Significance
Bunyamwera virus (BUNV) is the prototype of the Orthobunyavirus genus and Bunyaviridae family that contains important human and animal pathogens. The cleavage mechanism of orthobunyavirus glycoprotein precursor (GPC) and the host proteases involved have not been clarified. Here we found that NSm and Gc contain their own internal signal peptides, which mediate the GPC cleavage by host signal peptidase and signal peptide peptidase (SPP). Furthermore, the NSm domain-I plays an important postcleavage role in cell fusion. Our data clarified the implication of host proteases in the processing of the orthobunyavirus GPC. This work identifies SPP as a potential intervention target, and the knowledge we gained will benefit preventive strategies against other orthobunyavirus infections.
Keywords: Bunyavirus, Bunyamwera virus, glycoprotein precursor processing, signal peptidase, signal peptide peptidase
Abstract
The M genome segment of Bunyamwera virus (BUNV)—the prototype of both the Bunyaviridae family and the Orthobunyavirus genus—encodes the glycoprotein precursor (GPC) that is proteolytically cleaved to yield two viral structural glycoproteins, Gn and Gc, and a nonstructural protein, NSm. The cleavage mechanism of orthobunyavirus GPCs and the host proteases involved have not been clarified. In this study, we investigated the processing of BUNV GPC and found that both NSm and Gc proteins were cleaved at their own internal signal peptides (SPs), in which NSm domain I functions as SPNSm and NSm domain V as SPGc. Moreover, the domain I was further processed by a host intramembrane-cleaving protease, signal peptide peptidase, and is required for cell fusion activities. Meanwhile, the NSm domain V (SPGc) remains integral to NSm, rendering the NSm topology as a two-membrane-spanning integral membrane protein. We defined the cleavage sites and boundaries between the processed proteins as follows: Gn, from residue 17–312 or nearby residues; NSm, 332–477; and Gc, 478–1433. Our data clarified the mechanism of the precursor cleavage process, which is important for our understanding of viral glycoprotein biogenesis in the genus Orthobunyavirus and thus presents a useful target for intervention strategies.
The family Bunyaviridae contains >350 named viruses that are classified into the five genera Orthobunyavirus, Hantavirus, Nairovirus, Phlebovirus, and Tospovirus, making it one of the largest families of RNA viruses. Several members of the family are serious human pathogens, such as La Crosse virus and Oropouche virus (OROV) (Orthobunyavirus); Hantaan (HTNV) and Sin Nombre viruses (Hantavirus); Rift Valley fever virus (RVFV) and severe fever with thrombocytopenia syndrome virus (Phlebovirus); and Crimean-Congo hemorrhagic fever virus (CCHFV; Nairovirus) (1, 2). The characteristic features of bunyaviruses include a tripartite single-stranded RNA genome of negative- or ambi-sense polarity, cytoplasmic site of viral replication, and assembly and budding at membranes of the Golgi complex (1–3). Bunyamwera virus (BUNV), the prototype of both the family and the Orthobunyavirus genus, remains an important research model for many pathogens within this family.
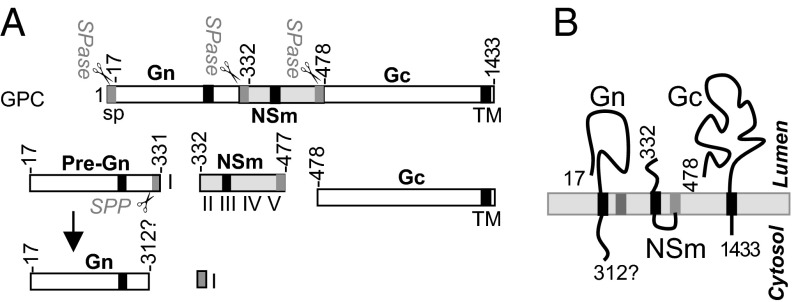
The medium (M) genomic RNA segment of orthobunyaviruses encodes the glycoprotein precursor (GPC; in order Gn-NSm-Gc) that is cotranslationally cleaved to yield the mature viral glycoproteins Gn and Gc and a nonstructural protein, NSm. Gn and Gc form viral spikes that play a crucial role in virus entry (1, 2). Both Gn and Gc are type I integral transmembrane proteins and form a heterodimer in the endoplasmic reticulum (ER) before trafficking to, and retention in, the Golgi compartment, where virus assembly occurs (2, 4, 5). Bunyavirus glycoproteins are fusogenic, and the fusion peptide is located on Gc, a class II fusion protein (6), but cell fusion requires the coexpression of both Gn and Gc glycoproteins (7). NSm, an integral membrane protein, comprises three hydrophobic domains (I, III, and V) and two nonhydrophobic domains (II and IV) (Fig. S1A), and its N-terminal domain (I) is required for BUNV replication (8).
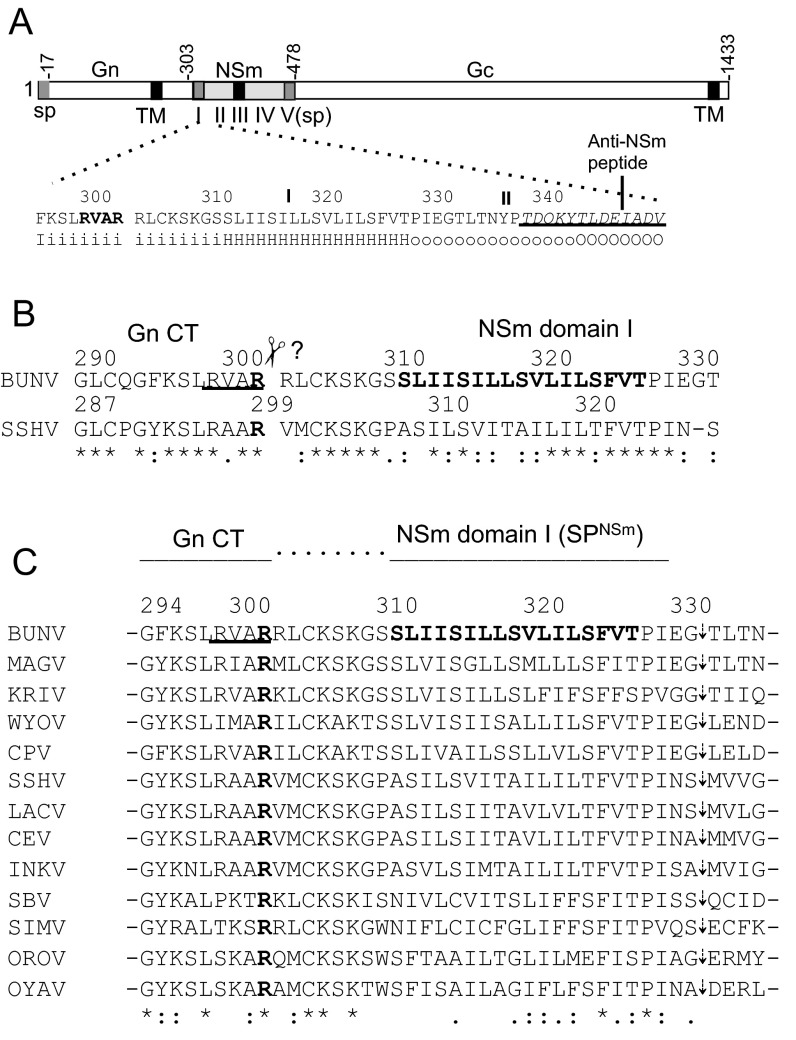
Fig. S1.
