Significance
In human females, one of the two X chromosomes is inactive (Xi) and adopts an unusual 3D conformation. The Xi chromosome contains superloops, large chromatin loops that are often anchored at the macrosatellite repeat DXZ4, and is partitioned into two large intervals, called superdomains, whose boundary lies at DXZ4. Here, we use spatial proximity mapping, microscopy, and genome editing to study the Xi. We find that superloops and superdomains are conserved across humans, macaque, and mouse. By mapping proximity between three or more loci, we show that superloops tend to occur simultaneously. Deletion of DXZ4 from the human Xi disrupts superloops, eliminates superdomains, and alters chromatin modifications. Finally, we show that a model in which CCCTC-binding factor (CTCF) and cohesin extrude chromatin can explain the formation of superloops and superdomains.
Keywords: X chromosome inactivation, inactive X chromosome, Hi‐C, CTCF, genome engineering
Abstract
During interphase, the inactive X chromosome (Xi) is largely transcriptionally silent and adopts an unusual 3D configuration known as the “Barr body.” Despite the importance of X chromosome inactivation, little is known about this 3D conformation. We recently showed that in humans the Xi chromosome exhibits three structural features, two of which are not shared by other chromosomes. First, like the chromosomes of many species, Xi forms compartments. Second, Xi is partitioned into two huge intervals, called “superdomains,” such that pairs of loci in the same superdomain tend to colocalize. The boundary between the superdomains lies near DXZ4, a macrosatellite repeat whose Xi allele extensively binds the protein CCCTC-binding factor. Third, Xi exhibits extremely large loops, up to 77 megabases long, called “superloops.” DXZ4 lies at the anchor of several superloops. Here, we combine 3D mapping, microscopy, and genome editing to study the structure of Xi, focusing on the role of DXZ4. We show that superloops and superdomains are conserved across eutherian mammals. By analyzing ligation events involving three or more loci, we demonstrate that DXZ4 and other superloop anchors tend to colocate simultaneously. Finally, we show that deleting DXZ4 on Xi leads to the disappearance of superdomains and superloops, changes in compartmentalization patterns, and changes in the distribution of chromatin marks. Thus, DXZ4 is essential for proper Xi packaging.
Mammalian females possess two copies of the X chromosome in each of their cells, whereas males possess one X and one Y chromosome. Ensuring that males and females exhibit similar expression levels for X-linked genes is accomplished by a process known as “X chromosome inactivation” (XCI) (1). XCI is initiated by the long noncoding RNA Xi-specific transcript (XIST) (2, 3), which triggers a cascade of events that repackage one X chromosome (called “Xi”) into facultative heterochromatin (4) but leave the other chromosome (called “Xa”) unaffected. XCI leads to stable silencing of most gene expression on the Xi chromosome throughout all subsequent somatic cell divisions (5, 6).
In addition, the Xi chromosome exhibits an unusual 3D conformation known as the “Barr body” (7–9). The mechanisms that bring about this conformation are largely unknown.
In previous work describing the structure of the Xi chromosome, we used DNA polymorphisms to assign in situ Hi-C reads to specific chromosomal homologs to create a diploid Hi-C map for human GM12878 cells (10). We showed that the Xi chromosome has a distinctive superstructure. First, it contains superdomains, which are unusually large contact domains (contiguous intervals of the genome in which all loci exhibit an enhanced probability of contact with one another). It also contains unusually large chromatin loops called “superloops.” Both superdomains and superloops can span dozens of megabases of the genome. We also showed that the macrosatellite element DXZ4 (11–13) is situated at the boundary of the superdomains (10) and is bound by CCCTC-binding factor (CTCF) exclusively on the Xi homolog (14). DXZ4 also lies at the anchor of superloops to XIST, FIRRE, LOC550643 (which we here dub the “inactive-X CTCF-binding contact element,” ICCE), and to a previously uncharacterized locus at ChrX:75350000–75400000 (which we refer to here as “x75”) (10, 15).
We also observed that this Xi superstructure coexists with compartmentalization. Compartmentalization refers to the fact that many genomes are partitioned into intervals belonging to a handful of types (called “subcompartments”), such that loci in intervals of the same subcompartment tend to colocalize during interphase. In a Hi-C contact map, these intervals [typically ∼300 kilobases (kb) long (10)] manifest as contact domains, i.e., as square domains of enhanced contact frequency along the diagonal, and the contacts between them give the map a “plaid” appearance. Each subcompartment is associated with a particular pattern of chromatin marks. Furthermore, the six known subcompartments can be assigned to one of two types, A or B, called “compartments.” Intervals that are in different subcompartments but are in the same compartment exhibit long-range contact patterns which are more similar than the contact patterns of intervals in entirely different compartments. The A and B compartments correspond closely with open and closed chromatin (10, 16).
In this study, we used contact mapping, microscopy, and genome engineering to study the superstructure of the Xi chromosome and the role of DXZ4.
Results
The Xi Chromosome Superstructure Is Conserved in Mouse and Rhesus Macaque.
