strains appear highly transformable in all growth phases (Fig. 4B). This suggests that a controlled loss of either Lpp0148 or RocR could be responsible for the transient development of competence observed in the wild-type strain. Interestingly, Western blot analysis of Lpp0148 shows that it is expressed in the exponential phase (OD600 < 0.9) before its expression steadily decreases at the midlog phase (OD600 of 1.7) and at the onset of the stationary phase (OD600 of 2.5) to become undetectable in the stationary phase (OD600 > 4) (Fig. 4C). As Lpp0148 is required to stabilize RocR, its reduced expression could impact the steady-state level of RocR. Northern blot analysis shows that RocR is indeed less expressed starting from an OD600 of 2.5 (Fig. 4C). The delay between the decrease of expression of RocR compared with that of Lpp0148 is consistent with the high half-life of RocR (Fig. 2C). The disappearance of Lpp0148 and RocR correlates with the detection of the comEA mRNA (Fig. 4C), which becomes highly stable at an OD600 of 2.5 (Fig. 4D). The data strongly support that competence development in L. pneumophila in the midlog phase is triggered by a programmed decreased expression of Lpp0148, which relieves the RocR-mediated silencing of genes encoding the DNA uptake system.
Fig. 4.
Lpp0148 and RocR control natural competence development. (A) The L. pneumophila Paris WT strain transiently develops competence during growth at 30 °C. Expression of comEA was followed using a comEA-gfp transcriptional fusion carried by pXDC91 (GFP/OD600, green line) and natural transformability (red circle, triangle, and square) determined at different time points during growth (OD600, blue line). Error bars on natural transformation efficiencies data represent SE. (B) Natural transformability during growth at 30 °C of the L. pneumophila Paris WT, lpp0148TAA, and ΔrocR strains. (C) Expression of Lpp0148 and RocR decrease at the onset of the transformability phase at 30 °C. Expression of Lpp0148 was analyzed by Western blot, and expression of comEA and RocR was determined by Northern blot analysis. A cross-reacting band and the 5S rRNA were used as loading controls for the Western blot and Northern blot, respectively. (D) comEA mRNA half-life determination by Northern blot analysis before (OD600 of 0.9) and during (OD600 of 2.5) the competence phase. Decay of comEA was followed after transcription was stopped with rifampicin at the indicated OD. The 5S rRNA was used as a loading control.
Lpp0148 Belongs to a Diverse Family of ProQ/FinO Domain-Containing Proteins.
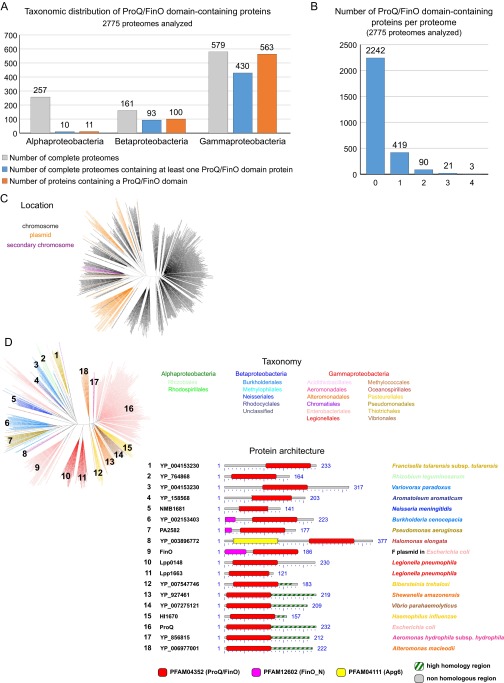
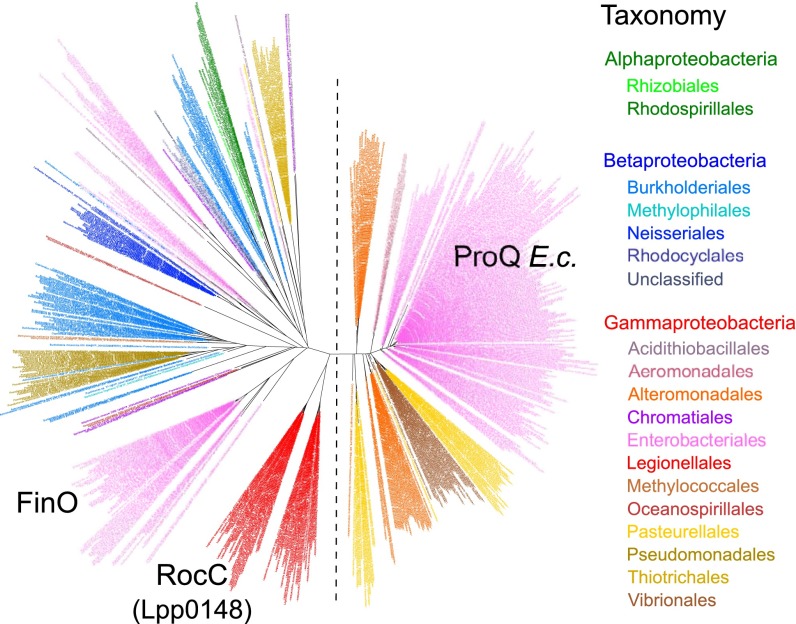
A systematic survey of 2,775 complete prokaryotic proteomes revealed the existence of 674 distinct proteins containing a ProQ/FinO domain (PF04352) (Dataset S1). Mostly found in Proteobacteria and mostly chromosome-encoded (Fig. S5C), they are widespread in Gammaproteobacteria (in 78% of species; Fig. S5A), with some species showing up to three or four homologs (Fig. S5B). The maximum likelihood phylogenetic tree of ProQ/FinO domain-containing proteins shows two distinct clusters: a compact cluster with short branches and a spread-out cluster with long branches (Fig. 5). Although the former (which includes the E. coli ProQ) is consistent with the current taxonomy, the latter presents intermixed sequences from beta- and gammaproteobacteria, which suggests a faster evolutionary rate and an impact of horizontal gene transfer (Fig. 5). This cluster is formed by ProQ/FinO domain-containing proteins with more diverse architecture and includes FinO and Lpp0148 (Fig. 5 and Fig. S5D). Broadly, the analysis shows that ProQ/FinO domain-containing proteins form a diverse family of RNA-binding proteins that may control various and unknown processes. Given that the L. pneumophila Lpp0148 specifically controls competence development, we propose to rename it RocC, for “Repressor of competence, RNA Chaperone.”
Fig. S5.
Phylogenetic analyses of ProQ/FinO domain-containing proteins (related to Fig. 5 and Dataset S1). (A) Taxonomic repartition of the 2,775 complete prokaryotic proteomes available at NCBI regarding the ProQ/FinO domain-containing proteins. (B) Number of ProQ/FinO domain-containing proteins per proteome in the 2,775 complete prokaryotic proteomes available at NCBI. (C) Distribution according to the encoding gene location. ProQ/FinO domain-containing proteins are colored in black, orange, or purple when their coding gene is located on the chromosome, a plasmid, or a secondary chromosome, respectively. (D) Architecture of the different ProQ/FinO domain-containing proteins. For each cluster (numbers on the tree), multiple members were aligned using BlastP, and conserved domains are shown. The C-terminal part of the proteins (after the ProQ/FinO domain) was aligned to extract the possible region of high similarity.
Fig. 5.
The ProQ/FinO domain-containing proteins are widespread in Proteobacteria. Shown are the phylogenetic analysis of ProQ/FinO domain-containing proteins (see also Dataset S1 and Fig. S5) and the maximum likelihood phylogenetic tree of the 674 protein sequences containing a ProQ/FinO domain found in 2,775 complete prokaryotic proteomes (80 amino acid positions were used). Colors correspond to taxonomic main lineages.