Schematic of BUNV GPC and mutations at RVAR motif and amino acid sequence alignment at the BUNV Gn-NSm junction. (A) BUNV GPC with positions of amino acid residues marking the predicted boundaries (Gn, NSm, and Gc) indicated. The amino acid sequence and the secondary structure prediction at the Gn–NSm junction (residues 295–350) are shown below. RVAR motif (residue 299–302) is in bold. The epitope to anti-NSm is underlined. The secondary structure is projected by using the HMMTOP server (www.enzim.hu/hmmtop) with H indicating hydrophobic transmembrane helix, i residue on inside, and O residues outside of transmembrane domains (TM/TMD). The NSm domains (I–V) (8) are shown under the NSm coding region. SP and TM are shown as gray and black boxes, respectively. (B) Alignment between BUNV and SSHV GPCs. The previously predicted last C-terminal residues are arginine (R) at residue 302 for BUNV and residue 299 for SSHV (in bold). (C) Amino acid sequence alignment of 13 orthobunyavirus GPCs (aligned to residues 294–335 of BUNV GPC). The alignment was created by using EMBL-EBI Clustal Omega program (www.ebi.ac.uk/Tools/msa/clustalo). The conserved arginine residues at residue 302 (for BUNV) are in bold. The GenBank accession numbers of orthobunyavirus GPCs used in alignment are as follows: BUNV (P04505), Maguari virus (MAGV; AAQ23639), Kairi virus (KRIV; ACV89517), Wyeomyia virus (WYOV; AGA54137), Cachoeira Porteira virus (CPV; AEZ35284), SSHV (ABX47014), La Crosse virus (LACV; AAB62804), California encephalitis virus (CEV; AAD53039), Inkoo virus (INKV; AAB93841), Schmallenberg virus (SBV; AGC93514), Douglas virus (DOUV; HE795091), Simbu virus (SIMV; YP_006590085), OROV (AGH07923), and Oya virus (OYAV; AGS56983).
Cleavage of BUNV GPC is mediated by host proteases, but the details of which proteases are involved and the precise cleavage sites have not been clarified. Experimental data on GPC processing have only been reported for snowshoe hare orthobunyavirus (SSHV); the C terminus of SSHV Gn was determined by C-terminal amino acid sequencing to be an arginine (R) residue at position 299 (9) (Fig. S1B). Based on alignments of several orthobunyavirus GPC sequences, it was suggested that Gn-NSm cleavage occurs at a similar position to that defined for SSHV (10). This arginine (302R for BUNV) appears conserved in GPCs of all orthobunyaviruses analyzed to date and for most of the viruses lies in the sequence R-V/A-A-R (Fig. S1C), which has been believed to be the site of Gn-NSm cleavage by furin-like proteases (11).
In eukaryotes, most secreted and membrane proteins contain cleavable N-terminal signal peptides (SPs), which are recognized by the signal recognition particles (SRPs) when nascent polypeptide chains emerged from the ribosome at the ER and translocate it into the ER lumen, where they are usually cleaved by cellular signal peptidases (SPases) (12). The embedded peptide remnant is usually subsequently released for degradation by the cellular SP peptidase (SPP) or SPP-like proteases, which belong to the family of intramembrane-cleaving aspartyl proteases (I-CliPs) (13–16). SPP is an ER-resident I-CliP (17) and is implicated in other important biological functions, such as in generating C-terminal peptides for MHC class I presentation (18) and human lymphocyte antigen E epitopes (19). SPP activities also involve the intramembrane cleavage of the core proteins of hepatitis C virus, GB virus B, and classical swine fever virus (family Flaviviridae) (20, 21).
To investigate the cleavage events of orthobunyavirus GPC, we used several approaches, including mutagenesis, virus assays, RNA interference, mass spectrometry (MS), and biological assays. We aimed to determine the cleavage sites between the boundaries of the mature proteins (e.g., Gn and NSm, and NSm and Gc). Our data revealed the implementation of the cellular SPase and SPP in the cleavage of BUNV GPC and clarified the mechanism of orthobunyaviruses GPC cleavage.
Results
Gn-NSm Cleavage Does Not Occur at the RVAR Motif by Furin-Like Proteases.
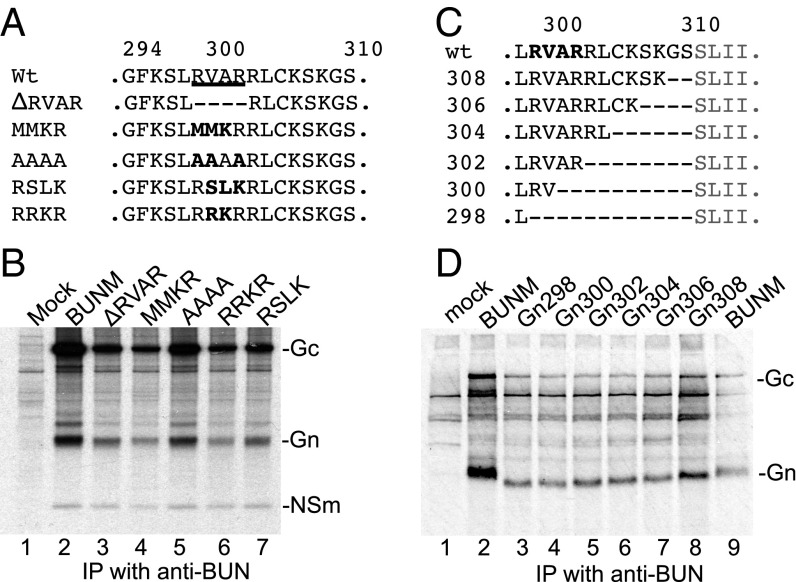
We first investigated whether the Gn-NSm cleavage takes place between residues 302R and 303R at the RVAR motif (Fig. S1). Five mutations were generated at this site in the M expression plasmid pTM1BUNM, including a deletion mutation (ΔRVAR) and four substitution mutations (MMKR, AAAA, RSLK, and RRKR) (Fig. 1A). These plasmids were transfected into BSR-T7/5 cells, and the radiolabeled viral proteins were immunoprecipitated with antiserum against BUNV particles (anti-BUN), followed by SDS/PAGE fractionation. Interestingly, like the wild-type (WT) BUNM control, all mutated GPCs were cleaved into Gn, NSm, and Gc (Fig. 1B), suggesting that this location is not a cleavage site by furin-like proteases. Furthermore, the furin inhibitor I (dec-RVKR-cmk; Calbiochem) had no effect on the BUNV GPC cleavage (Fig. S2A) and yields of virus produced in the presence of the drug over 30 h (Fig. S2B).
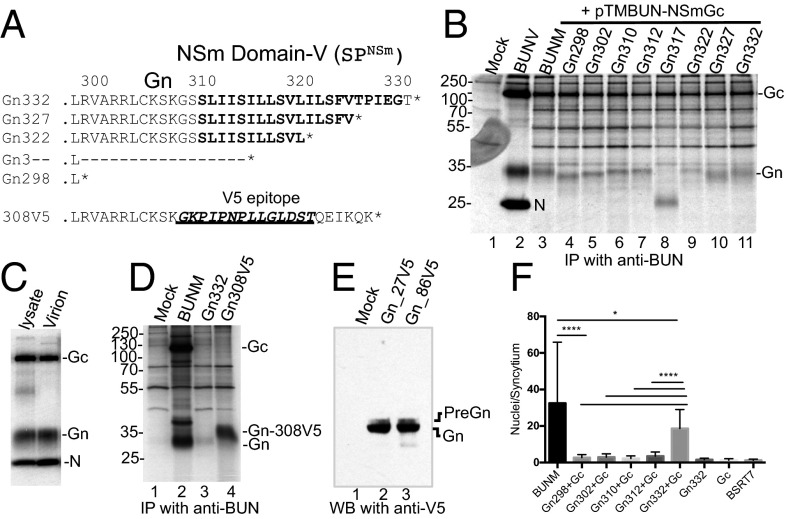
Fig. 1.
Gn-NSm does not cleave at the previously predicted RVAR site. (A) Substitution and internal deletions at RVAR motif. (B) Effect of deletion and substitution mutations on GPC cleavage. (C) Internal deletions between residues 298L and 311S at Gn-NSm junction. (D) The processing of mutant GPCs. Transfected BSRT7/5 cells were radiolabeled with [35S]methionine. The viral proteins were immunoprecipitated with anti-BUN and analyzed by SDS/PAGE. The position of the viral proteins is marked.
Fig. S2.
Effect of Furin inhibitor on BUNV infection. BUNV-infected Vero E6 cells (MOI of 0.01 pfu per cell) were incubated for 30 h in the presence of different concentrations (0, 12.5, 25, 50, and 100 µM) of furin inhibitor I (dec-RVKR-cmk, Calbiochem), and viral replication was evaluated by immunoprecipitation and virus titration. (A) Effect of the furin inhibitor I on GPC cleavage. Lane 2 is mock infected. (B) Yield of virus in the presence of Furin inhibitor I, the concentrations of which are similar to those that inhibit furin-mediated cleavage of the respiratory syncytial virus fusion protein (46).