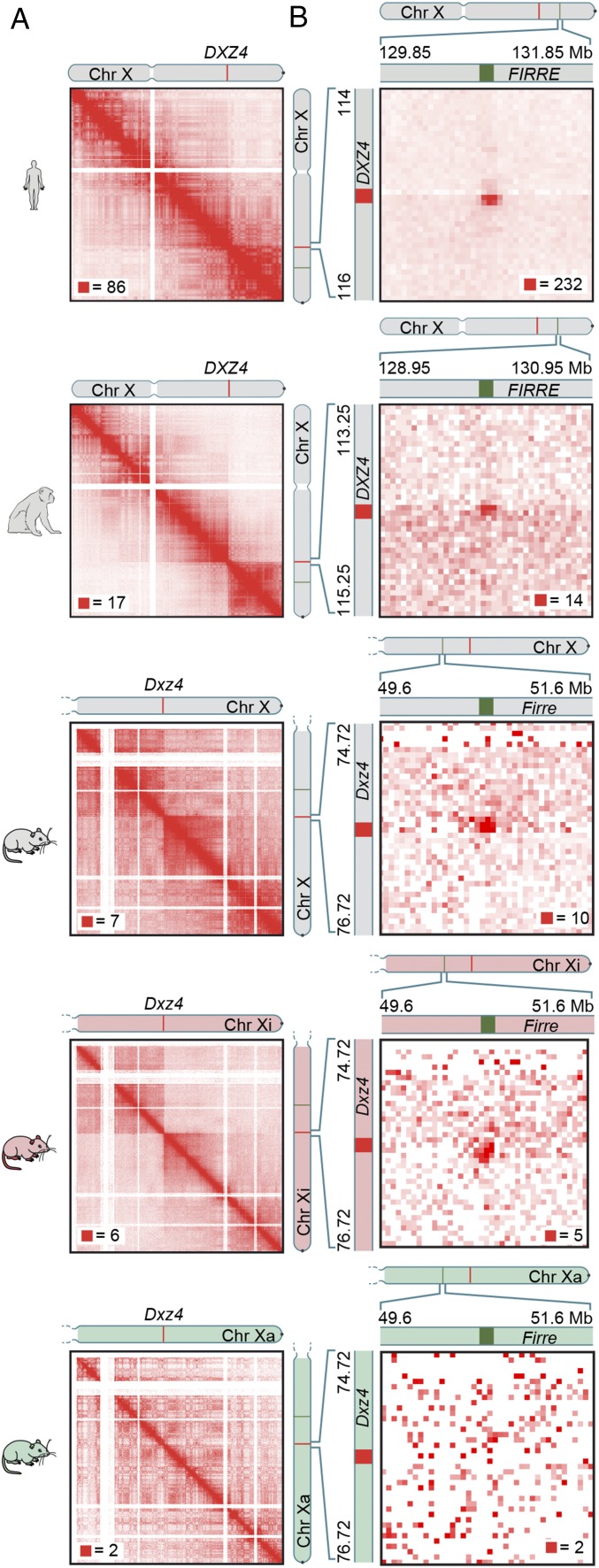
First, we explored whether the Xi chromosome superstructure is conserved among mammals by applying Hi-C to cells from mouse and rhesus macaque (SI Appendix, Table S1). For mouse, we studied the female Patski cell line (17). Because the cell line was derived from an interspecies hybrid cross (Mus musculus × Mus spretus), it has a high heterozygosity rate (1 in ∼64 bases) that facilitates assignment of a large fraction of alignable reads (49%) to particular chromosome homologs. Superdomains were apparent in the maps of the Xi chromosome but absent from maps of the Xa chromosome (Fig. 1A). Notably, despite extensive rearrangement of the murine X chromosome relative to its human counterpart, the boundary between the superdomains again occurred at Dxz4 [chrX: 75.7–75.8 megabases (Mb), mm10] (18). [While this manuscript was in preparation, this point was reported by multiple groups (19, 20).] As in human, we also observed superloops between Dxz4 and Firre, between Dxz4 and Xist, and between Dxz4 and x75 (Fig. 1B and SI Appendix, Fig. S1).
Fig. 1.
The Xi chromosome superstructure is conserved across human, rhesus macaque, and mouse. (A) Superdomains on the Xi chromosome are conserved across human, rhesus macaque, and mouse. The boundary of the superdomains lies at DXZ4 and its orthologs. In diploid Hi-C maps of mouse, the superdomain is seen only on the Xi chromosome. (Resolution: 100 kb.) For all contact maps, the color scale of each map goes from 0 (white) to red, whose value is given by the red square in each map. The chromosome icons are colored gray to indicate unphased maps of the X chromosome, in which data from both the Xi and Xa chromosomes are superimposed; they are colored red to indicate diploid Xi-only maps or green to indicate diploid Xa-only maps. The phased SNP calls used to generate homolog-specific maps are outlined in SI Appendix, Supplementary Materials and Methods. (B) A superloop forms between DXZ4 and FIRRE in human. Superloops are present at orthologous positions in rhesus macaque and mouse. In diploid Hi-C maps of mouse, the superloop is only seen on the Xi chromosome. (Resolution: 50 kb.)
However, there was one notable difference between maps of the human and murine Xi chromosomes. Compartment structure was almost entirely absent in the murine Xi chromosome: The Hi-C map of the Xi chromosome did not have a plaid appearance, and square domains were rarely seen along its diagonal. To check whether the absence of compartment structure is a feature of mice in general or of the Patski cell line in particular, we reanalyzed data from the two recent Hi-C studies of murine X chromosome inactivation mentioned above (19, 20). A map of the Patski cell line from ref. 19 confirms the lack of compartmentalization on the Xi chromosome in Patski cells. Similarly, in maps of murine fibroblasts (20), the compartment structure on the Xi chromosome was either attenuated or absent. However, maps of murine brain cells (19) exhibited clear compartmentalization on the Xi chromosome (SI Appendix, Fig. S12). Thus, compartmentalization of the Xi chromosome is present in some, but not all, murine cell types.
In situ Hi-C in female fibroblasts derived from rhesus macaque (AG08312; Coriell Institute) similarly revealed superdomains (Fig. 1A) with their boundary at the macaque DXZ4 locus (chrX: 114.2–114.3 Mb, rheMac2) (21) as well as a prominent superloop linking the macaque orthologs of DXZ4 and FIRRE (Fig. 1B).
Taken together, the contact maps of human, rhesus macaque, and mouse suggest that the Xi chromosome superstructure, including both superdomains and superloops, is widely conserved across eutherian mammals. Because DXZ4’s genomic context differs greatly in mouse and human, these findings support a functional role for DXZ4 in determining the placement of superloop anchors and of the superdomain boundary.
Mapping Contacts Among Three or More Loci Demonstrates That Superloops Tend to Occur Simultaneously, Leading to a Hub on the Xi Chromosome.
