Significance
Management of oxidative stress in plant chloroplasts involves signaling pathways to the nucleus that trigger stress response mechanisms. Yet, how oxidative stress is initially sensed in the chloroplast to activate accumulation of a stress signal remains enigmatic. We show that inactivation of a phosphatase, SAL1, by oxidative stress in chloroplasts controls accumulation of its substrate, as a plant stress signal. This regulatory mechanism is highly conserved across the plant kingdom and confers a second function to this metabolic enzyme as an oxidative stress sensor.
Keywords: retrograde signaling, redox regulation, stress sensing, chloroplast, drought stress
Abstract
Intracellular signaling during oxidative stress is complex, with organelle-to-nucleus retrograde communication pathways ill-defined or incomplete. Here we identify the 3′-phosphoadenosine 5′-phosphate (PAP) phosphatase SAL1 as a previously unidentified and conserved oxidative stress sensor in plant chloroplasts. Arabidopsis thaliana SAL1 (AtSAL1) senses changes in photosynthetic redox poise, hydrogen peroxide, and superoxide concentrations in chloroplasts via redox regulatory mechanisms. AtSAL1 phosphatase activity is suppressed by dimerization, intramolecular disulfide formation, and glutathionylation, allowing accumulation of its substrate, PAP, a chloroplast stress retrograde signal that regulates expression of plastid redox associated nuclear genes (PRANGs). This redox regulation of SAL1 for activation of chloroplast signaling is conserved in the plant kingdom, and the plant protein has evolved enhanced redox sensitivity compared with its yeast ortholog. Our results indicate that in addition to sulfur metabolism, SAL1 orthologs have evolved secondary functions in oxidative stress sensing in the plant kingdom.
Alleviating oxidative stress is a common challenge across evolution, occurring at the cellular, organellar, and systemic levels. In plant chloroplasts, drought and high-light (HL) stress induce production of reactive oxygen species (ROS) such as singlet oxygen (1O2) at photosystem II (PSII) and hydrogen peroxide (H2O2) as well as superoxide (O2−) at photosystem I (PSI) (1). There is also a shift from reducing to more oxidizing states in the redox poise of plastoquinone (PQ) and other stromal redox couples such as glutathione (GSH/GSSG). All of these changes are associated with adjustment of photosystem stoichiometry and chloroplastic metabolic enzymes by chloroplast-resident kinases (2) and redox-sensitive thioredoxins (TRXs) (3), respectively, as well as activation of signaling pathways for the induction of common and unique sets of nuclear genes (4, 5).
The nuclear transcriptional response to stress in chloroplasts is mediated by chemical signals emanating from the chloroplasts to the nucleus in a process called retrograde signaling (6). There are at least seven distinct retrograde signaling pathways responding to changes in chloroplastic ROS and redox state (7), including beta-cyclocitral for PSII-1O2 responses (8) and the PAP–XRN pathway which alters expression of 25% of the HL-associated transcriptome, many of which are ROS and redox associated (9). The unique gene sets which expression are induced by PSI ROS and changes in chloroplast redox poise are collectively referred to herein as plastid redox associated nuclear genes (PRANGs) (7); they include key and common stress marker genes such as ASCORBATE PEROXIDASE 2 (APX2) (10, 11) and ZAT10 (12) critical for acclimation. The nuclear regulators of PRANGs and the subsequent chloroplast-targeted stress responses, including induction of chloroplast antioxidant and redox regulation enzymes such as redoxin proteins, have been extensively elucidated for the different retrograde pathways (7, 12). Despite these advances, however, in all of the PRANG retrograde signaling pathways no chloroplastic sensor(s) of ROS and redox state has been conclusively identified (7). For instance, a previously hypothesized sensor kinase for the PQ redox state (2) has recently been reascribed to facilitate H2O2 production rather than redox sensing per se (13).
A substantial proportion of PRANGs are regulated by the phosphonucleotide, 3′-phosphoadenosine 5′-phosphate (PAP), which acts as a mobile chloroplast-to-nucleus stress retrograde signal (9). PAP accumulation is induced by drought and high-light stress, and the metabolite signal moves between the chloroplast, cytosol, and nucleus (9). PAP is produced by sulfotransferase-catalyzed sulfation reactions in secondary sulfur metabolism, which transfer activated sulfate from 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to various key acceptor molecules including peptides and hormones (14). This sulfate transfer generates PAP as a by-product that inhibits sulfotransferase activity and feedback-regulates overall sulfur flux (14, 15). During unstressed conditions, PAP is enzymatically degraded by the Arabidopsis thaliana SAL1 (AtSAL1) phosphatase in the chloroplast (9). AtSAL1 loss-of-function leads to constitutive PAP accumulation, up-regulation of PRANGs, increased stress tolerance, and altered sulfur metabolism (9, 15). Hence, SAL1 and PAP perform dual functions in sulfur metabolism (15) and stress signaling (9). The role of SAL1–PAP in chloroplast stress signaling is likely conserved beyond Arabidopsis to other members of the plant kingdom because transient silencing of SAL1 also enhanced stress tolerance in wheat (Triticum aestivum) (16).
PAP controlling PRANG expression during drought and HL has known degradation and production site(s) for the signal, a mechanism for signal movement, and a protein target for the signal (7, 9). However, similar to all other PRANG-regulating retrograde pathways, the mechanism by which chloroplast oxidative stress and redox state are initially sensed and transduced is unknown. Here we demonstrate that contrary to expectation for a metabolic enzyme, SAL1 can in and of itself act as a molecular sensor for oxidative stress. The switches for accumulation of the PAP chloroplast retrograde signal reside within the SAL1 protein, thereby providing a common site of perception of PSI ROS and redox couples for regulating PRANGs.
Results
Accumulation of the Stress Signal PAP Occurs via Oxidative Down-Regulation of AtSAL1 Activity.
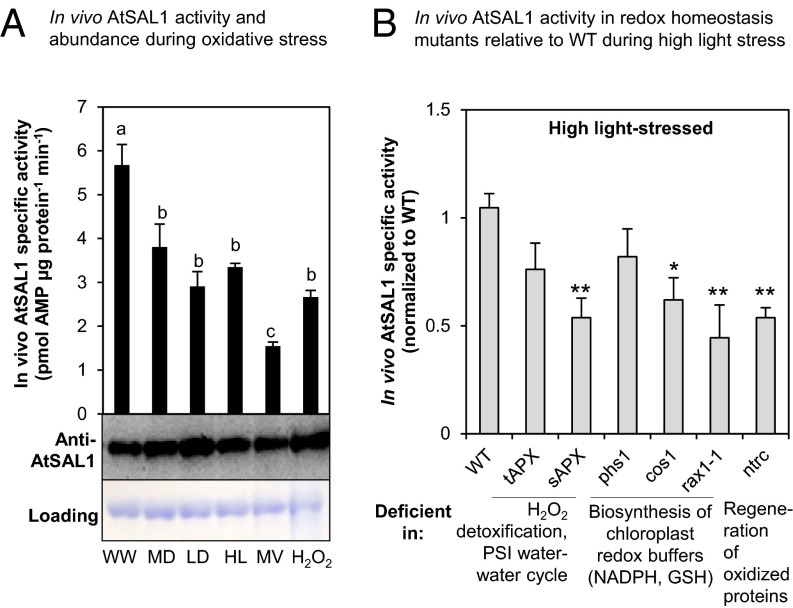
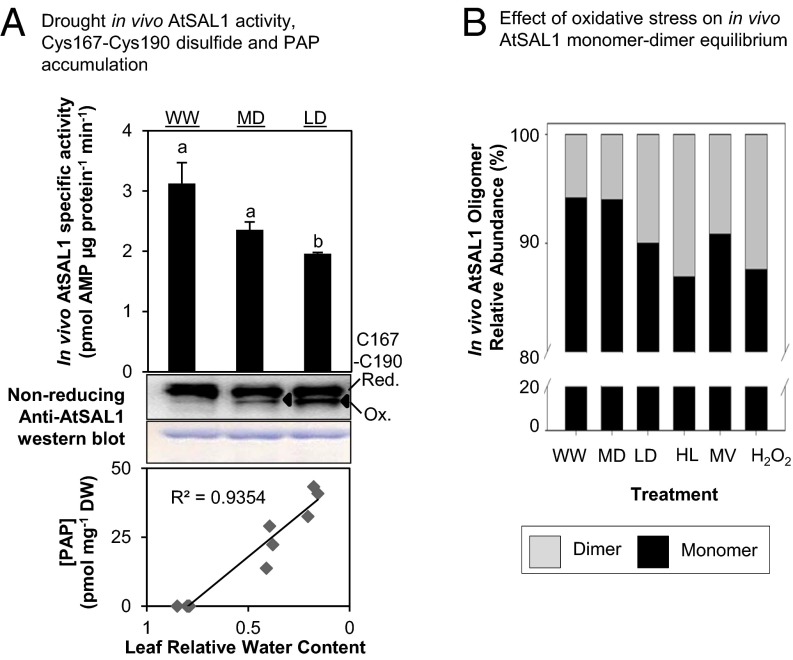
Given the multiple redox couples altered in the chloroplast in response to HL and drought (1, 14), we hypothesized that PAP accumulation during these stress conditions (9) can be regulated by AtSAL1 if the enzyme acted as a sensor of, and its activity regulated by, oxidative stress. Indeed, PSI-sourced ROS that invoke PRANG regulation such as H2O2 and O2− significantly lowered in vivo AtSAL1 activity (Fig. 1A). This was replicated in plants exposed to the abiotic stresses drought and HL that induce H2O2 and O2− formation (Fig. 1A). These treatments did not significantly alter AtSAL1 protein abundance, however (Fig. 1A), suggesting that the regulation of AtSAL1 activity in the chloroplast is therefore most likely via a posttranslational mechanism(s).
Fig. 1.
In vivo AtSAL1 activity is down-regulated by oxidative stress. (A) In vivo AtSAL1 activity is down-regulated by oxidative stress with negligible change in protein abundance (WW, well watered; MD, middrought; LD, late drought; HL, high light; MV, methyl viologen; H2O2, hydrogen peroxide). Activity was measured without any reducing agent, whereas protein electrophoresis and Western blotting were performed under reducing conditions for optimal protein transfer from gel to membrane. Similar results were obtained from two independent experiments. Means and SE for three to four biological replicates per treatment are shown. a, b, and c (P < 0.05) show significant differences between treatments. (B) In vivo AtSAL1 activity is sensitive to redox state. Disrupting redox homeostasis at photosystem I water–water cycle, ascorbate detoxification of ROS, cellular redox buffers, or regeneration of oxidized proteins (also see Table S1) results in significantly greater (*P < 0.05, **P < 0.01) down-regulation of AtSAL1 activity compared with WT under high-light stress. Some mutants show a trend of down-regulation in activity, but the differences were not significant. Means and SE for averaged relative activities of two biological replicates at three different concentrations of PAP per genotype are shown.
It is well established that in mutants deficient in regeneration of oxidized proteins, redox buffer homeostasis (NADPH/NADP+ and GSH/GSSG), or the water–water cycle that degrades PSI O2− and H2O2 via ascorbate (1), there are increased ROS accumulation, shifts in redox balance toward more oxidizing states, and /or deregulated PRANG expression (Table S1). In six of such mutants, in vivo AtSAL1 activity in response to HL-induced oxidative stress was further reduced by up to 50% relative to wild type (WT) (Fig. 1B).
Table S1.
Summary of redox homeostasis-deficient mutants used in Fig. 1
| Name | Gene identifier | Type of mutation | Biological pathway affected | Ref. |
| tAPX (thylakoidal ascorbate peroxidase) | AT1G77490 | amiRNA-knockdown | Detoxification of H2O2 in the chloroplast thylakoid and detoxification of superoxide by superoxide dismutase in water–water cycle at photosystem I (PSI). | 1 |
| sAPX (stromal ascorbate peroxidase) | AT4G08390 | T-DNA insertion, homozygous | Detoxification of H2O2 in the chloroplast stroma, where AtSAL1 is also located. | 1 |
| phs1 (photosensitive 1) | AT3G47390 | T-DNA insertion, homozygous | Involved in riboflavin and FAD synthesis. Mutant allele has increased oxidative stress due to reduced NADPH/NADP+ ratios and overproduction of ROS at PSI under high light. | 53 |
| cos1 (coronatine insensitive1 suppresor) | AT2G44050 | T-DNA insertion, heterozygous | Involved in the same metabolic pathway as phs1 (riboflavin and FAD synthesis). | 59 |
| rax1-1 (regulator of ascorbate peroxidase 2 1-1) | AT4G23100 | Point mutation, homozygous | Rate-limiting step of glutathione synthesis. GSH:GSSG redox ratio may be altered under oxidative stress. | 54 |
| ntrc (NAPDH-dependent thioredoxin reductase C) | AT2G41680 | T-DNA insertion, homozygous | Reduction of 2-Cys peroxiredoxins and redox control within the chloroplast. Deficiency causes hypersensitivity to abiotic stress. | 52 |
Loss-of-function mutants deficient in various aspects of redox homeostasis in the chloroplast were chosen for analysis of AtSAL1 activity under abiotic stress. The mutants are deficient in ROS detoxification, redox control at PSI, or chloroplast redox buffer pathways.