There exist eight residues between residues 302R and 311S at Gn-NSm junction (Fig. 1C and Fig. S1). To investigate whether these residues harbor the Gn-NSm cleavage site, we constructed six mutants that contain internal progressive deletions between residues 298L and 311S (Fig. 1C). As shown in Fig. 1D, all mutated precursors were properly cleaved. Moreover, the deletions resulted in the increased migration of Gn bands on the gel, with a relative shift corresponding to the number of amino acids removed (lanes 3–8), suggesting that these residues still belong to the Gn cytoplasmic tail (Gn CT) and that the Gn-NSm cleavage must occur at or within NSm domain I.
NSm Domain I Functions as an Internal SP.
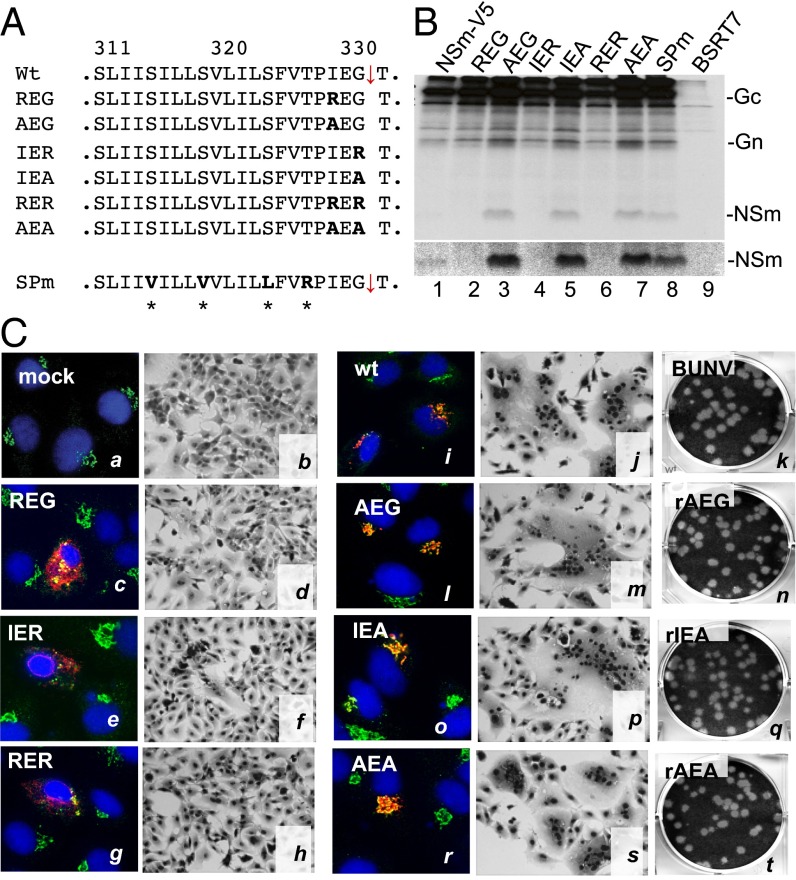
After excluding the Gn-NSm cleavage at motif RVAR302, we speculated that NSm domain I, a type II transmembrane domain (TMD), could function as an internal SP for NSm (SPNSm). By using the SignalP 4.1 server (www.cbs.dtu.dk/services/SignalP) (22), the domain was predicted as a cleavable internal SP that cleaves between residues 331G and 332T (Fig. 2A and Fig. S3A). It has been reported that the residues at the −3 and −1 positions relative to the SPase cleavage site are most critical for cleavage by the cellular SPase complex (23). Therefore, we generated a series of mutant GPCs, including six substitution mutations at the −3 and −1 positions and one substitution mutation in the core region of the domain (SPm) (Fig. 2A). When the residues at either −3 (329I) or −1 (331G), or at both positions, were changed to the charged arginine (R), the NSm protein was not detected (Fig. 2B, lanes 2, 4, and 6). However, substitution with alanine at these positions and substitution mutation within the domain (SPm) did not affect Gn-NSm cleavage (Fig. 2B, lanes 3, 5, 7, and 8) in that NSm was clearly seen. Furthermore, we purified NSm protein from the cells infected with recombinant virus, rBUNNSmV5, in which the V5-epitope was inserted in the NSm cytoplasmic domain (Fig. S3B), for MS analysis and confirmed that residue 332T is the first N-terminal residue of NSm (Fig. S3 C and D).
Fig. 2.
NSm domain-I acts as an internal SPNSm. (A) Mutations at −3 (329I) and −1 (331G) positions of the SPase cleavage site (on pTM1BUNM-NSmV5). (B) Effect of substitution mutations on NSm processing. The radiolabeled viral proteins were immunoprecipitated with anti-V5 antibody. (C) Effect of mutations on the Golgi targeting of the viral glycoproteins, cell fusion, and virus viability. For immunofluorescence assays, the transfected BSRT7/5 cells were stained with a mixture of anti-Gc MAb (M810, in red) and anti-GM130 (in green for the Golgi staining) antibodies and examined by confocal microscopy. Nuclei were stained in blue with DAPI. Cell fusion and virus rescue were performed as described in SI Materials and Methods.
Fig. S3.
MS analysis of BUNV NSm. (A) SP Prediction of NSm domain-I using SignalP 4.1 (www.cbs.dtu.dk/services/SignalP) (22) with cleavage between residues 331G and 332T. (B) The V5-tagged NSm (NSmV5). V5 epitope is inserted in the NSm cytoplasmic domain (IV) between residues 403 and 420. (C) Purification of NSm from cells infected with recombinant rBUNV-NSmV5. The V5-tagged NSm gel band was excised and subject to in-gel digestion and MS analysis. (D) MS/MS fragment pattern for the NSm N-terminal residues.
The effects of the mutations on glycoprotein function were also assessed with regard to the Golgi trafficking and cell fusion activities. Consistent with the above observation, the arginine substitutions at the −3 and −1 positions totally abolished Golgi targeting (Fig. 2C, c, e, and g) and cell fusion (Fig. 2C, d, f, and h), whereas alanine substitution had no effect on either Golgi colocalization (Fig. 2C, l, o, and r) or cell fusion (Fig. 2C, m, p, and s). When these mutations were tested for virus rescue, we were able to generate viruses from M-segment mutants containing alanine substitution mutations (rAEG, rIEA, and rAEA), but not from arginine substitution mutants. The rescued viruses showed similar plaque phenotypes to the WT control (Fig. 2C, n, q, and t). Together, our data confirmed that the NSm domain-I functions as an internal SPNSm.
Mapping the C Termini of Gn Protein.
After SPase cleavage of SPNSm at residue 332T, the SP was still attached to the upstream Gn CT (preGn). To define the Gn end, we constructed a series of individual Gn mutants that terminate between residues 298–332 (Fig. 3A). Consistent with the earlier results from internal deletions, the deletions from residues 312–298 resulted in the linear reduction in the molecular mass of Gn proteins (Fig. 3B, lanes 4–7), and Gn312 is comparable in size with WT Gn (comparing lanes 3 and 7), indicating that Gn likely ends at residue 312 or nearby residues. However, deletions in SPNSm did not cause a linear reduction in Gn molecular masses (Fig. 3B, lanes 8–10). It is noticeable that Gn317, which contains only five remnant hydrophobic residues at the C terminus, produced a smaller band (∼25 kDa), which we believed to be a degradation product (Fig. 3B, lane 8). We also compared the size of the intracellular and virion Gn proteins and found no difference (Fig. 3C), confirming that the intracellular and virion Gn proteins end at same position.
Fig. 3.
Mapping the C termini of Gn protein by mutagenesis. (A) Mutations in the Gn CT and NSm domain I (on pTM1BUN-Gn332). * represents the stop codon. (B) Effect of deletions on the migration shift of Gn protein. (C) Profile of the radiolabeled intracellular and virion proteins. (D) The expression of Gn332 (preGn) and Gn308V5. (E) WB analysis of V5-tagged Gn332 at residue 27 or 86. (F) Cell fusion assay on BSRT7/5 cells cotransfected with pTM1BUN-NSmGc (Gc) and one of the Gn mutants. *P < 0.05; ****P < 0.0001.
