Significance
The outer membrane of Gram-negative bacteria provides a barrier that allows these organisms to live in a variety of harsh environments. Here, we study the machine responsible for the insertion of integral membrane β-barrel proteins into this outer membrane. How this barrel assembly machine (Bam) functions is unknown because it has not been possible to characterize substrates in the process of folding. We slowed the assembly of a substrate and observe a partially folded state still bound to the Bam complex. These studies define how and where folding occurs and the roles that the two essential components of the Bam complex play. Understanding how Bam works could enable development of compounds that inhibit its function and kill Gram-negative bacteria.
Keywords: outer membrane, Bam complex, β-barrel, protein folding
Abstract
The assembly of β-barrel proteins into membranes is mediated by an evolutionarily conserved machine. This process is poorly understood because no stable partially folded barrel substrates have been characterized. Here, we slowed the folding of the Escherichia coli β-barrel protein, LptD, with its lipoprotein plug, LptE. We identified a late-stage intermediate in which LptD is folded around LptE, and both components interact with the two essential β-barrel assembly machine (Bam) components, BamA and BamD. We propose a model in which BamA and BamD act in concert to catalyze folding, with the final step in the process involving closure of the ends of the barrel with release from the Bam components. Because BamD and LptE are both soluble proteins, the simplest model consistent with these findings is that barrel folding by the Bam complex begins in the periplasm at the membrane interface.
The assembly of β-barrel membrane proteins into the outer membrane (OM) of Gram-negative bacteria, mitochondria, and chloroplasts is facilitated by conserved cellular machinery (1–4). The β-barrel assembly machine (Bam) folds and inserts integral membrane proteins into the OM of Gram-negative organisms (5). Bam is a five-protein complex consisting of the essential protein BamA, a β-barrel itself, and four lipoproteins, BamB, -C, -D, and -E, of which only BamD is essential (4–8). The Bam complex recognizes a large number of different substrates, but how each component catalyzes the folding and insertion of such structurally diverse substrates is unclear.
How β-barrels are assembled into membranes is not obvious. Where and how folding occurs is unclear because intermediates could contain both exposed polar amides and hydrophobic residues until the barrel has completed its fully hydrophobic exterior. By contrast, α-helical membrane proteins have internally satisfied hydrogen bonds, making stepwise assembly from stable secondary structural elements possible. Although Bam has been shown to accelerate membrane β-barrel assembly (9–11), the transient nature of folding intermediates has made accumulating such discrete species for characterization difficult (12–15). If structurally defined folding intermediates were to exist long enough for characterization, they could reveal crucial aspects of the folding process.
Here, we studied the assembly of an essential, slow-folding β-barrel, LptD. LptD is one of two components of the OM translocon that transports lipopolysaccharide to the cell surface (16–18). The other component, LptE, is a lipoprotein that forms a plug inside the LptD barrel (19–22). LptD also contains two disulfide bonds (23), and its assembly involves the formation of consecutive disulfide bonds that after barrel folding rearrange to form nonconsecutive disulfide bonds (24). The assembly of LptD is orders-of-magnitude slower (∼20 min versus seconds) than that of other barrel substrates (24–26). Because of the slow rate of folding and our ability to use oxidation state as a proxy for barrel folding, LptD is a prime candidate to capture folding intermediates.
We have used lptD mutations that further slow barrel assembly to trap substrate LptD on Bam. Characterization of this intermediate demonstrates that both essential components of the Bam complex, BamA and BamD, interact with the substrate and that the soluble lipoprotein LptE templates the formation of the LptD barrel. Because a significant amount of the LptD barrel enclosing LptE is formed before barrel closure while still interacting with the periplasmic portion of the Bam complex, we propose that barrel folding begins in the periplasm and that the last step in the assembly process is closure of the barrel with concomitant release from the Bam complex.
Results
Identification of a Substrate That Accumulates on the Bam Complex.