AtSAL1 Is Redox-Regulated by Dual Mechanisms of Intramolecular Disulfide Formation and Dimerization.
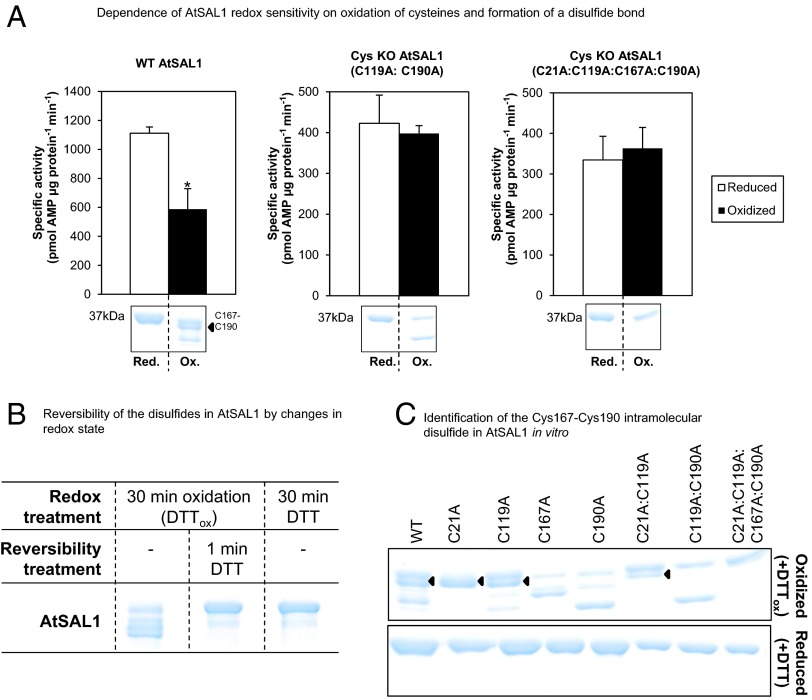
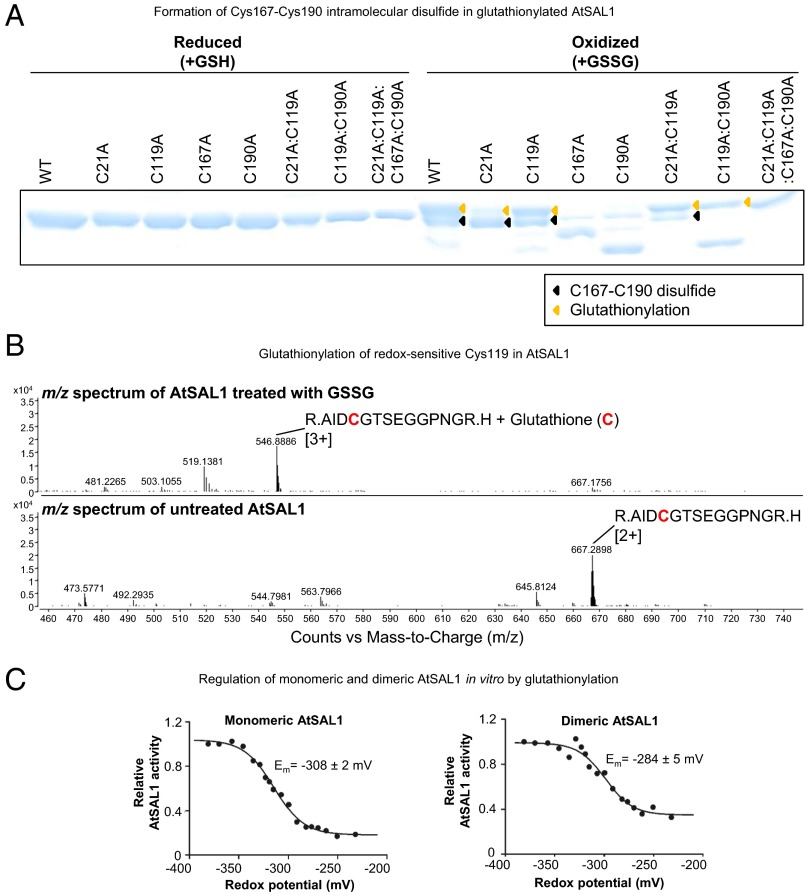
The suppression of in vivo AtSAL1 activity under oxidative stress conditions (Fig. 1A), coregulation of PRANG expression by photosynthetic ROS and PAP (4, 5, 9), and redox regulation of some sulfur metabolism enzymes in chloroplasts (14) led us to hypothesize that down-regulation of in vivo AtSAL1 activity is directly mediated by redox potential. Indeed, in vitro AtSAL1 activity is decreased under oxidizing conditions (Fig. 2A). The amino acid sequence of AtSAL1 includes four cysteine residues as potential targets for redox regulation (Cys21, Cys119, Cys167, and Cys190). When these cysteine residues were mutated to redox-insensitive alanine residues, total activity decreased 50%, but more importantly, oxidative down-regulation of AtSAL1 activity was lost (Fig. 2A). The down-regulation of AtSAL1 activity coincided with the appearance of multiple protein bands on SDS/PAGE (Fig. 2A, Bottom). Significantly, the multiple band patterns of oxidized AtSAL1 on SDS/PAGE were reversible upon addition of reducing agent (Fig. 2B); the altered migration of proteins with disulfide bond(s) due to intramolecular loop formation is well established (17, 18).
Fig. 2.
Regulation of AtSAL1 activity by redox state via its intramolecular Cys167–Cys190 disulfide. (A) Down-regulation of AtSAL1 activity by oxidation requires oxidation of cysteines because mutagenesis of cysteines to alanine in AtSAL1 abrogated redox sensitivity. The redox sensitivity correlates with a band directly beneath the full-length reduced protein (black arrow), which was determined to be a Cys167–Cys190 intramolecular disulfide (C). Vertical dashed lines indicate splicing and truncation of the gel shown in full in C. *P < 0.1. Activities of all proteins were assayed in the presence of 13.4 µM PAP. Means and SE of two independent experiments are shown. (B) Formation of disulfides in AtSAL1 by oxidation is rapidly reversed by returning the redox state to reducing conditions. Vertical dashed lines indicate splicing of the gel to show these three samples side by side; all samples were run on the same gel. (C) Determination of Cys–Cys disulfide pairs observed in WT AtSAL1 using cysteine to alanine substitution mutants of AtSAL1 under oxidation. Oxidized AtSAL1 proteins migrate at different rates to reduced AtSAL1 protein. The different Cys–Cys disulfide pairs were identified by cross-comparison with cysteine mutants: AtSAL1 containing a Cys167–Cys190 intramolecular disulfide (black triangles) migrates closest to reduced AtSAL1. The oxidized form is absent in all AtSAL1 mutants lacking either or both of Cys167 and Cys190. Other combinations such as the Cys21–Cys167 and Cys21–Cys190 disulfide did not correlate with the down-regulation of AtSAL1 activity by oxidation (A). These are likely nonspecifically formed during protein denaturation and SDS/PAGE. AtSAL1 containing the Cys167–Cys190 intramolecular disulfide is the only oxidized AtSAL1 species detected in endogenous plant protein samples pretreated with iodoacetamide to block reduced cysteines during protein extraction to prevent nonspecific disulfide formation (Fig. 6). Experiments were performed twice, with identical results.
Combinatorial analysis of Cys to Ala mutations facilitated identification of the Cys pairs involved in the dominant disulfide bonded bands (Fig. 2C). This revealed two important features of the oxidation/inactivation process: first, full oxidation of recombinant AtSAL1 was not possible, and only a fraction of the protein was converted to anomalously migrating oxidized bands; second, a specific, dominant band that was observed when the WT protein was oxidized was lost when either Cys167 or Cys190 was mutated to Ala, suggesting that they form a disulfide bond. Kinetic analysis of Cys to Ala mutations showed that when Cys119 or Cys190 was mutated to Ala, no loss of activity was observed in oxidizing conditions, suggesting that Cys167–Cys190 and Cys119 may be required for inactivation of AtSAL1 (Fig. 2A).
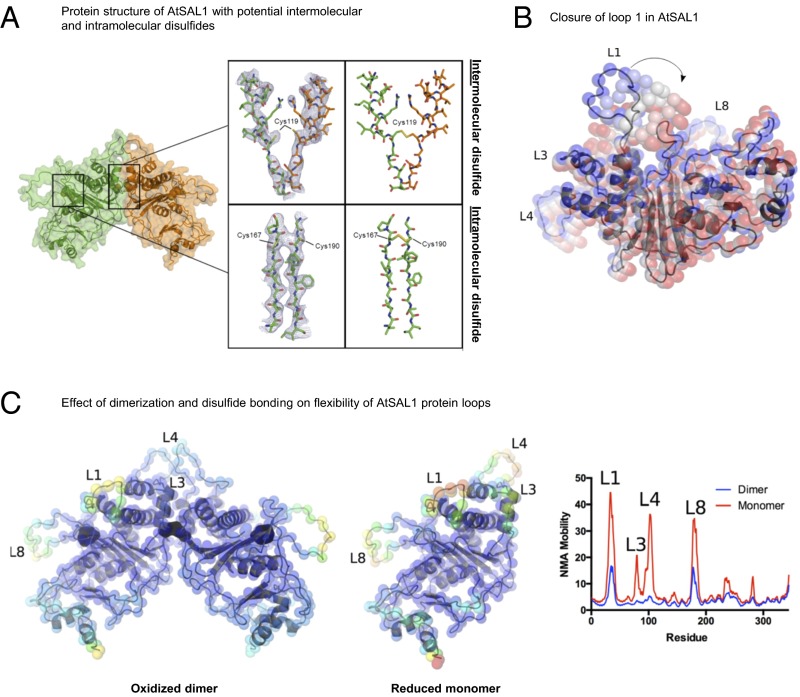
To investigate this redox sensing mechanism in more detail, we subsequently crystallized and solved the molecular structure of AtSAL1 to 3.05-Å resolution in its apo form (PDB 5ESY; Fig. 3 and Table S2). AtSAL1 is an α/β protein belonging to the carbohydrate phosphatase fold and superfamily (19) showing the closest structural homology to the yeast (Saccharomyces cerevisiae) PAP phosphatase ortholog, ScHAL2 [PDB 1KA1 (20), rmsd for Cα atoms = 2.4 Å, with amino acid sequence identity of 37% as calculated by the distance alignment (DALI) server (21)]. We found that AtSAL1 crystallized as a dimer; a crystallographic twofold interface was clearly visible and detected with the Protein Interfaces, Surfaces and Assemblies server (22). The dimer interface is centered on a symmetrical pair of Cys119 side chains from each monomer in the dimer (Fig. 3A), suggesting a role for an intermolecular disulfide in dimerization. Each monomer also contains a potential intramolecular Cys167–Cys190 disulfide pair located across adjacent beta strands (Fig. 3A); such a cross-strand disulfide is often a metastable switch used to control protein activity (23). Cys21 was not located near any potential disulfide bonding partners. Notably, none of the cysteine residues are located in the vicinity of the active site, suggesting that the regulation must be remote, or allosteric.
Fig. 3.
Structural basis for redox regulation of AtSAL1 activity. (A) (Left) Structural elucidation of AtSAL1 reveals a dimerization interface and three potentially redox-sensitive cysteine residues. (Middle) A view of the 2mFo-dFc map (blue lines, contoured at 1.0 σ) centered on Cys119 which is located at the interface between chain A (orange sticks) and chain B (green sticks) or a view of the 2mFo-dFc map centered on Cys167 and Cys190. (Right) The disulfide bonds present in an energy minimized model of the oxidized AtSAL1 dimer. (B) Closure of loop 1 of AtSAL1, as predicted by normal mode analysis (NMA) (31). The lowest-frequency normal mode is shown. Positions of Cα atoms are shown as colored spheres, from the crystal structure (blue) to the most closed conformation (red). (C) Dimerization and disulfide formation reduces the mobility of key loops (loops 1, 3, 4, and 8) in AtSAL1. Energy minimized models of the oxidized AtSAL1 dimer (Left) or the reduced AtSAL1 monomer (Middle) are colored according to mobility (blue indicating least mobile and red indicating most mobile); for details of energy minimization and normal mode analysis, see Materials and Methods. (Right) Plot of NMA mobility by residue for the oxidized dimer and reduced monomer.