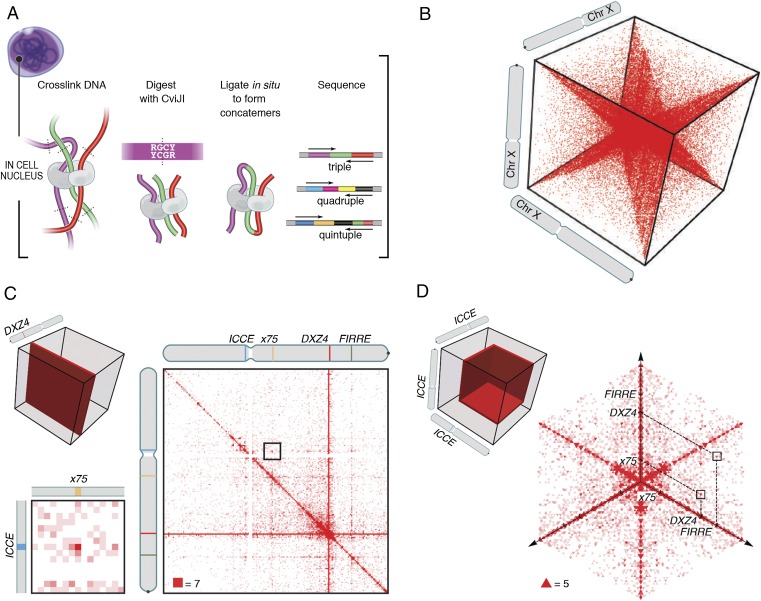
Second, we sought to characterize the structure formed by DXZ4 and other superloop anchors in greater detail. Specifically, we wondered whether superloops tend to occur simultaneously in the same cell, which would suggest the existence of a spatial hub on the Xi chromosome at which all superloop anchors colocate, or whether they occur in different cells. To explore this question, we re-examined our recently published Hi-C dataset, specifically the 21 billion reads that were derived from female human cell lines (10; see also ref. 22). Although most Hi-C reads bring together two nearby fragments, the procedure sometimes can result in the ligation of three or more nearby fragments in a nucleus. Such events make it possible to examine higher-order spatial relationships within a cell that cannot be interrogated from pairwise contacts. We searched the Hi-C data for reads containing sequences from three or more genomic loci. Although such reads were rare (∼1 in 2,056), the large size of the dataset enabled us to find more than 10 million unique “triples,” each exhibiting high-quality alignments (MAPQ ≥10) to three widely separated loci (all pairwise distances >20 kb) on the same chromosome. The data also contain about 4.6 million unique quadruples and 892 unique quintuples.
Because each triple (A-B-C) implies the proximity of three pairs of loci (A-B, A-C, and B-C), we assessed the quality of the triple contacts by creating matrices of the implied pairwise contacts. These matrices closely resembled matrices derived from ordinary, pairwise Hi-C data (Pearson’s r = 0.96 at 2.5 Mb) (SI Appendix, Fig. S2A), suggesting that the higher-order contacts observed using in situ Hi-C reflect genuine patterns of nuclear proximity.
We also developed a modified in situ Hi-C protocol using a restriction enzyme that digests chromatin into much finer fragments, designed to increase the proportion of reads containing three or more nearby fragments. The procedure, dubbed “COLA” (Concatemer Ligation Assay) uses CviJI, which cleaves RGCY sites between the G and C, producing short (average, 64 bp) blunt-end fragments that are ligated in situ (Fig. 2A). We sequenced 276 million paired-end reads from a COLA library from human GM12878 lymphoblastoid cells. Although the pairwise contact matrix closely resembled the one obtained from ordinary in situ Hi-C experiments (Pearson’s r = 0.82 at 2.5 Mb) (SI Appendix, Fig. S2A), the frequency of triples was 13 times higher (1 in 171). (The frequency was 43-fold higher than in dilution Hi-C experiments.) The COLA maps of GM12878 contributed an additional 2,272,943 unique triples, 251,048 unique quadruples, and 1,447 unique quintuples.
Fig. 2.
Concatemers produced by proximity ligation indicate simultaneous colocation of three or more loci. (A) In COLA, concatemers spanning three or more fragments are created by cutting chromatin using CviJI, followed by DNA–DNA proximity ligation in intact nuclei. (B) Contact triples visualized as a 3D contact tensor. Broad patterns of contacts can be revealed by slicing the tensor and examining the results at low resolution. (C) A planar cut of the contact tensor enables the examination of all triples containing DXZ4. Enlargement: Enrichment is seen in the plane at x75(–DXZ4)–FIRRE. (Resolution: 800 kb.) (D) A different cut makes it possible to study triples in the vicinity of ICCE. Superloop triples (ICCE–)DXZ4–FIRRE and (ICCE–)x75–FIRRE are highlighted. (Resolution: 2 Mb.)
Such higher-order contacts are naturally represented and visualized as an n-dimensional matrix, or tensor. Because the number of entries in this tensor is proportional to the size of the interrogated genome raised to the nth power, the tensor is extremely sparse. Features of chromosome organization manifest inside this tensor as n-dimensional shapes. For instance, the bright diagonal seen in 2D contact matrices naturally manifests as an n-dimensional hyperstar (Fig. 2B and SI Appendix, Fig. S2B).
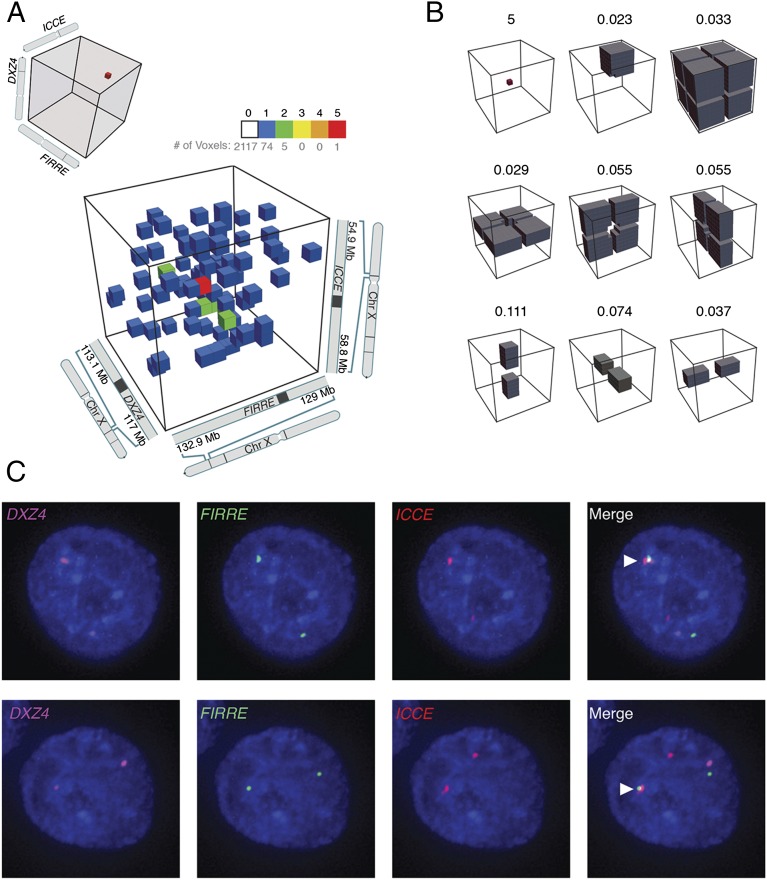
Because the 3D tensor was so sparse, we did not expect to see triples indicating simultaneous occurrence for pairs of ordinary loops, but we reasoned that we might see them for DXZ4–FIRRE and ICCE–DXZ4 superloops, because these anchors are so large (up to 300 kb) and thus produce many more fragments. Indeed, we found that the tensor contained a cluster of four triples corresponding to the triad of loci ICCE–DXZ4–FIRRE. (Because all three loci are tandem repeats, we included triples that did not align uniquely so long as all the possible alignments fell in the same locus.) Given the extreme sparsity of the contact tensor, the probability of even a single triple occurring at these three loci at random is <2% (based on locus size and pairwise distance between loci). The presence of four triples represents an enrichment of 300-fold over random expectation and is extremely unlikely (P = 1.4 × 10−9) (Fig. 2 C and D). The ICCE–DXZ4–FIRRE voxel was also very strongly enriched with respect to its local 3D neighborhood (Fig. 3 A and B and SI Appendix, Fig. S3). Similarly, we noticed a cluster of four triples at the triad ICCE–x75–DXZ4 (a 154-fold enrichment, P = 1.8 × 10−8) (Fig. 2 C and D and SI Appendix, Fig. S4).
