Significance
ATP, the fuel of life, is produced by a molecular machine consisting of two motors linked by a rotor. One motor generates rotation by consuming energy derived from oxidative metabolism or photosynthesis; the other uses energy transmitted by the rotor to put ATP molecules together from their building blocks ADP and phosphate. The anionic lipid cardiolipin, a component of bacterial membranes and the inner membranes of mitochondria where the machine is found, is an essential component of the enzyme. It interacts specifically, transiently, and repeatedly with the rotor of the machine, possibly lubricating its rotation or participating directly in the generation of rotation from the transmembrane proton motive force.
Keywords: mitochondria, ATP synthase, trimethyllysine, cardiolipin, molecular dynamics simulation
Abstract
The anionic lipid cardiolipin is an essential component of active ATP synthases. In metazoans, their rotors contain a ring of eight c-subunits consisting of inner and outer circles of N- and C-terminal α-helices, respectively. The beginning of the C-terminal α-helix contains a strictly conserved and fully trimethylated lysine residue in the lipid head-group region of the membrane. Larger rings of known structure, from c9-c15 in eubacteria and chloroplasts, conserve either a lysine or an arginine residue in the equivalent position. In computer simulations of hydrated membranes containing trimethylated or unmethylated bovine c8-rings and bacterial c10- or c11-rings, the head-groups of cardiolipin molecules became associated selectively with these modified and unmodified lysine residues and with adjacent polar amino acids and with a second conserved lysine on the opposite side of the membrane, whereas phosphatidyl lipids were attracted little to these sites. However, the residence times of cardiolipin molecules with the ring were brief and sufficient for the rotor to turn only a fraction of a degree in the active enzyme. With the demethylated c8-ring and with c10- and c11-rings, the density of bound cardiolipin molecules at this site increased, but residence times were not changed greatly. These highly specific but brief interactions with the rotating c-ring are consistent with functional roles for cardiolipin in stabilizing and lubricating the rotor, and, by interacting with the enzyme at the inlet and exit of the transmembrane proton channel, in participation in proton translocation through the membrane domain of the enzyme.
Cardiolipin is associated uniquely with energy-transducing membranes in mitochondria and eubacteria. In mitochondria, it is found in the inner membrane and is synthesized close to, or in, the inner leaflet (1) where most of it remains (2, 3). Cardiolipin consists of two 3-phosphatidyl groups linked by a glycerol bridge, and in bovine mitochondria, the four acyl chains have 18 carbon atoms with one or two unsaturated linkages (4). It has been proposed that under physiological conditions, the central hydroxyl and the two phosphates trap a proton in a resonance structure and that cardiolipin carries one net negative charge (5). However, re-evaluations of the pKa values of the phosphates indicate that under physiological conditions, the head-group of cardiolipin bears two negative charges (6, 7).
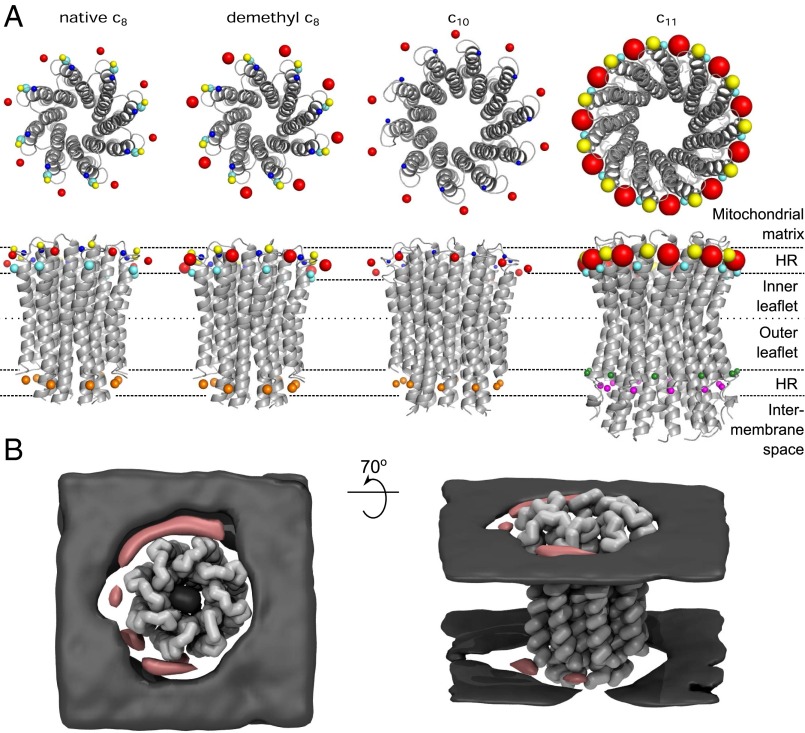
Cardiolipin and other phospholipids are essential components of active ATP synthases isolated from mitochondria (8–12). It has been suggested that cardiolipin acts to stabilize and lubricate the rotating c-ring (13) or to aid in proton transfer (5), but it is not known where or how cardiolipin binds to the enzyme. Excluding the regulatory protein IF1, the bovine ATP synthase complex is built from 28 polypeptide chains of 16 varieties (14). About 85% of a mosaic overall structure has been determined to atomic resolution by structural analysis of constituent domains (15–18), and an intact enzyme structure has been described at about 6-Å resolution (19). The enzyme consists of a spherical catalytic F1-domain in the matrix of the mitochondria, attached to the inner membrane domain by central and peripheral stalks. The central stalk is bound to a ring of c-subunits in the membrane domain, and together they constitute the enzyme’s rotor (13, 20). Each bovine c-subunit is folded into two transmembrane α-helices, and in each rotor-ring, the N- and C-terminal α-helices form concentric inner and outer circles, linked by eight loop regions exposed in the phospholipid head-group region on the matrix side of the inner membrane. Contacts between the loops and the central stalk add to the stability of the rotor. Vertebrate and probably all metazoan ATP synthases have c8-rings (21). Fungal ATP synthases contain c10-rings (20), various eubacterial enzymes have rings of c9, c11, c12, c13, and c15 (22–28), and the enzyme from spinach chloroplasts has a c14-ring (29). The turning of the rotor carries energy from the transmembrane proton-motive force generated by oxidative metabolism or photosynthesis to the catalytic domain, to energize the phosphorylation of ADP. The transmembrane path for protons involved in generating rotation is in the interface between the external surface of the c-ring and the membrane subunit a (30), which has not been resolved to high resolution, although a 4-Å X-ray structure of a bacterial enzyme with a c12-ring (25) and structures of the bovine and an algal enzyme determined by electron cryo-microscopy (19, 31) have revealed its rudiments. In the structures of intact ATP synthases (19, 25, 31), of F1-c-ring subcomplexes (13, 20), and of c-rings (22–24, 27–29, 32–35), there is no evidence of any bound phospholipid. However, the presence of a fully trimethylated and conserved lysine residue, in the phospholipid head-group region of c-subunits of metazoan ATP synthases, suggested that it could provide a site for cardiolipin to bind selectively, by impeding the binding of the head-group of other phospholipids (21). Therefore, here, we describe coarse-grain molecular dynamics simulations to examine how phospholipids interact with bovine c8-rings, with and without methyl groups, with the c10-ring in the ATP synthase from Saccharomyces cerevisiae (20) and the c11-ring from the Na+-dependent ATP synthase from Ilyobacter tartaricus (23), where the equivalent lysine residues are conserved but not methylated.
Results
Models of c-Rings in Lipid Bilayers.