Table S2.
Data collection and refinement statistics for crystal structure of AtSAL1 (PDB 5ESY)
| Space group | P61 |
| Data processing | |
| Cell dimensions (Å) a = b, c | 137.1, 74.6 |
| Resolution range, Å | 38.25–3.05 (3.24–3.05)* |
| Total number of reflections | 125243 (20978) |
| Number of unique reflections | 15341 (2522) |
| Completeness, % | 99.6 (92.8) |
| Mean I/σ(I) | 15.2 (2.2) |
| Wilson B factor, Å2 | 83.6 |
| CC1/2† | 0.999 (0.620) |
| Rmerge | 0.104 (1.11) |
| Refinement | |
| Rwork/Rfree, % | 22.7/25.8 (33.5/41.8) |
| Total number protein of atoms | 5082 |
| RMSD for bonds, Å | 0.004 |
| RMSD for angles, deg | 1.00 |
| Ramachandran favored, % | 86 |
| Ramachandran outliers, % | 1.9 |
| Clashscore | 12 |
| Average B factor, Å2 | 106.0 |
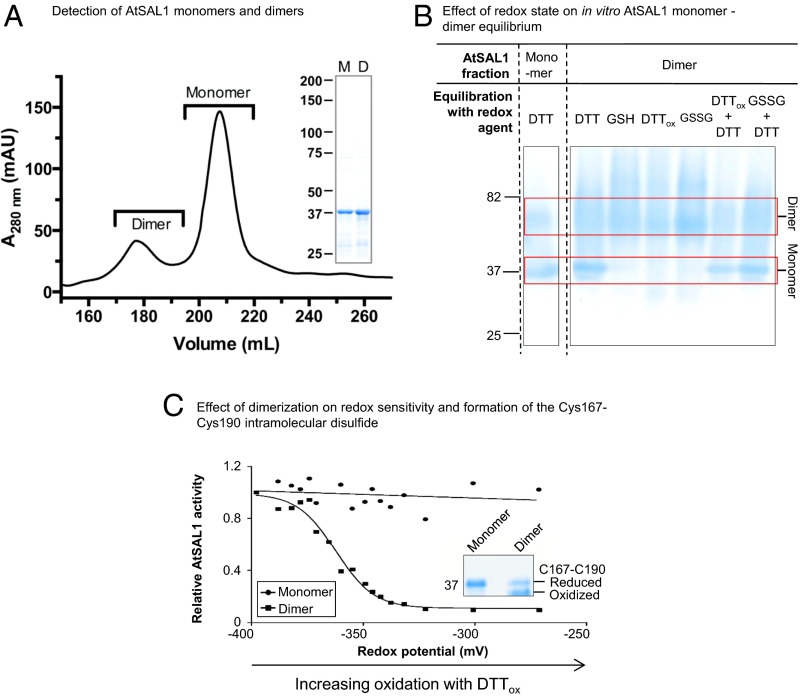
Although previously reported to be a monomer (24), we found that AtSAL1 exists in monomer–dimer equilibrium in solution through size exclusion chromatography (SEC) and SEC–multiangle laser light scattering (MALLS) (Fig. 4A and Fig. S1). Native PAGE analysis of recombinant AtSAL1 reveals that the dimeric fraction is stable under oxidizing conditions, owing to the presence of an intermolecular disulfide, but under sufficiently reducing conditions the protein returns to equilibrium between monomer and dimer (Fig. 4B). The monomeric and dimeric species displayed strikingly different redox sensitivity: whereas the monomer displayed high catalytic activity and is resistant to oxidation and inactivation, the dimer is less active under reducing conditions but is extremely sensitive to oxidation and is rapidly inactivated, with formation of disulfide bonds, including the intramolecular Cys167–Cys190 bond, in the presence of oxidized DTT (Fig. 4C and Table 1). Thus, it appears that dimerization is a precursor to oxidative inactivation and that the oligomeric equilibrium can be altered under oxidizing conditions through the formation of an intermolecular disulfide bridge. This result is consistent with the data presented in Fig. 2 showing that the mixed oligomeric species of AtSAL1 do not undergo full oxidation owing to the resistance of monomer to oxidation and that the Cys119 and Cys190 residues are involved in separate disulfide bonds that are both important for inactivation.
Fig. 4.
AtSAL1 is also regulated via dimerization involving Cys119. (A) AtSAL1 in monomer–dimer equilibrium detected in size exclusion chromatography of purified recombinant protein. Dimers were detected in at least three independent purification runs. (Inset) SDS/PAGE of monomeric and dimeric AtSAL1 indicating the proteins were of similar purity. Monomer and dimer masses were confirmed by SEC–MALLS (Fig. S1). (B) The monomer–dimer equilibrium can be shifted by an intermolecular disulfide under oxidizing conditions, thus increasing dimer abundance, or reduction of the disulfide by DTT dissociating the dimer. Whereas DTT is sufficient to achieve dimer separation, GSH is not. This is consistent with the relative redox potentials of these compounds: −264 mV at pH 7.4 for GSH compared with DTT (−360 mV) and the redox potential of disulfide bonds (ranging from −330 to −95 mV in thiol–disulfide oxidoreductases). Oxidation (DTTox, GSSG) increased dimer abundance to 100%, indicating formation of an intermolecular disulfide dimer under oxidation. Reversing the oxidation with reductant (DTTox + DTT and GSSG + DTT lanes) that breaks disulfide bonds shifts the equilibrium to monomer. The low resolution and fuzzy appearance of the higher MW bands are likely due to the type of gel (Tris-Glycine), lack of detergent (SDS), or reductant (DTT) that inhibits resolving native, folded proteins that are oxidized. Similar results were obtained in two independent experiments. (C) Under redox titration by DTTox in vitro, which induces formation of the Cys167–Cys190 disulfide, only dimeric AtSAL1 showed significant down-regulation of activity. Identical results were obtained from two independent experiments. (Inset) Dimerization is required for formation of the Cys167–Cys190 intramolecular disulfide that regulates AtSAL1 activity.
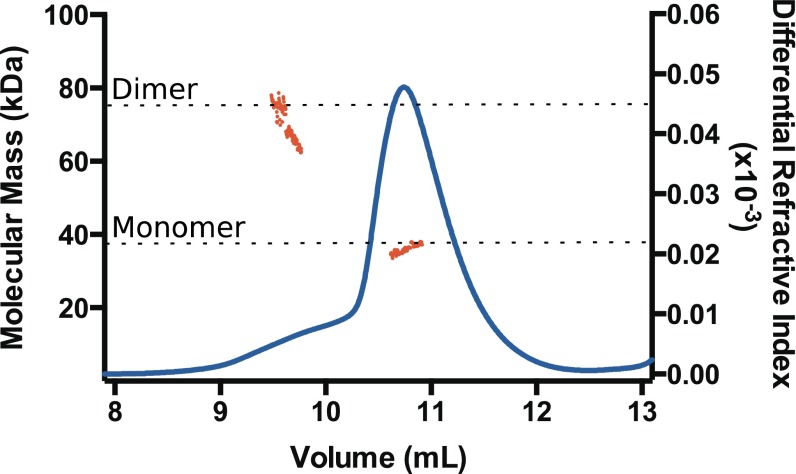
Fig. S1.
AtSAL1 exists in a monomer–dimer equilibrium. Purified AtSAL1 was exchanged into assay buffer and analyzed by MALLS following separation by size exclusion chromatography. Differential refractive index is shown as a blue line (right axis), and molecular mass derived from light scattering data is shown as red points (left axis). The expected masses of AtSAL1 monomer (37.5 kDa) and dimer (75 kDa) are indicated by dashed lines.
Table 1.
Enzyme kinetics under different redox states for two oligomeric forms of AtSAL1 and its orthologous rice protein, OsSAL1
| Kinetic parameters | Monomeric AtSAL1 | Dimeric AtSAL1 | OsSAL1 | |||
| Reduced | Oxidized | Reduced | Oxidized | Reduced | Oxidized | |
| kcat, min−1 | 123 ± 18 | 119 ± 13 | 24.4 ± 1.1 | 4.3 ± 1.3 | 44 ± 3.9 | 11 ± 1.5 |
| KM, µM PAP | 9.9 ± 3.3 | 8.3 ± 1.2 | 8.7 ± 5.2 | 4.2 ± 1.8 | 20 ± 6.9 | 61 ± 23.6 |
| kcat/KM, µM−1⋅min−1 | 12.4 | 14.2 | 2.8 | 1.0 | 2.2 | 0.2 |
Enzymatic assays were performed at 25 °C with 0.2 µg recombinant AtSAL1 in the presence of 5 mM DTT (reduced) or DTTox (oxidized). Results shown are means and SE from two independent experiments. For OsSAL1, activity of 0.2 µg recombinant protein was assayed at 25 °C in the presence of 20 mM GSH (reduced) or GSSG (oxidized). Experiment was performed twice, with similar results.
Given that dimerization appears to be essential for the intramolecular Cys167–Cys190 disulfide bond formation and that oxidation can inactivate AtSAL1, we sought to investigate the mechanisms that underlie these processes. Allosteric regulation of protein activity is commonplace (25) and frequently involves alteration of protein dynamics and trapping of proteins in inactive conformations (26–29). Thus, we performed molecular dynamics (MD) energy minimization to generate monomeric (reduced) and dimeric (oxidized) AtSAL1 models to investigate the effect of dimerization and the disulfide bonds on the protein structure and dynamics. Normal mode analysis of elastic network models (NMA-ENM) can effectively predict and analyze large-scale collective motions in proteins (30, 31) and was used here to investigate the effect of the Cys119–Cys119 and Cys167–Cys190 disulfide bridges and dimerization on the flexibility of AtSAL1.
As shown in Fig. 3B, loop 1 overhangs the active site in an open conformation; the equivalent loop to loop 1 in the AtSAL1 ortholog, ScHAL2, has been shown to close over the active site to stabilize substrate (20). In AtSAL1, NMA-ENM indicates that loop 1 fluctuates between open and closed conformations in a manner common to active site loops in almost all phosphatases (Fig. 3B) (32, 33), suggesting that this movement is likely to be important for catalytic activity. Loops 3 and 4 are located at the dimer interface and adjacent to loop 1, whereas loop 8 is also adjacent to loop 1.
The mobility of these loops in the oxidized dimer and reduced monomer are strikingly different, with loops 1, 3, 4, and 8 all being substantially rigidified through dimerization and disulfide bond formation (Fig. 3C). The dynamic coupling of these loops is also significantly different between the monomer and dimer, and these loops become significantly more hindered in the dimer (Fig. S2). These data also allow a plausible explanation for the resistance of the monomer to oxidation because the formation of the Cys167–Cys190 disulfide bond will require the two residues to be located in close proximity and to be relatively stable, which would be the case in the dimer but not the monomer. Thus, the decreased activity of AtSAL1 in the dimerized/oxidized state likely results from the rigidification of the active site loops, allosterically inhibiting the enzyme by preventing it from adopting conformations that are essential for activity and substrate binding. Indeed, both kcat and KM are affected by dimerization and oxidation (Table 1). This result is consistent with the current view of allosteric inhibitory regulation of proteins (28, 29) and provides a rapid means of reversible enzyme inactivation.
Fig. S2.
Prediction of coupled motions in AtSAL1. Covariance maps of Cα motions (74) calculated for the reduced AtSAL1 monomer and oxidized AtSAL1 dimer. Residues with coupled motions are shown in red, and residues with anticorrelated motions are shown in blue. The positions of the mobile loops described in Fig. 3C (loops 1, 3, and 8) are indicated.
Formation of the Intramolecular Disulfide Bond Controlling AtSAL1 Activity Can Be Mediated by the Chloroplast Redox Buffer GSH/GSSG.