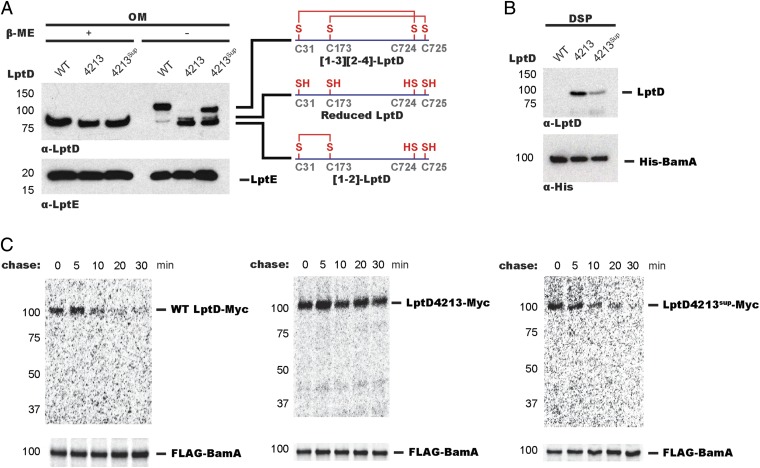
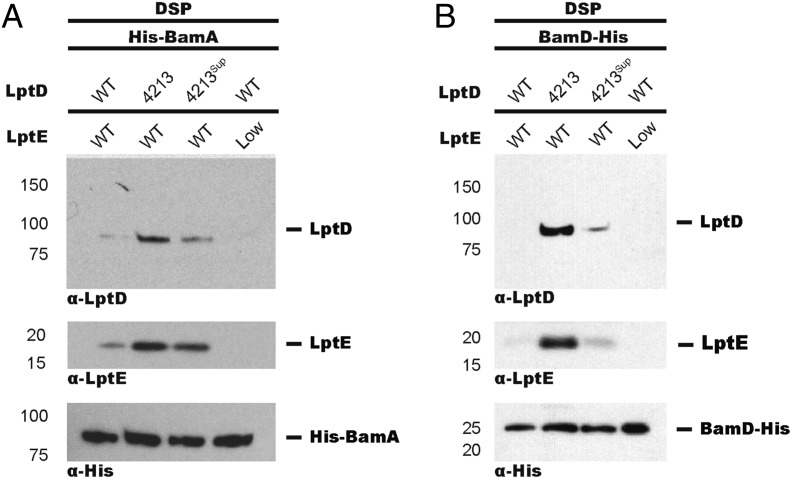
Oxidative folding of LptD in vivo involves the rearrangement of a form containing a disulfide bond between consecutive cysteines (designated [1,2]-LptD) to its mature form containing disulfide bonds between nonconsecutive cysteines (designated [1,3][2,4]-LptD for the order in which the cysteines appear in the primary sequence) (23, 24). More than 25 y ago, an lptD mutant allele was isolated, called lptD4213, which encodes a 23-amino acid deletion of an extracellular loop and confers OM assembly defects (16, 18–20). To further characterize this mutant protein, we isolated LptD from OMs of cultures expressing either WT LptD or LptD4213 (Fig. 1A). Whereas total levels of LptD were similar, LptD4213 primarily existed in a nonnative [1,2] disulfide-bonded form, which is an intermediate in the WT LptD assembly pathway (24). Because LptD4213 is membrane-associated but mostly remains in the [1,2] oxidation state, we surmised that it stalls at some point during assembly.
Fig. 1.
A mutant LptD substrate can be accumulated on Bam during folding. (A) LptD4213 accumulates as a nonfunctional disulfide-bonded species. LptD4213sup partially suppresses the LptD4213 assembly defect. LptD4213 migrates faster than WT LptD because it lacks 23 amino acids. OM fractions from wt (MC4100), lptD4213, and lptD4213sup strains were subject to α-LptD, α-LptE, and α-His immunoblot analyses. (B) LptD4213, but not WT LptD, accumulates on BamA at steady state. wt, lptD4213, and lptD4213sup strains expressing His-tagged BamA were cross-linked using DSP and affinity-purified. Adducts were identified by α-LptD and α-His immunoblot analyses after linker cleavage by β-ME. (C) WT LptD transiently interacts with BamA. LptD4213sup restores proper assembly of LptD4213 and mirrors the assembly of the native substrate. Cells expressing FLAG-tagged BamA and Myc-tagged WT LptD, LptD4213, or LptD4213sup were pulsed with [35S]-methionine and chased with cold methionine. Samples were analyzed by SDS/PAGE/autoradiography after DSP treatment, affinity purification, and cleavage of cross-linkers.