Discussion
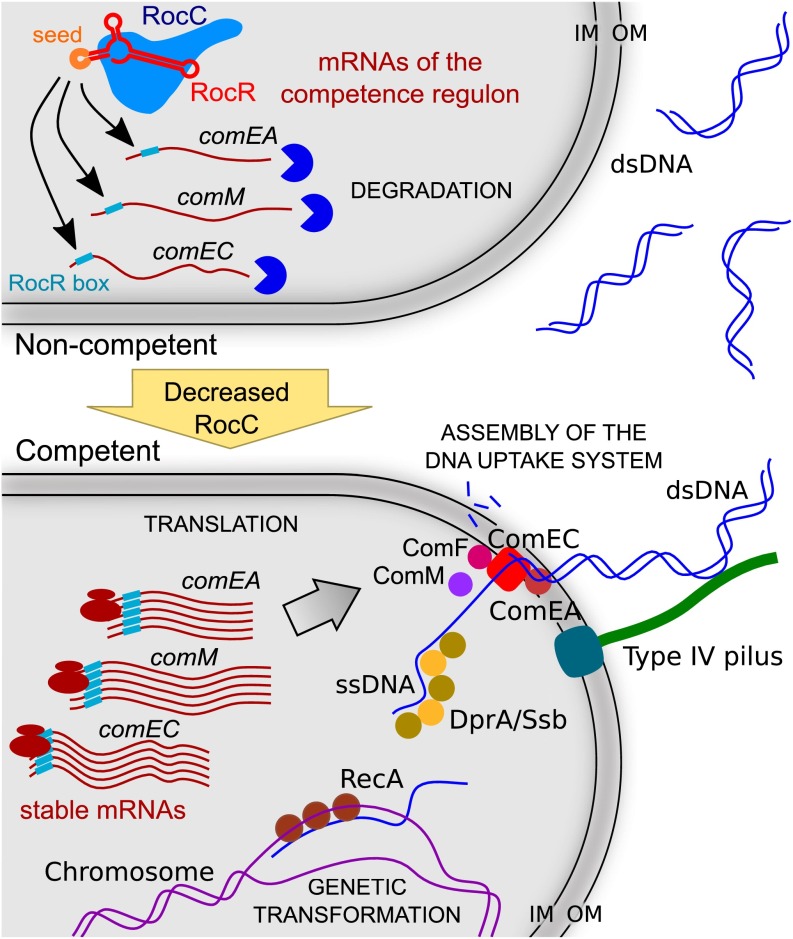
In stark contrast with all known regulatory mechanisms of competence, we report here that competence in L. pneumophila is directly controlled by the post-transcriptional repression of genes encoding the DNA uptake system (Fig. S6). This silencing system requires at least two components: a modular trans-acting sRNA and an RNA chaperone of non-constitutive expression.
Fig. S6.
Model of regulation of natural transformability in L. pneumophila. When cells are noncompetent, RocC stabilizes RocR and this ribonucleoprotein complex can bind the mRNAs of genes of the competence regulon via a conserved 6-nt sequence called RocR box and promote their degradation. Under competence-inducing conditions (for example, at the end of the exponential phase at 30 °C), RocC expression decreases. This triggers the destabilization of RocR and thus the stabilization of the mRNAs of the competence genes. These are then translated, the DNA uptake system is assembled, and horizontal gene transfer by natural genetic transformation can occur. IM, inner membrane; OM, outer membrane.
RocR is both the first described sRNA to directly control a competence regulon and the first identified trans-acting RNA partner of a ProQ/FinO-domain RNA chaperone. We propose that RocR interacts with the mRNAs of the DNA uptake system encoding genes by a base-pairing mechanism involving a 6-nt sequence, the RocR box, located upstream of the RBS and acting as a seed sequence (Fig. 3 and Fig. S6). Our results suggest that the SL1 and SL2 of RocR form an imperfect duplex with the targeted mRNAs, while the SL3 is engaged in direct interaction with RocC. Similarly to the 3′ end of FinP (27), the RocR SL3 is a hairpin–polyU tail that serves as a rho-independent transcriptional terminator and also specifically binds the ProQ/FinO domain of the protein. Thus, this structure may be a hallmark of sRNAs associated with ProQ/FinO-domain RNA chaperones. Although FinO only works with an antisense (i.e., cis-acting) sRNA, RocC interacts with a trans-acting sRNA, a function that was, until now, thought to be the exclusive property of the well-studied Hfq protein (28). This functional difference might come from the part of the protein outside the ProQ/FinO domain. Indeed, although this domain alone can bind RNA with higher affinity than the full-length protein (Fig. 1D and ref. 20), it is not sufficient to repress conjugation (20) or competence (Fig. 2 and Fig. S4). It was proposed that the flexible N-terminal region of FinO is necessary for sense–antisense RNA pairing (20). Although RocC lacks a similar N-terminal region, it carries an extended C-terminal domain that, despite being dispensable for RocR binding and stabilization, appears required for repression of competence (Fig. 1D, Fig. 2, and Fig. S4). We hypothesize that RNA remodeling activities to stimulate base-pairing of RocR with its mRNA targets lie within the C-terminal domain of RocC. Most importantly, RocC exemplifies that the world of RNA chaperones controlling trans-acting sRNAs extends beyond the well-documented Hfq protein (30). Chromosome-encoded ProQ/FinO-domain proteins should now be considered bona fide sRNA chaperones that play an important role in the biology of the cell.
We propose here a model in which RocC functions as the master regulator of competence development in L. pneumophila by directly controlling expression of the DNA uptake system (Fig. S6). Central to this activity is the regulated expression of RocC, which to our knowledge is the first example of control of a regulon by altering the expression of an RNA chaperone. As the specificity of the targeted regulon lies in a short sequence of the RocC-bound sRNAs, a few base changes may be sufficient to target a different regulon. This represents a way to rapidly evolve a conditional multitarget silencing strategy for disadvantageous processes. As intracellular bacteria thriving on environmental protozoans, Legionella species are continuously exposed to the DNA of their defunct eukaryotic hosts and their genome shows numerous acquired eukaryotic-like genes (31, 32). Although some acquired protozoan genes may prove beneficial for Legionella to hijack the cellular functions of its host, massive import of foreign DNA may also jeopardize chromosome integrity. The exceptionally conserved RocR sRNA in Legionella species (Fig. S3) may have emerged to limit the import of genetic material when Legionella is exposed to the potentially harmful foreign DNA released by its dead hosts.
Materials and Methods
Detailed protocols for all sections are described in SI Materials and Methods.
Bacterial Strains, Plasmids, and Oligonucleotides.
See SI Materials and Methods and Tables S3 and S4.
Table S3.