Fig. 3.
The ICCE–DXZ4 and DXZ4–FIRRE superloops tend to occur simultaneously, forming a hub on the Xi chromosome. (A) Examination of a small region from the 3D contact tensor of the X chromosome, centered on ICCE, DXZ4, and FIRRE, reveals a peak relative to the local neighborhood. Five contacts are seen in the (300 kb)3 voxel (i.e., 3D pixel) associated with simultaneous colocation of all three loci. There are more than 2,000 other (300 kb)3 voxels in the region shown; the number of contacts in each is indicated by the color. Of these voxels, five contain two contacts each, 74 contain one contact each, and more than 2,000 voxels contain no contacts. Note that because of the fixed bin width, the voxel size presented in this figure, (300 kb)3, is slightly larger than (and has slightly more contacts than) the exact volume defined by ICCE–DXZ4–FIRRE boundaries, which was used for the analyses in the main text. (B) The average frequency of contact in various local neighborhoods surrounding the ICCE–DXZ4–FIRRE peak. The peak is strongly enriched with respect to every model. (C) Representative examples of direct-labeled three-color DNA FISH in GM12878, a female cell line, showing collocation of ICCE–DXZ4–FIRRE. FISH signals overlay DAPI (blue) and are merged in the panel at the far right. The white arrowhead indicates a three-way overlap on one X chromosome.
To obtain independent evidence that DXZ4, FIRRE, and ICCE simultaneously colocalize on the Xi chromosome, we used 3D-FISH. When we examined X chromosomes in male lymphoblastoid cells (GM06992), we did not find a single case in which all three loci colocalized (out of 219 chromosomes examined). For X chromosomes in the female GM12878 cell line, we observed colocalization of all three loci on 17 of 222 X chromosomes (7.7%; P ≤ 0.0001, Fisher’s exact test), always on only one of the two alleles (SI Appendix, Table S2). Because females contain both Xa and Xi chromosomes, these FISH data suggest that all three loci overlap >15% of the time on the Xi chromosome (vs. 0% on the Xa chromosome) (Fig. 3C). Because typical pairs of nearby loci (<1 Mb) in the same loop tend to overlap less than 25% of the time when probed using 3D-FISH (10), an observed overlap frequency of >15% among three loci spread across a chromosome is extremely high.
Taken together, these findings suggest that the DXZ4, FIRRE, and ICCE loci on the Xi chromosome tend to colocate at a single spatial position, i.e., that they form a hub.
Xi Chromosome Subcompartments Are Closely Associated with Heterochromatin Type.
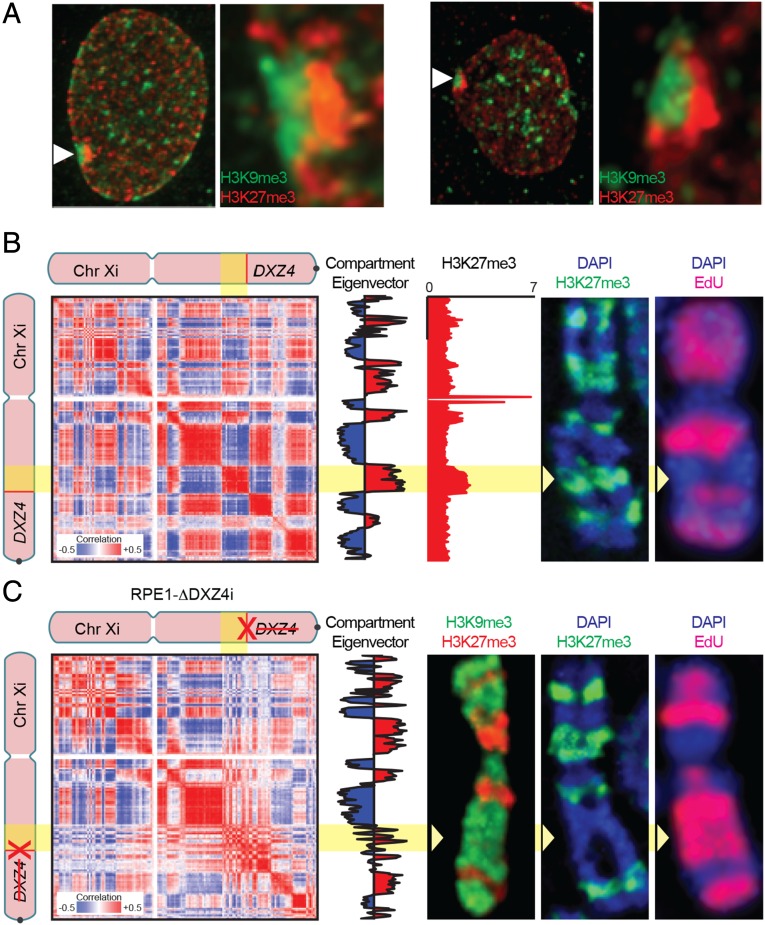
Third, we sought to study the relation between compartment structure and histone modifications on the Xi chromosome. For this purpose, we chose to use RPE1, a diploid retinal pigment epithelial cell line derived from a human female, because previous studies have characterized the distribution of histone modifications in this cell line. In particular, the Xi chromosome in RPE1 cells has been shown to have alternating multimegabase intervals marked by either histone H3 lysine 9 trimethylation (H3K9me3) or histone H3 lysine 27 trimethylation (H3K27me3); these large intervals are visible as distinct bands during metaphase, and the two types of intervals tend to colocalize in the nucleus during interphase (23). We confirmed these results by performing immunofluorescence using antibodies for H3K9me3 and H3K27me3 during both interphase (Fig. 4A) and metaphase (Fig. 4B). To map precisely the genomic intervals decorated by each mark during interphase, we examined RPE1 ChIP-Seq data for H3K27me3 (Fig. 4B) (24). We observed a correspondence between the ChIP-Seq signal seen during interphase and the immunofluorescence pattern seen during metaphase, supporting the suggestion that the extent of each mark does not change significantly between the two states (25).