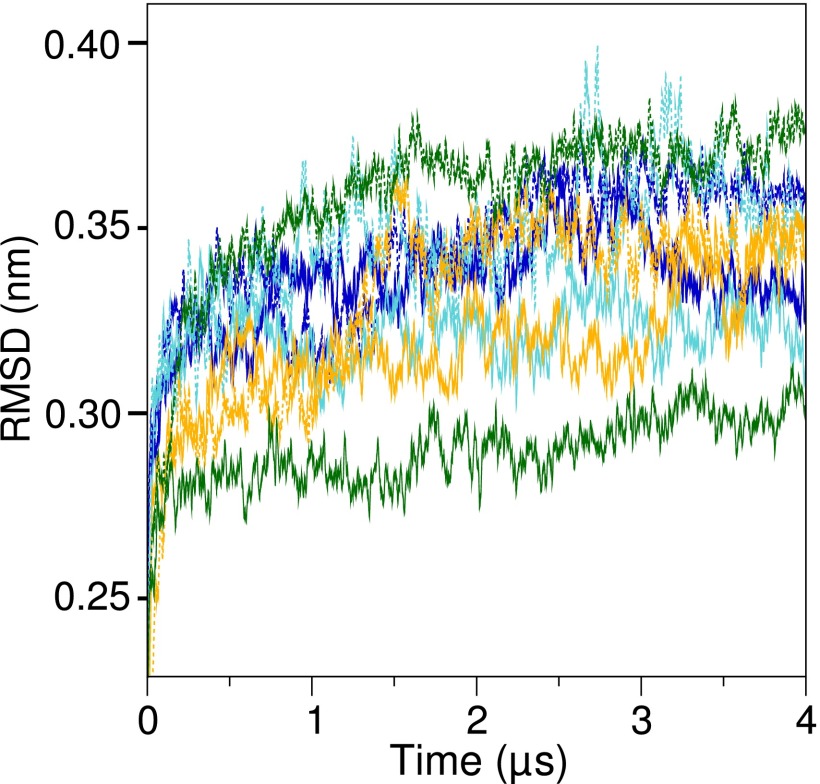
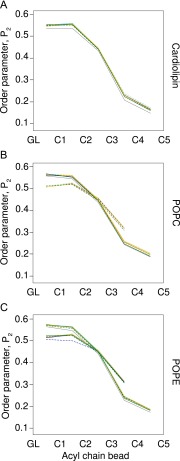
The structures of c-rings at the start and end of the molecular dynamics simulations were essentially the same, with RMSDs of less than 0.35 nm between initial and final structures superimposed by backbone beads (Fig. S1). However, c11-rings were the most stable, probably because intersubunit interactions occurred between Lys-50 residues and Asp-52 residues in adjacent c-subunits (23). Sometimes, in the c10-ring, the orthologous residues were sufficiently close to allow the formation of equivalent bonds, whereas the orthologous residues Lys-43 and Gln-45 in the c8-ring were too far apart. The formation of bilayers in the liquid phase was confirmed by the order parameters of the lipids, which were greatest in the lipid head-group region, diminishing down the acyl chain toward zero at the distal ends (Fig. S2).
Fig. S1.
Stability of simulated c-rings. Stabilities of c-rings were assessed during simulation production runs from the RMSD from the starting structure of each c-ring model. The RMSDs were calculated every 1.2 ns over a 50-point running average. Their values are shown as solid and dashed lines corresponding to the two simulations of each ring. Blue, trimethylated native c8-ring; cyan, demethylated c8-ring; orange, c10-ring; green, c11-ring.
Fig. S2.
Melting of lipid chains during simulations. Lipid disorder was calculated from the order parameters of the acyl chains of (A) cardiolipin, (B) POPC, and (C) POPE. The simulations were performed twice, shown as solid and dashed lines. The lines corresponding to different simulations are colored as follows: gray, no c-ring; dark blue, native c8-ring; cyan, demethylated c8-ring; orange, c10-ring; green, c11-ring. The order parameters for cardiolipin were averaged over all four chains; they are shown separately for phosphatidyl POPC and POPE that have chains of differing lengths.
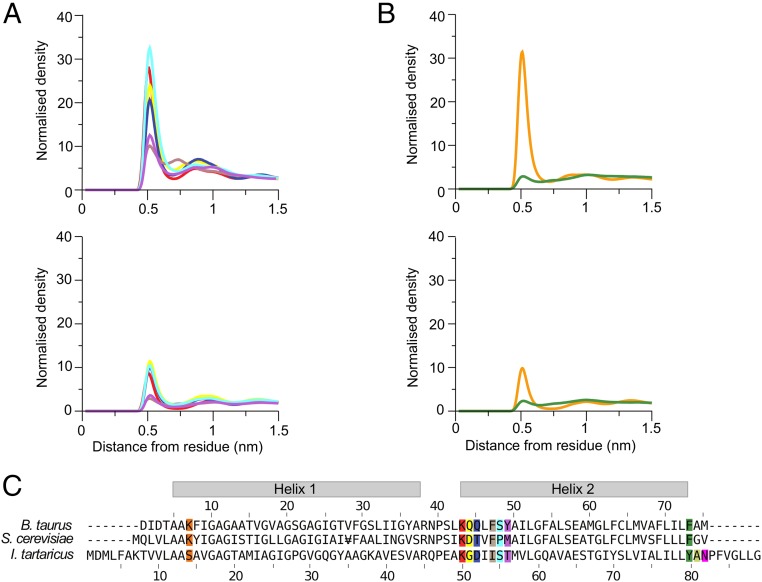
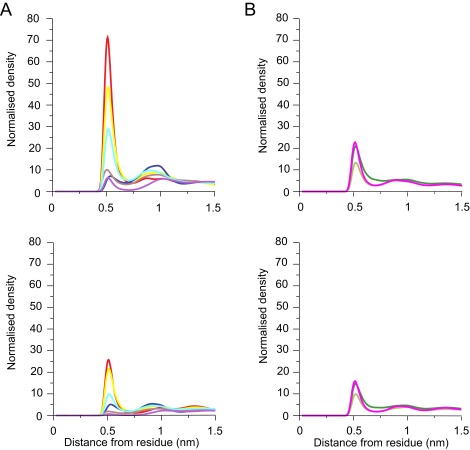
The lipids became organized into an annulus in direct contact with the c-ring, and the next concentric layer, with maxima in the radial distribution functions of lipid head-groups at 0.6 and 0.9 nm (Fig. 1). Cardiolipin remained longer in the inner annulus than phosphatidyl lipids. Once a cardiolipin entered the lipid annulus in either leaflet, it remained bound to the c-ring longer than phosphatidyl lipids, which tended to touch fleetingly and then diffuse back into the bulk membrane.
Fig. 1.
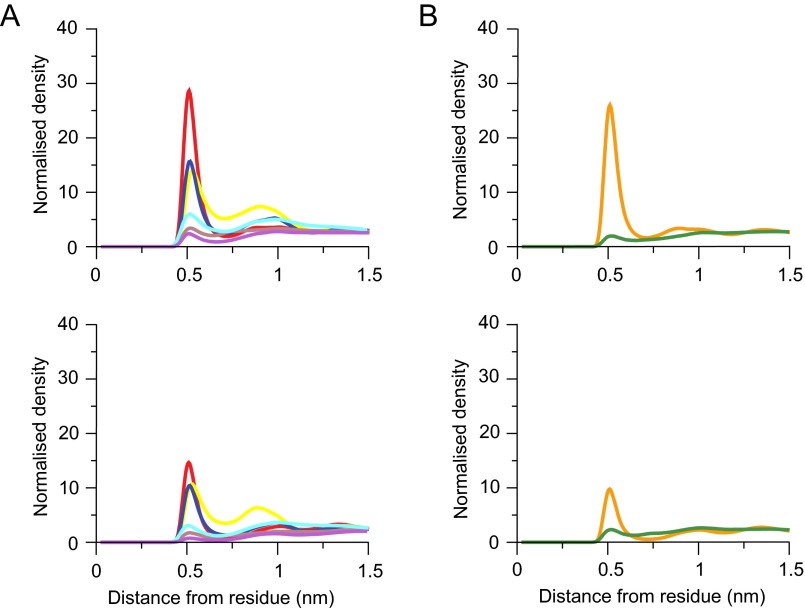
Interactions between lipids and amino acids in native c8-rings. (A and B) Average radial distribution functions from two independent simulations for lipids in the inner and outer leaflets of the membrane, respectively. Upper and Lower correspond to interactions with cardiolipin and with POPC and POPE, respectively. Their colors correspond to specific amino acids in the bovine c-subunit, as defined in C. (C) Alignment of sequences of c-subunits from cows, S. cerevisiae, and I. tartaricus. Residues interacting with lipids are colored. For the radial distribution curves for the demethylated c8-rings and c10- and c11-rings from S. cerevisiae and I. tartaricus, respectively, see Figs. S4, S6, and S8.
Interactions of Lipids with the Native c8-Ring.
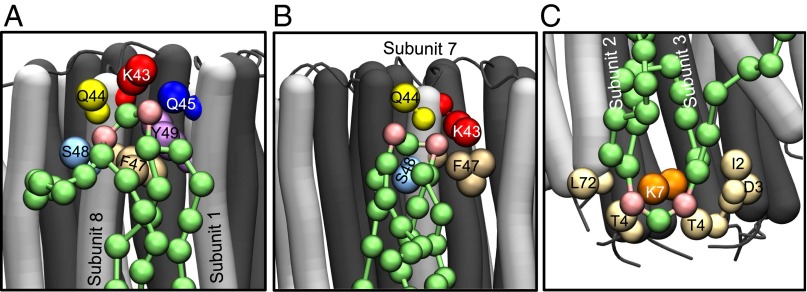
The densities of radial distribution functions of the phosphate moieties of cardiolipin (Fig. 1) were greatest around the side-chains of TM-Lys-43, Gln-44, Gln-45, and Ser-48 in the inner leaflet of the membrane (Fig. 1A). Likewise, the densities of phosphate groups of phosphatidyl lipids were greatest around the same residues, but much lower than those of cardiolipin (Fig. 1A). In the outer leaflet of the membrane, there was a high density of cardiolipin around residue Lys-7, which is at the beginning of the N-terminal α-helix in the vicinity of the lipid head-groups. The densities of phosphatidyl lipid head-groups were much lower (Fig. 1B).