Bacterial strains and plasmids used in this study
| Strains/plasmids | Relevant genotype | Reference |
| Strains | ||
| L. pneumophila | ||
| JR32 WT | Philadelphia-1 derivative; SmR; r− m+ | (44) |
| JR32 Δlpp0148 | JR32; Δlpp0148 | This study |
| JR32 ΔrocR | JR32 ΔrocR | This study |
| JR32 ΔrocR::MK | JR32 ΔrocR::(lacIq, Ptac-mazF, nptII); KanR | This study |
| JR32 rocRm3 | JR32 rocRm3 | This study |
| JR32 rocRm4 | JR32 rocRm4 | This study |
| JR32 rocRm5 | JR32 rocRm5 | This study |
| LELA3825 | JR32; lpp0148::(Tn903dIIlacZ::nptII); KanR | (34) |
| LELA3825G | JR32; lpp0148::(Tn903dIIlacZ::aacC1); GentR | This study |
| Paris WT | Paris Outbreak isolate CIP107629 | CNR Lyon |
| Paris lpp0148TAA | Paris; lpp0148TAA | (22) |
| Paris ΔrocR | Paris; ΔrocR | This study |
| Paris ΔrocR::MK | Paris; ΔrocR::(lacIq, Ptac-mazF, nptII); KanR | This study |
| E. coli | ||
| DH5α, λpir | F−, supE44, ΔlacU169 (ΦlacZΔM15), recA1, endA1, hsdR17, thi-1, gyrA96, relA1, λpir lysogen | Laboratory strain collection |
| BL21(DE3) | F−, ompT, gal, dcm, lon, hsdSB(rB- mB-), λ[DE3 (lacI, lacUV5-T7 gene 1, ind1, sam7, nin5)] | Laboratory strain collection |
| Plasmids | ||
| pET-15b_Lpp0148aa1–230 | pET-15b(+) derivative; IPTG-inducible production of His-Lpp0148aa1–230 (Paris); AmpR | This study |
| pET-47b_Lpp0148aa1–230 | pET-47b(+) derivative; IPTG-inducible production of His-(HRV-3C)-Lpp0148aa1–230 (Paris); KanR | This study |
| pGEM-ihfB::Kan | pGEM-T Easy; ihfB::nptII; AmpR, KanR | (22) |
| pGEM-MK | pGEM-T Easy; lacIq, Ptac-mazF, nptII; AmpR, KanR | This study |
| pGEM-SK | pGEM-T Easy; sacB-mazF, nptII; AmpR, KanR | This study |
| pGEMPKD4 | pGEM-T Easy; FRT- nptII -FRT; AmpR, KanR | This study |
| pGEX-6P-1_Lpp0148aa1–126 | pGEX-6P-1 derivative; IPTG-inducible production of GST-(HRV-3C)-Lpp0148aa1–126 (Paris); AmpR | This study |
| pKD4 | pANTS-γ; FRT- nptII -FRT; AmpR, KanR | (35) |
| pLLA21 | pX3; ProcR-rocR*, cat; CmR | This study |
| pLLA27 | pXDC91 with mutated RocR box (m3: TttAAC); ΔmobA; PcomEAm3-comEA_gfp+, cat; CmR | This study |
| pLLA28 | pXDC91 with mutated RocR box (m4: TCCttC); ΔmobA; PcomEAm4-comEA_gfp+, cat; CmR | This study |
| pLLA29 | pXDC91 with mutated RocR box (m5: ctacAC); ΔmobA; PcomEAm5-comEA_gfp+, cat; CmR | This study |
| pX3 | RSF1010 derivative (replicative in L. pneumophila); ΔmobA; gfp+ (no promoter), cat; CmR | (12) |
| pX3_148 | pX3; Plpp0148-lpp0148†, cat; CmR | This study |
| pX3_148-9 | pX3; Plpp0148-lpp0148_lpp0149‡, cat; CmR | This study |
| pXDC46 | pGEM-T Easy; Tn903dIIlacZ::aacC1; AmpR, GentR | (34) |
| pXDC91 | RSF1010 derivative (replicative in L. pneumophila); ΔmobA; PcomEA-comEA§_gfp+, cat; CmR | (12) |
Abbreviations of antibioresistance (XxxR) are as follows: Amp, ampicillin; Cm, chloramphenicol; Gent, gentamycin; Kan, kanamycin; Sm, streptomycin.
Locus 3384431–3384710 from Paris strain.
Locus 165011–165943 from Paris strain.
Locus 165011–167014 from Paris strain.
Locus 889182–889694 from JR32 strain.
Table S4.
Oligonucleotides used in this study
| Oligonucleotides | Sequence, 5′ to 3′* | Restriction site | Detected RNA | Amplified RNA |
| Cloning primers | ||||
| catsac-F | CGACTCACTATAGGGCGAATTGGGCC | |||
| catsac-R | CATATGCCACCGACCCGAGCAAAC | |||
| comElacZ-P1 | CGTTGTATATTAATCATCGAAGCGTG | |||
| comElacZ-P4 | GCGGTTGTTTCTCCGATGAGAGTAC | |||
| LA51_up-lppnc0692-F | GGTTAGCACTCGTCAATCAC | |||
| LA52_up-lppnc0692-R-tail-maz | GGCCCAATTCGCCCTATAGTGAGTCGAAAATAAATATCTCATACTC | |||
| LA53_down-lppnc0692-F-tail-maz | GGGTTTGCTCGGGTCGGTGGCATATGCCTGAAATCCTGACAAGAGC | |||
| LA54_down-lppnc0692-R | GCCGGATTGCATCATTACCC | |||
| LA55-up-lppnc0692-R+tail LA56 | TAGAGAGCTCTTGTCAGGATTTCAGGTCTCATACTCCACTAATTAATG | |||
| LA56-down-lppnc0692-F | CCTGAAATCCTGACAAGAGCTCTCTA | |||
| LA61_rocR-F-NotI | GTAAGCGGCCGCCTCATTGTGATGCGGGAGAG | NotI | ||
| LA62_rocR-R-XmaI | CTTACCCGGGGCTCTTGTCAGGATTTCAGG | XmaI | ||
| LA68_RocRbox_m3-F | CTCTGGAACGAGCCGTTTAGTttAACAATTAGGAGAAATTTATG | |||
| LA69_RocRbox_m3-R | CATAAATTTCTCCTAATTGTTaaACTAAACGGCTCGTTCCAGAG | |||
| LA70_RocRbox_m4-F | CTCTGGAACGAGCCGTTTAGTCCttCAATTAGGAGAAATTTATG | |||
| LA71_RocRbox_m4-R | CATAAATTTCTCCTAATTGaaGGACTAAACGGCTCGTTCCAGAG | |||
| LA72_RocRbox_m5-F | CTCTGGAACGAGCCGTTTAGctacACAATTAGGAGAAATTTATG | |||
| LA73_RocRbox_m5-R | CATAAATTTCTCCTAATTGTgtagCTAAACGGCTCGTTCCAGAG | |||
| LA79_RocR_m4-R | ACACATAGCAAGTCCttCCTCGCAAAATAAATATC | |||
| LA80_RocR_m4-F | GATATTTATTTTGCGAGGaaGGACTTGCTATGTGT | |||
| LA81_up-rocR-F | TGGGCTTGAAGGCGATCAAC | |||
| LA82_down-rocR-F-tail-maz | GGGTTTGCTCGGGTCGGTGGCATATGTCCTAAAACCCTGACATGAA | |||
| LA85_RocR_m3-R | ACACATAGCAAGTttAACCTCGCAAAATAAATATC | |||
| LA86_RocR_m3-F | GATATTTATTTTGCGAGGTTaaACTTGCTATGTGT | |||
| LA87-up-rocR-R+tail LA88 | TCCTAAAACCCTGACATGAACTCTCT | |||
| LA88-down-rocR-F | TCCTAAAACCCTGACATGAACTCTCT | |||
| LA91_RocR_m5-R | ACACATAGCAAGctacACCTCGCAAAATAAATATC | |||
| LA92_RocR_m5-F | GATATTTATTTTGCGAGGTgtagCTTGCTATGTGT | |||
| lpp0148_P1 | GCCGTTTTAAATCGGCCAGAAAG | |||
| lpp0148_P4 | CTTCCCCCTAAAAATCAGGATGTC | |||
| lpp0148pX3_F | CTCTGCGGCCGCATTTTGCTAATTGGATACAAACCAAAAG | NotI | ||
| lpp0148pX3_R | ATCACCCGGGTTACTCTGTTGTTTCCTTTTTATCTTCTG | XmaI | ||
| lpp0149pX3_R | ATCACCCGGGCTAAGGTTGTTTTAAAATCATAAAGTAAGC | XmaI | ||
| lpp0148RM_F | CAATAGACTAAACACAATAAGGGTACCAG | |||
| lpp0148RM_P2 | TCTGGTACCCTTATTGTGTTTAGTCTATTG | |||
| lpp0148RM_P3 | GAACTAAGGAGGATATTCATATGGACCATGGCTGTTTATTTCTCCATTATGGGCTG | |||
| lpp0148RM_R | GGAACTTCGAAGCAGCTCCAGCCTACACAATCTGCTGAAAATAATGTCGCTATTCGC | |||
| MazF-R | CATATGCCACCGACCCGAGCAAACCCGAAGAAGTTGTCCATATTGGCCAC | |||
| MazFk7-F | CGACTCACTATAGGGCGAATTGGGCCGCTTTCCAGTCGGGAAACCTG | |||
| pKD4_P1 | GATTGTGTAGGCTGGAGCTGCTTCG | |||
| pKD4_P2 | GCCATGGTCCATATGAATATCCTCC | |||
| proQ15b-F | CCGGATTAATATGAGAAAGCAGGCGCTGCAC | AseI | ||
| proQ15b-R | AGCCGGATCCTTACTCTGTTGTTTCCTTTTTCTCTTCTG | BamHI | ||
| rocc_paris_pet47b+_fwd_1 | CTAGTCCCCGGGATGAGAAAGCAGGCGCTG | SmaI | ||
| rocc_paris_pet47b+_rev_230 | GACTAGGCGGCCGCTTACTCTGTTGTTTCCTTTTTATCTTCT | NotI | ||
| rocc-pgex6p1-fwd_1 | CTAGTCGGATCCATGAGAAAGCAGGCGCT | BamHI | ||
| rocc-pgex6p1-rev_126 | GACTAGGCGGCCGCTTACTTTTCCACGCGTTTTTTAATTTTC | NotI | ||
| Probes for Northern blot (biotinylated in 5′) | ||||
| comEA-NB2 | CTACAAAACGCTGCCCTATACCCTTGACTTCCGCAAGCTCTTCCAGAG | comEA | ||
| LA59_rocR-small-NB | TGTGTCGCCAATTGACCCAGTGGAATGACACATAGCAAGTCCAACCTC | RocR | ||
| RT-qPCR primers | ||||
| lpp0872-RT-F2 | CGGATGTGTATTCTTTAACAGGC | comEA | ||
| lpp0872-RT-R2 | GCTGCCCTATACCCTTGACT | comEA | ||
| lpptmRNA-RT-F1 | CCGCTTATCGGTATCGAATC | tmRNA (control) | ||
| lpptmRNA-RT-R1 | CGCAAGTCCTCTGCCTTTAG | tmRNA (control) |
Relevant restriction sites are underlined. The RocR box sequence is written in bold, with the nucleotides introducing mutations in lowercase. The seed sequence of RocR is highlighted in gray, with the nucleotides introducing mutations in lowercase. The notation Pxxx is used to designate the Promoter of the gene xxx. For transcriptional fusion, an underscore (_) is used (e.g., comEA_gfp).