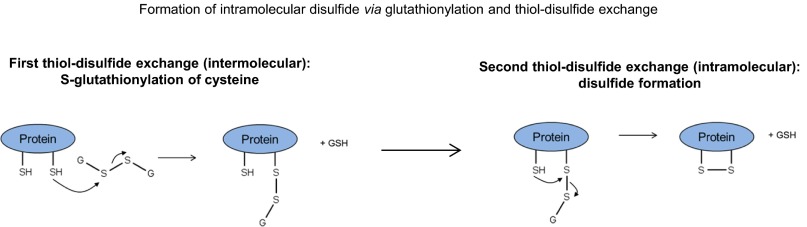
We then investigated whether in vitro AtSAL1 inactivation by the redox regulatory mechanisms shown in Fig. 3 can be induced by in vivo redox couples present in the chloroplast, such as glutathione (GSH). During oxidative stress, GSH can be oxidized to oxidized glutathione (GSSG). GSSG is known to glutathionylate cysteine residues of chloroplast proteins to regulate their activity (3), and it promotes formation of an intramolecular disulfide bond between proximal cysteine residues via thiol–disulfide exchange (34) (Fig. S3). Therefore, we tested whether glutathionylation may also induce formation of the Cys167–Cys190 bond in AtSAL1. The GSSG-treated AtSAL1 was able to form the Cys167–Cys190 disulfide in all recombinant proteins containing both residues (Fig. 5A). Additionally, decreased activity in GSSG-treated AtSAL1 correlated with glutathionylation of Cys119 and Cys190 as detected by mass spectrometry (Fig. 5B). Critically, glutathionylation down-regulated activity in both monomeric and dimeric AtSAL1 samples (Fig. 5C). The redox titration of both monomeric and dimeric AtSAL1 with GSH/GSSG yielded redox midpoint potentials (Em) close to the physiological glutathione redox potential (−317 ± 8 mV) in Arabidopsis chloroplasts (35) (Fig. 5C). Therefore, two redox processes can decrease AtSAL1 activity: the first involving dimerization and intermolecular disulfide bonding and the second a dimerization-independent process involving glutathionylation by the chloroplast redox couple GSH/GSSG. Both mechanisms result in the formation of the Cys167–Cys190 intramolecular disulfide and down-regulation of AtSAL1 activity.
Fig. S3.
Formation of an intramolecular disulfide via a thiol–disulfide exchange initiated by glutathionylation of a cysteine residue. See ref. 34 for review of this mechanism.
Fig. 5.
AtSAL1 can be regulated by glutathionylation at redox-sensitive cysteines. (A) Glutathionylation of AtSAL1 with oxidized glutathione (GSSG; yellow arrows) results in formation of the intramolecular C167–C190 disulfide (black arrows), presumably via the thiol–disulfide exchange mechanism (34). Identical results were obtained in two independent experiments. (B) Observed shift in mass consistent with cysteine glutathionylation in AtSAL1 treated with GSSG compared with untreated AtSAL1. A representative m/z spectrum for Cys119 is shown. Charge is indicated in brackets. (C) Both monomeric and dimeric AtSAL1 are sensitive to glutathionylation, with decrease in activity in redox titration with GSH/GSSG (a less negative potential is more oxidizing). The redox midpoint potential (Em) was close to physiological GSH/GSSG redox potential of Arabidopsis chloroplasts (35). Although dimeric AtSAL1 activity only decreased to 40% under fully oxidizing conditions compared with 10% for monomeric AtSAL1, the basal activity of dimeric AtSAL1 is already significantly lower than monomeric AtSAL1 under the same redox state (Table 1). Measurements were performed twice.
In Vitro Redox Regulatory Mechanisms of AtSAL1 Are Recapitulated in Vivo.
If the SAL1–PAP pathway is a primary regulator of PRANGs and PAP concentrations are controlled by redox regulation of SAL1 as hypothesized, then the multiple redox mechanisms regulating AtSAL1 in vitro should be recapitulated in vivo in response to chloroplast redox cues that initiate PRANG regulation. We tested this hypothesis by analyzing AtSAL1 activity, and intramolecular disulfide formation and dimerization, when photosynthetic ROS and the chloroplast redox state were manipulated in various ways.
First, we observed in vivo formation of the characteristic Cys167–Cys190 disulfide band in AtSAL1 during drought stress. In vivo abundance of the Cys167–C190 disulfide bonded form of AtSAL1 progressively increased in correlation with decreasing AtSAL1 activity and increasing PAP accumulation in leaves of drought-stressed Arabidopsis (Fig. 6A). Second, across the multiple abiotic stress treatments that increase abundance of PSI-sourced ROS and lead to reduction of in vivo AtSAL1 activity shown in Fig. 1A, the proportion of dimeric AtSAL1 in oxidatively stressed Arabidopsis leaves increased relative to control (Fig. 6B). This is in agreement with the observed Cys119 intermolecular disulfide-mediated dimerization under oxidative conditions (Fig. 4B).
Fig. 6.
AtSAL1 is redox-regulated via intramolecular disulfide formation and dimerization in vivo, and it is sensitive to the chloroplast redox state. (A) Down-regulation of AtSAL1 activity and concomitant PAP accumulation correlates with formation of the Cys167–Cys190 intramolecular disulfide (black triangles) in endogenous AtSAL1 during drought stress. Means and SE are shown for n = 4 biological replicates for well-watered and n = 3 for drought. In contrast to Fig. 1A, leaf protein extracts were blocked with iodoacetamide, and then protein electrophoresis and Western blotting were performed under nonreducing conditions to visualize the Cys167–Cys190 disulfide. Loading control was Coomassie Blue staining. Similar results were obtained in two independent experiments. (B) The monomer–dimer equilibrium of AtSAL1 in vivo is shifted in favor of the dimer during oxidative stress, suggesting formation of the Cys119–Cys119 intermolecular disulfide to stabilize the dimer. Total leaf protein pooled from four biological replicates per treatment was resolved on Native-PAGE and immunoblotted, and the relative quantities of dimeric to monomeric AtSAL1 were estimated by image analysis on ImageJ. WW, well watered; MD, middrought; LD, late drought; HL, high light; MV, methyl viologen; H2O2, hydrogen peroxide.
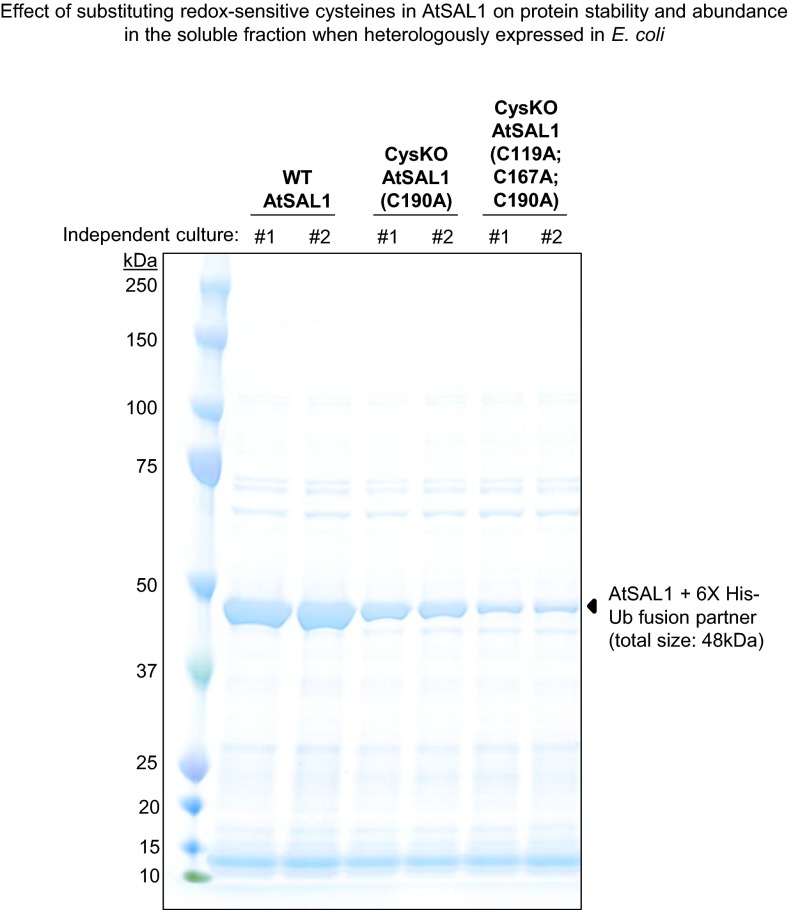
To complement the in vivo results shown in Fig. 6, either one of two approaches can demonstrate that redox regulation of AtSAL1 enables PAP accumulation and PRANG expression. First, AtSAL1 Cys–Ala mutants that are redox-insensitive in vitro can be expressed in Arabidopsis; however, abundance of the Cys–Ala proteins was lower than WT when expressed in Escherichia coli (Fig. S4). It is well established in plants that point mutations, let alone four mutations in a single gene, can affect a variety of protein characteristics, including in vivo stability, activity, and/or protein–protein interactions (36). Indeed, two other SAL1 point mutations, alx8 and hos2, affect protein stability and activity, respectively, in a temperature-dependent manner (37, 38).
Fig. S4.
Cys–Ala mutations negatively affect AtSAL1 protein stability and abundance. The yield of soluble AtSAL1 protein is drastically decreased when the redox-sensitive Cys residues were mutagenized to Ala. The SDS/PAGE gel shows semipurified recombinant WT or mutated AtSAL1 proteins after a soluble protein fraction from 7 mL of induced E. coli cells was incubated with Ni-NTA beads in a 1.5 mL Eppendorf tube, washed with 20 mM imidazole, and the bound AtSAL1+6X His–Ub fusion proteins (black triangles) eluted with 250 mM imidazole. The abundance of soluble AtSAL1 protein decreased with increasing number of Cys–Ala substitutions. The negative effect of the Cys–Ala mutations was reproducible in two independent transformed bacterial colonies.
A second approach involves analyzing the same gene across many species to identify strong conservation of the same characteristic, which would indicate strong evolutionary selection to maintain this function (39, 40). The SAL1–PAP pathway for PRANG regulation and stress tolerance is functional across dicotyledonous (9, 37) and monocotyledonous plants (16). Therefore, if AtSAL1 is genuinely redox-regulated in vivo, then the cysteine residues conferring in vitro redox sensitivity in AtSAL1 should be strongly conserved in evolution beyond Arabidopsis to other plant species, and distantly related orthologs should also show redox-sensitive biochemical activity.
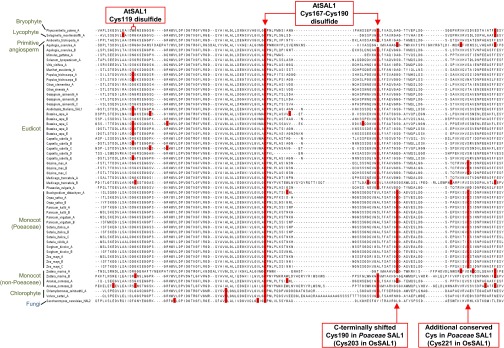
We found that the redox-responsive cysteine residues in AtSAL1 are indeed highly conserved (Fig. S5). The Cys167–C190 intramolecular disulfide pair is strongly conserved across the representative bryophyte, chlorophyte, early angiosperm, eudicot, and monocot species examined. The Cys119 residue that mediates the intermolecular dimerization is less conserved but is still present in 33% of eudicot SAL1 orthologs and in two monocot proteins. In the distantly related Poaceae SAL1 orthologs including the Oryza sativa SAL1 (OsSAL1) protein, the position of the conserved AtSAL1 Cys190 is C-terminally shifted by seven amino acids. These proteins possess an additional conserved cysteine residue (Fig. S5).
Fig. S5.
Conservation of redox-sensitive cysteines in SAL1 orthologs in plants. The Cys119 involved in dimerization of AtSAL1 is moderately conserved in eudicots and non-Poaceae monocot plants but is absent in other lineages. The intramolecular disulfide pair Cys167–Cys190 is invariant across bryophytes, lycophytes, primitive angiosperm, eudicots, and non-Poaceae monocots. Cys190 is C-terminally shifted by seven amino acids in Poaceae monocot plants. Additionally, another cysteine is strongly conserved in the Poaceae family of monocots (Cys221 in OsSAL1; Fig. 7). In contrast to plant SAL1 orthologs (green), the fungal SAL1 ortholog in Saccharomyces cerevisiae HAL2 (blue) lacks Cys119 and Cys190.