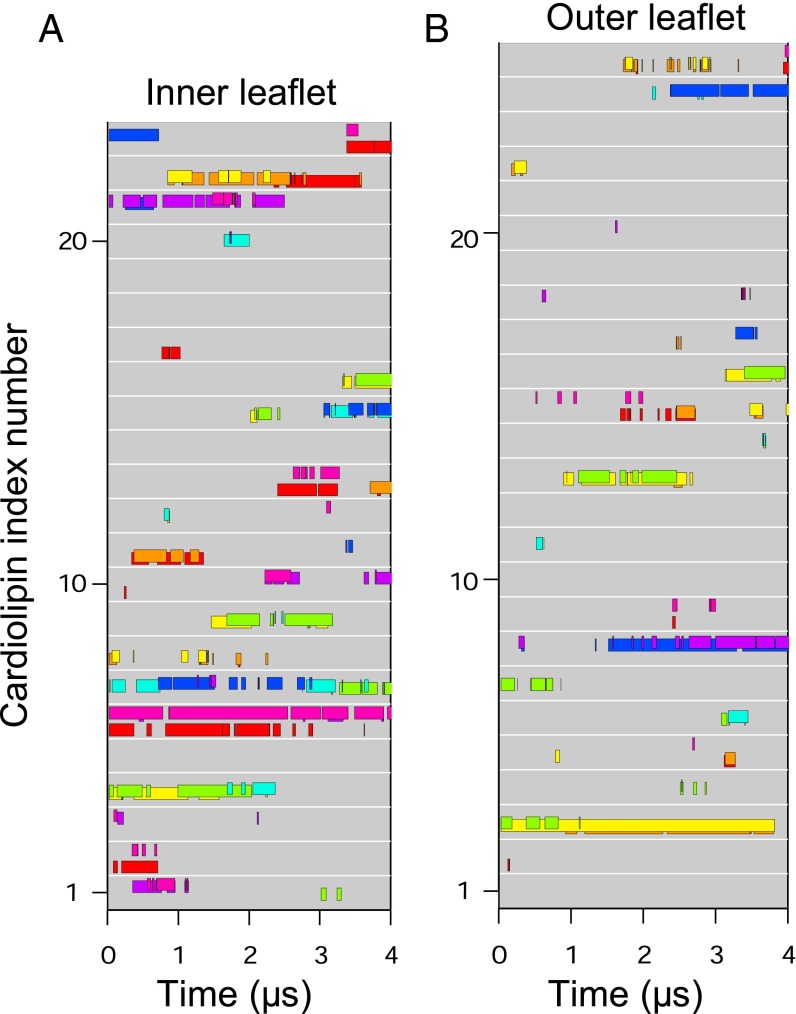
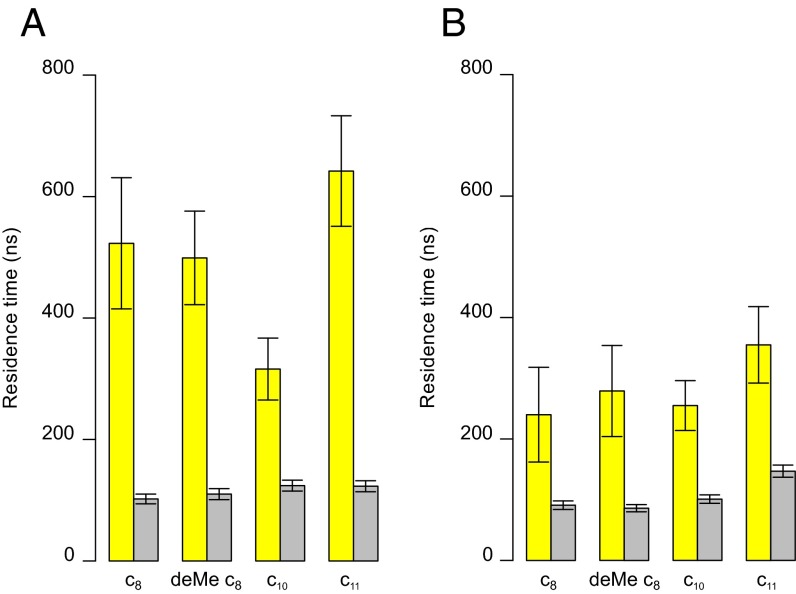
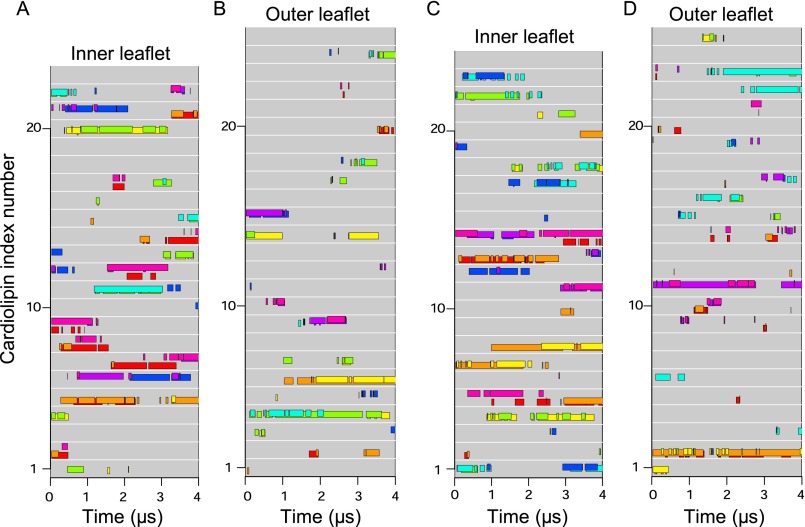
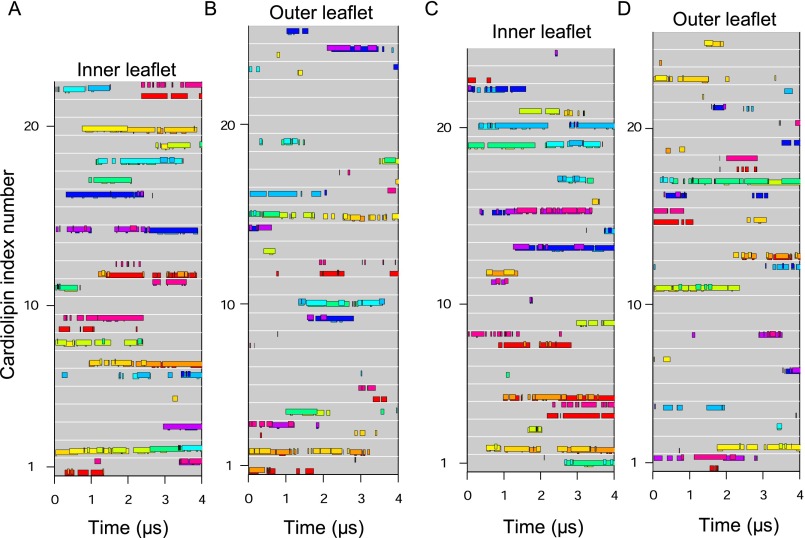
During simulations, several cardiolipins became bound to the native c8-ring simultaneously, clustered around residues TM-Lys-43 and Lys-7 (Fig. 2 A and B). A typical cardiolipin would move into the lipid annulus surrounding the c-ring, binding via its head-group in one of two modes (Fig. 3 A and B and Movie S1). In one mode, the cardiolipin spanned between two adjacent C-terminal α-helices with its acyl chains lying against their surfaces; in the other it bound to a single c-subunit. In both modes, one phosphate of cardiolipin interacted with residues Ser-48 and Gln-44 in one c-subunit and the other with either Gln-45 or TM-Lys-43 in the same subunit or its immediate neighbor. In the outer leaflet of the membrane, a cardiolipin phosphate usually became bound to Lys-7 in a single c-subunit via one or both of its phosphates (Fig. 3C). Although some cardiolipin molecules were bound to c-subunits, others diffused around the surface of the c-ring (Fig. 4 and Fig. S3). For example, in Fig. 4, cardiolipin 22 of the inner leaflet became bound first to c-subunits s2 and s3 and then to subunits s1 and s2. More cardiolipins were bound simultaneously in the inner leaflet than in the outer leaflet (Fig. 4). The residence times of cardiolipin in the inner and outer leaflets of the membrane were 500 and 220 ns, respectively, and for phosphatidyl lipids in the inner leaflet, 100 ns (Fig. 5).
Fig. 2.