Natural Transformability.
Transformation ability was determined by incubating bacterial cultures with a non-replicative plasmid containing a kanamycin-resistance cassette that can recombine with the chromosome and produces kanamycin-resistant transformants. Following incubation, serial dilutions were plated on nonselective and selective (i.e., with kanamycin) medium. Transformation frequency is the ratio of the number of cfus counted on selective versus nonselective media.
Gene Expression Analysis and Expression Profiling by RNA-Seq.
Total RNA from bacterial cultures was extracted (22), and Northern blot analysis was performed as previously described (12). RNA-seq analysis was performed on DNase-treated RNA samples from bacterial cultures grown to an OD600 of 0.8 at 37 °C. Strand-specific cDNA libraries were sequenced on a HiSeq 2000 instrument (ArrayExpress accession no. E-MTAB-4094). Enriched transcripts were determined using DESeq2 (33).
RIP-Seq of RocC-Bound RNAs.
Exponentially growing cells were fixed with formaldehyde, lysed, and incubated with protein A magnetic beads coated with affinity-purified antibodies directed against Lpp0148. Bound RNAs were extracted with a tri-reagent solution, and strand-specific cDNA libraries were prepared following a 3′-end polyadenylation and a 5′-end RNA adapter ligation and sequenced on HiSeq 2000.
mRNA Decay and RNA Half-Life Determination.
Cultures at an OD600 of 0.8 were treated with 100 μg/mL of rifampicin to stop transcription. RNA was extracted at the time points indicated in the figure legends. Transcript levels were analyzed by Northern blot (RocR, comEA) and real-time quantitative PCR (RT-qPCR) (comEA).
EMSAs.
Lpp0148 1–230 and Lpp0148 1–126 were affinity-purified with a 6×His N-terminal tag that was subsequently removed by cleavage with the HRV-3C protease. RNAs were synthesized by in vitro transcription in reactions containing α-[32P]ATP and purified. Binding reactions were carried out for 30 min at room temperature and directly loaded onto polyacrylamide gels run at 4 °C.
Phylogenetic Analysis.
A survey of 2,775 complete prokaryotic proteomes was conducted, and the retrieved homologs were aligned. The resulting alignment was trimmed to 80 positions defining the ProQ/FinO domain and used to infer a global phylogeny.
SI Materials and Methods
Bacterial Strains and Growth Conditions.
The L. pneumophila strains in this study are derived from the Paris clinical isolate (Outbreak isolate CIP107629) or the JR32 strain, a restriction-deficient and streptomycin-resistant derivative of the Philadelphia-1 strain. These strains (see Table S3) were grown in liquid media ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered yeast extract (AYE) or on solid media ACES-buffered charcoal yeast extract (CYE) plates at 30 °C or 37 °C. Liquid cultures were performed in a 13-mL tube containing 3 mL of medium or in 200-mL erlenmeyer flasks containing 50 mL of medium in a shaking incubator (Minitron, Infors HT) at 200 rpm. Alternatively, strains were grown in 100 μL AYE in 96-well plates placed inside a temperature-controlled plate reader (Tecan Infinite F200 PRO). The plate was submitted to intermittent shaking (100 rpm, 60 s every 7 min), and growth (OD600) and GFP signal (480 nm/520 nm) were monitored every 15 min. When appropriate, chloramphenicol (Cm), gentamicin (Gent), IPTG (isopropyl β-d-1-thiogalactopyranoside), and X-gal (5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside) were used at 5 μg/mL, 10 μg/mL, 500 µM, and 20 µg/mL, respectively. Kanamycin (Kan) was used at 15 μg/mL or 50 µg/mL for Paris or JR32 derivatives, respectively.
E. coli strains (see Table S3) were cultivated in LB medium with shaking or on LB–agar plates at 37 °C. When appropriate, kanamycin and chloramphenicol were used at 50 μg/mL and 25 μg/mL, respectively.
Recombinant DNA Techniques, Oligonucleotides, and DNA Isolation.
The plasmids (see Construction of Plasmids and Strains) and oligonucleotides (purchased from Eurogentec) used in this study are listed in Tables S3 and S4, respectively. For transformation of replicative plasmids in L. pneumophila, electroporation was performed as previously described (13). Enzymes were purchased from New England Biolabs and were used as described by the manufacturer. For PCR amplification, PrimeSTAR Max (TaKaRa) was used. Isolation of genomic DNA from L. pneumophila was performed using the Wizard Genomic DNA Purification Kit (Promega). Plasmid preparations from E. coli and L. pneumophila were realized using the QIAprep Spin Miniprep Kit (QIAGEN) or the E.Z.N.A. Plasmid Mini Kit I (Omega Bio-tek).
Natural Transformation of L. pneumophila.
Natural transformation of L. pneumophila strains was realized as follows. The strains were streaked on CYE solid medium from the –80 °C frozen stock culture and incubated for 3 d at 37 °C. The strains were then restreaked on a new CYE plate and incubated overnight at 37 °C to obtain exponentially growing cells. Bacteria were resuspended to an OD600 of 1 (∼109 cells per mL) in 3 mL AYE. We centrifuged 2× 1 mL of cell suspension for 3 min at 5,000 × g in a table-top microcentrifuge, and each pellet was resuspended in 50 µL of AYE with or without transforming DNA. Each suspension was spotted on a new CYE plate and left to dry. The plate was incubated overnight at 30 °C. Each spot was resuspended in 200 µL AYE. Tenfold serial dilutions were then plated on nonselective medium and selective medium. Plates were incubated for 72 h at 37 °C, and cfu counting was performed. Transformation frequency is the ratio of the number of cfus counted on selective medium divided by the number of cfus counted on nonselective medium. For routine transformation efficiency determination, the DNA used was 2 µg (for 109 cells) of pGEM-ihfB::KanR, a plasmid containing a kanamycin-resistance cassette (KanR) inserted in the ihfB gene of L. pneumophila (22). As this plasmid is non-replicative in L. pneumophila, the internalized molecules recombine with the chromosome via a double crossover allowing the integration of the KanR cassette in the ihfB locus. Transformants were selected on CYE + Kan.