Fig. 4.
Deletion of DX4 disrupts compartmentalization, distribution of histone marks, and replication timing. (A) Immunofluorescence showing the distribution of H3K9me3 (green, indirect immunofluorescence) and H3K27me3 (red, direct immunofluorescence) in wild-type RPE1 cells at interphase. White arrowheads indicate the location of the Xi chromosome that is expanded in the panels to the right, showing the corresponding nuclei. (B) A correlation matrix derived from the contact map of the wild-type RPE1 Xi chromosome reveals two distinct long-range contact patterns, indicating two subcompartments (first column, numbered left to right). These patterns are reflected in its principal eigenvector, which is shown to the right of the matrix (second column). The color of the eigenvector indicates its sign and thus the long-range pattern exhibited by the corresponding locus (Resolution: 500 kb.) One of these subcompartments correlates well with H3K27me3 ChIP-Seq in RPE1 (third column), as well as with a representative metaphase Xi chromosome showing the distribution of H3K27me3 (green, indirect immunofluorescence) merged with DAPI (blue) in RPE1 (fourth column), and with a representative metaphase Xi chromosome showing the pattern of EdU incorporation (red) merged with DAPI (blue) (fifth column). The yellow arrowheads indicate the H3K27me3 band between x100 and DXZ4 that replicates earlier in S-phase. (C) A correlation matrix derived from the contact map of the RPE1-ΔDXZ4i Xi chromosome (first column); its principal eigenvector (second column); a representative RPE1-ΔDXZ4i metaphase Xi chromosome showing the distribution of H3K9me3 (green, indirect immunofluorescence) and H3K27me3 (red, direct immunofluorescence) (third column); a representative metaphase Xi chromosome showing the distribution of H3K27me3 (green, indirect immunofluorescence) merged with DAPI (blue) (fourth column); and a representative metaphase Xi chromosome showing the pattern of EdU incorporation (red) merged with DAPI (blue) (fifth column). The compartment interval between x100 and DX74 is compromised, and corresponding changes are seen in metaphase histone mark distribution and in replication timing.
We then used in situ Hi-C to create a diploid contact map of the X chromosome in RPE1 cells. As in our previous high-resolution in situ Hi-C maps from human females, the Hi-C map exhibited superdomains and superloops on the Xi chromosome but not on the Xa chromosome. Like our previous diploid human map, and unlike our diploid murine map of Patski cells, the Xi chromosome in RPE1 cells also exhibited extensive compartmentalization. Compartmentalization patterns vary among cell lines, but we observed that RPE1 cells had several long compartment intervals on the Xi chromosome that were not present on the Xa chromosome, notably a contiguous 15-Mb stretch from a locus at 100 Mb (denoted “x100”) all the way to DXZ4 (Fig. 4B). Strikingly, the compartment intervals on the Xi chromosome, reflected in the first principal component of the observed/expected matrix, correspond closely to the alternating intervals of H3K27me3 and H3K9me3 on the Xi chromosome seen by ChIP-Seq and by immunofluorescence (Fig. 4B and SI Appendix, Fig. S5A). For example, the 15-Mb region between x100 and DXZ4 corresponded to a contiguous H3K27me3 interval in all 151 Xi chromosomes examined (Fig. 4B and SI Appendix, Fig. S5A) (25).
Loss of DXZ4 on the Xi Chromosome Disrupts Superloops, Eliminates Superdomains, and Influences Heterochromatin Identity.
Fourth, we explored the effect of deleting DXZ4 on the structures characterized above. We designed a pair of transcription activator-like effector nucleases (TALENs) (a class of nucleases with a customizable DNA-binding motif) that target two unique inverted repeats flanking the DXZ4 locus (SI Appendix, Fig. S6A). Simultaneous cutting at both ends of the locus resulted in loss of the intervening DNA, including DXZ4 (SI Appendix, Fig. S6B). We isolated clones of RPE1 cells that lacked DXZ4 on either the Xi (RPE1-ΔDXZ4i) or Xa (RPE1-ΔDXZ4a) chromosome (SI Appendix, Fig. S7). We defined the precise extent of the deletions by sequencing the deletion product (SI Appendix Fig. S8). [We created additional RPE1-ΔDXZ4i clones using a dual-guide, CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats-CRISPR associated 9)-based genome-editing strategy (SI Appendix, Fig. S6C).]