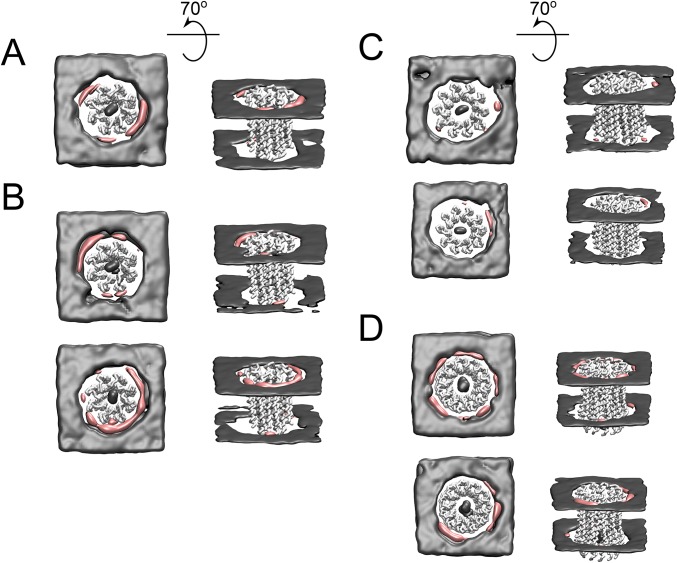
Locations of lipid head-groups around c-rings shown both from protein-centric (A) and lipid-centric (B) viewpoints. In the upper and lower lines of A, views of c-rings from, respectively, the mitochondrial matrix side of the membrane and in the plane of the bilayer (HR, head-group region). The comparative densities of the head-groups of cardiolipin around side chain beads of selected residues are shown by the relative sizes of the spheres. For each residue, the density is the maximum point on their radial distribution function. The colors of beads and amino acid residues in Fig. 1 correspond. For aligned sequences of c-subunits, see Fig. 1C. (B) Equivalent time-averaged densities of cardiolipin phosphate groups (pink) and POPC and POPE phosphate groups (gray) for the first repeat simulation of the native c8 ring. Densities for other simulations are shown in Fig. S11. The view is from the mitochondrial matrix side (Left) and with this view rotated behind the plane of the page by 70° (Right). The densities demonstrate the preferred position of cardiolipin proximal to the c-ring and the diffuse density of POPC and POPE.
Fig. 3.
Modes of binding of cardiolipin to trimethylated c8-rings. The N- and C-terminal α-helices of c-subunits are dark and light gray, respectively. Cardiolipin molecules are green, with pink phosphate groups. The large colored spheres represent the coarse grain beads for specific amino acids, as indicated, lying within 0.7 nm of the phosphate beads of cardiolipin. (A and B) Head-group region of cardiolipin in the inner leaflet of the membrane bound, respectively, to two adjacent c-subunits (subunits 8 and 1) and to a single c-subunit (the relative frequency of these two binding modes is shown in Fig. S12). (C) A cardiolipin molecule in the outer leaflet of the membrane bound to a single c-subunit.
Fig. 4.
Binding of individual cardiolipin molecules to discrete c-subunits in the trimethylated native c8-ring. (A and B) Data are shown for cardiolipin molecules in the inner and outer leaflets of the membrane, respectively. Each gray line represents a cardiolipin molecule, and colored bars along the line indicate when a cardiolipin phosphate was bound to a specific c-subunit, indicated by the color of the bar. The eight c-subunits, s1–s8, are colored as follows: s1, red; s2, orange; s3, yellow; s4, green; s5, cyan; s6, blue; s7, purple; s8, magenta.
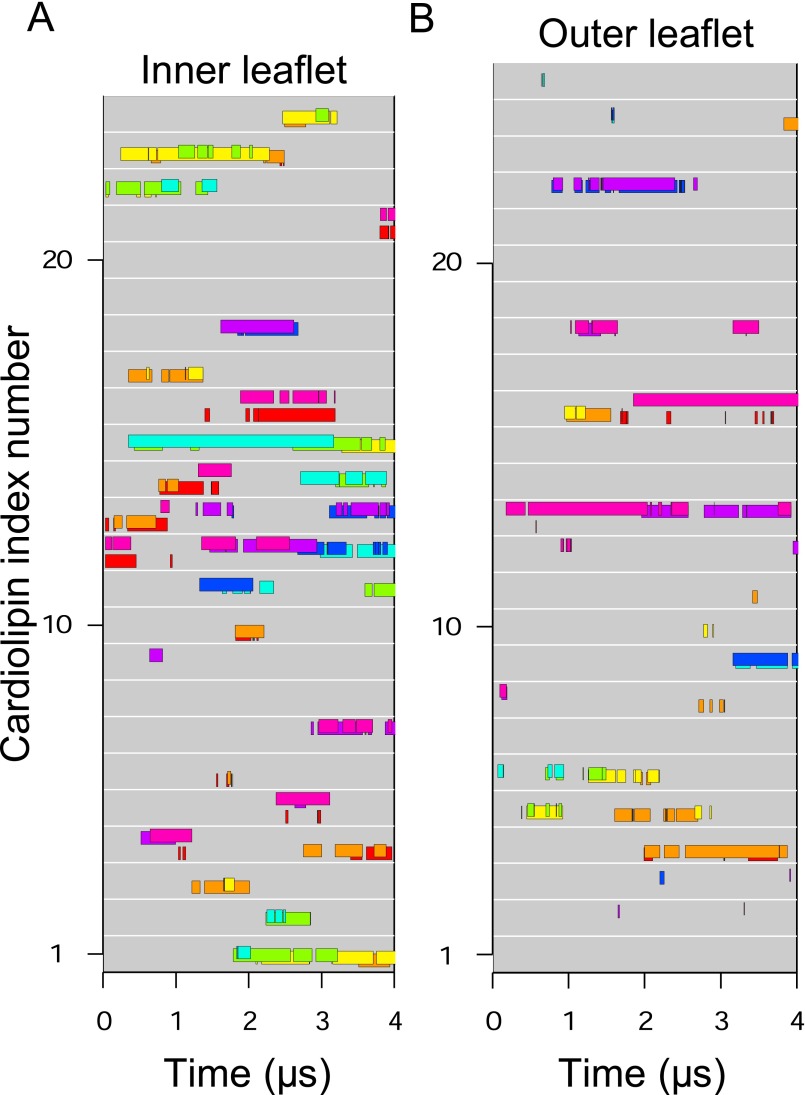
Fig. S3.
Binding of individual cardiolipin molecules to discrete c-subunits during a second simulation of the trimethylated native c8-ring. (A and B) Data are shown for cardiolipin molecules in the inner and outer leaflets of the membrane, respectively. Each gray line represents a cardiolipin molecule, and colored bars along the line indicate when a cardiolipin phosphate was bound to a specific c-subunit, indicated by the color of the bar. The eight c-subunits, s1–s8, are colored as follows: s1, red; s2, orange; s3, yellow; s4, green; s5, cyan; s6, blue; s7, purple; s8, magenta.
Fig. 5.
Residence times for lipids in association with c-rings. They correspond to the duration of binding of a lipid to any part of a c-ring. (A and B) Values for the inner and outer leaflets of the membrane, respectively, averaged over both simulations of each c-ring. The yellow and gray bars correspond to residence times of cardiolipin and POPC/POPE, respectively, in simulations containing cardiolipin, POPC and POPE. Error bars were obtained by bootstrapping.
Fig. S11.
Equivalent time-averaged densities of cardiolipin phosphate groups (pink) and POPC and POPE phosphate groups (gray) for the second repeat simulation of the native c8-ring (A), both repeats of the demthylated c8-ring (B), both repeats of the c10-ring (C), and both repeats of the c11-ring (D). The view is from the mitochondrial matrix side (Left) and with this view rotated behind the plane of the page by 70° (Right).
Fig. S12.
Relative frequency of cardiolipin phosphate binding modes in the inner leaflet. The bar graph shows the percentage of cardiolipin phosphates interacting with a single subunit (pale pink bars) as opposed to two subunits simultaneously (magenta bars) for each c-ring simulated.
The Demethylated c8-Ring.
In silico demethylation of Lys-43 increased the interaction of the cardiolipin head-group with this residue (Fig. 2 and Fig. S4), but the binding site and the residence times were unchanged (Figs. 2 and 5 and Fig. S5). The density of cardiolipin phosphates was greatest around residue Lys-43, and significant densities were observed around Ser-48, Gln-44, and Gln-45 (Fig. 2 and Fig. S4A). The densities of cardiolipin phosphates around residues Gln-44, Gln-45, and Ser-48 were similar in trimethylated and demethylated c8-rings, but the density around demethylated Lys-43 was nearly twice that observed around TM-Lys-43, reflecting the weaker Lennard Jones interaction between terminal trimethyllysine side chain bead (modified from the bead type in coarse-grained lysine) and lipid phosphates. Demethylation of Lys-43 had no effect on the interactions of cardiolipin in the outer leaflet with the c8-ring (Fig. 2).
Fig. S4.