Alternatively, transformability can be tested during growth in liquid medium (3). Exponentially growing cells on plate were used to inoculate 50 mL AYE at a starting OD600 of 0.05 and were incubated at 30 °C with shaking. When the culture reached the OD to be tested, the transformability test was done as follows. A volume of culture containing 109 cells was collected two times, and each was pelleted by centrifugation. Each pellet was resuspended in 200 µL AYE, and 2.5 µg of pGEM-ihfB::KanR was added in one tube, with the other tube being the control without DNA. Both tubes were incubated for 30 min standing at 30 °C to allow the DNA to enter the cells, and then 9 µL of Dnase I (1 U/µL; Sigma) and 791 µL AYE were added. Tubes were incubated for 2 h at 37 °C with shaking to stop DNA entry and to allow expression of the resistance gene. Tenfold serial dilutions were then plated on nonselective medium and selective medium. Plates were incubated for 72 h at 37 °C, and cfu counting was performed.
Construction of Plasmids and Strains.
JR32 Δlpp0148.
To construct this lpp0148 deletion mutant, the locus was amplified from the JR32 WT chromosome by PCR using the primer pair lpp0148_P1/lpp0148_P4 (4 kb). The PCR product was then cut by SphI (two sites: after nucleotides 181 and 346 of the lpp0148 coding sequence). The two obtained fragments were purified and ligated together. The ligation mixture was used to transform LELA3825 [JR32; lpp0148::(Tn903dIIlacZ, KanR)] (34). The bacterial suspension was plated on CYE, overlaid with Xgal, and screened for colonies presenting a white color (lac− cells). Colonies were tested for sensitivity to kanamycin, and the deletion in lpp0148 was then confirmed by PCR and sequencing.
LELA3825G [JR32; lpp0148::(Tn903dIIlacZ, GentR)].
The kanamycin resistance gene (nptII) inserted in lpp0148 of LELA3825 (34) was swapped for a gentamycin resistance gene (aacCI) by using the pXDC46 as described previously (34). Transformants were selected on CYE + Gent and verified for kanamycin sensitivity. The integration of GentR at the correct locus was verified by PCR. This strain was then transformed with pXDC91 to be used to screen for hypercompetent lpp0148 mutants from the random mutagenesis.
JR32 ΔrocR and Paris ΔrocR.
Markerless mutants of rocR were constructed in two steps, taking advantage of the counterselectable MK cassette. This MK cassette bears a kanamycin resistance gene and the toxin-encoding mazF gene under the control of an IPTG-inducible promoter (lacIq, Ptac-mazF, nptII) and was cloned into the pGEM-T Easy vector (Promega) to create plasmid pGEM-MK.
The first step in constructing the ΔrocR mutants was to insert this cassette in the chromosome, replacing rocR (strains ΔrocR::MK). To do so, the upstream and downstream regions of rocR were amplified with primers carrying 30-nt sequences complementary to ends of the MK cassette. The upstream (PCR A) and downstream (PCR C) regions were assembled to the MK cassette (PCR B, 3.2 kb amplified from plasmid pGEM-MK with primer pair MazFk7-F/MazF-R) by PCR overlap extension and used for natural transformation. For Paris ΔrocR::MK, PCR A (LA51_up-lppnc0692-F/LA52_up-lppnc0692-R-tail-maz, 1.4 kb) and PCR C (LA53_down-lppnc0692-F-tail-maz/LA54_down-lppnc0692-R, 1.5 kb) were amplified from Paris WT chromosome. For JR32 ΔrocR::MK, PCR A (LA81_up-rocR-F/LA52_up-lppnc0692-R-tail-maz, 2 kb) and PCR C (LA82_down-rocR-F-tail-maz/LA54_down-lppnc0692-R, 1.5 kb) were amplified from JR32 WT chromosome. The overlapping PCRs were transformed in the WT strains, and transformants were selected on CYE + Kan and tested for sensitivity to IPTG. Integration of the MK cassette at the correct locus was verified by PCR.
To obtain the markerless mutants, a second step was performed as follows. The upstream and downstream regions of rocR were amplified again but with primers carrying a 30-nt tail sequence that makes the 3′ end of the upstream region complementary to the 5′ end of the downstream region (PCR D and PCR E). These PCRs were assembled by PCR overlap extension and used to transform the ΔrocR::MK strains. For Paris ΔrocR, PCR D (LA51_up-lppnc0692-F/LA55-up-lppnc0692-R+tail LA56, 1.5 kb) and PCR E (LA56-down-lppnc0692-F/LA54_down-lppnc0692-R, 1.4 kb) were amplified from the Paris WT chromosome. For JR32 ΔrocR::MK, PCR D (LA81_up-rocR-F/LA87-up-rocR-R+tail LA88, 2 kb) and PCR E (LA88-down-rocR-F/LA54_down-lppnc0692-R, 1.5 kb) were amplified from the JR32 WT chromosome. Transformants were selected on CYE + IPTG and tested for sensitivity to kanamycin. Proper deletion of rocR was verified by PCR and sequencing.
Complementation of lpp0148 and rocR deletions.
For complementation of Paris lpp0148TAA, plasmids pX3_148 and pX3_148-9 were created as follows. Loci 165011–165943 (lpp0148) and 165011–167014 (lpp0148_lpp0149) were amplified from the Paris WT chromosome using primer pairs lpp0148pX3_F/lpp0148pX3_R (955 bp) and lpp0149pX3_R (2 kb), respectively. These fragments were cloned into the pX3 plasmid [RSF1010 derivative, replicative in E. coli and L. pneumophila (12)] open with NotI and XmaI. The ligation mixture was transformed into E. coli DH5α, and λpir and transformants were selected on LB plates containing chloramphenicol. Plasmids were verified by PCR and sequencing. They were then used to transform L. pneumophila (by electroporation followed by selection on CYE + Cm).
For complementation of Paris ΔrocR, plasmid pLLA21 (i.e., pX3_rocR) was created as follows: The rocR locus (3384431–3384710) of Paris WT was amplified by PCR using primer pair LA61_rocR-F-NotI/LA62_rocR-R-XmaI (302 bp). This fragment was cloned into the pX3 plasmid open with NotI and XmaI. The ligation mixture was transformed into E. coli DH5α, and λpir and transformants were selected on LB plates containing chloramphenicol. The plasmid was verified by PCR and sequencing and then used to transform L. pneumophila (by electroporation followed by selection on CYE + Cm).
Testing RocR box functionality: Creation of plasmids pLLA27, pLLA28, and pLLA29.
To assess the impact of mutations in the RocR box on the recognition of an mRNA by the complex Lpp0148/RocR, plasmid pXDC91 (PcomEA_comEA_gfp) was submitted to site-directed mutagenesis. PCRs were done using primer pairs designed to mutate the RocR box (TCCAAC) on pXDC91: LA68_RocRbox_m3-F/LA69_RocRbox_m3-R for pLLA27 (with RocR box m3, TttAAC), LA70_RocRbox_m4-F/LA71_RocRbox_m4-R for pLLA28 (with RocR box m4, TCCttC), and LA72_RocRbox_m5-F/LA73_RocRbox_m5-R for pLLA29 (with RocR box m5, ctacAC). PCR products were then digested by DpnI to remove the parental pXDC91 and transformed in E. coli DH5α λpir. Transformants were selected on LB plates with chloramphenicol. Plasmids were verified by sequencing and then used to transform L. pneumophila JR32 WT (electroporation followed by selection on CYE + Cm).
Creation of RocR mutants that recognize the mutated RocR box of pLLA27, pLLA28, and pLLA29.