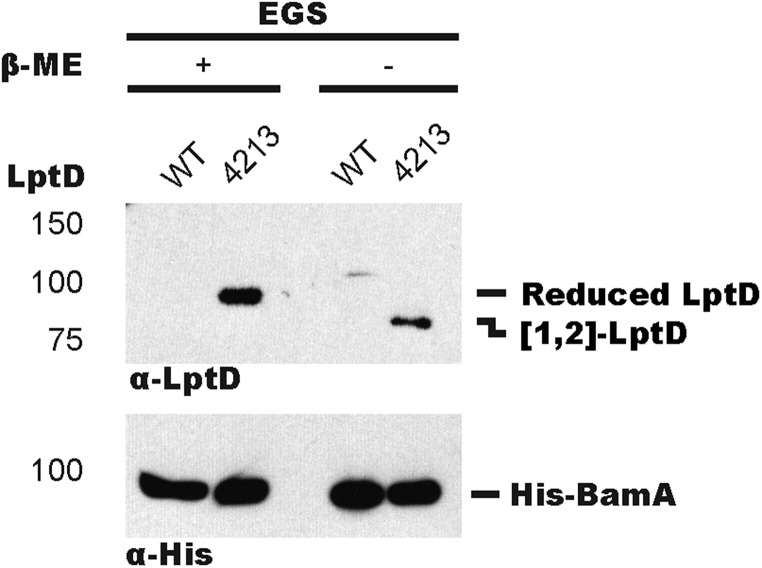
To determine where LptD4213 stalls along the assembly pathway, we used a chemical cross-linking strategy using the amine-reactive cross-linker dithiobis(succinimidyl proprionate) (DSP) to capture transient protein–protein interactions. Cells expressing His-tagged BamA (Fig. S1) were treated with DSP and affinity-purified (Fig. S2). After linker cleavage, we observed accumulation of LptD4213, but not of WT LptD, on BamA (Fig. 1B). Because WT LptD does not accumulate on BamA, we concluded that mature [1,3][2,4]-LptD does not stably associate with BamA. To determine the oxidation state of the LptD4213 cross-linked adduct, we used the same cross-linking strategy except with the chemical cross-linker ethylene glycol bis(succinimidylsuccinate) (EGS), which can be cleaved under nonreducing conditions to preserve disulfide bond configuration. We found that LptD4213 accumulated on BamA in the [1,2] disulfide-bonded configuration (Fig. S3). These results established that LptD4213 stalls on BamA with an oxidation state characteristic of a folding intermediate.
Fig. S1.
Strains containing mutagenized plasmids exhibit no change in OM content. [1,2]-LptD accumulates at steady state when LptE levels are limiting and when LptD is defective. Samples were subject to α-LptD, α-LptE, and α-His immunoblot analyses after OM extraction in wt, lptD4213, lptD4213sup, and LptE-limiting strains expressing His-tagged BamA (A) and His-tagged BamD (B).
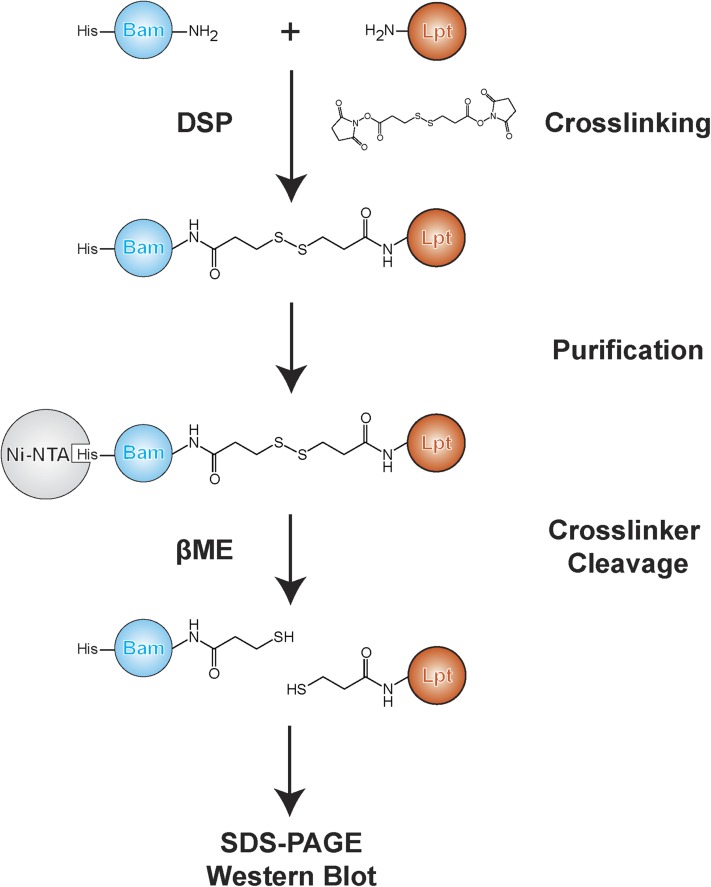
Fig. S2.
Experimental workflow for chemical cross-linking in vivo. Cells expressing a His-tagged Bam component are treated with DSP to capture transient protein–protein interactions. Cross-linked adducts are purified by Ni-NTA and then treated with reducing agent to cleave the cross-linker before analysis by Western blot.
Fig. S3.
LptD4213 stalls on BamA as the [1,2] disulfide bonded species. LptD4213 stalls on BamA in an intermediate disulfide-bonded state. wt and lptD4213 strains expressing His-tagged BamA were cross-linked using EGS and affinity-purified. Adducts were identified by α-LptD and α-His immunoblot analyses after cleavage of EGS by hydroxylamine.