Interactions between lipids and residues in demethylated c8-rings. Radial distribution functions were calculated for each lipid type with amino acid residues in the demethylated c8-rings. The values are the averages taken from two independent simulations. (A and B) Radial distribution functions of lipids in the inner and outer leaflets of the membrane, respectively. In each part, Upper and Lower correspond to interactions with cardiolipin and interactions with POPC and POPE, respectively. The color of each curve corresponds to a specific amino acid residue in the bovine c-subunit, as defined in Fig. 1C.
Fig. S5.
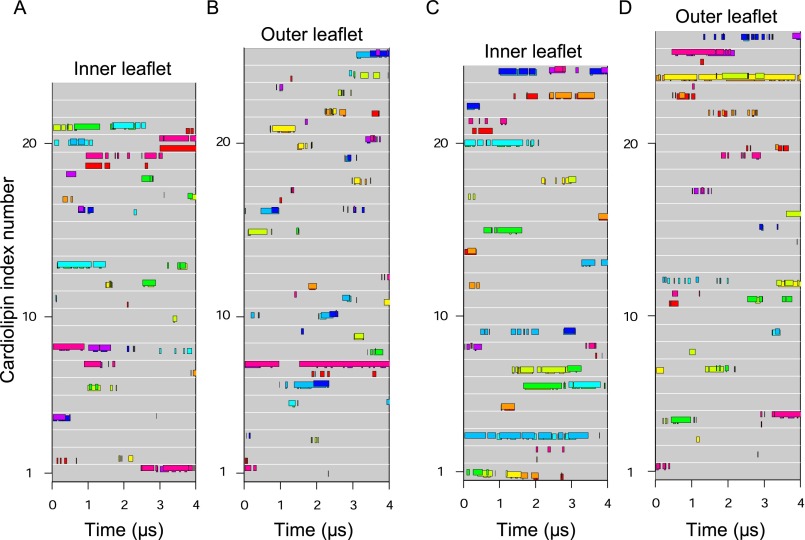
Individual cardiolipins bound to distinct c-subunits in inner and outer leaflets during both simulations of the demethylated c8-ring. (A and B) First simulation. (C and D) Second simulation. Data are shown for cardiolipin molecules in the inner (A and C) and outer (B and D) leaflets of the membrane. Each gray line represents a cardiolipin, and colored bars along the line indicate when a cardiolipin phosphate bound to a c-subunit, as indicated by bar color. The c-subunits s1–s8 are colored as follows: s1, red; s2, orange; s3, yellow; s4, green; s5, cyan; s6, blue; s7, purple; s8, magenta.
Binding of Lipids to c10- and c11-Rings.
In the c10-rings, cardiolipin phosphates in the inner leaflet of the membrane bound mostly to residue Lys-44 (equivalent to bovine Lys-43) in a single c-subunit (Figs. S6 and S7). The head-groups of cardiolipin molecules in the outer leaflet were bound to Lys-8 (Fig. S6 and Fig. 2), which is equivalent to Lys-7 in bovine c-subunit (Fig. 1C). The phosphatidyl lipids were present at a lower density around the c10-ring than cardiolipin (Fig. S6), and the greatest concentration of their head-groups in the inner leaflet of the membrane was around residues Lys-44, Asp-45 and Thr-46 (Fig. 2 and Fig. S6A), and, in the outer leaflet, around residue Lys-8 (Fig. 2 and Fig. S6B). The residence time of the cardiolipin in the inner leaflet of the membrane (300 ns) was slightly longer than in the outer leaflet (220 ns), whereas the values for the phosphatidyl lipids, 100 ns, were similar in both the inner and outer leaflets (Fig. 5).
Fig. S6.
Interactions between lipids and residues in c10-rings. Radial distribution functions were calculated for each lipid type with amino acid residues in the c10-rings. The values are the averages taken from two independent simulations. (A and B) Radial distribution functions of lipids in the inner and outer leaflets of the membrane, respectively. In each part, Upper and Lower correspond to interactions with cardiolipin and interactions with POPC and POPE, respectively. The color of each curve corresponds to a specific amino acid residue in the bovine c-subunit, as defined in Fig. 1C.
Fig. S7.
Individual cardiolipins bound to distinct c-subunits in inner and outer leaflets during both simulations of the c10-ring. (A and B) First simulation. (C and D) Second simulation. Data are shown for cardiolipin molecules in the inner (A and C) and outer (B and D) leaflets of the membrane. Each gray line represents a cardiolipin, and colored bars along the line indicate when a cardiolipin phosphate bound to a c-subunit, as indicated by bar color. The c-subunits s1–s10 are colored as follows: s1, red; s2, orange; s3, yellow; s4, pale green; s5, green; s6, cyan; s7, light blue; s8, dark blue; s9, purple; s10, magenta.
With c11-rings, cardiolipin phosphates were bound around residue Lys-50 (equivalent to bovine Lys-43) for long periods, with appreciable additional density around residues Gly-51 and Ser-55 (Fig. 2 and Fig. S8A). There was also significant density for cardiolipin phosphates about 1 nm from Asp-52, corresponding to the second coordination sphere. The region of the c11-ring in contact with the outer leaflet of the membrane has no lysine residues in the head-group region with which cardiolipin might interact. However, there was some density of cardiolipin and phosphatidyl lipids around residues Tyr-80 and Asn-82 (Fig. S8B). The residence time for cardiolipins in the inner and outer leaflets of the membrane were ca. 600 and 300 ns, the highest values observed in the simulations (Fig. 5). Moreover, at any instant, several cardiolipins were bound to the c11-ring (Fig. S9).
Fig. S8.
Interactions between lipids and residues in c11-rings. Radial distribution functions were calculated for each lipid type with amino acid residues in the c11-rings. The values are the averages taken from two independent simulations. (A and B) Radial distribution functions of lipids in the inner and outer leaflets of the membrane. In each part, Upper and Lower correspond to interactions with cardiolipin and interactions with POPC and POPE, respectively. The color of each curve corresponds to a specific amino acid residue in the bovine c-subunit, as defined in Fig. 1C.
Fig. S9.
Individual cardiolipins bound to distinct c-subunits in inner and outer leaflets during both simulations of the c11-ring. (A and B) First simulation. (C and D) Second simulation. Data are shown for cardiolipin molecules in the inner (A and C) and outer (B and D) leaflets of the membrane. Each gray line represents a cardiolipin, and colored bars along the line indicate when a cardiolipin phosphate bound to a c-subunit, as indicated by bar color. The c-subunits s1–s11 are colored as follows: s1, red; s2, dark orange; s3, pale orange; s4, yellow; s5, pale green; s6, green; s7, cyan; s8, light blue; s9, dark blue; s10, purple; s11, magenta.
Discussion
Role of Cardiolipin in Mitochondrial Enzymes.
Bound cardiolipin molecules influence the stabilities and activities of transport proteins and enzyme complexes in the inner membranes of mitochondria and may help to stabilize interactions between respiratory enzyme complexes organized in supercomplexes (36). In some instances, the mode of binding of cardiolipin to proteins has been defined structurally (37). In the ADP/ATP translocase, a cardiolipin is bound between two adjacent transmembrane α-helices, probably providing additional stability (38); in cytochrome c oxidase, a cardiolipin spans between two monomeric complexes, evidently helping to stabilize the dimer (39); and in the cytochrome bc1 complex, a cardiolipin molecule bound in the vicinity of a proton uptake pathway leading from the matrix side of the membrane to the site where quinone reduction takes place, may have a role in proton translocation through the inner membrane (40). In simulations with complexes III and IV (41, 42) and the ADP/ATP translocase, cardiolipin molecules became and remained bound to the same sites occupied by cardiolipin in their crystal structures.
In contrast, the requirement for phospholipids, including cardiolipin, for a fully functional ATP synthase is well established, and it has been proposed that about four cardiolipin molecules are bound on average to each bovine enzyme complex (43). Cardiolipin has been found to be entrapped inside the annulus of the K-ring of a V-type ATPase (44). However, no phospholipids have been detected yet in structures of intact ATP synthases, or in those of F1-c-ring and c-ring subcomplexes, but the fairly harsh conditions for purifying them would be likely to remove any bound lipids. Although the enzyme from the α-proteobacterium, Paracoccus denitrificans, has been purified with associated endogenous phospholipids (45), the resolution of the current structure is insufficient to determine whether or not any lipids are bound specifically (25).
Binding of Cardiolipin to c-Rings.