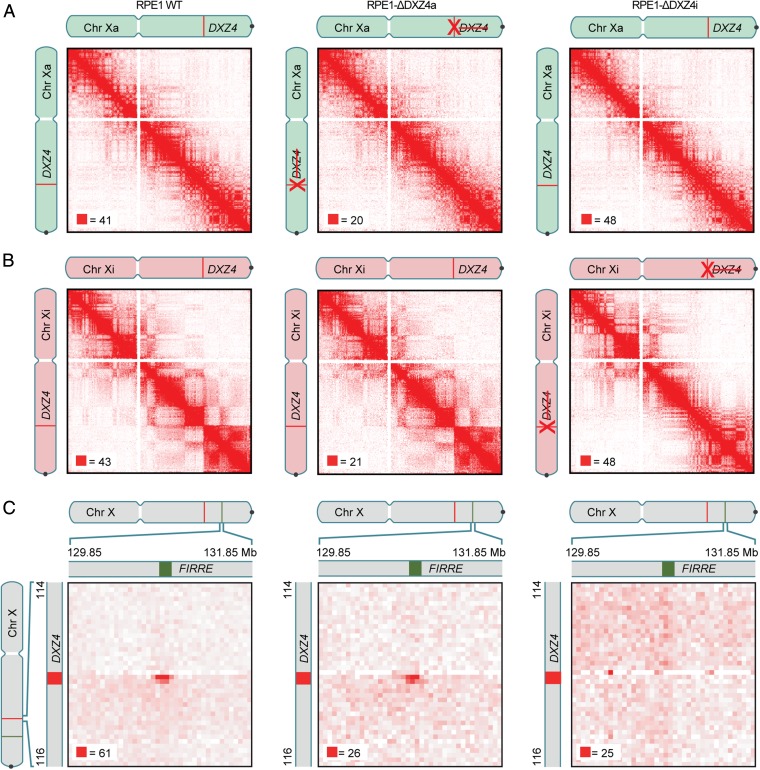
We then used in situ Hi-C to create diploid contact maps of both mutant cell lines (RPE1-ΔDXZ4i and RPE1-ΔDXZ4a) and compared these maps to the maps of wild-type RPE1 cells generated above (Fig. 5 A and B). RPE1-ΔDXZ4a mutants exhibited the same superdomains and superloops on the Xi chromosome seen in wild-type RPE1 cells. In contrast, the Xi chromosome in RPE1-ΔDXZ4i mutants lacked both the superdomains and the superloops anchored at DXZ4 (Fig. 5 B and C). Notably, RPE1-ΔDXZ4i continued to exhibit superloops not anchored at DXZ4 (SI Appendix, Fig. S9), suggesting that superloops form independently of one another; we confirmed this observation by using FISH to probe the FIRRE–ICCE superloop [anchor overlap frequency in wild-type RPE1 cells: 7 of 114 Xi chromosomes (6%); in RPE1-ΔDXZ4a: 7 of 107 chromosomes (7%); and in RPE1-ΔDXZ4i: 2 of 102 chromosomes (2%)].
Fig. 5.
Deletion of DXZ4 eliminates the Xi chromosome superstructure. (A) Maps of the Xa chromosome in RPE1 (Left), RPE1-ΔDXZ4a (Center), and RPE1-ΔDXZ4i (Right) cells. Compartmentalization is seen. Superstructure is absent. The chromosome icons are colored gray to indicate unphased maps of the X chromosome, in which data from both Xi and Xa chromosomes are superimposed; they are colored red to indicate diploid Xi-only maps; and they are colored green to indicate diploid Xa-only maps. The phased SNP calls used to generate homolog-specific maps are outlined in the SI Appendix. (B) Maps of the Xi chromosome in RPE1 (Left), RPE1-ΔDXZ4a (Center), and RPE1-ΔDXZ4i (Right) cells exhibit compartmentalization with unusually long compartment intervals. Superdomains are present in wild-type RPE1 cells and remain after DXZ4 is deleted on the Xa chromosome but not after DXZ4 is deleted on the Xi chromosome. (Resolution: 500 kb.) (C) The DXZ4–FIRRE superloop is present in wild-type RPE1 (Left) and in RPE1-ΔDXZ4a (Center) cells but disappears in RPE1-ΔDXZ4i cells (Right). (Resolution: 50 kb.)
When we examined the large contiguous compartment interval between x100 and DXZ4 that is present on the Xi chromosome in wild-type RPE1 cells, we found that deletion of DXZ4 on the Xi chromosome (but not on the Xa chromosome) disrupted the interval’s compartmentalization pattern, histone modifications, and replication timing. The single-compartment interval was replaced by small alternating compartment intervals (Fig. 4C). Consistent with the observation that changes in compartmentalization are associated with changes in histone modification patterns (10, 16), the contiguous interval of H3K27me3 across this interval was either significantly diminished or completely absent in three independent RPE1-ΔDXZ4i clones (clone 1 frequency: 29.1%, n = 148; clone 2 frequency: 23.8%, n = 172; clone 3 frequency: 1.0%, n = 97; P = <0.0001 Fisher’s exact test) (Fig. 4B and SI Appendix, Fig. S5B), and instead the interval was decorated by H3K9me3 (Fig. 4B and SI Appendix, Fig. S5C). By using 5-ethynyl-2′-deoxyuridine (EdU) labeling, we found that DNA in this interval also replicates much later in RPE1-ΔDXZ4i cells than in the wild-type RPE1 cells (Fig. 4 B and C and SI Appendix, Fig. S5D). What accounts for the variability between these independent clones is unclear. It is not caused by the extent of the deletion or the mode of targeting, because precisely the same DNA sequence is removed in clones 2 and clone 3 using CRISPR-Cas9 (SI Appendix, Figs. S6 and S8). Although highly speculative, it is possible that the point during the cell cycle at which the deletion occurred could impact maintenance of heterochromatin, or chromatin band variability could simply reflect cloning of cells from cell populations that show variation in Xi heterochromatin (25).
Loss of DXZ4 on the Xi Chromosome Does Not Have a Major Effect on Xi Transcription.
In contrast to these findings, we observed that loss of DXZ4 has no major effect on overall transcription of the Xi chromosome. RNA-Seq showed expression changes exceeding twofold in only 10 of 982 genes after false-discovery rate correction (SI Appendix, Table S3). The altered genes had no obvious significance. Similarly, RNA-FISH of 12 genes on the Xi chromosome revealed no consistent changes across RPE1-ΔDXZ4i clones (SI Appendix, Fig. S10 and Table S4).
A Model in Which CTCF and Cohesin Extrude Chromatin Can Explain the Formation of Superloops and Superdomains.
The results described above show that DXZ4 plays an important role in the nuclear architecture of the Xi chromosome. A key question is how DXZ4 brings about the presence of superloops and superdomains on the Xi chromosome. The DXZ4 locus typically comprises between 10 and 100 copies of a repeat element containing a CTCF motif (14). These CTCF motifs are the only sequences in DXZ4 showing evidence of strong purifying selection (18). DXZ4 on the Xi chromosome is known to be hypomethylated and to bind CTCF, whereas on the Xa chromosome it is hypermethylated and does not bind CTCF (14).