To determine if the stalled LptD4213 is still folding-competent, we used a previously described intragenic suppressor of lptD4213, lptD4213sup, which harbors both the 23-codon deletion and a mutation that changes Asn-274 to Ile (27). OM fractions from strains expressing WT LptD, LptD4213sup, or LptD4213 (Fig. 1A) displayed no difference in LptD levels, suggesting that the N274I change in LptD4213sup does not affect protein stability or expression. However, we observed a significant increase in population of the mature [1,3][2,4] species in strains expressing LptD4213sup with respect to those expressing LptD4213 (Fig. 1A). Because total LptD levels remained constant, the N274I change in LptD4213sup appeared to facilitate conversion of the stalled [1,2]-LptD4213 intermediate to the mature [1,3][2,4] species. Consistent with this finding, we observed a corresponding decrease in cross-linking of LptD4213 to BamA (Fig. 1B). Because a single amino acid change increased the amount of properly oxidized LptD4213 at the expense of the species cross-linked to BamA, we concluded that the [1,2]-LptD4213 on Bam is an assembly intermediate.
The absence of a strong cross-link between WT LptD and BamA at steady state could reflect an interaction time too short for a cross-link to form, rather than a fundamental difference in the folding pathway of WT LptD compared with LptD4213. That is, WT LptD cross-linked weakly to BamA simply because [1,2]-LptD proceeds rapidly to mature [1,3][2,4]-LptD, which does not associate with Bam. Using pulse-labeling, we previously showed [1,2]-LptD is detectable only in the first 10 min after a cold methionine chase (24). To determine if we could observe short-lived intermediates of WT LptD on Bam, we examined the time-dependence of cross-linking in pulse-labeled cells expressing WT LptD or LptD4213. In cells expressing WT LptD, we detected a time-dependent cross-link between WT LptD and BamA, which appeared immediately after the pulse and disappeared 20 min into the chase (Fig. 1C). However, in cells expressing LptD4213, we observed substantial accumulation of LptD4213 on BamA immediately after the pulse that remained high throughout the chase (Fig. 1C).
To assess whether LptD4213 follows the same folding pathway as WT LptD, we examined the cross-linking profile of LptD4213sup. The time course showed immediate formation of a transient, cross-linked intermediate that decreased rapidly, mirroring the profile observed with WT LptD rather than that observed with LptD4213 (Fig. 1C). Because the residue altered by the suppressor (N274I) faces into the hydrophobic core of the OM and not the barrel lumen (19, 20), we assume no gross structural changes between [1,2]-LptD4213 and LptD4213sup. The intragenic suppressor has altered folding kinetics such that LptD4213 can now be converted to the native oxidation state. Because a single amino acid substitution allows LptD4213 to continue to functional product, we inferred that the stalled LptD4213 complex is a putative folding intermediate that could reveal crucial aspects of β-barrel assembly.