To confirm the subsequent processing of SPNSm upon SPase cleavage, we constructed a mutated Gn (Gn308V5) with domain I replaced with nonhydrophobic V5 epitope and a further six residues from the Gc CT (QEIKQK) (Fig. 3A). It is worth mentioning that the unprocessed Gn332 (preGn) would be similar in size to Gn308V5 (35.33 vs. 35.12 kDa). As anticipated, Gn308V5 ran at a higher molecular mass than WT Gn and the processed Gn332 (Fig. 3D), indicating that SPNSm was further processed from preGn. The preGn was visible by Western blot (WB) analysis of V5-tagged Gn proteins (Gn-27V5 and -86V5) as a faint band above the predominant processed Gn protein (Fig. 3E). Cell fusion assays with Gn mutants (in coexpression with Gc) indicated that the extensive syncytia were formed only from cells coexpressing Gc and Gn332 (PreGn), and any deletions in SPNSm diminished the cell fusion (Fig. 3F), suggesting that the liberated SPNSm is required in cell fusion.
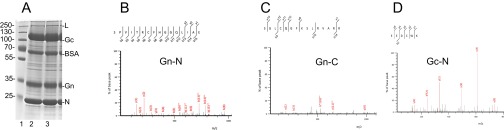
MS analysis of virion Gn protein identified 17S as the first residue of the processed Gn protein (Fig. S4B), but was unable to confirm the Gn end, with the furthest C-terminal residue mapped to 303R (Fig. S4C and Table S1), similar to the previously determined SSHV Gn end (9). It is probably because of the fact that the newly identified Gn C-terminal residues (303–312) are rich in positively charged arginine and lysine residues being targets by trypsin-like proteases (24). The terminal residues identified by MS are summarized in Table S1.
Fig. S4.
MS analysis of BUNV Gn and Gc. (A) Coomassie Blue-stained viral proteins of purified virus particles. (B–D) MS/MS fragment patterns for the terminal residues at Gn N terminus (B), Gn C-terminal region (C), and Gc N terminus (D).
Table S1.
Summary of N- and C-terminal amino acids of Gn, NSm, and Gc proteins identified by MS
| Protein | Enzyme | Amino acid sequence and position |
| Gn N-terminal | GluC + PMSF | 17SPVITRCFHGGQLIAE |
| Gn C-terminal | GluC + PMSF | SGLCQGFK 296 |
| SGLCQGFKS297 | ||
| SGLCQGFKSLR299 | ||
| SGLCQGFKSLRVAR302 | ||
| SGLCQGFKSLRVARR303 | ||
| AspN | DRMRMHRESGL291 | |
| Tryp | ESGLCQGFK296 | |
| ESGLCQGFKSLR299 | ||
| NSm N-terminal | GluC | 332TLTNYPTDQK |
| NSm C-terminal | GluC | CGFCTCGLLEDPEGVVVHK441 |
| Gc N-terminal | Tryp | 478EEDCWK |
| Gc C-terminal | Tryp | LYLQEIK1431 |
GluC, Glutamyl endopeptidase; AspN, Peptidyl-Asp metalloendopeptidase; Tryp, trypsin.
NSm Domain V Functions as an Internal Noncleavable SPGc.
Our previous study suggested that NSm domain-V functions as a SPGc (8). Indeed, the deletion of the domain abolished Gc processing because no Gc protein was detected from the mutated BUNV GPCs that contained whole or partial internal deletion in the domain (Fig. 4A, lanes 4–6). To study whether the domain undergoes any further processing as SPNSm, we compared the size of C-terminal truncated NSm proteins (based on pTmBUNGn-NSmV5) (Fig. 4B). The deletions resulted in the increased migration shift of NSm bands (Fig. 4C, lanes 4–8). NSm476 that contains the intact domain V was identical in size with the parental NSmV5 (comparing lanes 9 and 10), indicating that domain V is not further cleaved. Furthermore, no size change was noticeable when the domain V was swapped by either nonhydrophobic residues from EGFP (residues 1–20) or hydrophobic SP of HTNV (strain 76–118) Gn protein (SPHGn, residues 1–19) (Fig. 4D), whereas NSm456 that lacks the domain V was obviously smaller (Fig. 4D, lane 6), providing corroborating evidence that SPGc remains integral to the mature NSm. This finding renders the topology of mature NSm as a two-membrane-spanning protein (residues 332–477) that consists of the ectodomain (II), TMD (III), endoplasmic loop (IV), and C-terminal type-II TMD (V) (Fig. 4E).
Fig. 4.
NSm domain V functions as a noncleavable SPGc. (A) Deletion of domain V abolished the Gc processing. (B) Mutations at NSm domain-V. * represents the stop codon. (C) Effect of deletions on the migration shift of the V5-tagged NSm protein. (D) WB analysis of V5-tagged NSm and its mutants. (E) The revised topology of NSm protein.
Requirement of SPNSm for Cell Fusion and Virus Replication.
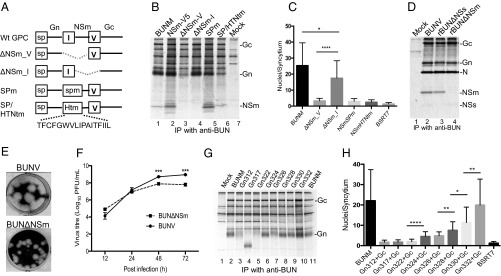
To further investigate the role of SPNSm in virus replication, we made four mutant GPC constructs (Fig. 5A) and compared the impact of mutations in promoting cell fusion and virus viability. All four mutated GPCs were cleaved into Gn and Gc, and also NSm, from the BUNM-SPmNSm and -SPNSm/HTNtm (Fig. 5B). A cell fusion assay revealed that only the GPC-∆NSm_I, which contained deletion of whole mature NSm (residues 332–477) but retained SPNSm for Gc translocation, produced extensive syncytial formation (Fig. 5C). For the other three GPC mutants, the domain V, SPmNSm, and SPNSm/HTNtm functioned properly as SPGc for GPC processing, but these mutated GPCs failed to cause cell fusion. Consistently, from GPC-∆NSm_I we rescued a recombinant virus (rBUN∆NSm) that did not express NSm protein (Fig. 5D, lane 4). The rBUN∆NSm was attenuated, evidenced by smaller plaque phenotype (Fig. 5E) and reduced virus yield at late infection (Fig. 5F). Because BUNV NSm is one of the key factors in the Golgi-associated “virus factory,” the depletion of the protein likely affects the virus assembly (8, 25, 26). The same strategy was used to create a viable recombinant OROV lacking NSm (27).
Fig. 5.
Requirement of SPNSm for GPC processing, cell fusion, and virus replication. (A) Schematic showing either deletion of NSm or mutations in SPNSm. (B) Effect of mutations on the GPC cleavage. (C) Cell fusion on BSRT7/5 cells transfected with BUNM mutants. (D–F) The protein profile (D), plaque phenotype (E), and growth curve (F) of the recombinant virus lacking NSm (rBUN∆NSm). (G) Effect of deletion mutations in SPNSm on the migration shift of Gn protein. (H) Cell fusion on BSRT7/5 cells cotransfected with pTM1BUN-NSmGc and one of the Gn mutants. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To characterize further SPNSm, we assessed the effect of the deletion mutations in the domain on GPC processing and cell fusion. As we mentioned early, deletions in SPNSm did not lead to a linear reduction in the molecular mass of Gn proteins (Fig. 5G). The Gn bands from Gn324 to Gn332 are comparable in size with WT Gn (Fig. 5G, lanes 6–11), but further deletion toward Gn312 (in the case of Gn317 and Gn322) would interrupt the proper processing of Gn protein (Fig. 5G, lanes 4 and 5), suggesting that the processing requires the stable SP TMD structure. When these mutants were tested in cell fusion, we noticed that even removal of two residues from SPNSm (Gn330) had a significant impact on syncytia formation and that further deletions diminished the extent of cell fusion (Fig. 5H). The data together indicated that, in addition to the role as SP, the liberated SPNSm has postcleavage function.
Involvement of SPP in the Processing of BUNV GPC.