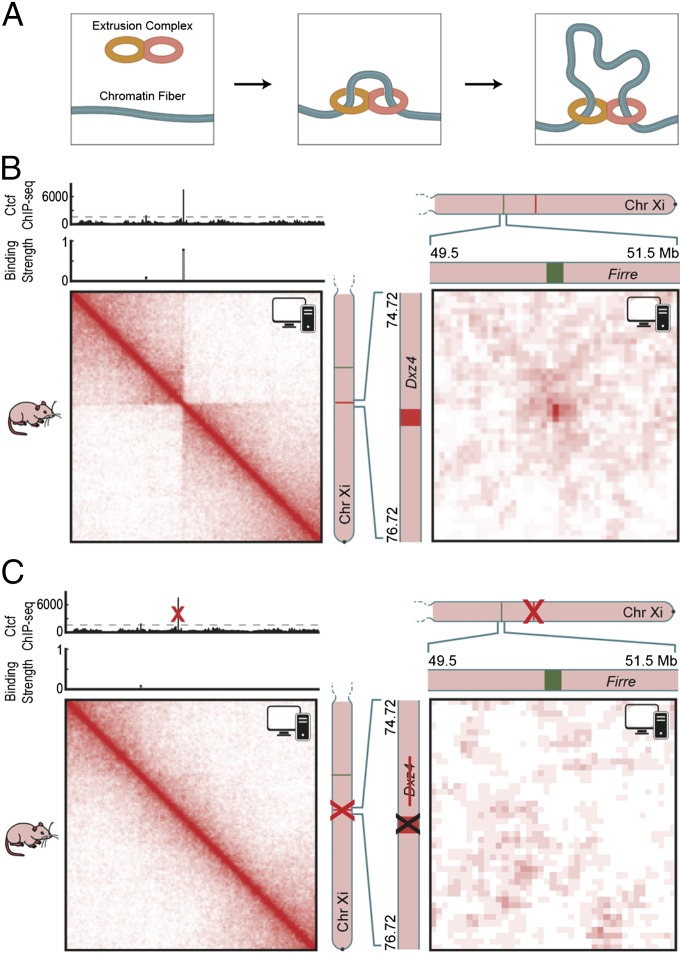
Recently, we proposed a model in which CTCF and the ring-shaped cohesin complex mediate the formation of chromatin loops and domains via extrusion (26; also see refs. 27–29). In this model, an extrusion complex comprising two DNA-binding subunits is loaded onto chromatin at a single genomic position. The subunits slide along chromatin in opposite directions, thereby extruding a loop. Each subunit stops when it arrives at a CTCF site in the appropriate orientation, which serves as an “anchor” (Fig. 6A). When both subunits have stopped, a “persistent loop,” i.e., a loop that is present for a protracted period, is formed between the anchor sites. Simulations showed that these extruded loops naturally give rise to domains and that knowledge of the CTCF-bound sites is sufficient to predict, with reasonable accuracy, the results of Hi-C experiments.
Fig. 6.
(A) In our physical model of loop formation by extrusion (26), an extrusion complex comprising two DNA-binding subunits is loaded onto chromatin. The subunits form a loop by sliding in opposite directions. When they arrive at an anchor site, they have a probability of binding and thus halting the extrusion process. (B) We generated an ensemble of Patski Xi chromosome configurations using the extrusion model, with anchor-binding probabilities drawn from Ctcf ChIP-Seq data in Patski. We calculated a contact map from this ensemble. (Left) A superdomain boundary is formed at Dxz4. (Right) A superloop forms between Dxz4 and Firre. The icon in the upper right corner of each map indicates that these maps were generated in silico, using physical simulations, rather than via experiment. (C) Simulating the deletion of Dxz4 leads to the disappearance of the superdomain (Left) and superloop (Right).
Although the model above assumed that extrusion complexes stop completely at appropriately oriented CTCF sites, similar results are obtained if the complexes undergo partial arrest for a period of time (SI Appendix, Fig. S11). If so, the model also would account for the presence of superloops and superdomains at loci on the Xi chromosome that contain a large number of CTCF-binding sites: Extrusion complexes would be sequestered at such loci for a much longer period than at typical CTCF sites. In contrast, these structures would not be seen on the Xa chromosome, where DXZ4 is packaged into constitutive heterochromatin (14) and does not bind CTCF and therefore would not arrest the extrusion complex.
To test this hypothesis, we modeled the mouse Xi chromosome as a long homopolymer containing CTCF-binding sites, whose positions were derived from Patski Ctcf ChIP-Seq data (30). We used molecular dynamics simulations to examine the behavior of this polymer in a solvent containing extrusion complexes and used the resulting ensemble to generate a contact map for the Xi chromosome (Fig. 6B). We found that the contact map was partitioned into two superdomains whose boundary lay at Dxz4. It also clearly exhibited the Dxz4–Firre superloop. When we simulated the effects of deleting Dxz4, these features disappeared (Fig. 6C). Thus, Hi-C data for the Xi chromosome are consistent with a model in which DXZ4 brings about the formation of superdomains and superloops by serving as a tandem anchor during loop extrusion.
Discussion
We find that the position of DXZ4 with respect to superloop anchors and the superdomain boundary on the Xi chromosome is broadly conserved across human, rhesus macaque, and mouse; that DXZ4, FIRRE, ICCE, and possibly other loci simultaneously colocate to form a superhub comprising superloop anchors on the Xi chromosome; and that the deletion of DXZ4 broadly disrupts the chromatin structure of the Xi chromosome by eliminating superdomains, disrupting superloops anchored at DXZ4, and modifying the distribution of chromatin marks and compartments.
Note that, consistent with our findings, a simultaneous study from another group has shown that deletion of Dxz4 from the mouse Xi also results in loss of superdomain structure (31).
Interestingly, when we deleted DXZ4, we did not observe a pronounced effect on either silencing or escape, perhaps because we deleted DXZ4 after the induction of XCI. It is possible that a different transcriptional phenotype would be observed if DXZ4 were deleted before the induction of XCI. At the same time, the broad changes in replication timing and chromatin marks are both consistent with a transition to constitutive heterochromatin, suggesting that DXZ4 strongly influences the chromatin environment on the Xi chromosome. Given the influence of H3K9me3 on the reprogramming of somatic cells to pluripotency (32), it is conceivable that DXZ4 may facilitate that process during X chromosome reactivation (33–35).
Finally, we show that the behavior of DXZ4, which contains numerous CTCF-binding sites, resembles the behavior of individual CTCF sites but produces features—loops, domains, histone mark patterns, and compartmentalization—at the whole-chromosome scale. Despite the larger feature size, the observations appear to be consistent with extrusion as an underlying mechanism, suggesting that genomes may exhibit similar folding principles across a wide range of scales.
Materials and Methods
For the COLA protocol, five million cells were cross-linked for 10 min with 1% formaldehyde. Nuclei were permeabilized. DNA was digested with CviJI in CviJI reaction buffer and ligated with T4 DNA ligase. The library was enriched for ligation products after proximity ligation using size selection on an agarose gel. The library was prepared for sequencing with the Illumina platform. The full protocol can be found in SI Appendix, COLA Protocol.