BamA and BamD Interact with Substrate Throughout the Assembly Process.
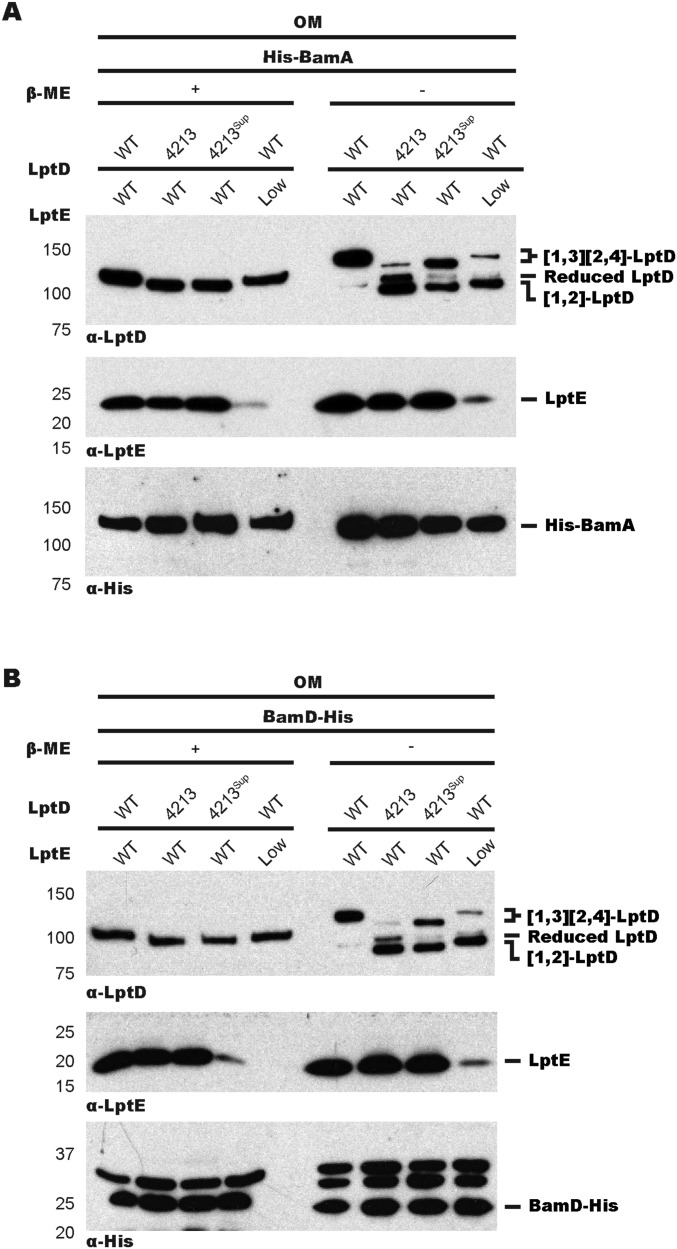
Proper assembly of the lipopolysaccharide translocon requires the lipoprotein LptE, which forms a plug for the LptD barrel (22, 28). Biochemical and genetic data have suggested that LptD and LptE may interact during folding on Bam (21, 29). To determine if LptD4213 arrests on BamA with LptE, we performed similar chemical cross-linking experiments to those described above to detect LptE. We observed increased accumulation of LptE on BamA in cells expressing LptD4213 with respect to those expressing WT LptD (Fig. 2A). Consistent with LptD4213 representing a putative folding intermediate, LptD4213sup exhibited lower levels of both LptD and LptE accumulated on BamA.
Fig. 2.
LptE is required for engagement of LptD with Bam. LptD4213 stalls on the Bam complex with LptE. WT LptD does not accumulate on BamA or BamD when LptE levels are limiting. Samples were subjected to α-LptD, α-LptE, and α-His immunoblot analyses after DSP cross-linking, affinity purification, and cleavage of cross-linkers in wt, lptD4213, lptD4213sup, and LptE-limiting strains expressing His-tagged BamA (A) or His-tagged BamD (B).
Because Bam also contains an essential lipoprotein, BamD (5, 7), we tested if this protein, like BamA (30–33), is involved in the assembly of barrel substrates (34, 35). We performed the same cross-linking experiments as for BamA except in cells expressing His-tagged BamD (Fig. S1). As with BamA, we observed an increase in cross-linking of both LptD and LptE to BamD in cells expressing LptD4213 compared with those expressing WT LptD and LptD4213sup (Fig. 2B). Therefore, we have captured a stalled substrate on the Bam complex, containing BamA, BamD, LptD, and LptE.
LptD Requires LptE to Fold on the Bam Complex.
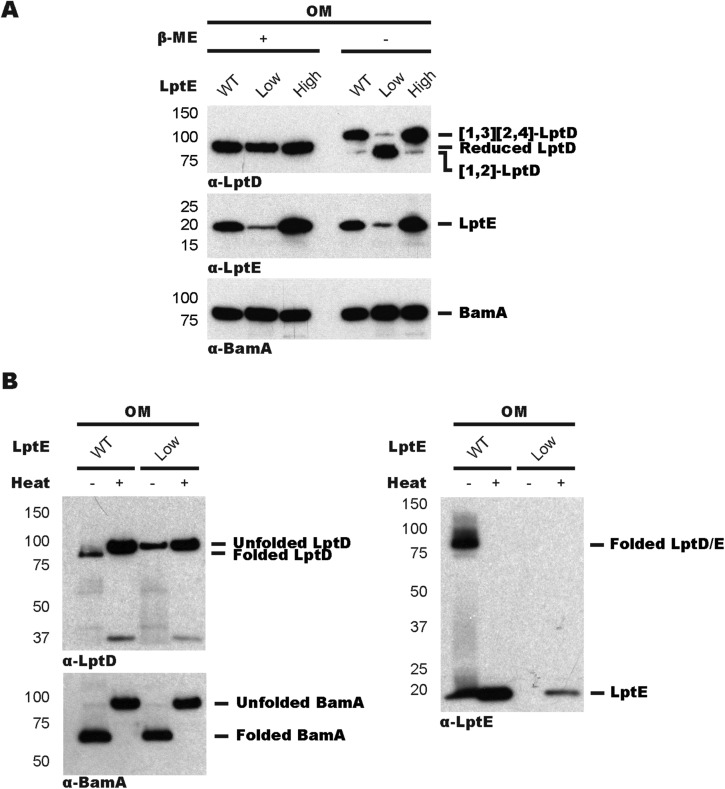
Identification of an LptD4213/E stalled substrate on Bam revealed that LptE is present before folding is complete. To ask if LptD can assemble on Bam without its plug, we constructed a strain in which we could regulate the levels of LptE and monitored the resulting disulfide configuration of WT LptD. When LptE was limiting, we observed no changes in total LptD levels but detected an accumulation of unfolded [1,2]-LptD (Fig. S4), which was expected because the LptD oxidative rearrangement is LptE-dependent (23, 24). We then investigated if this unfolded [1,2]-LptD interacts with Bam using the same cross-linking strategy described above. Analysis of whole-cell lysates revealed no accumulation of WT LptD on BamA (Fig. 2A) or on BamD (Fig. 2B) under LptE-limiting conditions. We conclude that LptD does not assemble independently of LptE on Bam.