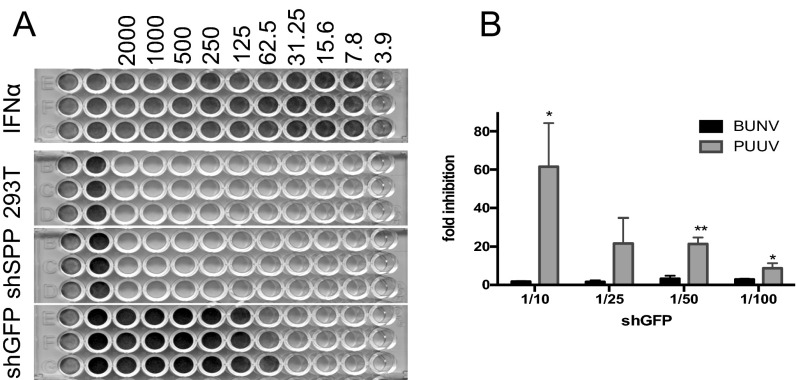
Because SPP is the ER-resident I-Clip, we suspect that SPP is probably involved in the further processing of SPNSm. To address the issue, we generated three lentiviruses that express small hairpin RNAs (shRNAs)—two specific to human SPP mRNA and one to EGFP (shGFP) as negative control. Both shSPP1 and shSPP2 were able to inhibit the SPP expression in the transduced A549 cells (shSPP2 showed a better silencing effect) (Fig. 6A). We then examined the impact of SPP knockdown on BUNV replication after low-multiplicity infection of A549V cells [multiplicity of infection (MOI) of 0.01 pfu per cell]. WB analysis showed that the detection of BUNV N was delayed by 24 h in shSPP-silenced cells compared with shGFP control (Fig. 6B), and the virus titer in SPP-knockdown cells was >10-fold lower than controls across the infection period (Fig. 6C). This finding indicates the likely involvement of SPP in BUNV replication. To investigate whether SPP is implicated in replication of other bunyaviruses, we infected the transduced A549V cells with SBV (Orthobunyavirus genus), RVFV (Phlebovirus genus), and Puumala virus (PUUV; Hantavirus genus), and their N proteins and virus titers were determined by WB and plaque assay. A significant inhibitory effect of SPP silencing was observed for SBV (Fig. 6 D and E). For RVFV, inhibition was noticeable, but to a lesser extent than BUNV and SBV (not statistically significant) (Fig. 6 F and G). Significant inhibition of PUUV replication was observed for PUUV replication in SPP-knockdown cells (Fig. 6 H and I), but PUUV growth was inhibited to a greater extent in shGFP control cells than in SPP-knockdown cells, as evidenced by the N protein detection in SPP-knockdown cells, but not in shGFP control at 72 h postinfection (p.i.) (Fig. 6H, lane 7 at bottom row of long exposure) and significantly lower virus titer in shGFP control (Fig. 6I). This pattern for PUUV is largely due to antiviral activity that we found present in shGFP lentivirus preparation (Fig. S5A), for which PUUV is more sensitive to the inhibitory effect on virus replication than BUNV to the inhibition effect (Fig. S5B).
Fig. 6.
SPP KO affects BUNV infection. (A) SPP knockdown in A549 cells. (B and C) WB analysis (B) and growth kinetics (C) of BUNV infection in shRNA expressing A549V cells. (D and E) Effects of SPP knockdown on SBV infection. (F and G) Effects of SPP knockdown on RVFV infection. (H and I) Effects of SPP knockdown on PUUV infection. The transduced A459V cells were infected with virus (MOI of 0.01). At each time point, the supernatants were harvested for virus titration, and cell lysates were collected for WB. The relevant proteins were probed with antibodies against SPP, tubulin (T), or viral N proteins. (J) Effect of SPP knockdown on BUNV spreading. Transduced Huh7 cells were infected with rBUNGc-EGFP (MOI of 0.01) and incubated until time points 10, 24, and 48 h p.i., as indicated. Cells were stained with anti-GM130 and examined by confocal microscopy. EGFP-tagged Gc and virus particles were shown in green; GM130 were stained in red, and the nuclei were stained in blue with DAPI. *P < 0.05; **P < 0.01; ****P < 0.0001.
Fig. S5.
shGFP lentivirus contains antivirus activity. (A) Biological IFN assay. A549/Npro cells were treated with series-diluted IFN-α (pbI assay science), lentivirus, or cell medium for 24 h and then challenged with EMCV (at MOI of 0.1) for 48 h or until obvious CPE occurred. Cells were fixed and stained with 4% formaldehyde/0.1% crystal violet solution. (B) Comparison of BUNV and PUUV infections responsive to shGFP lentivirus. A549/Npro cells were treated with different dilutions of shGFP lentivirus and infected with either BUNV or PUUV at MOI of 0.1. The result was shown as fold inhibition of virus titers between the induced and noninduced cells. *P < 0.05; **P < 0.01.
The inhibitory effect of SPP knockdown on BUNV infection was also visualized by using a recombinant virus (rBUNGc-EGFP) with EGFP fused to Gc (3). Naïve Huh7 cells and cells expressing shSPP2 were infected with rBUNGc-EGFP (MOI of 0.01), and cells were examined at 10, 24, and 48 h p.i. by fluorescence microscopy. Production of EGFP-tagged Gc—and hence production of progeny virus particles—was observed in naïve cells at 10 h p.i., and virus spread to adjacent cells was clearly evident at 24 h p.i. All cells were infected by 48 h p.i. (Fig. 6J, a–c). In SPP-knockdown cells, the EGFP-tagged Gc was not observed until 24 h p.i., and the spreading to neighboring cells was still limited at 48 h p.i. (Fig. 6J, d–f).
Discussion
The cleavage between BUNV Gn and NSm has long been thought to occur at the amino acid motif RV/AAR, which is conserved in several orthobunyaviruses (10, 11) and fits the minimum furin cleavage site (RXXR) (28). However, because the furin-like proprotein convertases process substrates in the lumen of the Golgi complex and endosome or at the cell surface (29), it is unlikely that the RxxR motif in the Gn CT can be accessed by these proteases. Moreover, some members of the genus Orthobunyavirus, such as Wyeomyia virus, SBV, and OROV, lack the RxxR motif (Fig. S1C). In fact, we have proven that the motif and the downstream eight residues (residues 303–310) are still part of Gn CT. In the Bunyaviridae family, the furin-like protease is involved in the GPC processing of CCHFV (Nairovirus genus) for generating a 38-kDa NSm protein, whereas the CCHFV furin site is located at the ectodomain of pre-Gn protein (30).
By using mutagenesis and MS analysis, we confirmed that the NSm domain-I is SPNSm which is cleaved by SPase at residue 332T of mature NSm. The residual SPNSm, which is still linked to the upstream Gn CT (as preGn), is further processed from Gn C terminus by the ER-resident SPP. The implication of SPP in BUNV GPC process is validated by our observations as follows: (i) The further processing of NSm domain-I (SPNSm) upon SPase cleavage; (ii) the detection of preGn by WB analysis of V5-tagged Gn protein; and (iii) inhibition of BUNV and SBV infection in SPP-knockdown cells. We also assessed the impact of SPP knockdown on two other bunyaviruses, RVFV (Phlebovirus) and PUUV (Hantavirus). SPP knockdown had no significant inhibitory effect on RVFV infection, whereas it inhibited PUUV infection. However, because PUUV replicated less efficiently in the shGFP-induced cells, we were unable to draw a clear conclusion. Some lentivirus expressing shRNAs can trigger IFN activation (31), and the effect of siRNA on innate immunity is sequence- and structure-related (32). It should be mentioned that the coding strategies and sizes of products encoded by M segments of the viruses in the family are very divergent, and thus it is plausible that the precursor processing differs from genus to genus.
Besides its role as a SP, we provide evidence that the liberated SPNSm has postcleavage function in cell fusion. We speculate that the liberated SPNSm is likely incorporated into the virion by interacting with one of the viral glycoproteins upon cleavage by SPP, which is probably required for that interaction. However, we were unable to find the peptide from the purified virus particles by MS analyses, perhaps because of the technical challenges of the small size and hydrophobicity of the domain. Another possibility is that the Gn CT is modified during the processing of SPNSm by SPP and that modification might be crucial for glycoprotein activities. Whatever the cases, it seems that the sequence specificity of SPNSm is important. Several SPs of viral proteins have postcleavage functions. For instance, SPGP-C of lymphocytic choriomeningitis virus and Junín virus (Arenaviruses) precursor glycoproteins C (GP-C) is an essential structural component of mature virions and is required in glycoprotein maturation, cell fusion, and virus infectivity (33–35).