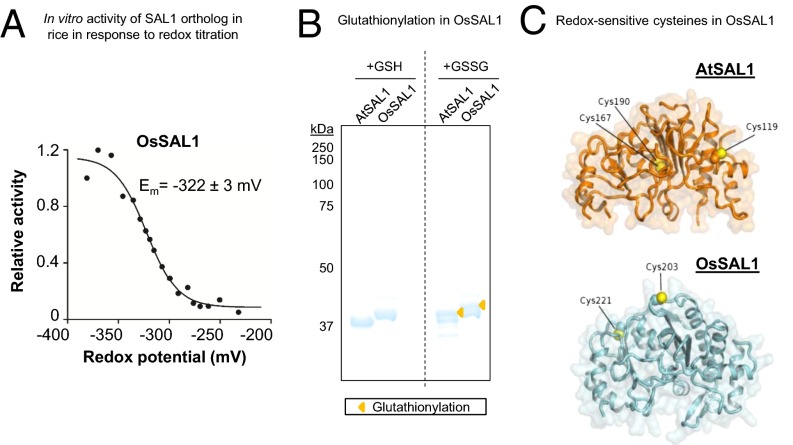
We investigated OsSAL1 in detail, given that it has been shown to have activity against PAP (41), and monocots are estimated to have diverged from dicots 140–150 Mya (42). Conserved similarities in redox sensitivity between dicot AtSAL1 and monocot OsSAL1 would therefore be functionally and evolutionarily significant. As was observed for AtSAL1, OsSAL1 activity is inhibited by oxidation (Table 1), and redox titration of the protein shows a physiologically relevant Em (Fig. 7A). OsSAL1 conformation can also be modified by glutathionylation (Fig. 7B). Homology modeling of OsSAL1 reveals that both the strongly conserved cysteine residues are surface-exposed (Fig. 7C), which may potentiate redox regulation.
Fig. 7.
Biochemical and structural evidence for conservation of redox sensitivity in a rice SAL1 ortholog. (A) Redox titration on OsSAL1 shows that the protein is redox sensitive and has a redox midpoint potential (Em) in the physiologically relevant range. A less negative potential is more oxidizing. (B) Oxidation of AtSAL1 and OsSAL1 with GSSG similarly result in glutathionylation of the proteins, increasing their apparent molecular weight when resolved on nonreducing SDS/PAGE (yellow triangles). Vertical dashed lines indicate splicing and truncation of the gel to remove additional lanes not relevant to this result. (C) Comparison between redox-sensitive cysteine residues detected in structures of AtSAL1 and modeling of OsSAL1. Unlike AtSAL1 which contains both surface-exposed and intramolecular disulfide cysteines, OsSAL1 is predicted to contain surface exposed cysteines (marked in yellow). Both Cys203 and Cys221 of OsSAL1 are strongly conserved in Poaceae SAL1 orthologs (Fig. S5).
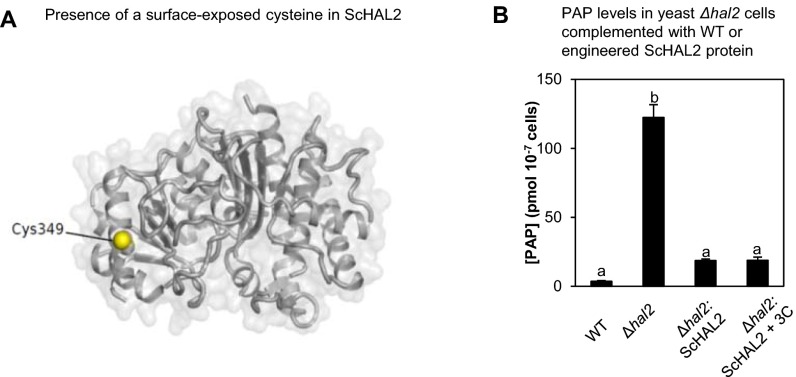
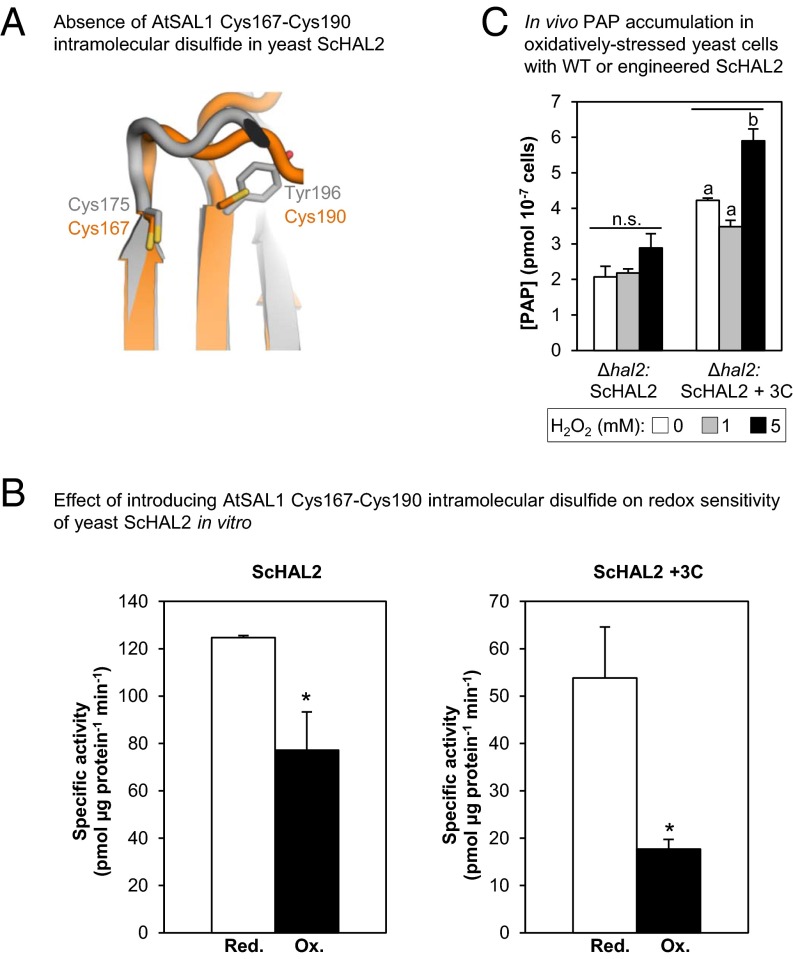
Finally, a corollary to the results described above is that if a SAL1 ortholog lacks the redox-responsive cysteine residues, then introduction of these residues should enhance redox sensitivity in the new protein. The activity of the yeast (Saccharomyces cerevisiae) ortholog, ScHAL2, shows some redox sensitivity to glutathionylation (Table S3), possibly due to presence of a surface-exposed cysteine (Fig. S6A). However, ScHAL2 lacks the three cysteines at positions structurally equivalent to those of AtSAL1, including the Cys119 side chain that promotes dimerization (Fig. S5) and the Cys167–Cys190 intramolecular disulfide (Fig. 8A). This suggests that the redox-sensing mechanisms regulating AtSAL1 activity in plants (Figs. 1–6) are absent in yeast and may have evolved in plants for chloroplast redox sensing and PRANG regulation. We therefore introduced three additional cysteine residues (Thr21Cys, Phe127Cys, and Tyr196Cys) into ScHAL2 (ScHAL2+3C).
Table S3.
Effect of introducing AtSAL1 Cys167-Cys190 disulfide pair on redox sensitivity of ScHAL2
| Protein | [PAP], µM | Specific activity, pmol per µg protein per min | |||
| Reduced | Oxidized | Decrease in activity, % | Average decrease in activity, % | ||
| ScHAL2 | 3.35 | 124 ± 0.9 | 77 ± 16 | 38 | 46 ± 4.8a |
| 13.4 | 414 ± 66 | 220 ± 31 | 46 | ||
| 26.8 | 597 ± 150 | 270 ± 35 | 54 | ||
| ScHAL2+3C | 3.35 | 53 ± 10 | 17 ± 2 | 67 | 67 ± 1.4b (P < 0.005 relative to ScHAL2) |
| 13.4 | 72 ± 24 | 25 ± 5 | 65 | ||
| 26.8 | 81 ± 29 | 24 ± 0.8 | 70 | ||
Enzymatic assays were performed with 0.2 µg recombinant protein in the presence of 20 mM GSH (reduced) or GSSG (oxidized) at 30 °C. Means and SE from two independent experiments are shown. The decrease in activity for ScHAL2+3C by oxidation is significantly greater (a, b; P < 0.005) than that in WT ScHAL2.
Fig. S6.
WT and engineered ScHAL2. (A) Presence of a surface-exposed Cys349 in the ScHAL2 crystal structure (1KA1), which may explain the redox-sensitive activity observed in Fig. 8B. (B) The WT ScHAL2 and ScHAL2+3C proteins are equally active in vivo when expressed in yeast cells deficient in ScHAL2 (Δhal2) because both proteins complement PAP levels in Δhal2 to similar levels, albeit still about 10-fold higher than WT. Similar results were obtained in two independent experiments.
Fig. 8.
Enhancement of redox sensitivity in yeast ScHAL2 by introduction of the AtSAL1 intramolecular disulphide. (A) Structural alignment-guided introduction of the intramolecular disulfide from AtSAL1 (orange) into yeast ScHAL2 (gray) by the Tyr176Cys mutation. Thiol groups are indicated in yellow. (B) Introduction of additional disulfide in ScHAL2+3C results in increased redox sensitivity in vitro compared with WT ScHAL2. Means and SE from two independent experiments for specific activity at 3.35 µM PAP are shown. For full results, see Table S3. Asterisks indicate significant differences (P < 0.05). (C) Introduction of additional disulfide in ScHAL2+3C increased redox sensitivity and PAP accumulation in vivo when expressed in yeast Δhal2 cells under mild oxidative stress. Significant differences are indicated by a and b (P < 0.05). Error bars indicate SE; n = 3 independent cultures for all experiments. n.s., no significant difference.
Phe127Cys is the equivalent of AtSAL1 Cys119 that promotes dimerization, whereas Tyr196Cys introduces a potential disulfide pair in the same position as the critical intramolecular disulfide that regulates activity of the dimeric AtSAL1 (Fig. 8A). Indeed, ScHAL2+3C showed significantly greater redox sensitivity compared with WT ScHAL2 in vitro (Fig. 8B and Table S3). When expressed in vivo, the engineered ScHAL2+3C was as efficient as WT ScHAL2 in degrading PAP under unstressed conditions (Fig. S6B). Critically, introduction of the intramolecular disulfide also increases redox sensitivity in vivo compared with the WT form: yeast Δhal2 overexpressing ScHAL2+3C significantly accumulated PAP when challenged with mild H2O2 stress, whereas those overexpressing WT ScHAL2 did not (Fig. 8C). Thus, addition of the cysteine residues alone is sufficient to induce enhanced redox sensitivity to the yeast SAL1 ortholog.
Discussion
Redox Regulation of AtSAL1 Involves Multiple Structural Mechanisms.
A diverse range of proteins are specifically regulated by disulfide formation (23). The Cys167–Cys190 intramolecular disulfide in AtSAL1 is intriguing because it occurs across two antiparallel beta strands immediately adjacent to one another, at the base of a hairpin loop connecting these strands (Fig. 3A). Formation of a disulfide across adjacent beta strands is a form of cross-strand disulfide (CSD) that has been termed “forbidden disulfides” (23) because they disobey the established rules of protein stereochemistry (43, 44) and introduce strain into the protein structure that may be energetically and structurally unfavorable (44). However, recent findings indicate that strain in local areas of a protein are tolerated for regulation of protein function (23). An increasing number of proteins have been characterized that contain CSDs; in most cases, these disulfides regulate function (23). These examples include proteins involved in chloroplast redox control such as TRXs, which have a canonical CSD-containing motif that is strongly conserved across evolution (23, 45). CSD formation directly blocks catalytic cysteines in TRXs (46, 47), whereas the CSD decreases protein flexibility in AtSAL1 (Fig. 3).
Formation of the intramolecular Cys167–Cys190 disulfide in response to oxidizing conditions is dependent upon dimerization (Fig. 4C), which stabilizes the protein conformation (Fig. 3). The dimerization interface between AtSAL1 monomeric subunits is relatively small (Fig. 3A), which may result in a relatively weak or transient interaction in vivo and explain the monomer–dimer equilibrium in solution under reducing conditions, until the interface is locked together through the Cys119–Cys119 intermolecular disulfide bonding during oxidative stress. The inactivation of both monomeric and dimeric AtSAL1 by GSH/GSSG, and formation of the Cys167–Cys190 disulfide by glutathionylation via thiol–disulfide exchange (Fig. 5), reveals an additional mechanism for redox regulation of AtSAL1.
AtSAL1 Redox Regulation Allows Both Metabolic Control of Sulfur Assimilation and Oxidative Stress Signaling.