Fig. S4.
Limiting LptE levels causes LptD assembly defects. (A) Regulation of LptE levels by an arabinose-inducible promoter. Samples were subject to α-LptD, α-LptE, and α-LptE immunoblot analyses after outer membrane extraction in wt and LptE-limiting strains. LptE-low and LptE-high samples were taken with or without the addition of 0.2% arabinose, respectively. (B) Unfolded [1,2]-LptD accumulates when LptE is limiting. OM fractions from wt and LptE-limiting strains were subject to subject to α-LptD, α-LptE, and α-BamA immunoblot analyses under reducing conditions. Samples were heated for 10 min at 100 °C as indicated.
The LptD4213 Intermediate on the Bam Complex Is a Stable Partially Folded Barrel.
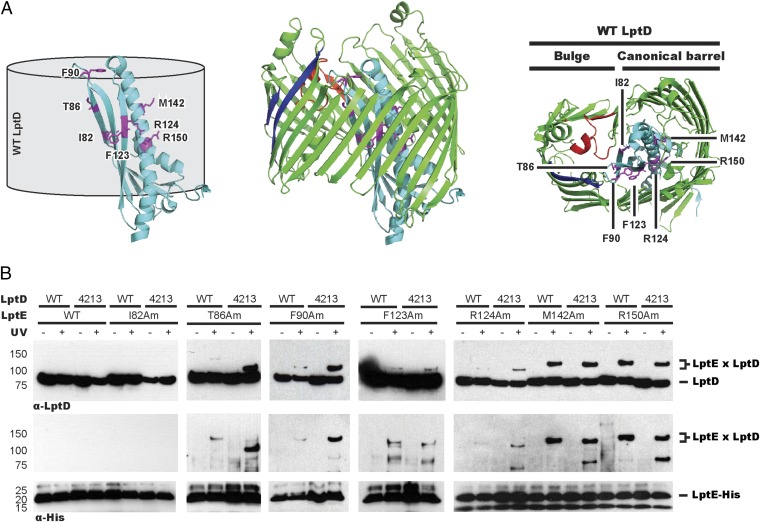
We have previously used a site-specific in vivo photo–cross-linking strategy to identify residues on multiple faces of LptE that interact with the lumen of the LptD barrel (Fig. 3A) in a properly assembled LptD/E translocon (21). We wondered if we could use photo–cross-linking at these positions to better characterize the LptD4213 stalled on Bam. We compared the cross-linking profiles of WT LptD and LptD4213 when the UV photo–cross-linker parabenzoyl-l-phenyalanine (pBPA) (36) was introduced into His-tagged LptE at seven positions. As expected, we observed cross-links between six residues in LptE with WT LptD (Fig. 3B). The same six residues in LptE also cross-link with LptD4213, but several of these residues differed in their cross-linking intensity compared with WT LptD (Fig. 3B).
Fig. 3.
The LptD substrate accumulated on Bam is a rudimentary barrel plugged by LptE. (A) Specific sites in LptE (magenta) cross-link to the lumen of LptD (green). LptE T86 interacts with an extracellular loop that is deleted in LptD4213 (red). Images were generated using the structure of LptD/E from Shigella flexneri (PDB ID code 4Q35). The N-terminal soluble domain and the L11 loop have been removed for clarity. (B) In vivo photo–cross-linking of LptE to LptD4213 or WT LptD. wt or lptD4213 strains both harboring the amber suppression system and expressing His-tagged LptE pBPA derivatives were either left untreated or irradiated with UV. Adducts were identified after affinity purification by α-LptD and α-His immunoblot analyses under reducing conditions.
The intensity of cross-linking at residues M142, R150, and F123 was similar in both the WT and mutant strains. LptD does not form a canonical barrel, but instead forms a bulge where its N and C termini come together to close the barrel (Fig. 3A) (19, 20). M142 and R150 interact with the LptD barrel on the side opposite the bulge (Fig. 3A). Because these residues in LptE form cross-links of similar intensity to both WT LptD and LptD4213, this region of LptD4213/E has already adopted its final, folded conformation.