To obtain the RocR mutants, the JR32 ΔrocR::MK strain was transformed by a mutated rocR locus constructed as follows. The upstream and downstream regions of rocR were amplified so that the upstream 3′ end and the downstream 5′ end are complementary to each other and create a mutated seed sequence. These PCRs were assembled by PCR overlap extension and used to transform the ΔrocR::MK strain. For JR32 rocRm3, PCR F (LA81_up-rocR-F/LA85_RocR_m3-R, 2 kb) and PCR G (LA86_RocR_m3-F/LA54_down-lppnc0692-R, 1.6 kb) were amplified from JR32 WT chromosome. For JR32 rocRm4, PCR H (LA81_up-rocR-F/LA79_RocR_m4-R, 2 kb) and PCR I (LA80_RocR_m4-F/LA54_down-lppnc0692-R, 1.6 kb) were amplified from the JR32 WT chromosome. For JR32 rocRm5, PCR N (LA81_up-rocR-F/LA91_RocR_m5-R, 2 kb) and PCR P (LA92_RocR_m5-F/LA54_down-lppnc0692-R, 1.6 kb) were amplified from JR32 WT chromosome. Transformants were selected on CYE + IPTG and tested for sensitivity to kanamycin. The rocR locus was verified by PCR and sequencing.
Random Mutagenesis of lpp0148 in the L. pneumophila JR32 Strain.
To isolate mutations of Lpp0148 that would impair its function, we first amplified its ORF from the JR32 WT strain using primer pair lpp0148RM_F/lpp0148RM_R and used it as a template for random mutagenesis with the GeneMorph Random Mutagenesis Kit (Agilent Technologies). The obtained mutated products (PCR B*, 782 bp) were then incorporated in a longer DNA fragment that was used for natural transformation of L. pneumophila. To do so, the upstream (PCR A, 1.8 kb) and downstream (PCR D, 1.6 kb) regions of lpp0148 were amplified from JR32 WT chromosome using primer pairs lpp0148_P1/lpp0148RM_P2 and lpp0148RM_P3/lpp0148_P4, respectively. A kanamycin resistance cassette was also obtained so as to fit between the mutated lpp0148 ORF and the downstream region. This KanR cassette [cloned into the pGEM-T Easy vector (Promega) from the pKD4 plasmid (35)] was amplified from the pGEMPKD4 plasmid using primer pair pKD4_P1/pKD4_P2 (PCR C, 1.5 kb). PCR A, B*, C, and D were assembled by PCR overlap extension and used to transform the strain LELA3825G, pXDC91. This strain is a lpp0148::GentR JR32 derivative with the comEA_gfp reporter on a CmR plasmid. Transformants were selected on CYE + Cm + Kan, and bright green colonies (hypercompetent mutants) were picked (60 clones). These were isolated and checked for gentamycin sensitivity (34 of the 60 clones were KanR GentS). Then the presence of Lpp0148 was ascertained by Western blot in the KanR GentS clones (Lpp0148 was detectable in 33 of the 34 clones), and the lpp0148 locus was sequenced. We obtained 19 original mutants, among which were 10 single nonsynonymous mutations and three single nucleotide deletions causing a frameshift and a truncation of the C-terminal part of the protein.
Western Blot Analysis and Immunodetection.
Cells were grown in AYE + Cm and were harvested at an OD600 of ∼0.8 by centrifugation at 5,000 × g for 5 min. The pellet was resuspended directly in 125 mM Tris·HCl, pH 6.8, 2% (wt/vol) SDS, 15% (vol/vol) glycerol, 0.005% Bromophenol Blue, and 100 mM β-mercaptoetanol and incubated for 5 min at 85 °C. Samples were then separated by SDS/PAGE. Next, proteins were transferred to a PVDF membrane on a Trans-Blot Turbo Transfer System (Bio-Rad). Lpp0148 was detected with polyclonal anti-Lpp0148 antibodies and anti-rabbit IgG and HRP-conjugated antibodies (Sigma), and revelation was done using the ECL system (Thermo Scientific) according to the manufacturer’s instructions. Luminescence signals were acquired using an imaging workstation equipped with a charge-coupled device camera (Thermo Scientific).
Gene Expression Analysis by Northern Blot.
Total RNA from bacterial cultures was extracted according to a previously described procedure (22). Briefly, cells were fixed in ice-cold methanol and lysed in RNAsnap buffer (18 mM EDTA, 0.025% SDS, 1% 2-mercaptoethanol, 95% formamide). RNA was then extracted using a tri-reagent solution (acid guanidinium thiocyanate–phenol–chloroform) and isopropanol-precipitated. Northern blot was performed using a denaturing Tris/Borate/EDTA (TBE)–urea 6% acrylamide gel. One to two micrograms of total RNA in denaturing buffer were loaded per lane and run in TBE buffer. RNA was transferred to a nylon membrane (IMMOBILON-NY+; Millipore Corporation) by electrophoretic transfer in 0.5× TBE buffer. RNA was cross-linked to the membrane by UV irradiation. Membranes were hybridized at 42 °C with 5 nM of a 5′-biotinylated oligonucleotide probe (Table S4) in ULTRAhyb Ultrasensitive Hybridization Buffer (Ambion) and then washed according to ULTRAhyb’s instructions. Membranes were developed using HRP-conjugated streptavidin and enhanced luminol substrate (Chemiluminescent Nucleic Acid Detection Module; Pierce). Luminescence signals were acquired using an imaging workstation equipped with a charge-coupled device camera (Thermo Scientific). Ethidium-bromide staining of 5S ribosomal RNA was used to check for equal loading of the lanes.
Flow Cytometry.
Fresh cultures grown for 12 h on CYE + Cm plates were resuspended in PBS at an OD600 of 1. We mixed 450 µL of cell suspension with 50 µL of formaldehyde, and 0.5 µL of 10 mg/mL FM4-64 was added. Cells were incubated for 30 min at room temperature. Then cells were washed and resuspended in 500 µL of PBS. GFP levels (Ex 488 nm, Em 530/30 nm) of 5.105 FM4-64–positive particles (Ex 488 nm, Em 640LP) were measured on 1,000× diluted samples by an Attune Acoustic Focusing Cytometer (Applied Biosystems).
Global Gene Expression Profiling by RNA-Seq.
Total RNA from bacterial cultures grown to an OD600 of 0.8 at 37 °C was extracted as described earlier. RNA samples were treated with DNase I and purified on silica-based columns (Zymo Research). Strand-specific cDNA libraries were prepared from Paris WT and lpp0148TAA mutant RNA using the ScriptSeq v2 kit (Epicentre) and sequenced on an HiSeq 2000 platform (Illumina) with single-read 50-bp assay. Enriched transcripts were determined using the Bioconductor DESeq2 package (33) with the R statistical software, version 3.2.2. Appropriate normalization of count matrix by sequencing depth, excluding outliers based on the Cook’s distance and independent filtering to gain statistical power regarding the Benjamini and Hochberg multiple testing correction method, was performed before downstream analysis. Of note, the DEseq2 method of moderated estimation of fold change tends to deflate the ratios compared with conventional raw fold change determination methods. Data were visualized with the Artemis Genome Browser (36). RNA-seq data were deposited to the MINSEQE-compliant public ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/) (accession no. E-MTAB-4094) and brokered to the European Nucleotide Archive (www.ebi.ac.uk) (accession no. ERP013398).
Purification of His-Lpp0148 for Specific Antibody Production and Antibody Purification.