The flux of sulfur in plants is regulated partly via redox control of key enzymes in the sulfur assimilation pathway (48–50). Oxidative stress is expected to increase sulfur flux into sulfur reduction for production of the redox buffer GSH/GSSG because two key enzymes in this primary branch of sulfur metabolism, APS reductase (APR) and Glutamate-Cysteine Ligase (GCL), are more active when oxidized (49, 50). Conversely, PAP biosynthesis in the parallel, secondary pathway should be down-regulated by oxidative stress because enzymatic synthesis of the PAP precursor, PAPS, by APS kinase (APK) is decreased by oxidation (48).
Within the present study we show that AtSAL1 is significantly less active when oxidized (Figs. 1–6), thus providing a mechanism for the 30-fold accumulation of PAP seen in WT plants during drought (9), without necessitating increased sulfur allocation into secondary sulfur metabolism and PAP synthesis. We also show that metabolites from one sulfur metabolism branch can influence biosynthesis of metabolites in the parallel pathway because GSH/GSSG directly regulates AtSAL1 activity (Fig. 5) and hence PAP levels. Significantly, the redox midpoint potential (Em) of AtSAL1 (−308 ± 2 mV for monomer and −284 ± 5 mV for dimer at pH 7.5; Fig. 5C) overlaps with those determined for the oxidation-activated primary sulfur metabolism (GSH/GSSG) enzymes (−330 ± 10 mV at pH 8.0 for APR and −318 ± 11 mV at pH 7.0 for GCL) (49, 50) and oxidation-inhibited secondary sulfur metabolism (PAPS/PAP) APK enzymes (−286 ± 18 mV at pH 7.5) (48). Therefore, the same redox state in chloroplasts can concomitantly regulate multiple sulfur metabolism enzymes including AtSAL1. This could allow coordination of flux through the sulfur pathway for redox buffering (GSH/GSSG) concomitant to stress signaling (PAP).
The redox-responsive cysteine residues in AtSAL1 are conserved in plant species that lack glucosinolate biosynthesis, which constitute a major sink for sulfur (and source of PAP) in Arabidopsis and its relatives in the Brassicaceae (14) (Fig. S5). Indeed, the redox regulation was conserved in OsSAL1 (Fig. 7), despite O. sativa being monocotyledonous and lacking glucosinolates. Another indication that the SAL1–PAP pathway functions independently of glucosinolates is that it also mediates stress tolerance in wheat (16). Therefore, the redox regulation of SAL1 can be uncoupled from sulfur metabolism.
SAL1 Acts as a General Redox Sensor in the Chloroplast for Retrograde Signaling and PRANG Regulation.
Our results indicate that SAL1 activity in plants is sensitive to the overall redox state of the chloroplast and not to a specific stimulus or sensor protein (Fig. 1). The SAL1 protein is sensitive to ROS production and redox state of PSI, where O2− is produced and detoxified via Superoxide Dismutase (SOD)-mediated dismutation to H2O2, which is further detoxified by thylakoidal and stromal APXs (tAPX and sAPX). Deficiency in sAPX compromises H2O2 degradation and concomitantly increases O2− abundance at PSI due to SOD inhibition (1); this increases the effect of HL stress on suppression of AtSAL1 activity (Fig. 1B). Indeed, direct induction of O2− production at PSI by methyl viologen inhibited AtSAL1 (Fig. 1A). Perturbing multiple aspects of chloroplast redox homeostasis also influenced AtSAL1 activity (Fig. 1B). This includes the major oxidized protein regeneration pathway in chloroplasts involving NADPH-dependent Thioredoxin Reductase C (NTRC) (51) and the redox buffers NADPH /NADP+ and GSH/GSSG. Mutants in these pathways are unsurprisingly hypersensitive to oxidative stress (52, 53) but also deregulated in PRANG expression during stress (54). The latter probably reflects the influence of redox poise on AtSAL1 activity and PAP accumulation.
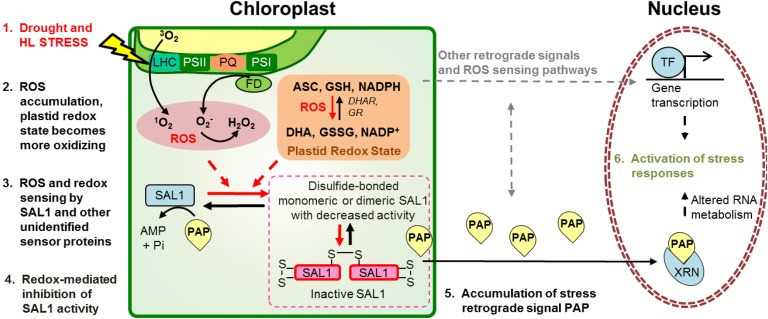
The convergence of photosynthetic ROS and redox cues on SAL1 has key implications for stress retrograde signaling and regulation of PRANGs. Our results here demonstrating the coupling of SAL1 activity and PAP accumulation to chloroplastic redox poise present this enzyme as an unconventional oxidative stress sensor and hub for the convergence of H2O2, O2−, and redox cues in the chloroplast that alter nuclear gene expression (4, 6, 7). That is, the primary function of SAL1 is not to sense ROS and redox state, yet its sensitivity to these chloroplast cues enables them to be sensed by the nucleus via PAP, providing capacity for fine-tuning responses (Fig. 9). Such a hypothesis does not preclude parallel pathways that also sense or respond to ROS or redox state in the chloroplast. For example, recent evidence shows that projections from chloroplasts called stromules increase in abundance during stress and can enable transport of ROS from chloroplasts to the nucleus (55, 56). The extent to which PAP and other ROS communication pathways overlap is a subject of future research.
Fig. 9.
SAL1 as an oxidative stress sensor in the chloroplast for regulation of PAP-mediated retrograde signaling. Oxidative stresses (drought and HL) invoke physiological and biochemical changes in chloroplasts, such as ROS accumulation and altered redox poise. These are sensed by SAL1 via redox-mediated inhibition of SAL1 activity, enabling PAP accumulation. Stress responses, including transcription of stress-responsive genes, are activated. ROS sensors may also control other HL retrograde signals, such as β-cyclocitral or MEcPP, or use stromules; their identity and interaction with SAL1-PAP remain to be determined (gray arrows).
The conserved secondary redox sensing by SAL1 suggests that these enzymes may be considered “moonlighting” proteins (57). Many moonlighting proteins are evolutionarily ubiquitous enzymes that have secondary functions in diverse processes including metabolism and disease (57). To our knowledge, SAL1 would constitute the first moonlighting oxidative stress sensor described in plants. The sensitivity of the sensor may be fine-tuned differently across evolution to fulfill kingdom-specific functions because Arabidopsis and rice SAL1 appear more responsive to ROS and redox state than yeast HAL2 due to presence of the intramolecular Cys167–Cys190 disulfide (Figs. 3A and 8 B and C and Table S3).
Whether chloroplast communication necessitates additional layers of complexity in regulation of SAL1 in plants requires further elucidation. Regardless, there is precedent for the evolution of increased redox sensitivity in PAP/PAPS metabolism in plants. For instance, the adenosine 5′-phosphosulfate kinase (APK) enzyme catalyzing PAPS production evolved increased redox sensitivity in the transition from cyanobacteria to higher plants (58). It is also fascinating that WT ScHAL2 shares some conservation of redox sensitivity (Fig. 8 and Table S3), albeit to a lesser extent, with AtSAL1 despite lacking the AtSAL1 redox-responsive cysteine residues. Instead, ScHAL2 may be regulated via a different surface-exposed cysteine residue (Fig. S6). It may be that PAP phosphatases in other kingdoms are redox-regulated via analogous mechanisms targeting different cysteine residues, and this will be interesting to explore.
In summary, transient elevation of PAP levels as an oxidative stress signal in plants is coupled to the redox state perceived by SAL1. That at least four different ROS/redox couples (H2O2, O2−, GSH/GSSG, and DTTred/DTTox) regulate SAL1 enables a fine-tuning of SAL1 activity and thus retrograde signaling to communicate the different fluctuations of chloroplast ROS balance/redox poise in response to environmental stimuli. It is intriguing that a similarity in regulation of SAL1 orthologs relates more to the secondary function of redox sensing and stress signaling than sulfation and raises the question as to which function was the primary driver for evolutionary conservation of this protein. Our results suggest that dual-function SAL1 orthologs may be uniquely positioned as single-component integrators of sensing and signaling of aspects of oxidative stress in plants.
Materials and Methods
Plant Material and Growth Conditions.
Arabidopsis seeds were germinated on soil and kept at 4 °C for 3 d to synchronize germination. Plants were grown at 100–150 µmol photons per m2 per s, 12-h photoperiod, 21–23 °C, and 50–55% humidity. Five-week-old plants were used for all stress assays (SI Text). T-DNA insertion lines for the redox homeostasis mutants sapx (1), ntrc (52), phs1 (53), and cos1 (59) (Table S1) were obtained from the Arabidopsis Biological Resource Centre. The amiRNA silencing line for tAPX (1) was provided by Christophe Laloi, Aix-Marseille Universite, Marseille, France, and rax1-1 (54) was obtained from Phil Mullineaux, University of Essex, Essex, United Kingdom (Table S1).
Protein Purification from Biological Samples.
Arabidopsis leaf native proteins were extracted using a protocol modified from Murguia et al. (60) (SI Text). Native proteins were kept on ice and used immediately in Clear-Native PAGE and activity assays, as described later.
For detection of the Cys167–Cys190 intramolecular disulfide in endogenous AtSAL1, native proteins were incubated with 10 mM iodoacetamide in the dark for 1 h to prevent oxidation of any cysteines that were reduced in vivo. Leaf protein was then precipitated in TCA/acetone, washed twice with cold acetone, and resuspended in solubilisation buffer (9 M urea, 4% (wt/vol) CHAPS, 1% (wt/vol) DTT, and 35 mM Tris base) before SDS/PAGE and Western blotting (SI Text).
Recombinant Protein Purification.
Recombinant WT AtSAL1 protein was expressed in Escherichia coli BL21 DE3 cells (New England BioLabs) and purified by standard metal affinity purification (SI Text). The purified recombinant proteins were stored in storage buffer [50 mM Tris⋅HCl, pH 8.0; 150 mM NaCl; 20 mM KCl; 1 mM MgCl2; and 15% (vol/vol) glycerol] at −80 °C.
Protein Gel Electrophoresis.
Recombinant protein, 0.5–1 µg, was incubated in degassed storage buffer [50 mM Tris⋅HCl, pH 8.0; 150 mM NaCl; 50 mM KCl; 1 mM MgCl2; and 15% (vol/vol) glycerol] in the presence of either 5 mM DTTred (reducing conditions) or 5 mM DTTox (trans-4,5-Dihydroxy-1,2-dithiane; oxidizing) for 1 h at RT, then resolved by SDS/PAGE and stained as described in SI Text. To visualize effects of GSH/GSSG, proteins were incubated in degassed storage buffer containing 20 mM GSH or 20 mM GSSG, then run on gels and stained as above.
For Clear-Native PAGE, proteins were incubated with redox agents as above but resuspended in native sample loading buffer [100 mM Tris⋅HCl, 10% (vol/vol) glycerol, 0.0025% bromophenol blue, pH 8.6] and resolved on a 3–12% Novex NativePAGE gel (Life Technologies) in native running buffer (25 mM Tris, 192 mM Glycine, pH 8.3) without denaturing agents.
Activity Assays.
Recombinant protein activity against PAP was assayed by incubating 0.2 µg protein in degassed activity buffer (100 mM Tris⋅Mes, pH 7.5, and 1 mM Mg acetate) in presence of either reducing or oxidizing equivalents of DTTred/DTTox (5 mM) or GSH/GSSG (20 mM) for 1 h at 25 °C, then increasing concentrations of PAP were added to a final volume of 150 µL and initial activity assayed at 25 °C (AtSAL1 and OsSAL1) or 30 °C (ScHAL2). The reaction was stopped by flash-freezing in liquid N2. AMP produced from degradation of PAP was quantified using the method for derivatization and detection of adenosines via high-performance liquid chromatography (9). All Michaelis–Menten kinetics parameters were calculated using GraphPad Prism (GraphPad Software Inc.). For redox titration of activity, SAL1 protein was incubated as above with different ratios of DTTred:DTTox (final total concentration: 5 mM) or GSH:GSSG (final concentration: 20 mM). Redox midpoint potential, Em, was calculated by fitting titration data to the Nernst equation using GraphPad Prism:
and
with an RT/F of 25.7 mV and n = 2 (49, 61).
For activity of AtSAL1 in native protein extracts from Arabidopsis, 10 µg of total native protein extract was incubated in the same activity buffer as above with increasing concentrations of PAP at 25 °C without any redox agents.