In contrast, the other three pBPA substitutions in LptE resulted in markedly different cross-linking intensities to WT LptD compared with LptD4213. Cross-links from residues 90 and 124 in LptE were much stronger to LptD4213 than to WT LptD (Fig. 3B), implying a closer association of LptE with the C terminus of LptD in LptD4213 than in mature WT LptD/E. Residue 86 showed the largest difference in cross-linking, forming a strong cross-link to LptD4213, but a very weak cross-link to mature LptD/E. Residue 86 points toward the N-terminal region of LptD that is involved in barrel closure (19, 20) and likely interacts with the region of LptD that is deleted in LptD4213 (Fig. 3A). The difference in mobility observed for the cross-linked WT and LptD4213 species is consistent with cross-linking to a different site. These results indicate that LptD4213 is wrapped around LptE; although a large portion of the barrel resembles its final folded form, the bulge region has not yet completed folding.
Barrel Folding Precedes Closure and Release from the Bam Complex.
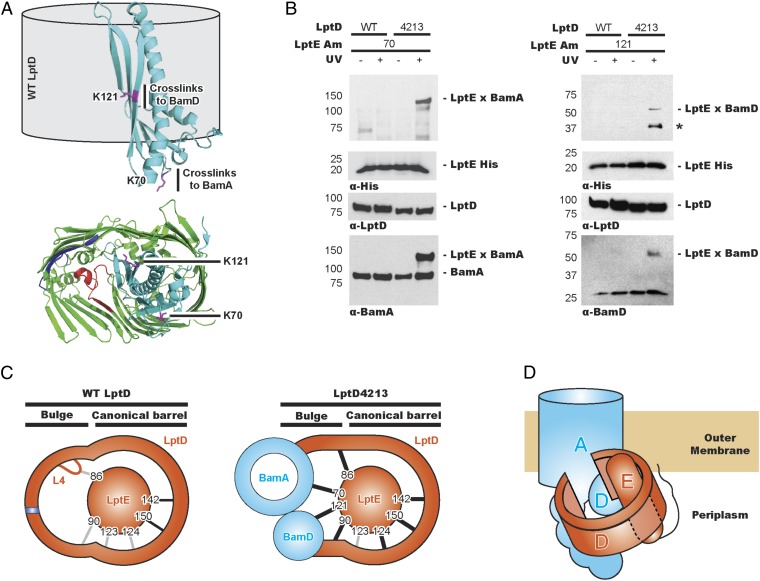
Our observation that LptE plugs the LptD4213 barrel (Fig. 3), yet can cross-link to Bam (Fig. 2), led us to ask what sites in LptE contact Bam. Assuming that DSP cross-links lysine side chains, we replaced those lysines in LptE that are not facing the canonical barrel region of LptD (Fig. 4A) with pBPA. When pBPA replaced K70 and K121, we observed no cross-links to WT LptD (Fig. 4B). However, in cells expressing LptD4213, we observed the appearance of high-molecular weight adducts from positions K70 and K121 in LptE to BamA and BamD, respectively (Fig. 4B). Residue K121 in LptE points into the LptD lumen and is protected from proteolysis by the barrel in the final folded form of LptD/E (Fig. 4A) (22). A cross-link can only form from LptE to BamD if the LptD barrel is still open. Therefore, we propose that LptD4213 exists largely as a plugged barrel in which the ends of the barrel have not closed (Fig. 4C).
Fig. 4.
The LptD substrate accumulated on Bam is an open barrel. (A) LptE K121 is protected by, but does not directly interact with, LptD. LptE K79 is exposed to the periplasmic space. (B) LptE displays LptD4213-dependent cross-links to Bam. Bands marked with an asterisk have not been identified. (C) LptE displays differential cross-linking profiles to WT LptD (Left) and LptD4213 (Right). Strong and weak cross-links are marked by gray and black lines, respectively. (D) Side view model of Bam-assisted LptD folding.
Discussion
In this study, we have used the essential β-barrel, LptD, as a substrate to study Bam-mediated assembly in vivo. We have isolated a partially folded LptD intermediate that is plugged by its lipoprotein LptE and stabilized by interactions with BamA and BamD. A number of β-hairpins have properly aligned because we observe cross-links identical to those found in the fully folded barrel (Fig. 3). However, the barrel remains open because we can observe a cross-link from LptE K121 to BamD in this intermediate (Fig. 4), even though K121 should be fully enclosed in the mature LptD barrel (19, 20). It is worth noting that K121 is found in a region of LptE that is functionally relevant for barrel assembly. Previously, we found that a 6-bp deletion in lptE that changes residues 116–120 from YPISA to YRA causes defects in LptD assembly (29). Importantly, missense mutations in bamA suppress this deletion. We argue here that this region of LptE forms important interactions with BamD during folding, and if these cannot form, then mutations in bamA change how Bam handles LptD/E to overcome the problem.