Lpp0148 (full size, amino acids 1–230) from L. pneumophila Paris was cloned into the NdeI and BamHI restriction sites of pET-15b (Novagen) after amplification with primer pair proQ15b-F/proQ15b-R. Ligation products were transformed into E. coli BL21(DE3) cells. For overexpression, a 100-mL cell culture was grown for 3 h at 37 °C to an OD600 between 0.4 and 0.8 and induced with IPTG at a final concentration of 1 mM. The temperature was reduced to 30 °C, and cell cultures were pelleted after a further 3 h of growth, flash-frozen in liquid nitrogen, and stored at –80 °C. The cell pellet was resuspended at 4 °C in 5 mL of lysis buffer (50 mM sodium phosphate, pH 7.5, 150 mM NaCl, 1 mM PMSF, and 1 mM β-mercaptoethanol). Cells were lysed by French press three times at room temperature, and the lysate was centrifuged for 30 min at 10,000 × g at 4 °C. The cleared cell lysate was then mixed with 200 µL of HisPur Ni-NTA Magnetic Beads (Thermo Scientific) pre-equilibrated in lysis buffer and incubated with gentle mixing for 2 h at 4 °C. The beads were washed five times with 1 mL of 50 mM sodium phosphate, pH 7.5, 150 mM NaCl, 20 mM imidazole, 1 mM PMSF, and 1 mM β-mercaptoethanol. Elution of His6-Lpp0148 was done with 500 µL of 50 mM sodium phosphate, pH 7.5, 150 mM NaCl, 300 mM imidazole, 1 mM PMSF, and 1 mM β-mercaptoethanol. Eluted protein was concentrated by dialysis using Zeba Spin Desalting Columns (7K molecular weight cut-off, 10 mL; Thermo Scientific) against 100 mM sodium phosphate and 150 mM NaCl, pH 7.5. Glycerol was added to a final concentration of 40% solution, and the purified protein solution was stored at –20 °C.
The purified His6-Lpp0148 was used to produce polyclonal rabbit antibodies (Covalab). Obtained serum was purified by affinity using the AminoLink Plus Immobilization Kit (Thermo Scientific) with bound His6-Lpp0148, depleted of nonspecific reactive antibodies by using the AminoLink Plus Immobilization Kit (Thermo Scientific) coated with a lysate from the Paris lpp0148TAA mutant strain, and concentrated by centrifugation on a Microsep 30K Omega, Red (Pall Corporation).
RIP-Seq of Lpp0148-Bound RNAs.
Exponentially growing bacterial cultures were fixed with 1% formaldehyde for 30 min. Pelleted bacterial cells were resuspended in lysis buffer (50 mM Hepes–KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate) and sonicated at 4 °C. Lysates were incubated with protein A magnetic beads (Dynabeads, Invitrogen) coated with rabbit-raised affinity-purified antibodies directed against Lpp0148. Following the washing steps, RNA was eluted from the magnetic beads by extraction with a tri-reagent solution (acid guanidinium thiocyanate–phenol–chloroform) and isopropanol-precipitated. RNA samples were treated with DNase I and concentrated on silica-based columns (Zymo Research). Strand-specific cDNA libraries were prepared following 3′-end polyadenylation and 5′-end RNA adapter ligation and sequenced on a HiSeq 2000 platform (Illumina) with single-read 75-bp assay. Data were visualized and the number of reads per feature extracted using Artemis Genome Browser (36). Read counts were normalized using voom methodology (37), and the statistical significance of the obtained enrichment was calculated as previously described (38).
mRNA Decay and RNA Half-Life Determination.
Bacterial cultures at an OD600 of 0.8 were treated with 100 μg/mL of rifampicin to stop transcription. RNA was extracted as described above at the time points indicated in the figure legends. Transcript levels were detected by Northern blot analysis (RocR, comEA) as previously described (22). For RT-qPCR analysis, traces of genomic DNA were removed by a DNase I treatment and RNA was purified on a silica-based column (Zymo Research). RNA (2 μg) was reverse-transcribed with RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific) and used as a template for real-time PCR with Premix Ex Taq (Tli* RNase H Plus) (TaKaRa) on LC480 instruments (Roche). Data were analyzed with the ΔCt method using tmRNA (encoded by the lpptmRNA1 gene) as a reference. Amounts of mRNA were expressed relative to time t = 0. Data were fit to a first-order exponential decay with the Qtiplot software.
Lpp0148 Cloning, Expression, and Purification for Biochemistry Analyses.
Lpp0148aa1–230 (full size) from L. pneumophila Paris was cloned into the SmaI and NotI restriction sites of pET-47b(+) (Novagen) after PCR amplification with primer pair rocc_paris_pet47b+_fwd_1/rocc_paris_pet47b+_rev_230. Ligation products were transformed into E. coli BL21(DE3) cells. For overexpression, a 2 L cell culture was grown for 3 h at 37 °C to an OD600 between 0.4 and 0.8 and induced with IPTG at a final concentration of 1 mM. The temperature was reduced to 30 °C, and cell cultures were pelleted after a further 3 h of growth, flash-frozen in liquid nitrogen, and stored at –80 °C. The pellet was resuspended at 4 °C in 40 mL of lysis buffer (50 mM Hepes, pH 7.3, 0.5 M NaCl, 10 mM Imidazole, 40 mg lysozyme, and 300 μL Halt protease inhibitors mixture; Thermo Scientific). Cells were lysed by sonication (Branson Digital Sonifier 450) and the cleared cell lysate loaded onto 10 mL of HisPur Ni-NTA Superflow Agarose (Thermo Scientific). The column was washed with 90 mL of lysis buffer, and Lpp0148 was cleaved from the hexa-histidine tag with an overnight incubation of HRV-3C protease (Sigma-Aldrich H9916). Eluted protein was concentrated and further purified on a Superdex 75 16/60 size-exclusion column equilibrated in 150 mM NaCl and 50 mM Hepes, pH 7.3.
Lpp0148aa1–126 was cloned into pGEX-6P-1 (GE Healthcare Life Sciences) using restriction sites BamHI and NotI after PCR amplification with primer pair rocc-pgex6p1-fwd_1/rocc-pgex6p1-rev_126. Ligation products were transformed into E. coli BL21(DE3) cells. Overexpression and lysis were performed as for Lpp0148aa1–230 but with no imidazole in the lysis buffer. Cleared lysate was incubated on 10 mL of Glutathione Superflow Agarose (Pierce 25238) for 2 h at 4 °C, washed with 10 column volumes of lysis buffer, and incubated overnight with HRV-3C protease. Cleaved protein was eluted, concentrated, and purified on a Superdex 75 16/60 size-exclusion column equilibrated in 150 mM NaCl and 50 mM Hepes, pH 7.3.
EMSA.
RNAs used in gel mobility shift assays were prepared by T7 transcription from synthetic DNA templates, in reactions containing α-[32P]ATP, from synthetic DNA templates and were purified by 15% denaturing PAGE:
RocR → 5′ GCGAGGUUGGACUUGCUAUGUGUCAUUCCACUGGGUCAAUUGGCGACACACUGAUUGGCCCUUUCU 3′,
5′ RocR → 5′ GCGAGGUUGGACUUGCUAUGUGUCAUUCCAC 3′, and
3′ RocR → 5′ UGGGUCAAUUGGCGACACACUGAUUGGCCCUUUCU 3′.
Binding reactions [50–100 × 103 cpm 32P-labeled RNAs and 0–16 µM full length (amino acids 1–230) or N-terminal (amino acids 1–126) Lpp0148 in 2 mM MgCl2, 2 units RNaseOut, 50 mM NaCl, 25 mM Hepes, pH 7.3] were carried out for 30 min at room temperature and directly loaded onto 10% native Tris–glycine (wt/vol) polyacrylamide gels run at 4 °C. Gels were visualized by exposure to a phosphor screen (Molecular Dynamics) that was scanned by a Typhoon PhosphorImager (GE HealthCare) and analyzed using ImageQuant Software (Molecular Dynamics).
Phylogenetic Analysis.
A survey of the 2,775 complete prokaryotic proteomes available at the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov) was conducted to identify ProQ/FinO domain-containing proteins. We used two different sequence similarity-based approaches. First we applied an iterative BLASTP procedure (39) using different seeds. In addition, we performed an HMM profile-based search with the HMMSEARCH program from the HMMER package (40) and the ProQ/FinO protein domain (Pfam accession no. PF04352). The 474 retrieved homologs were aligned using MAFFT version 7.037b with the accurate option l-INS-i (41). The resulting alignment was trimmed to 80 positions with BMGE version 1.1 with the BLOSUM30 model (42) and used to infer a maximum likelihood phylogeny of ProQ/FinO domain-containing proteins with FASTTREE v.2 (43) with the wag cat 4 gamma options. The branch robustness of the resulting tree was estimated using SH-like supports with FASTTREE. The location of the ProQ/FinO domains in the 474 retrieved homologs was determined with the program HMMSCAN from the HMMER package (40).