Crystallization, Data Collection, and Refinement.
AtSAL1 crystals were grown by vapor diffusion in hanging drops. Crystals formed at a protein concentration of 20 mg⋅mL−1 in 20–30% (wt/vol) PEG 2000-MME, 0.2 M (NH4)2SO4, and 0.1 M Hepes, pH 8.0–8.6. Diffraction data were collected at the Australian Synchrotron at the MX2 beamline (λ = 0.9537 Å). The resolutions limits of the data were assessed on the basis of the significance of the CC1/2 at the P = 0.001 level (62, 63). Diffraction data were integrated using XDS (64) and scaled using SCALA from the CCP4 program suite (65). The crystals belonged to the P61 space group, and significant merohedral twinning was identified by xtriage (66) (twin operator h,-h-k,-l). Phases were obtained by molecular replacement in Phaser (67) using the yeast ortholog of SAL1 (PDB 1QGX) (20) as the search model. The crystallographic asymmetric unit contained two copies of SAL1. For both chains, the entire 346 residue polypeptide of SAL1 was modeled. Twin refinement (h,-h-k,-l), with noncrystallographic symmetry (NCS restraint sigma 2.5°, restraint limit 15°) and automatically generated secondary structure restraints, was completed with phenix.refine (66, 68).
Simulation of the Oxidized AtSAL1 Dimer.
The intermolecular (Cys119–Cys119) and intramolecular (Cys167–Cys190) disulfides were modeled using the GROMACS package (69) in conjunction with the GROMOS 53a7 force field for condensed phases (70). To prepare the model, the crystallographic coordinates of the AtSAL1 dimer were immersed in a cubic shaped box of solvent with a minimum of 1 nm between the protein and the box edge. The simple point charge model was used to represent water (71), and the protein’s charge was neutralized by the addition of sodium ions. Electrostatic energy was calculated using the particle mesh Ewald (PME) method (72), and cutoff distances for the calculation of van der Waals and Coulomb interactions were set at 0.9 and 1.4 nm, respectively. To model the disulfide bonds between Cys167–Cys190 and Cys119–Cys119, the AtSAL1 topology file was edited to include a bond between the sulfur atoms of the cysteines. The resulting system was energy minimized via steepest descent to the limit of machine precision. During energy minimization the disulfide bond length decreased to an appropriate length (2 ± 0.2 Å). For comparison the reduced AtSAL1 monomer was modeled. To prepare the model, the crystallographic coordinates of each monomer in the AtSAL1 asymmetric unit were energy minimized. The same procedure was used to energy minimize the monomeric structure as the dimeric structure, except disulfide bonds (Cys167–Cys190) were not included in the monomer. To obtain an estimate of the flexibility of the energy minimized structures, the structures were submitted to the iMOD normal mode analysis webserver (73). To analyze the coupling of AtSAL1 motions, the covariance of Cα motions was plotted (74).
SI Materials and Methods
Stress Treatments on Plants.
Drought stress treatment was performed in a randomized manner as previously described (9, 37). For other oxidative stress treatments, leaf disks were floated on 0.1% Tween and exposed either to 1,000 µmol photons per m2 per s white light (high light) or to 500 mM H2O2, 100 µM methyl viologen (O2− generator), or untreated (control) under standard light intensity for 4 h.
Yeast Growth Conditions and Stress Treatments.
All yeast (Saccharomyces cerevisiae) strains used were isogenic with the haploid BY4741 background and were cultured at 30 °C in yeast synthetic drop-out medium (SD) with the omission of leucine where necessary for plasmid maintenance. The Δhal2 strain was complemented with WT ScHAL2 and ScHAL2+3C cDNA sequences overexpressed under the glyceraldehyde-3-phosphate dehydrogenase promoter in the pJR1138 vector (75) using a standard lithium acetate transformation protocol. These strains were designated Δhal2:ScHAL2 and Δhal2:HAL2+3C, respectively. For oxidative stress treatments, cultures were grown till early exponential phase, then H2O2 (1 or 5 mM final concentration) was added. Cells were grown for a further 12 h before harvest for PAP quantification.
Cloning, Cysteine Knockout, and Knock-in Mutagenesis.
The mature full-length WT AtSAL1 cDNA was amplified from WT Arabidopsis leaf cDNA using proofreading polymerase and cloned into the pGEM 3zf(+) vector. Mutagenesis was then carried out using different combinations of primers containing point mutations to change the individual cysteine codons into alanine, with the QuikChange II Site-Directed Mutagenesis kit (Stratagene) as per manufacturer’s instructions. All mutations were confirmed by Sanger sequencing. The resulting mutated AtSAL1 sequences were then cloned into either pHUE (76) or prSETb (Life Technologies) and resequenced to confirm presence of mutation(s), and recombinant proteins were purified as described below. For introduction of additional cysteines into ScHAL2 to produce ScHAL2+3C, the mutated sequence was synthesized and directly cloned into prSETb (Life Technologies).
Recombinant Protein Purification.
The mature full-length AtSAL1 cDNA (excluding the transit peptide sequence) was expressed as an AtSAL1–Ubiquitin fusion protein in the pHUE expression vector under IPTG induction and purified using Ni-NTA His-Bind Resin (Novagen) according to manufacturer’s instructions. AtSAL1–Ubiquitin was then digested with the deubiquitinylating enzyme Usp2 in a digestion buffer (50 mM Tris⋅HCl, pH 8.0; 150 mM NaCl; 20 mM KCl; and 2 mM β-mercaptoethanol) and repurified through the Ni-NTA resin to yield the mature AtSAL1 protein of 95% purity as assayed by SDS/PAGE. The mature AtSAL1 protein was further purified into monomeric and dimeric fractions by size exclusion chromatography (SEC) on a HiLoad 26/60 Superdex-200 SEC column (Life Technologies). The monomeric (eluting between 195 and 220 mL) and dimeric (eluting between 170 and 195 mL) fractions obtained were concentrated by filter centrifugation, quantified by Bradford assay, and stored in storage buffer [50 mM Tris⋅HCl, pH 8.0; 150 mM NaCl; 20 mM KCl; 1 mM MgCl2; and 15% (vol/vol) glycerol] at −80 °C. Mutagenized variants of AtSAL1 and WT OsSAL1 proteins were purified essentially as described above but were expressed with an N-terminal TEV protease recognition sequence in prSETb (Life Technologies) instead of pHUE, and the fusion protein was digested with TEV protease instead of Usp2 to yield the mature protein.
Protein Quantification.
All recombinant proteins and Arabidopsis protein extracts were quantified using the Bradford assay (77). Ten microliters of protein (prediluted 1:4 in water) were incubated with 240 µL of 1× Bradford reagent dye (Bio-Rad) in clear 96-well plates at room temperature for 15 min, then absorbance was measured at 595 nm. Three technical replicates for each protein sample were quantified. BSA (Sigma Aldrich) at a range of concentrations (0–1.2 mg⋅mL−1) were used as standards.
Size Exclusion Multiple Angle Laser Light Scattering.
Purified AtSAL1 protein was dialyzed against AtSAL1 assay buffer without reducing agent. The protein was then applied to a WTC-030S5 size exclusion column (Wyatt Technologies) equilibrated with AtSAL1 assay buffer. Multiangle laser light scattering (Dawn8+; Wyatt Technologies) and differential refractive index (Optilab Rex; Wyatt Technologies) of the eluted protein were monitored, and data were analyzed using ASTRA (Wyatt Technologies).
SDS/PAGE.
To recombinant proteins incubated with redox agents at RT for 1 h, NuPAGE LDS 4× sample loading buffer (Life Technologies) was added to a final concentration of 1×. Samples were heated at 70 °C for 10 min, cooled on ice, and then resolved on a 10% Bis-Tris NuPAGE gel with Mops buffer (Life Technologies) at 150 V for 2.5 h without addition of reducing agents or antioxidants. The protein bands were visualized by staining with GelCode Blue Safe Stain (Thermo Scientific) according to manufacturer’s instructions.
Native Protein Purification from Arabidopsis.
Approximately 100 mg of either control or stressed Arabidopsis leaves were frozen in liquid nitrogen and ground to a fine powder with a 1/8′′ steel ball with the TissueLyzer II (Qiagen). Native proteins were resuspended in ice-cold 50 mM Tris⋅HCl, pH 7.5, supplemented with 1% PVP 360 and 1× Roche Protease Inhibitor Mixture (Roche). Cellular debris was removed by centrifugation at 4 °C, and the proteins in the supernatant were quantified by Bradford assay.
Immunoblotting.
Five micrograms total Arabidopsis leaf protein were resolved by SDS/PAGE as described above, in absence of additional redox agents, then electrotransferred to a PVDF membrane and probed with a 1:1,000 dilution of polyclonal antibodies raised against AtSAL1 (Catalog Number AS07 256; Agrisera) as described previously (37). After three washes with PBST, the blot was incubated with 1:10,000 dilution of HRP-conjugated goat anti-rabbit IgG for 10 min. After a further three washes with PBST, the membrane was developed with the Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare).
Mass Spectrometry.
Gel bands were manually excised for identification by mass spectrometry. Excised gel bands were destained and digested with trypsin as previously described (78), with the modification of using 55 mM iodoacetamide but omitting DTT to avoid loss of glutathione. Tryptic peptides were extracted in 80% ACN/0.1% formic acid, dried under vacuum, and resuspended in 5% (vol/vol) ACN and 0.1% formic acid before MS analysis using liquid chromatography connected to an accurate-mass Quadrupole Time-of-Flight MS (Agilent 6550) equipped with a Chip Cube ion source as described previously (79). MS/MS spectra analysis and database searching was performed as described (80) using the following modifications. The results were searched against the TAIR10 database for A. thaliana protein sequences (33,621 protein sequences) using the Mascot ver. 2.3 search engine (Matrix Science). The following parameters were selected: MS error tolerance of ±100 ppm, MS/MS error tolerance of ±0.5 Da, semitryptic peptides, 1 maximum missed cleavage tolerated, variable modifications oxidation (M), carboxymethyl (C), glutathione (C), and oxidation (C); finally, the instrument was set to ESI-Q-ToF. In addition to MS/MS identification, all data files were searched for the presence of cysteine-containing peptides, as well as glutathionylated peptides. Within the Mass Hunter qualitative software (Agilent), the extracted ion chromatogram was extracted for all peptides of interest.
Amino Acid Alignment.
SAL1 amino acid sequences from the plant kingdom were retrieved from the Phytozome database (www.phytozome.net). Mature amino acid sequences (after manual removal of confirmed and putative transit peptides) of SAL1 orthologs in plants were aligned using Clustal Omega with default parameters and visualized in JalView.
Quantification of Adenosines.
Total adenosines were extracted with 0.1 M HCl, derivatized with chloroacetaldehyde, and quantified fluorometrically after HPLC fractionation as previously described (9). PAP quantification was performed using calibration curves constructed from authentic standards.
Statistical Analyses.
For all experiments where three or more sample groups were present, analysis of variance was used to test for significant differences (P < 0.05). Where only two sample groups of interest were compared, the two-sample Student’s t test was used. All statistical analyses were carried out using GraphPad Instat 3.0 (GraphPad Software) or R3.08 (www.r-project.org).
Acknowledgments
We thank Prof. Ian Dawes (University of New South Wales) for providing the yeast strains, as well as Dr. Derek Collinge, Mr. Diep Ganguly, and Miss Nay Chi Khin [Australian National University (ANU)] for technical support. We thank Dr. Paul D. Carr for assistance with crystallization and helpful discussions. We received financial support from the Australian Research Council Centre of Excellence in Plant Energy Biology (CE140100008) and scholarships to K.X.C. [Endeavour International Postgraduate Research Scholarship (IPRS) and ANU] and S.Y.P. (ANU). This research was also supported by the Wellcome Trust (Project Grant 092283, to W.A.) and the European Commission (Marie Curie Fellowship 625451, to J.W.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank (PDB ID code 5ESY).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604936113/-/DCSupplemental.