Cardiolipin, in strong preference to phosphatidyl lipids, became bound to native c8-rings at specific sites around TM-Lys-43 on the side of the membrane next to the mitochondrial matrix, and around Lys-7 on the opposite side (Fig. 2). Also, the residence times for cardiolipin around these sites and elsewhere in the lipid annulus next to the c-ring were longer than those for phosphatidyl lipids (Fig. 5). However, there was a greater tendency for cardiolipin molecules to be bound in the vicinity of TM-Lys-43 than of Lys-7. At any instant, three or four cardiolipins were bound to c-rings in the inner leaflet, similar to estimates of the number of cardiolipin molecules bound per ATP synthase complex (43). Demethylation of Lys-43 increased the density of the head-groups of cardiolipin molecules, but had little effect on their residence times. Increase in the ring size to c10 also had little effect, but increasing the ring to c11 increased both the density of cardiolipin molecules and their residence times (Figs. 3 and 4). Again, cardiolipin became bound in two modes via equivalent amino acids in the head-group regions of the lipid bilayer (Figs. 1C, 2, and 3), and it tended to diffuse around the rings (Fig. 4 and Figs. S3, S5, S7, and S9).
These simulations show that the regions of the c-subunit in contact with the head-group regions of the inner membranes of mitochondria have evolved to attract cardiolipin molecules preferentially over phosphatidyl lipids. Lysine residues, or arginine residues in some bacteria, and their positive charges are intrinsic components of many of these binding sites, although the positive charge is not an absolute requirement (Fig. S10). Irrespective of the size of the ring, and the methylation status of Lys-43 in the c8-rings, the site in the inner leaflet is more attractive than the site in the outer leaflet, and given the asymmetric distribution of cardiolipin molecules in the inner membranes of mitochondria, the site around Lys-43 is more significant than the site around Lys-7. However, in the c8-rings, the selectivity in the inner leaflet is not, as proposed (21), endowed by the complete trimethylation of Lys-43. Rather, trimethylation has little effect on the attraction of cardiolipin. The complete trimethyation of Lys-43 of the c-subunit of ATP synthase has been found uniquely throughout all classes of metazoans that have been examined, and it probably persists throughout the approximately two million species in this kingdom of life. It is an intrinsic feature of ATP synthases with c8-rings, which according to current models experience the greatest rotational torque of all c-rings, during catalysis. However, the exact role of the modification remains obscure.
Fig. S10.
Alignments of sequences of c-subunits. (A and B) Selected sequences from various eukaryotic classes and sequences of c-subunits where the structure of the c-ring has been characterized, respectively. The positions of the N- and C-terminal α-helices are shown. Conserved and substituted residues are black and red, respectively. The conserved glutamate involved in proton translocation is highlighted in blue, the lysine residues at the beginning of the C-terminal α-helix are purple, and alanine residues that influence the symmetry of the ring are green.
Role of Cardiolipin Molecules in ATP Synthase.
The sites where cardiolipins are bound permanently to complexes III and IV and ADP/ATP translocase are characterized by having positively charged and other polar residues in the region of the lipid head-group, in a groove in their surfaces where the head-groups and acyl chains can nestle. The sites of interaction of cardiolipin with c-rings also have positively charged and polar residues in the lipid head-group region, but in contrast, the convex surface of the c-ring provides no associated grooves where the phosphate groups can associate stably. Thus, cardiolipin molecules interact repeatedly, but transiently, with the c-ring via two belts of continuous binding sites formed by the repeating pattern of polar and basic residues in each c-subunit, one on the inner leaflet exposed part of the protein, and the other on the outer one, and then they diffuse away without being affected by rotation. In an active ATP synthase rotating at 100 Hz, one rotation of the c-ring takes 10 ms, and so during the ∼500 ns of interaction with a cardiolipin, the ring turns about 0.02°. At any given instant, there are approximately three to four cardiolipins interacting with the c-ring; the cumulative effect of these repetitive, but brief, specific interactions could contribute to the stability of the ring, and may also lubricate its passage through the lipid bilayer. In the integral ATP synthase complex, the c-ring and the a subunit are in intimate contact (25), leaving about 70% of the external surface of the c-ring exposed to the bilayer and available for these transient interactions. It is unlikely that any cardiolipin molecules can bind, even transiently, in this tight interface region.
Another possible function for cardiolipin in the ATP synthase, discussed before, is that its negative charges might participate directly in proton translocation through the membrane by trapping a proton in a resonance structure, leaving it with one negative charge overall (5). However, the re-estimated values of the pKa values of cardiolipin are consistent with each cardiolipin carrying two negative charges at physiological pH values, negating the proton-trap model. Nonetheless, proton transport along the membrane surface between proton pumps and ATP synthase has recently been shown to enhance ATP production by ATP synthase (46), and the negative charges of cardiolipin could still be intrinsic components of the transmembrane proton pathway. In the current structural models of the bacterial and bovine ATP synthases (19, 25, 31), the a subunit contains a bundle of four α-helices tilted at about 30° to the plane of the membrane. This bundle appears to provide two polar half channels. In mitochondria, one of them would lead from the intermembrane space, carrying protons to the essential glutamate residue in the middle of the C-terminal α-helix of a c-subunit in the a-c-ring interface. It has been proposed that once neutralized, this carboxyl residue moves by Brownian motion in a counter clockwise direction generating the first element of a rotary cycle. More negatively charged glutamates are brought successively to the proton half channel and neutralized by protons, providing further incremental rotary steps. Eventually, after being rotated through the lipid bilayer the neutralized glutamates are brought successively into a second polar half-channel in the subunit a–c-ring interface, where they reionise, regenerating the negatively charged carboxylate, and releasing the proton into the half-channel leading to the matrix of the mitochondrion (30). It is conceivable that the negative charges of cardiolipin molecules bound around TM-Lys-43 near to the exit of this second half-channel could be accepting protons from the ATP synthase and then be dispersing them to respiratory complexes. Such a mechanism would avoid the local accumulation of positive charges around the proton exit half channel. Similarly, in the outer leaflet of the inner mitochondrial membrane they could be bringing protons to the inlet channel by associating briefly in the vicinity of Lys-7. Although this mechanism can apply to both bacterial and mitochondrial ATP synthases, it cannot apply to the chloroplast enzyme, as chloroplast membranes are devoid of cardiolipin, However, they do contain the anionic lipid, sulfoquinovosyl diacylglceride, which may substitute functionally for cardiolipin (47).
Limitations and Future Directions for Simulation Work.
The current simulations, with a length of 2 × 4 μs, are short in comparison with simulations performed by Arnarez et al. of cytochrome bc1 and cytochrome c oxidase with cardiolipin (41, 42). However, the length of simulations here is justified as the symmetry of the c-ring means that the sampling is enhanced and the cardiolipin exchange at the c-ring is fast. The coarse-grained approach has the advantage of accessing timescales appropriate to observe protein–lipid interactions, with enough molecular detail to elicit meaningful results. However, secondary structure is fixed during simulations, thus preventing the observation of detailed conformational changes at protein–lipid and protein–protein interfaces. All-atom simulations of the system would provide an excellent complement to the results presented here.
Materials and Methods
Coarse-Grained Models of Proteins and Lipids.
Coordinates of c-rings were taken from the bovine F1-c8 (13), the S. cerevisiae F1-c10 (20), and the I. tartaricus c11 complexes (23) (Protein Data Bank ID codes 2XND, 2XOK and 2WGM, respectively). Each structure was converted to a coarse-grained model with MARTINI scripts (48). Parameters for POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylehanolamine) and POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine) were taken from MARTINI (49). In those for cardiolipin (50–52), each phosphate bead had a negative charge as previously (52), and in recognition of its reassessed charged state (6). Each cardiolipin model had four 18:1 acyl groups.
Coarse-Grained Simulations.
The simulations and the analysis of trajectories are described in SI Materials and Methods.
SI Materials and Methods
Conversion of Protein Structures to Coarse-Grained Models.
Transmembrane α-helices were modeled with α-helical backbone beads. In c10- and c11-rings, the region linking the two transmembrane α-helices in each c-subunit contains a short α-helix, which was modeled as such. The glutamate involved in ion translocation in the middle of the C-terminal α-helix of c-subunits was modeled in the protonated state. A coarse-grained model of TM-Lys was based on lysine and choline where the inner side chain apolar C2 bead replaced the polar C3 bead of unmodified lysine in MARTINI. The trimethylamino group was assigned a Q0 bead. The bond length between the two side-chain beads was increased from 0.28 nm in lysine to 0.34 nm in TM-Lys.
Models of Lipid Bilayers.
Two hydrated bilayer models were created by simulating their self-assembly from lipids and water with GROMACS (53), and the MARTINI force field for lipids (49) and proteins (48). Each model contained 56 POPC, 44 POPE, and 25 cardiolipin molecules, plus 375 water beads, corresponding to the composition of the inner mitochondrial membrane in porcine hearts and in rat and Guinea pig livers (54). The first system was self-assembled into bilayers during a 2.4-μs simulation, with pressure coupled isotropically and with the Berendsen barostat. Then the bilayers were simulated for 60 ns with semi-isotropic pressure coupling, but otherwise as before. A bilayer of 250 lipids was created by pasting together two copies of smaller self-assembled bilayers and simulated for a further 60 ns with semi-isotropic pressure coupling. The second bilayer was created by simulating this first bilayer for a further 1.2 μs.