Based on our findings, we propose a model for the process and topology of orthobunyavirus glycoproteins (Fig. 7A). All three proteins, in precursor form, contain their own SPs. The N-terminal SPGn targets the nascent Gn polypeptide through the ER membrane, where it is cleaved by SPase at residue 17S. The internal SPNSm mediates the translocation of the nascent NSm chain into the ER membrane, and it is subsequently cleaved at residue 332T by SPase from the NSm. SPNSm is further processed by SPP to free the Gn CT. NSm domain-V/SPGc translocates the nascent Gc chain to the ER membrane and is cleaved at residue 478E to separate the mature NSm and the nascent Gc chain. However, unlike SPNSm, the domain-V/SPGc is not further processed and remains as the C-terminal domain of mature NSm. The updated topology model of mature Gn, NSm, and Gc proteins is illuminated in Fig. 7B. Gn and Gc proteins are type I transmembrane proteins, and NSm is a two-membrane-spanning protein.
Fig. 7.
The model of BUNV GPC (Gn-NSm-Gc) cleavage. (A) Schematic showing BUNV GPC processing. (B) Topology of Gn, Gc, and NSm proteins. See Discussion for details.
This study revealed a dimension for SPP in virus replication. The knowledge will benefit vaccine development and help identify new antiviral drugs against pathogenic virus infections caused by viruses in the family.
Materials and Methods
The materials and methods are described in SI Materials and Methods. They include cells and viruses, antibodies, plasmids and mutagenesis, transfection of cells, metabolic radiolabeling and immunoprecipitation, immunofluorescence staining, BUNV glycoprotein fusion assay, virus rescue by reverse genetics, virus infection, titration and purification, preparation of BUNV glycoprotein Gn, Gc, and NSm proteins; MS analysis, SPP knockdown by lentivirus shRNA; and WB.
Data were expressed as the mean and SD. The P value and statistical significance of difference was analyzed by using unpaired t tests with GraphPad 6 software. *P < 0.05, significant; **P < 0.01, very significant; ***P < 0.001, extremely significant.
SI Materials and Methods
Cells and Viruses.
A549, A459V (36), A549-NPro (37), Vero E6, BHK-21, HEK 293T, Huh7, and BSR-T7/5 (38) cells were maintained as described (4). BUNV, SBV, RVFV (strain MP12), and PUUV (strain CG1820) were used as representative strains for genera of Orthobunyavirus, Phlebovirus, and Hantavirus in SPP knockdown assays. rBUNGc-EGFP is a recombinant BUNV with the truncated Gc tagged by EGFP (3).
Antibodies.
MAb810, a monoclonal antibody against BUNV Gc, anti-BUN, and anti-NSm, the rabbit antisera against BUNV particles and NSm peptide TDQKYTLDEIADVLQA (residues 338–353 of BUNV GPC), have been described (26). Rabbit anti-BUN N, anti-SBV N, and anti-RVFV N antisera were raised against nucleoproteins of BUNV, SBV, and RVFV, respectively (39–41). A rabbit antiserum against GM130, a cis-Golgi matrix protein was provided by M. Lowe, University of Manchester, Manchester, United Kingdom. Other antibodies were purchased commercially: rabbit polyclonal antibody against SPP (anti-SPP) (Abcam); mouse monoclonal antibody against the V5 epitope (anti-V5) (Serotec); goat anti-rabbit antibody conjugated with FITC (Sigma); and goat anti-mouse antibody conjugated with Cy5 (Chemicon International).
Production of Anti-PUU N. The 6His-tagged PUUV N protein was expressed in BL21-CodonPlus (DE3)-RP Escherichia coli (Agilent Technologies) under IPTG induction at 18–20 °C for 20 h and purified with Ni–NTA resin, and its identity was confirmed by MS. The purified PUU N was used for generating rabbit antisera against PUUV N protein (Eurogentec).
Plasmids and Mutagenesis.
Plasmids that generate full-length BUNV antigenome RNA transcripts, pT7riboBUNL(+), TVT7R-BUNM(+), and pT7riboBUNS(+), or express the full-length GPC, pTM1BUNM, have been described (8, 42); pT7riboBUNM-NSmV5(+) and pTM1BUNM-NSmV5 contain the V5 epitope (GKPIPNPLLGLDST) inserted between residues 403 and 420 of the NSm coding region (Fig. S3B). The substitution and internal deletion mutations in BUNV GPC (in the backbone of either pTM1BUNM for high expression or TVT7RBUNM for virus rescue) were generated by PCR-directed mutagenesis on the appropriate parental templates.
A series of individual Gn expression plasmids (pTM1BUNGn298 to Gn332) were derived from pTM1BUNM (Fig. 3A), and Gn-NSm expression plasmids (pTM1BUNGn-NSmV5-453-476) were derived from pTM1BUNM-NSmV5 (Fig. 4B). Two mutated Gn-NSm constructs (pTmBUNGnNSmV5-SPGc/SPHGn and pTmBUNGnNSmV5-SPGc/eGFP1–20) were created by replacement of the domain V with either SP of HTNV Gn protein (residues 1–19) or N-terminal 20 residues of eGFP (Fig. 4B). Three mutated GPC constructs (pTM1BUNMΔ446–476, Δ457–476, and Δ467–476) contain internal deletions in the coding region of NSm domain IV and V). pTM1BUN-NSmGc expresses NSm and Gc proteins (residues 309–1433). pTmBUNM∆NSm_V and pTmBUNMΔNSm_I contain internal deletions in NSm coding region, in which ∆NSm_V has deletion of NSm domains I–IV (residues 311–456) except domain V (SPGc) and pTmBUNMΔNSm_I has deletion of NSm domains II–V (residues 332–477) except domain I (SPNSm) as SP for Gc. pTMBUNM-SPNSm/HTNtm has 17 residues of SPNSm (residues 311–327) swapped with those of HTNV Gn TMD (TFCFGWVLIPAITFIIL, residues 490–506) (Fig. 5A). pTmBUNM-SPmNSm contains four point mutations in the coding region of NSm domain I (Fig. 2A). All of the constructs were confirmed by DNA sequencing.
Transfection of Cells.
BSRT7/5 cells were grown in 35-mm dishes for immunoprecipitation experiments, in 12-well plates for cell fusion assay, or on 13-mm glass coverslips for immunofluorescence assays. Cells were transfected with the mixtures of plasmid DNA and transfection reagent (Lipofectamine 2000 Transfection Reagent; Life Technologies) and incubated at 37 °C for 24 h.
Metabolic Radiolabeling and Immunoprecipitation.
At 24 h post transfection or p.i., cells were labeled with [35S]methionine (PerkinElmer) for 3 h. For labeling of virus particles, the infected BHK21 cells grown in 175-cm2 flask were labeled with [35S]methionine for 6 h p.i., and virus particles were purified by ultracentrifugation (4). Cells were then lysed on ice in 300 µL of nondenaturing radioimmunoprecipitation assay (RIPA) buffer [50 mM Tris⋅HCl (pH 7.4), 1% Triton X-100, 300 mM NaCl, and 5 mM EDTA] containing a mixture of cOmplete protease inhibitors (Roche). BUNV proteins were immunoprecipitated with anti-BUN, -NSm, or -V5 antibodies conjugated to Dynabeads Protein A (Novex; Life Technologies). The beads bound to immune complexes were washed with RIPA buffer containing 0.1% Triton X-100 and with cold PBS, and bound viral proteins were analyzed by SDS/PAGE under reducing conditions.
Immunofluorescence Staining.
The transfected or infected cells were fixed with 4% (vol/vol) formaldehyde/2% (wt/vol) sucrose/PBS and permeabilized with 0.2% Triton X-100 in PBS before staining with specific primary antibodies and secondary antibody conjugates. Cells were examined by using a Zeiss LSM confocal microscope.
BUNV Glycoprotein Fusion Assay.