Detailed descriptions of all procedures, including assembly of TALENs and CRISPR/Cas9 constructs, isolation and characterization of deletion mutants, the in situ Hi-C procedure and analysis pipeline, including that of higher-order contacts, DNA and RNA FISH, RNA-Seq, immunofluorescence, and details on extrusion simulations can be found in SI Appendix, Supplementary Materials and Methods, as can the sources for all reagents and antibodies.
Supplementary Material
Acknowledgments
We thank C. Schatschneider for discussions regarding statistical tests; I. Bochkov for discussions and assistance with design; R. Didier for assistance with flow cytometry; Cheng-Zhong Zhang and David Pellman for access to RPE1 single-cell sequencing data; H. F. Willard for providing the RPE1 Xa somatic cell hybrid; Christine Disteche for providing the Patski cell line; Feng Zhang for use of pX458; and Sigrid Knemeyer for assistance with the design and preparation of figures. The Center for Genome Architecture thanks Janice, Robert, and Cary McNair for support. This work was supported by National Institute of General Medical Science Grant R01GM073120 (to B.P.C.); a National Science Foundation (NSF) Graduate Research Fellowship under Grants DGE0946799 and DGE1144152 (to M.H.H.); NIH New Innovator Award 1DP2OD008540-01, NIH 4D Nucleome Grant U01HL130010, NSF Physics Frontier Center Grant PHY-1427654, National Human Genome Research Institute (NHGRI) Center for Excellence for Genomic Sciences Grant HG006193, an NVIDIA Research Center Award, an IBM University Challenge Award, a Google Research Award, Cancer Prevention Research Institute of Texas Scholar Award R1304, a McNair Medical Institute Scholar Award, and the President’s Early Career Award in Science and Engineering (to E.L.A.); and NHGRI Grant HG003067 (to E.S.L.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The Hi-C and RNA-Seq sequence data reported in this paper have been deposited at the GEO database, www.ncbi.nlm.nih.gov/geo/ (accession no. GSE71831).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609643113/-/DCSupplemental.
References
- 1.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Brockdorff N, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71(3):515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 3.Brown CJ, et al. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 4.Dixon-McDougall T, Brown C. The making of a Barr body: The mosaic of factors that eXIST on the mammalian inactive X chromosome. Biochem Cell Biol. 2016;94(1):56–70. doi: 10.1139/bcb-2015-0016. [DOI] [PubMed] [Google Scholar]
- 5.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 6.Schulz EG, Heard E. Role and control of X chromosome dosage in mammalian development. Curr Opin Genet Dev. 2013;23(2):109–115. doi: 10.1016/j.gde.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163(4148):676–677. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- 8.Naughton C, Sproul D, Hamilton C, Gilbert N. Analysis of active and inactive X chromosome architecture reveals the independent organization of 30 nm and large-scale chromatin structures. Mol Cell. 2010;40(3):397–409. doi: 10.1016/j.molcel.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teller K, et al. A top-down analysis of Xa- and Xi-territories reveals differences of higher order structure at ≥ 20 Mb genomic length scales. Nucleus. 2011;2(5):465–477. doi: 10.4161/nucl.2.5.17862. [DOI] [PubMed] [Google Scholar]
- 10.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giacalone J, Friedes J, Francke U. A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat Genet. 1992;1(2):137–143. doi: 10.1038/ng0592-137. [DOI] [PubMed] [Google Scholar]
- 12.Schaap M, et al. Genome-wide analysis of macrosatellite repeat copy number variation in worldwide populations: Evidence for differences and commonalities in size distributions and size restrictions. BMC Genomics. 2013;14:143. doi: 10.1186/1471-2164-14-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tremblay DC, Moseley S, Chadwick BP. Variation in array size, monomer composition and expression of the macrosatellite DXZ4. PLoS One. 2011;6(4):e18969. doi: 10.1371/journal.pone.0018969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chadwick BP. DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res. 2008;18(8):1259–1269. doi: 10.1101/gr.075713.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horakova AH, Moseley SC, McLaughlin CR, Tremblay DC, Chadwick BP. The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome. Hum Mol Genet. 2012;21(20):4367–4377. doi: 10.1093/hmg/dds270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lingenfelter PA, et al. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat Genet. 1998;18(3):212–213. doi: 10.1038/ng0398-212. [DOI] [PubMed] [Google Scholar]
- 18.Horakova AH, et al. The mouse DXZ4 homolog retains Ctcf binding and proximity to Pls3 despite substantial organizational differences compared to the primate macrosatellite. Genome Biol. 2012;13(8):R70. doi: 10.1186/gb-2012-13-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng X, et al. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 2015;16:152. doi: 10.1186/s13059-015-0728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minajigi A, et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349(6245):aab2276. doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLaughlin CR, Chadwick BP. Characterization of DXZ4 conservation in primates implies important functional roles for CTCF binding, array expression and tandem repeat organization on the X chromosome. Genome Biol. 2011;12(4):R37. doi: 10.1186/gb-2011-12-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ay F, et al. Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C. BMC Genomics. 2015;16:121. doi: 10.1186/s12864-015-1236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chadwick BP, Willard HF. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci USA. 2004;101(50):17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nozawa RS, et al. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat Struct Mol Biol. 2013;20(5):566–573. doi: 10.1038/nsmb.2532. [DOI] [PubMed] [Google Scholar]
- 25.Chadwick BP. Variation in Xi chromatin organization and correlation of the H3K27me3 chromatin territories to transcribed sequences by microarray analysis. Chromosoma. 2007;116(2):147–157. doi: 10.1007/s00412-006-0085-1. [DOI] [PubMed] [Google Scholar]
- 26.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA. 2015;112(47):E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alipour E, Marko JF. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40(22):11202–11212. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fudenberg G, et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15(9):2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nasmyth K. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- 30.Berletch JB, et al. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015;11(3):e1005079. doi: 10.1371/journal.pgen.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heard E, Dekker J, Structural organization of the inactive X chromosome in the mouse. Nature, in press. [DOI] [PMC free article] [PubMed]
- 32.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151(5):994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mak W, et al. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303(5658):666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- 34.Ohhata T, Wutz A. Reactivation of the inactive X chromosome in development and reprogramming. Cell Mol Life Sci. 2013;70(14):2443–2461. doi: 10.1007/s00018-012-1174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303(5658):644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.