Coarse-Grained Simulations.
Two coarse-grained simulations of each c-ring were run. Their starting points were generated by putting the c-ring into the two pre-equilibrated bilayers. Lipids were packed around the c-rings by the Inflategro method (55). The central cavities of c8-rings were filled with four POPC molecules, two on each side of the bilayer, and the larger cavities of the c10- and c11-rings were occupied by two POPC molecules in the inner leaflet of the membrane and four in the outer one.
Coarse-grained simulations of proteins and lipids were carried out with versions 4.5.3 and 4.5.4 of the GROMACS molecular dynamics package (56), and with the MARTINI force field (48, 49) for proteins, lipids, and water. During simulations, the temperature was maintained at 296 K and the pressure at 1 bar with a compressibility of 3 × 10−5 bar−1; periodic boundaries were applied; a time step of 30 fs was used; a dielectric constant of 15.0 was applied to electrostatic interactions; and nonbonded interactions were evaluated with a switch function between 0.0–1.2 nm for electrostatic interactions and 0.9–1.2 nm for van der Waals interactions. To equilibrate lipid and water molecules around each c-ring in equilibration simulations, water and lipids of each model were energy minimized before simulations. During equilibration simulations, beads of the c-ring were restrained with a force constant of 10,000 kJ⋅mol−1⋅nm−2. Equilibration simulations were carried out in three stages: first, as a constant volume and temperature ensemble (NVT) for 6 ns by using the Berendsen thermostat and then as a constant pressure and temperature ensemble (NPT) with the Parrinello-Rahman barostat with a time constant of 12 ps. A model was considered to be equilibrated with respect to pressure when its box dimensions had stabilized, usually within 600 ns. In a final stage of equilibration, the system was simulated for 120 ns with only the protein backbone beads restrained. In production simulations, two simulations of 4 μs were carried out with each c-ring by using the different equilibrated models. The simulations were performed under the NPT ensemble with coordinates and velocities of beads recorded every 30 ps for further analysis. Temperatures were coupled separately for lipids, protein, and water with the Nosé–Hoover thermostat with a coupling constant of 1.2 ps. The pressure was coupled semi-isotropically with the Parrinello–Rahman barostat with a coupling constant of 12 ps.
The use of the MARTINI coarse-grained parameterization in GROMACS allowed the time taken to run simulations to be reduced, and the dynamics were faster than in atomistic simulations. A conversion factor of four was used to convert raw simulation times into real times (57).
Analysis of Simulation Trajectories.
Data were analyzed with the R statistical package (58). Graphs were prepared with R and Grace (plasma-gate.weizmann.ac.il/Grace/). Coarse-grained molecular structures and simulation trajectories were visualized with VMD (59), Bendix (60), and MacPyMol (61). Fig. 2B was prepared using the VolMap tool in VMD (59) with a resolution of 0.2 nm. Radial distribution functions measure the variation of the density of beads as a function of distance from a reference bead relative to their average density in the system. They show whether lipid beads are more or less likely to be found around certain protein residues. They were calculated with GROMACS tools for lipids with respect to their distance from protein residues over simulations. Radial distribution functions of c-rings were an average over all c-subunits in a ring for simulations of both simulations of a c-ring and were smoothed using a running average over 0.05 nm. An interaction cutoff distance was defined as the location of the minimum between the first and second peaks of radial distribution functions. Particles were considered to be interacting when they were within the cutoff distance.
Protein–lipid distances were calculated from coordinates of simulations collected every 1.2 ns. The “lipid in a cage” motion was removed by smoothing trajectories with a moving window of 30 ns (41, 42). A lipid bead within 0.6 nm of any bead of the c-ring was considered to be in contact, and its head-group was considered to be interacting if at least one phosphate bead interacted with a protein bead. A whole lipid was considered to be interacting if at least four lipid beads interacted with a c-ring bead. The lipid (head-group) interaction time was the length of time a lipid (head-group) interacted continuously with the c-ring.
To characterize the dynamic behavior of lipid types, residence times were determined for the phosphate beads of lipid head-groups and for the lipid as a whole. The residence time, θ, of a lipid is defined as the average time that a single lipid continuously interacts with the surface of the c-ring. The residence time was obtained from the normalized survival time-correlation function, σ(t)
where T is the total simulation time, and Nj is the total number of a given lipid type with nonzero interaction time (41, 42, 62, 63). The function ρj(v, v + t) took the value 1 if lipid j interacted continuously with the protein from time v to time v + t (inclusive) and 0 otherwise. The value of v ran from 0 − T ns in steps of 1 ns, and values of σ(t) were determined at 1-ns intervals from 0 to T ns. σ(t) was normalized by dividing by σ(0), and so the survival time-correlation function is 1 at t = 0. The normalized time–correlation function was modeled as a single exponential function, with the rate parameter 1/θ
The value of θ was determined by fitting values of σ(t) to an exponential curve using the nonlinear least squares fitting function (nls) in the R statistical package.
Supplementary Material
Acknowledgments
We thank Drs. M. Dahlberg and S.-J. Marrink for discussion of the parameterization of coarse-grain cardiolipin and trimethyllysine, respectively, Dr. T. B. Walpole for preparing Fig. S10, and Mr. M. G. Montgomery for help in the design of other figures. We thank Prof. M. Wikström for comments on the manuscript. This work was funded by the intramural programme of the Medical Research Council (MRC) via Programme U105663150 (to J.E.W.). A.L.D. was a recipient of an MRC research studentship.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 8568.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608396113/-/DCSupplemental.
References
- 1.Schlame M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res. 2008;49(8):1607–1620. doi: 10.1194/jlr.R700018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krebs JJR, Hauser H, Carafoli E. Asymmetric distribution of phospholipids in the inner membrane of beef heart mitochondria. J Biol Chem. 1979;254(12):5308–5316. [PubMed] [Google Scholar]
- 3.de Kroon AI, Dolis D, Mayer A, Lill R, de Kruijff B. Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim Biophys Acta. 1997;1325(1):108–116. doi: 10.1016/s0005-2736(96)00240-4. [DOI] [PubMed] [Google Scholar]
- 4.Schlame M, Brody S, Hostetler KY. Mitochondrial cardiolipin in diverse eukaryotes. Comparison of biosynthetic reactions and molecular acyl species. Eur J Biochem. 1993;212(3):727–735. doi: 10.1111/j.1432-1033.1993.tb17711.x. [DOI] [PubMed] [Google Scholar]
- 5.Haines TH, Dencher NA. Cardiolipin: A proton trap for oxidative phosphorylation. FEBS Lett. 2002;528(1-3):35–39. doi: 10.1016/s0014-5793(02)03292-1. [DOI] [PubMed] [Google Scholar]
- 6.Olofsson G, Sparr E. Ionization constants pKa of cardiolipin. PLoS One. 2013;8(9):e73040. doi: 10.1371/journal.pone.0073040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sathappa M, Alder NN. The ionization properties of cardiolipin and its variants in model bilayers. Biochim Biophys Acta. 2016;1858(6):1362–1372. doi: 10.1016/j.bbamem.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kagawa Y, Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. IX. Reconstruction of oligomycin-sensitive adenosine triphosphatase. J Biol Chem. 1966;241(10):2467–2474. [PubMed] [Google Scholar]
- 9.Kopaczyk K, Asai J, Allmann DW, Oda T, Green DE. Resolution of the repeating unit of the inner mitochondrial membrane. Arch Biochem Biophys. 1968;123(3):602–621. doi: 10.1016/0003-9861(68)90181-1. [DOI] [PubMed] [Google Scholar]
- 10.Pitotti A, Contessa AR, Dabbeni-Sala F, Bruni A. Activation by phospholipids of particulate mitochondrial ATPase from rat liver. Biochim Biophys Acta. 1972;274(2):528–535. doi: 10.1016/0005-2736(72)90198-8. [DOI] [PubMed] [Google Scholar]
- 11.Santiago E, López-Moratalla N, Segovia JF. Correlation between losses of mitochondrial ATPase activity and cardiolipin degradation. Biochem Biophys Res Commun. 1973;53(2):439–445. doi: 10.1016/0006-291x(73)90681-5. [DOI] [PubMed] [Google Scholar]
- 12.Laird DM, Parce JW, Montgomery RI, Cunningham CC. Effect of phospholipids on the catalytic subunits of the mitochondrial F0.F1-ATPase. J Biol Chem. 1986;261(31):14851–14856. [PubMed] [Google Scholar]