BSR-T7/5 cells, grown on 12-well plates, were transfected with 1 μg of either pTM1BUNM or one of the mutant M expression plasmids. At 24 h after transfection, cells were treated with low-pH medium (pH 5.3) for 15 min, and then the medium was replaced by Glasgow's MEM supplemented with 10% FCS. After incubation for 4 h at 37 °C, the extent of cell fusion was observed after Giemsa staining. Cells that contained more than three nuclei were considered as syncytia, and the nuclei in 30 syncytia in total were counted. The fusion is represented as average nuclei per syncytium (n = 30).
Virus Rescue by Reverse Genetics.
Rescue experiments were performed essentially as described with minor modifications (42). Subconfluent monolayers of BSR-T7/5 cells grown in T25 were transfected with a mixture of plasmids comprising 1.0 μg each of pT7riboBUNL(+), pT7riboBUNS(+), and TVT7RBUNM(+) or one of the M cDNA mutants, and 7.5 µL of Lipofectamine 2000 (Life Technologies) in 0.7 mL of Opti-MEM medium (Life Technologies). At 4 h after transfection, 4 mL of growth medium was added, and incubation continued at 33 °C until cytopathic effect (CPE) was evident.
Virus Infection, Titration, and Purification.
Cells seeded in dishes or flasks were infected at an MOI of 0.01 or 1.0 pfu per cell. The inoculum was removed after 1 h, and cells were washed with PBS to remove the unattached viruses. Supernatants were harvested at the indicated time points. BUNV and SBV were titrated by plaque assay on BSRT7/5 cells and RVFV on BHK21 cells. PUUV was titrated on Vero E6 cells by using the Avicel immunofocus assay. Briefly, the virus-infected cells were covered with 0.6% Avicel (FMC BioPolymer) overlay medium and incubated for 7 d, fixed with 4% formaldehyde/PBS, and permeabilized with 0.5% Triton X-100 and 20 mM glycine/PBS. Cell monolayer was blocked with 4% skimmed milk, reacted with anti-PUUN, and probed with peroxidase-conjugated anti-rabbit IgG. The foci were developed after incubation with True Blue peroxidase substrate (KPL). The purification of BUNV particles was described (4).
Preparation of BUNV Glycoprotein Gn, Gc, and NSm Proteins.
BUNV NSm protein was purified from lysate of the rBUNV-NSmV5–infected cells by using anti-V5 Affinity gel (biotool.com). Briefly, BHK-21 cells were infected with rBUNV-NSmV5, and cells were harvested 48 h p.i. when cells showed obvious CPE. The cell pellet was lysed with nondenaturing RIPA buffer and mixed with anti-V5 Affinity bead. The V5-tagged NSm protein (NSmV5) was separated by 15% Tris–glycine SDS/PAGE (Fig. S3C). For preparation of BUNV Gn and Gc proteins, the purified BUNV particles were separated by 12.5% Tris–glycine SDS/PAGE and stained with Coomassie Brilliant Blue G-250 solution (Fig. S4A). Gn and Gc bands were excised for MS analysis.
MS Analysis.
Protein bands were cut into 1-mm cubes, which were then subjected to in-gel digestion with glutamyl endopeptidase (GluC) (NEB), AspN (NEB), or trypsin (Promega) by a ProGest Investigator in-gel digestion robot (Genomic Solutions) using standard protocols (43). Briefly, the gel cubes were destained by washing with acetonitrile and subjected to reduction and alkylation before digestion with endoproteinase at 37 °C. The peptides were then extracted with 10% formic acid and concentrated down to 20 μL by using a SpeedVac (ThermoSavant). In some GluC digestions of Gn protein, PMSF (1 mM) was added to prevent any trypsin-like proteolytic cleavage. The peptides were then separated on an Acclaim PepMap 100 C18 trap and an Acclaim PepMap RSLC C18 column (ThermoFisher Scientific), using a nanoLC Ultra 2D plus loading pump and nanoLC as-2 autosampler (Eksigent). The peptides were eluted with a gradient of increasing acetonitrile, containing 0.1% formic acid (5–40% acetonitrile in 5 min, 40–95% in a further 1 min, followed by 95% acetonitrile to clean the column, before reequilibration to 5% acetonitrile). The eluate was sprayed into a TripleTOF 5600+ electrospray tandem mass spectrometer (Sciex) and analyzed in information-dependent acquisition mode, performing cycles of 250 ms of MS followed by 100-ms tandem MS (MS/MS) analyses on the 15 most intense peaks seen by MS. The MS/MS data file generated via the “Create mgf file” script in PeakView (Sciex) was analyzed by using the Mascot algorithm (Matrix Science) against an internal database to which the amino acid sequence of the BUNV GPC (GenBank accession no. P04505) had been added, with GluC, AspN, or trypsin as the cleavage enzyme and carbamidomethyl as a fixed modification of cysteines and methionine oxidation and deamidation of glutamines and asparagines as a variable modifications, followed by an “error tolerant” search, to look for peptides where only one end conforms to the enzyme specificity criteria, to delineate the C-terminal peptide sequence.
SPP Gene (HM13) Knockdown by Lentivirus shRNA.
shRNA cDNAs specific for human SPP (nucleotides 777–795 for shSPP1 and 1,034–1052 for shSPP2; GenBank accession no. AJ420895) and eGFP (nucleotides 441–463; GenBank accession no. U57607) as control were cloned into pLKO.1 puro (Addgene). Lentivirus stocks were prepared in HEK 293T cells by transfection of constructs: pLKO, pMD-G (pVSVG), a plasmid expressing the vesicular stomatitis virus glycoprotein (44), and pCMV∆R8.91 (pCMV-R), a plasmid expressing the gag/pol rev genes of HIV-1 (45), with Lipofectamine 2000 (Life Technologies). shRNA expression in A549 or A549V cells was achieved by transducing cells with shRNA-expressing lentiviruses for 48 h in the presence of 8 µg/mL polybrene (Santa Cruz Biotechnology).
Western Blotting (WB).
Lysates of plasmid-transfected or virus-infected cells were prepared by the addition of 100 µL of NuPAGE LDS sample buffer (Life Technologies) containing 1 µL of Benzonase nuclease (Merck Millipore). Proteins were separated on 4–12% Bis–Tris protein gel (Life Technologies) and transferred to a Nitrocellulose Blotting Membrane (GE Healthcare). The membrane was probed with appropriate antibodies and signals detected by chemiluminescence.
Acknowledgments
This paper is dedicated to the memory of our colleague Richard M. Elliott who died on June 5, 2015 whilst this work was ongoing. We thank Drs. Klaus K. Conzelmann (Ludwig-Maximilians-Universität München), David Jackson, Richard Randall (University of St. Andrews), and Martin Lowe (University of Manchester) for providing reagents used in this work; Drs. Friedemann Weber (Justus-Liebig-Universität Gießen) and Alain Kohl (University of Glasgow) for critical reading of the manuscript; and Angela Elliott, Edward Dornan, and Dr. Ye Liu for technical support. This study was supported by Wellcome Trust Grant 099220/B/12/Z (to R.M.E.) and Grant 094476/Z/10/Z that funded the purchase of the TripleTOF 5600 mass spectrometer at the Biomedical Sciences Research Complex (BSRC) of University of St. Andrews.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603364113/-/DCSupplemental.