References
- 1.Asada K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- 2.Pesaresi P, et al. Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell. 2009;21(8):2402–2423. doi: 10.1105/tpc.108.064964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michelet L, et al. Glutathionylation of chloroplast thioredoxin f is a redox signaling mechanism in plants. Proc Natl Acad Sci USA. 2005;102(45):16478–16483. doi: 10.1073/pnas.0507498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gadjev I, et al. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;141(2):436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laloi C, et al. Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104(2):672–677. doi: 10.1073/pnas.0609063103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chi W, Sun X, Zhang L. Intracellular signaling from plastid to nucleus. Annu Rev Plant Biol. 2013;64(64):559–582. doi: 10.1146/annurev-arplant-050312-120147. [DOI] [PubMed] [Google Scholar]
- 7.Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu Rev Plant Biol. 2016;67:25–53. doi: 10.1146/annurev-arplant-043015-111854. [DOI] [PubMed] [Google Scholar]
- 8.Ramel F, et al. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA. 2012;109(14):5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estavillo GM, et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23(11):3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM. Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell. 1997;9(4):627–640. doi: 10.1105/tpc.9.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karpinski S, et al. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science. 1999;284(5414):654–657. doi: 10.1126/science.284.5414.654. [DOI] [PubMed] [Google Scholar]
- 12.Rossel JB, et al. Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell. 2007;19(12):4091–4110. doi: 10.1105/tpc.106.045898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tikkanen M, Gollan PJ, Suorsa M, Kangasjärvi S, Aro EM. STN7 operates in retrograde signaling through controlling redox balance in the electron transfer chain. Front Plant Sci. 2012;3:277. doi: 10.3389/fpls.2012.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan KX, Wirtz M, Phua SY, Estavillo GM, Pogson BJ. Balancing metabolites in drought: the sulfur assimilation conundrum. Trends Plant Sci. 2013;18(1):18–29. doi: 10.1016/j.tplants.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Lee BR, et al. Effects of fou8/fry1 mutation on sulfur metabolism: Is decreased internal sulfate the trigger of sulfate starvation response? PLoS One. 2012;7(6):e39425. doi: 10.1371/journal.pone.0039425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manmathan H, Shaner D, Snelling J, Tisserat N, Lapitan N. Virus-induced gene silencing of Arabidopsis thaliana gene homologues in wheat identifies genes conferring improved drought tolerance. J Exp Bot. 2013;64(5):1381–1392. doi: 10.1093/jxb/ert003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Després C, et al. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell. 2003;15(9):2181–2191. doi: 10.1105/tpc.012849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood MJ, Andrade EC, Storz G. The redox domain of the Yap1p transcription factor contains two disulfide bonds. Biochemistry. 2003;42(41):11982–11991. doi: 10.1021/bi035003d. [DOI] [PubMed] [Google Scholar]
- 19.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247(4):536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 20.Albert A, et al. X-ray structure of yeast Hal2p, a major target of lithium and sodium toxicity, and identification of framework interactions determining cation sensitivity. J Mol Biol. 2000;295(4):927–938. doi: 10.1006/jmbi.1999.3408. [DOI] [PubMed] [Google Scholar]
- 21.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Wouters MA, Fan SW, Haworth NL. Disulfides as redox switches: From molecular mechanisms to functional significance. Antioxid Redox Signal. 2010;12(1):53–91. doi: 10.1089/ars.2009.2510. [DOI] [PubMed] [Google Scholar]
- 24.Peltier JB, et al. The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol Cell Proteomics. 2006;5(1):114–133. doi: 10.1074/mcp.M500180-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Changeux J-P, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308(5727):1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 26.Li M, Smith CJ, Walker MT, Smith TJ. Novel inhibitors complexed with glutamate dehydrogenase: Allosteric regulation by control of protein dynamics. J Biol Chem. 2009;284(34):22988–23000. doi: 10.1074/jbc.M109.020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer AG., 3rd Enzyme dynamics from NMR spectroscopy. Acc Chem Res. 2015;48(2):457–465. doi: 10.1021/ar500340a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guarnera E, Berezovsky IN. Allosteric sites: Remote control in regulation of protein activity. Curr Opin Struct Biol. 2016;37:1–8. doi: 10.1016/j.sbi.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508(7496):331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bahar I, Rader AJ. Coarse-grained normal mode analysis in structural biology. Curr Opin Struct Biol. 2005;15(5):586–592. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López-Blanco JR, Aliaga JI, Quintana-Ortí ES, Chacón P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42(Web Server issue):W271–W276. doi: 10.1093/nar/gku339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiesmann C, et al. Allosteric inhibition of protein tyrosine phosphatase 1B. Nat Struct Mol Biol. 2004;11(8):730–737. doi: 10.1038/nsmb803. [DOI] [PubMed] [Google Scholar]
- 33.Choe JY, Fromm HJ, Honzatko RB. Crystal structures of fructose 1,6-bisphosphatase: Mechanism of catalysis and allosteric inhibition revealed in product complexes. Biochemistry. 2000;39(29):8565–8574. doi: 10.1021/bi000574g. [DOI] [PubMed] [Google Scholar]
- 34.Foyer CH, Noctor G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011;155(1):2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maughan SC, et al. Plant homologs of the Plasmodium falciparum chloroquine-resistance transporter, PfCRT, are required for glutathione homeostasis and stress responses. Proc Natl Acad Sci USA. 2010;107(5):2331–2336. doi: 10.1073/pnas.0913689107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartlett ME, Whipple CJ. Protein change in plant evolution: Tracing one thread connecting molecular and phenotypic diversity. Front Plant Sci. 2013;4:382. doi: 10.3389/fpls.2013.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson PB, et al. The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J. 2009;58(2):299–317. doi: 10.1111/j.1365-313X.2008.03780.x. [DOI] [PubMed] [Google Scholar]
- 38.Xiong L, Lee H, Huang R, Zhu JK. A single amino acid substitution in the Arabidopsis FIERY1/HOS2 protein confers cold signaling specificity and lithium tolerance. Plant J. 2004;40(4):536–545. doi: 10.1111/j.1365-313X.2004.02225.x. [DOI] [PubMed] [Google Scholar]
- 39.Studer RA, Dessailly BH, Orengo CA. Residue mutations and their impact on protein structure and function: Detecting beneficial and pathogenic changes. Biochem J. 2013;449(3):581–594. doi: 10.1042/BJ20121221. [DOI] [PubMed] [Google Scholar]
- 40.Mularoni L, Ledda A, Toll-Riera M, Albà MM. Natural selection drives the accumulation of amino acid tandem repeats in human proteins. Genome Res. 2010;20(6):745–754. doi: 10.1101/gr.101261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng Z, Verma DPS. A rice HAL2-like gene encodes a Ca(2+)-sensitive 3‘(2’),5′-diphosphonucleoside 3‘(2’)-phosphohydrolase and complements yeast met22 and Escherichia coli cysQ mutations. J Biol Chem. 1995;270(49):29105–29110. doi: 10.1074/jbc.270.49.29105. [DOI] [PubMed] [Google Scholar]
- 42.Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol. 2004;58(4):424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- 43.Richardson JS. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 44.Thornton JM. Disulphide bridges in globular proteins. J Mol Biol. 1981;151(2):261–287. doi: 10.1016/0022-2836(81)90515-5. [DOI] [PubMed] [Google Scholar]
- 45.Collet J-F, Messens J. Structure, function, and mechanism of thioredoxin proteins. Antioxid Redox Signal. 2010;13(8):1205–1216. doi: 10.1089/ars.2010.3114. [DOI] [PubMed] [Google Scholar]
- 46.Jeng MF, et al. High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure. 1994;2(9):853–868. doi: 10.1016/s0969-2126(94)00086-7. [DOI] [PubMed] [Google Scholar]
- 47.Qin J, Clore GM, Gronenborn AM. The high-resolution three-dimensional solution structures of the oxidized and reduced states of human thioredoxin. Structure. 1994;2(6):503–522. doi: 10.1016/s0969-2126(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 48.Ravilious GE, Nguyen A, Francois JA, Jez JM. Structural basis and evolution of redox regulation in plant adenosine-5′-phosphosulfate kinase. Proc Natl Acad Sci USA. 2012;109(1):309–314. doi: 10.1073/pnas.1115772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hicks LM, et al. Thiol-based regulation of redox-active glutamate-cysteine ligase from Arabidopsis thaliana. Plant Cell. 2007;19(8):2653–2661. doi: 10.1105/tpc.107.052597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bick J-A, et al. Regulation of the plant-type 5′-adenylyl sulfate reductase by oxidative stress. Biochemistry. 2001;40(30):9040–9048. doi: 10.1021/bi010518v. [DOI] [PubMed] [Google Scholar]
- 51.Nikkanen L, Rintamäki E. Thioredoxin-dependent regulatory networks in chloroplasts under fluctuating light conditions. Philos Trans R Soc Lond B Biol Sci. 2014;369(1640):20130224. doi: 10.1098/rstb.2013.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serrato AJ, Pérez-Ruiz JM, Spínola MC, Cejudo FJ. A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem. 2004;279(42):43821–43827. doi: 10.1074/jbc.M404696200. [DOI] [PubMed] [Google Scholar]
- 53.Ouyang M, et al. The photosensitive phs1 mutant is impaired in the riboflavin biogenesis pathway. J Plant Physiol. 2010;167(17):1466–1476. doi: 10.1016/j.jplph.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 54.Ball L, et al. Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell. 2004;16(9):2448–2462. doi: 10.1105/tpc.104.022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brunkard JO, Runkel AM, Zambryski PC. Chloroplasts extend stromules independently and in response to internal redox signals. Proc Natl Acad Sci USA. 2015;112(32):10044–10049. doi: 10.1073/pnas.1511570112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caplan JL, et al. Chloroplast stromules function during innate immunity. Dev Cell. 2015;34(1):45–57. doi: 10.1016/j.devcel.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huberts DH, van der Klei IJ. Moonlighting proteins: An intriguing mode of multitasking. Biochim Biophys Acta. 2010;1803(4):520–525. doi: 10.1016/j.bbamcr.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 58.Herrmann J, Nathin D, Lee SG, Sun T, Jez JM. Recapitulating the structural evolution of redox regulation in adenosine 5′-phosphosulfate kinase from cyanobacteria to plants. J Biol Chem. 2015;290(41):24705–24714. doi: 10.1074/jbc.M115.679514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao S, et al. COS1: An Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence. Plant Cell. 2004;16(5):1132–1142. doi: 10.1105/tpc.020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murguía JR, Bellés JM, Serrano R. A salt-sensitive 3‘(2’),5′-bisphosphate nucleotidase involved in sulfate activation. Science. 1995;267(5195):232–234. doi: 10.1126/science.7809627. [DOI] [PubMed] [Google Scholar]
- 61.Hutchison RS, Ort DR. Measurement of equilibrium midpoint potentials of thiol/disulfide regulatory groups on thioredoxinactivated chloroplast enzymes. Methods in Enzymol. 1995;252:220–228. doi: 10.1016/0076-6879(95)52025-2. [DOI] [PubMed] [Google Scholar]
- 62.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336(6084):1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diederichs K, Karplus PA. Better models by discarding data? Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1215–1222. doi: 10.1107/S0907444913001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pronk S, et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmid N, et al. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J. 2011;40(7):843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 71.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B, editor. The Jerusalem Symposia on Quantum Chemistry and Biochemistry. Vol 14. Springer; Dordrecht, Netherlands: 1981. pp. 331–342. [Google Scholar]
- 72.Essmann U, et al. A smooth particle mesh Ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- 73.Lopéz-Blanco JR, Garzón JI, Chacón P. iMod: Multipurpose normal mode analysis in internal coordinates. Bioinformatics. 2011;27(20):2843–2850. doi: 10.1093/bioinformatics/btr497. [DOI] [PubMed] [Google Scholar]
- 74.Ichiye T, Karplus M. Collective motions in proteins: A covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins. 1991;11(3):205–217. doi: 10.1002/prot.340110305. [DOI] [PubMed] [Google Scholar]
- 75.Yalovsky S, et al. Plant farnesyltransferase can restore yeast Ras signaling and mating. Mol Cell Biol. 1997;17(4):1986–1994. doi: 10.1128/mcb.17.4.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Catanzariti A-M, Soboleva TA, Jans DA, Board PG, Baker RT. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 2004;13(5):1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 78.Grassl J, et al. Analysis of the budding yeast pH 4-7 proteome in meiosis. Proteomics. 2010;10(3):506–519. doi: 10.1002/pmic.200900561. [DOI] [PubMed] [Google Scholar]
- 79.Grassl J, Pružinská A, Hörtensteiner S, Taylor NL, Millar AH. Early events in plastid protein degradation in stay-green Arabidopsis reveal differential regulation beyond the retention of LHCII and chlorophyll. J Proteome Res. 2012;11(11):5443–5452. doi: 10.1021/pr300691k. [DOI] [PubMed] [Google Scholar]
- 80.Lee CP, Eubel H, O’Toole N, Millar AH. Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol Cell Proteomics. 2008;7(7):1297–1316. doi: 10.1074/mcp.M700535-MCP200. [DOI] [PubMed] [Google Scholar]