- 13.Watt IN, Montgomery MG, Runswick MJ, Leslie AGW, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci USA. 2010;107(39):16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker JE. The ATP synthase: The understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41(1):1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 15.Gibbons C, Montgomery MG, Leslie AG, Walker JE. The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. Nat Struct Biol. 2000;7(11):1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- 16.Dickson VK, Silvester JA, Fearnley IM, Leslie AGW, Walker JE. On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 2006;25(12):2911–2918. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gledhill JR, Montgomery MG, Leslie AGW, Walker JE. How the regulatory protein, IF(1), inhibits F(1)-ATPase from bovine mitochondria. Proc Natl Acad Sci USA. 2007;104(40):15671–15676. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rees DM, Leslie AGW, Walker JE. The structure of the membrane extrinsic region of bovine ATP synthase. Proc Natl Acad Sci USA. 2009;106(51):21597–21601. doi: 10.1073/pnas.0910365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou A, et al. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife. 2015;4(e10180):e10180. doi: 10.7554/eLife.10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stock D, Leslie AG, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286(5445):1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 21.Walpole TB, et al. Conservation of complete trimethylation of lysine-43 in the rotor ring of c-subunits of metazoan adenosine triphosphate (ATP) synthases. Mol Cell Proteomics. 2015;14(4):828–840. doi: 10.1074/mcp.M114.047456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preiss L, et al. Structure of the mycobacterial ATP synthase Fo rotor ring in complex with the anti-TB drug bedaquiline. Sci Adv. 2015;1(4):e1500106. doi: 10.1126/sciadv.1500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science. 2005;308(5722):659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 24.Meier T, Ferguson SA, Cook GM, Dimroth P, Vonck J. Structural investigations of the membrane-embedded rotor ring of the F-ATPase from Clostridium paradoxum. J Bacteriol. 2006;188(22):7759–7764. doi: 10.1128/JB.00934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales-Rios E, Montgomery MG, Leslie AGW, Walker JE. Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 Å resolution. Proc Natl Acad Sci USA. 2015;112(43):13231–13236. doi: 10.1073/pnas.1517542112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier T, et al. A tridecameric c ring of the adenosine triphosphate (ATP) synthase from the thermoalkaliphilic Bacillus sp. strain TA2.A1 facilitates ATP synthesis at low electrochemical proton potential. Mol Microbiol. 2007;65(5):1181–1192. doi: 10.1111/j.1365-2958.2007.05857.x. [DOI] [PubMed] [Google Scholar]
- 27.Matthies D, et al. The c13 ring from a thermoalkaliphilic ATP synthase reveals an extended diameter due to a special structural region. J Mol Biol. 2009;388(3):611–618. doi: 10.1016/j.jmb.2009.03.052. [DOI] [PubMed] [Google Scholar]
- 28.Pogoryelov D, Yildiz O, Faraldo-Gómez JD, Meier T. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat Struct Mol Biol. 2009;16(10):1068–1073. doi: 10.1038/nsmb.1678. [DOI] [PubMed] [Google Scholar]
- 29.Vollmar M, Schlieper D, Winn M, Büchner C, Groth G. Structure of the c14 rotor ring of the proton translocating chloroplast ATP synthase. J Biol Chem. 2009;284(27):18228–18235. doi: 10.1074/jbc.M109.006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Junge W, Sabber D, Engelbrecht S. ATP-synthesis. Rotatory catalysis by F-ATPase: Real-time recording of intersubunit rotation. Berichte der Bunsengesellschaft für Phys Chemie. 2014;100(12):2014–2019. [Google Scholar]
- 31.Allegretti M, et al. Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase. Nature. 2015;521(7551):237–240. doi: 10.1038/nature14185. [DOI] [PubMed] [Google Scholar]
- 32.Symersky J, et al. Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation. Nat Struct Mol Biol. 2012;19(5):485–491, S1. doi: 10.1038/nsmb.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preiss L, et al. The c-ring stoichiometry of ATP synthase is adapted to cell physiological requirements of alkaliphilic Bacillus pseudofirmus OF4. Proc Natl Acad Sci USA. 2013;110(19):7874–7879. doi: 10.1073/pnas.1303333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulz S, et al. A new type of Na(+)-driven ATP synthase membrane rotor with a two-carboxylate ion-coupling motif. PLoS Biol. 2013;11(6):e1001596. doi: 10.1371/journal.pbio.1001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pogoryelov D, et al. Microscopic rotary mechanism of ion translocation in the F(o) complex of ATP synthases. Nat Chem Biol. 2010;6(12):891–899. doi: 10.1038/nchembio.457. [DOI] [PubMed] [Google Scholar]
- 36.Claypool SM, Koehler CM. The complexity of cardiolipin in health and disease. Trends Biochem Sci. 2012;37(1):32–41. doi: 10.1016/j.tibs.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palsdottir H, Hunte C. Lipids in membrane protein structures. Biochim Biophys Acta. 2004;1666(1-2):2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 38.Pebay-Peyroula E, et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426(6962):39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 39.Shinzawa-Itoh K, et al. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26(6):1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lange C, Nett JH, Trumpower BL, Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001;20(23):6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnarez C, Mazat JP, Elezgaray J, Marrink SJ, Periole X. Evidence for cardiolipin binding sites on the membrane-exposed surface of the cytochrome bc1. J Am Chem Soc. 2013;135(8):3112–3120. doi: 10.1021/ja310577u. [DOI] [PubMed] [Google Scholar]
- 42.Arnarez C, Marrink SJ, Periole X. Identification of cardiolipin binding sites on cytochrome c oxidase at the entrance of proton channels. Sci Rep. 2013;3(1263):1263. doi: 10.1038/srep01263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eble KS, Coleman WB, Hantgan RR, Cunningham CC. Tightly associated cardiolipin in the bovine heart mitochondrial ATP synthase as analyzed by 31P nuclear magnetic resonance spectroscopy. J Biol Chem. 1990;265(32):19434–19440. [PubMed] [Google Scholar]
- 44.Zhou M, et al. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science. 2011;334(6054):380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morales-Rios E, et al. Purification, characterization and crystallization of the F-ATPase from Paracoccus denitrificans. Open Biol. 2015;5(9):150119. doi: 10.1098/rsob.150119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nilsson T, et al. Lipid-mediated protein-protein interactions modulate respiration- driven ATP synthesis. Sci Rep. 2016;6:24113. doi: 10.1038/srep24113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haines TH. A new look at cardiolipin. Biochim Biophys Acta. 2009;1788(10):1997–2002. doi: 10.1016/j.bbamem.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Monticelli L, et al. The MARTINI coarse-grained force field: Extension to proteins. J Chem Theory Comput. 2008;4(5):819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 49.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: Coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111(27):7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 50.Dahlberg M, Maliniak A. Mechanical properties of coarse-grained bilayers formed by cardiolipin and zwitterionic lipids. J Chem Theory Comput. 2010;6(5):1638–1649. doi: 10.1021/ct900654e. [DOI] [PubMed] [Google Scholar]
- 51.Dahlberg M. Polymorphic phase behavior of cardiolipin derivatives studied by coarse-grained molecular dynamics. J Phys Chem B. 2007;111(25):7194–7200. doi: 10.1021/jp071954f. [DOI] [PubMed] [Google Scholar]
- 52.Dahlberg M, Marini A, Mennucci B, Maliniak A. Quantum chemical modeling of the cardiolipin headgroup. J Phys Chem A. 2010;114(12):4375–4387. doi: 10.1021/jp9110019. [DOI] [PubMed] [Google Scholar]
- 53.Van Der Spoel D, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 54.Daum G. Lipids of mitochondria. Biochim Biophys Acta. 1985;822(1):1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 55.Kandt C, Ash WL, Tieleman DP. Setting up and running molecular dynamics simulations of membrane proteins. Methods. 2007;41(4):475–488. doi: 10.1016/j.ymeth.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 56.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 57.Marrink SJ, de Vries AH, Mark AE. Coarse grained model for semiquantitative lipid simulations. J Phys Chem B. 2004;108(2):750–760. [Google Scholar]
- 58.R Development Core Team 2008. R: A Language and Environment for Statistical Computing. Available at https://www.R-project.org. Accessed October 1, 2010.
- 59.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 60.Dahl ACE, Chavent M, Sansom MSP. Bendix: Intuitive helix geometry analysis and abstraction. Bioinformatics. 2012;28(16):2193–2194. doi: 10.1093/bioinformatics/bts357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schrödinger 2013. The PyMOL molecular graphics system, Version 1.7.4. Available at pymol.org. Accessed January 15, 2013.
- 62.Rocchi C, Bizzarri AR, Cannistraro S. Water dynamical anomalies evidenced by molecular-dynamics simulations at the solvent-protein interface. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1998;57(3):3315–3325. [Google Scholar]
- 63.Periole X, Rampioni A, Vendruscolo M, Mark AE. Factors that affect the degree of twist in beta-sheet structures: A molecular dynamics simulation study of a cross-beta filament of the GNNQQNY peptide. J Phys Chem B. 2009;113(6):1728–1737. doi: 10.1021/jp8078259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.