References
- 1.Plyusnin A, Elliott RM, editors. Bunyaviridae: Molecular and Cellular Biology. Caister Academic; Norfolk, U.K.: 2011. [Google Scholar]
- 2.Elliott RM, Schmaljohn CS. Bunyaviridae. In: Knipe DM, Howley PM, editors. Fields Virology. 6th Ed. Wolters Kluwer; Philadelphia: 2013. pp. 1244–1282. [Google Scholar]
- 3.Shi X, van Mierlo JT, French A, Elliott RM. Visualizing the replication cycle of bunyamwera orthobunyavirus expressing fluorescent protein-tagged Gc glycoprotein. J Virol. 2010;84(17):8460–8469. doi: 10.1128/JVI.00902-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi X, Brauburger K, Elliott RM. Role of N-linked glycans on bunyamwera virus glycoproteins in intracellular trafficking, protein folding, and virus infectivity. J Virol. 2005;79(21):13725–13734. doi: 10.1128/JVI.79.21.13725-13734.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowden TA, et al. Orthobunyavirus ultrastructure and the curious tripodal glycoprotein spike. PLoS Pathog. 2013;9(5):e1003374. doi: 10.1371/journal.ppat.1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garry CE, Garry RF. Proteomics computational analyses suggest that the carboxyl terminal glycoproteins of Bunyaviruses are class II viral fusion protein (beta-penetrenes) Theor Biol Med Model. 2004;1:10. doi: 10.1186/1742-4682-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi X, Kohl A, Li P, Elliott RM. Role of the cytoplasmic tail domains of Bunyamwera orthobunyavirus glycoproteins Gn and Gc in virus assembly and morphogenesis. J Virol. 2007;81(18):10151–10160. doi: 10.1128/JVI.00573-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi X, et al. Requirement of the N-terminal region of orthobunyavirus nonstructural protein NSm for virus assembly and morphogenesis. J Virol. 2006;80(16):8089–8099. doi: 10.1128/JVI.00579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fazakerley JK, et al. Organization of the middle RNA segment of snowshoe hare Bunyavirus. Virology. 1988;167(2):422–432. [PubMed] [Google Scholar]
- 10.Briese T, Rambaut A, Lipkin WI. Analysis of the medium (M) segment sequence of Guaroa virus and its comparison to other orthobunyaviruses. J Gen Virol. 2004;85(Pt 10):3071–3077. doi: 10.1099/vir.0.80122-0. [DOI] [PubMed] [Google Scholar]
- 11.Elliott RM, Blakqori G. Molecular biology of Orthobunyaviruses. In: Plyusnin A, Elliot RM, editors. Bunyaviridae. Caister Academic; Norfolk, U.K.: 2011. pp. 1–39. [Google Scholar]
- 12.Auclair SM, Bhanu MK, Kendall DA. Signal peptidase I: Cleaving the way to mature proteins. Protein Sci. 2012;21(1):13–25. doi: 10.1002/pro.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golde TE, Wolfe MS, Greenbaum DC. Signal peptide peptidases: A family of intramembrane-cleaving proteases that cleave type 2 transmembrane proteins. Semin Cell Dev Biol. 2009;20(2):225–230. doi: 10.1016/j.semcdb.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemberg MK. Intramembrane proteolysis in regulated protein trafficking. Traffic. 2011;12(9):1109–1118. doi: 10.1111/j.1600-0854.2011.01219.x. [DOI] [PubMed] [Google Scholar]
- 15.Voss M, Schröder B, Fluhrer R. Mechanism, specificity, and physiology of signal peptide peptidase (SPP) and SPP-like proteases. Biochim Biophys Acta Biomembranes. 2013;(12):2828–2839. doi: 10.1016/j.bbamem.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 16.Weihofen A, Martoglio B. Intramembrane-cleaving proteases: Controlled liberation of proteins and bioactive peptides. Trends Cell Biol. 2003;13(2):71–78. doi: 10.1016/s0962-8924(02)00041-7. [DOI] [PubMed] [Google Scholar]
- 17.Friedmann E, et al. SPPL2a and SPPL2b promote intramembrane proteolysis of TNFalpha in activated dendritic cells to trigger IL-12 production. Nat Cell Biol. 2006;8(8):843–848. doi: 10.1038/ncb1440. [DOI] [PubMed] [Google Scholar]
- 18.Oliveira CC, et al. New role of signal peptide peptidase to liberate C-terminal peptides for MHC class I presentation. J Immunol. 2013;191(8):4020–4028. doi: 10.4049/jimmunol.1301496. [DOI] [PubMed] [Google Scholar]
- 19.Lemberg MK, Bland FA, Weihofen A, Braud VM, Martoglio B. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J Immunol. 2001;167(11):6441–6446. doi: 10.4049/jimmunol.167.11.6441. [DOI] [PubMed] [Google Scholar]
- 20.McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002;21(15):3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bintintan I, Meyers G. A new type of signal peptidase cleavage site identified in an RNA virus polyprotein. J Biol Chem. 2010;285(12):8572–8584. doi: 10.1074/jbc.M109.083394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 23.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evnin LB, Vásquez JR, Craik CS. Substrate specificity of trypsin investigated by using a genetic selection. Proc Natl Acad Sci USA. 1990;87(17):6659–6663. doi: 10.1073/pnas.87.17.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fontana J, López-Montero N, Elliott RM, Fernández JJ, Risco C. The unique architecture of Bunyamwera virus factories around the Golgi complex. Cell Microbiol. 2008;10(10):2012–2028. doi: 10.1111/j.1462-5822.2008.01184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lappin DF, Nakitare GW, Palfreyman JW, Elliott RM. Localization of Bunyamwera bunyavirus G1 glycoprotein to the Golgi requires association with G2 but not with NSm. J Gen Virol. 1994;75(Pt 12):3441–3451. doi: 10.1099/0022-1317-75-12-3441. [DOI] [PubMed] [Google Scholar]
- 27.Tilston-Lunel NL, Acrani GO, Randall RE, Elliott RM. Generation of recombinant oropouche viruses lacking the nonstructural protein NSm or NSs. J Virol. 2015;90(5):2616–2627. doi: 10.1128/JVI.02849-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3(10):753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11(5):367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez AJ, Vincent MJ, Erickson BR, Nichol ST. Crimean-congo hemorrhagic fever virus glycoprotein precursor is cleaved by Furin-like and SKI-1 proteases to generate a novel 38-kilodalton glycoprotein. J Virol. 2006;80(1):514–525. doi: 10.1128/JVI.80.1.514-525.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34(3):263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 32.Whitehead KA, Dahlman JE, Langer RS, Anderson DG. Silencing or stimulation? siRNA delivery and the immune system. Annu Rev Chem Biomol Eng. 2011;2(1):77–96. doi: 10.1146/annurev-chembioeng-061010-114133. [DOI] [PubMed] [Google Scholar]
- 33.Schrempf S, Froeschke M, Giroglou T, von Laer D, Dobberstein B. Signal peptide requirements for lymphocytic choriomeningitis virus glycoprotein C maturation and virus infectivity. J Virol. 2007;81(22):12515–12524. doi: 10.1128/JVI.01481-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bederka LH, Bonhomme CJ, Ling EL, Buchmeier MJ. Arenavirus stable signal peptide is the keystone subunit for glycoprotein complex organization. MBio. 2014;5(6):e02063. doi: 10.1128/mBio.02063-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.York J, Romanowski V, Lu M, Nunberg JH. The signal peptide of the Junín arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1-G2 complex. J Virol. 2004;78(19):10783–10792. doi: 10.1128/JVI.78.19.10783-10792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Killip MJ, et al. Deep sequencing analysis of defective genomes of parainfluenza virus 5 and their role in interferon induction. J Virol. 2013;87(9):4798–4807. doi: 10.1128/JVI.03383-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hale BG, et al. CDK/ERK-mediated phosphorylation of the human influenza A virus NS1 protein at threonine-215. Virology. 2009;383(1):6–11. doi: 10.1016/j.virol.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73(1):251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber F, Dunn EF, Bridgen A, Elliott RM. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology. 2001;281(1):67–74. doi: 10.1006/viro.2000.0774. [DOI] [PubMed] [Google Scholar]
- 40.Elliott RM, et al. Establishment of a reverse genetics system for Schmallenberg virus, a newly emerged orthobunyavirus in Europe. J Gen Virol. 2013;94(Pt 4):851–859. doi: 10.1099/vir.0.049981-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brennan B, Li P, Elliott RM. Generation and characterization of a recombinant Rift Valley fever virus expressing a V5 epitope-tagged RNA-dependent RNA polymerase. J Gen Virol. 2011;92(Pt 12):2906–2913. doi: 10.1099/vir.0.036749-0. [DOI] [PubMed] [Google Scholar]
- 42.Lowen AC, Noonan C, McLees A, Elliott RM. Efficient bunyavirus rescue from cloned cDNA. Virology. 2004;330(2):493–500. doi: 10.1016/j.virol.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 44.Naldini L, Blömer U, Gage FH, Trono D, Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93(21):11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15(9):871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 46.Sugrue RJ, Brown C, Brown G, Aitken J, McL Rixon HW. Furin cleavage of the respiratory syncytial virus fusion protein is not a requirement for its transport to the surface of virus-infected cells. J Gen Virol. 2001;82(Pt 6):1375–1386. doi: 10.1099/0022-1317-82-6-1375. [DOI] [PubMed] [Google Scholar]