Supplementary Material
Acknowledgments
L.A. and X.C. thank Chloé Vallantin for excellent technical support and Christine Gaspin, Unité de Mathématiques et Informatique Appliquées de Toulouse, Toulouse, France, for helpful discussions on sRNA-mRNA duplex predictions. L.A. is supported by a postdoctoral fellowship from the Fondation pour la Recherche Médicale (SPF20130526652). C.B.-A. is a member of the Institut Universitaire de France and is funded by the “Ancestrome” project (ANR-10-BINF-01-01). Work in the A.M.M. laboratory is supported by the Natural Sciences and Engineering Research Council of Canada (NSERC). Work in the M.G. laboratory is supported by the Canadian Institutes of Health Research (CIHR114975). Work in the X.C. laboratory is supported by a CNRS–INSERM ATIP-Avenir grant and Sanofi. This work was performed within the framework of the LABEX ECOFECT (ANR-11-LABX-0048) of Université de Lyon, within the program “Investissements d’Avenir” (ANR-11-IDEX-0007) operated by the French National Research Agency (ANR).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-seq data were deposited to the MINSEQE-compliant public ArrayExpress database, https://www.ebi.ac.uk/arrayexpress (accession no. E-MTAB-4094) and brokered to the European Nucleotide Archive, www.ebi.ac.uk (accession no. ERP013398).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601626113/-/DCSupplemental.
References
- 1.Johnston C, Martin B, Fichant G, Polard P, Claverys J-P. Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat Rev Microbiol. 2014;12(3):181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 2.Smith JM, Dowson CG, Spratt BG. Localized sex in bacteria. Nature. 1991;349(6304):29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 3.Anderson MT, Seifert HS. Opportunity and means: Horizontal gene transfer from the human host to a bacterial pathogen. MBio. 2011;2(1):e00005–e00011. doi: 10.1128/mBio.00005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Overballe-Petersen S, et al. Bacterial natural transformation by highly fragmented and damaged DNA. Proc Natl Acad Sci USA. 2013;110(49):19860–19865. doi: 10.1073/pnas.1315278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seitz P, Blokesch M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev. 2013;37(3):336–363. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2(3):241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 7.Provvedi R, Dubnau D. ComEA is a DNA receptor for transformation of competent Bacillus subtilis. Mol Microbiol. 1999;31(1):271–280. doi: 10.1046/j.1365-2958.1999.01170.x. [DOI] [PubMed] [Google Scholar]
- 8.Draskovic I, Dubnau D. Biogenesis of a putative channel protein, ComEC, required for DNA uptake: Membrane topology, oligomerization and formation of disulphide bonds. Mol Microbiol. 2005;55(3):881–896. doi: 10.1111/j.1365-2958.2004.04430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mortier-Barrière I, et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007;130(5):824–836. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 10.Attaiech L, et al. Role of the single-stranded DNA-binding protein SsbB in pneumococcal transformation: Maintenance of a reservoir for genetic plasticity. PLoS Genet. 2011;7(6):e1002156. doi: 10.1371/journal.pgen.1002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone BJ, Kwaik YA. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol. 1999;181(5):1395–1402. doi: 10.1128/jb.181.5.1395-1402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charpentier X, Kay E, Schneider D, Shuman HA. Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J Bacteriol. 2011;193(5):1114–1121. doi: 10.1128/JB.01146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchrieser C, Charpentier X. Induction of competence for natural transformation in Legionella pneumophila and exploitation for mutant construction. Methods Mol Biol. 2013;954:183–195. doi: 10.1007/978-1-62703-161-5_9. [DOI] [PubMed] [Google Scholar]
- 14.Sexton JA, Vogel JP. Regulation of hypercompetence in Legionella pneumophila. J Bacteriol. 2004;186(12):3814–3825. doi: 10.1128/JB.186.12.3814-3825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunte HJ, Crane RA, Culham DE, Richmond D, Wood JM. Protein ProQ influences osmotic activation of compatible solute transporter ProP in Escherichia coli K-12. J Bacteriol. 1999;181(5):1537–1543. doi: 10.1128/jb.181.5.1537-1543.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerr CH, Culham DE, Marom D, Wood JM. Salinity-dependent impacts of ProQ, Prc, and Spr deficiencies on Escherichia coli cell structure. J Bacteriol. 2014;196(6):1286–1296. doi: 10.1128/JB.00827-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghetu AF, Gubbins MJ, Frost LS, Glover JN. Crystal structure of the bacterial conjugation repressor finO. Nat Struct Biol. 2000;7(7):565–569. doi: 10.1038/76790. [DOI] [PubMed] [Google Scholar]
- 18.Arthur DC, et al. Mapping interactions between the RNA chaperone FinO and its RNA targets. Nucleic Acids Res. 2011;39(10):4450–4463. doi: 10.1093/nar/gkr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mark Glover JN, et al. The FinO family of bacterial RNA chaperones. Plasmid. 2015;78:79–87. doi: 10.1016/j.plasmid.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Arthur DC, et al. FinO is an RNA chaperone that facilitates sense-antisense RNA interactions. EMBO J. 2003;22(23):6346–6355. doi: 10.1093/emboj/cdg607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Biesen T, Frost LS. The FinO protein of IncF plasmids binds FinP antisense RNA and its target, traJ mRNA, and promotes duplex formation. Mol Microbiol. 1994;14(3):427–436. doi: 10.1111/j.1365-2958.1994.tb02177.x. [DOI] [PubMed] [Google Scholar]
- 22.Juan P-A, Attaiech L, Charpentier X. Natural transformation occurs independently of the essential actin-like MreB cytoskeleton in Legionella pneumophila. Sci Rep. 2015;5:16033. doi: 10.1038/srep16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berka RM, et al. Microarray analysis of the Bacillus subtilis K-state: Genome-wide expression changes dependent on ComK. Mol Microbiol. 2002;43(5):1331–1345. doi: 10.1046/j.1365-2958.2002.02833.x. [DOI] [PubMed] [Google Scholar]
- 24.Peterson SN, et al. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol. 2004;51(4):1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 25.Redfield RJ, et al. A novel CRP-dependent regulon controls expression of competence genes in Haemophilus influenzae. J Mol Biol. 2005;347(4):735–747. doi: 10.1016/j.jmb.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Sahr T, et al. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol. 2012;9(4):503–519. doi: 10.4161/rna.20270. [DOI] [PubMed] [Google Scholar]
- 27.Jerome LJ, Frost LS. In vitro analysis of the interaction between the FinO protein and FinP antisense RNA of F-like conjugative plasmids. J Biol Chem. 1999;274(15):10356–10362. doi: 10.1074/jbc.274.15.10356. [DOI] [PubMed] [Google Scholar]
- 28.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136(4):615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kessler A, et al. The Legionella pneumophila orphan sensor kinase LqsT regulates competence and pathogen-host interactions as a component of the LAI-1 circuit. Environ Microbiol. 2013;15(2):646–662. doi: 10.1111/j.1462-2920.2012.02889.x. [DOI] [PubMed] [Google Scholar]
- 30.Gottesman S, Storz G. RNA reflections: Converging on Hfq. RNA. 2015;21(4):511–512. doi: 10.1261/rna.050047.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cazalet C, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36(11):1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 32.de Felipe KS, et al. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187(22):7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charpentier X, Faucher SP, Kalachikov S, Shuman HA. Loss of RNase R induces competence development in Legionella pneumophila. J Bacteriol. 2008;190(24):8126–8136. doi: 10.1128/JB.01035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rutherford K, et al. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16(10):944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 37.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eddy SR. Accelerated profile HMM searches. PLOS Comput Biol. 2011;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price MN, Dehal PS, Arkin AP. FastTree 2--Approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadosky AB, Wiater LA, Shuman HA. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun. 1993;61(12):5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.