Characterization of a partially folded barrel trapped on the Bam complex allows us to propose a model for how Bam catalyzes the assembly of membrane β-barrel proteins (Fig. 4D). Folding initiates when BamA and BamD interact with the N- and C-terminal strands of unfolded LptD, drawing the barrel around the soluble LptE plug. Favorable interactions between LptE and LptD, including the L4 loop that contains the 23 amino acids deleted in LptD4213, facilitate barrel closure and release from BamA/D. LptD4213 accumulates on Bam as an open barrel because the deletion in LptD4213 removes critical interactions between LptD and LptE that permit barrel closure. We hypothesize that LptE not only templates barrel folding, but also triggers substrate release through an interaction with the L4 loop that brings the N and C termini of LptD together and away from Bam.
Not all β-barrel membrane proteins have a plug, but some of the largest ones do contain one that is part of the same polypeptide as the barrel (37). Although smaller barrels cannot accommodate folded domains within their lumens, some contain polypeptides that may be involved in barrel assembly (38, 39). Because bringing the N- and C-terminal strands together is not as entropically challenging for smaller barrels, folding may be less dependent on a structural plug. If plugs also act as release factors, as proposed here, then small barrels would have an analogous mechanism for promoting release from Bam.
Our model explains why Bam-catalyzed folding is more efficient than the uncatalyzed insertion of β-barrels into membranes (9). Biophysical studies suggest that the first step in uncatalyzed β-barrel assembly involves pairing of the N and C termini, possibly facilitated by a molten state at the membrane interface (14, 40). Pairing of the termini dramatically restricts conformational and rotational degrees-of-freedom to allow alignment of the β-hairpins. Once enough β-hairpins have arranged to form a well-defined barrel, spontaneous membrane insertion occurs (41–43). In our model of the catalyzed process, BamA/D interacts with the unfolded substrate, presumably also to restrict conformational freedom, templating formation of the β-sheet. We conclude that Bam, rather than changing the overall assembly mechanism, accelerates the intrinsic folding pathway.
Our data are also in agreement with the uncatalyzed mechanism of folding with respect to where folding occurs. In the uncatalyzed process, folding begins outside of the membrane (43). Because BamD is found at the membrane interface rather than in the membrane, Bam-catalyzed folding must also occur at this interface (Fig. 4D). In fact, BamD and LptE are both soluble periplasmic proteins anchored to the OM by lipid tails, and LptD folds around LptE. The simplest model, consistent with our results, is that folding likely begins in the periplasm at the membrane interface before membrane insertion.
Materials and Methods
Strains and Growth Conditions.
Strains and plasmids are provided in Tables S1 and S2, respectively. Unless otherwise noted, cultures were grown at 37 °C and supplemented with the appropriate antibiotics and amino acids.
Table S1.
Strains used
Table S2.
Plasmids used
| Plasmids | Description | Source |
| pET23/42 | pET23a(+) with multiple cloning sites of pET24a(+), PT7-dependent expression vector | (28) |
| pET23/42lptE-His | Encodes full-length LptE with a C-terminal His8 tag | (24) |
| pET23/42lptEI82Am-His | pET23/42lptE-His with I82Amber | (21) |
| pET23/42lptET86Am-His | pET23/42lptE-His with T86Amber | (21) |
| pET23/42lptEF90Am-His | pET23/42lptE-His with F90Amber | (21) |
| pET23/42lptEF123Am-His | pET23/42lptE-His with F123Amber | (21) |
| pET23/42lptER124Am-His | pET23/42lptE-His with R124Amber | (21) |
| pET23/42lptEM142Am-His | pET23/42lptE-His with M142Amber | (21) |
| pET23/42lptER150Am-His | pET23/42lptE-His with R150Amber | (21) |
| pET23/42lptEK70Am-His | pET23/42lptE-His with K70Amber | Present study |
| pET23/42lptEK121Am-His | pET23/42lptE-His with K121Amber | Present study |
| pET9a | Cloning vector; PT7-dependent expression vector | Novagen |
| pET9aHis-BamA | Encodes full-length BamA with a N-terminal His8 tag | Present study |
| pET9aBamD-His | Encodes full-length BamD with a C-terminal His8 tag | Present study |
| pET9aFLAG-BamA | Encodes full-length BamA with a N-terminal FLAG3 tag | Present study |
| pET9aBamD-FLAG | Encodes full-length BamD with a C-terminal FLAG3 tag | Present study |
| pBBR1MCS | Cloning vector; PT7 dependent expression vector | (48) |
| pBBR1MCSlptE | Encodes full-length LptE | Present study |
| pSup-BpaRS-6TRN | Encodes an orthogonal tRNA and aminoacyl-tRNA synthetase permitting ribosomal incorporation of pBPA at TAG stop codons | (49) |
In Vivo Chemical Cross-Linking.
Cross-linking experiments are based on techniques previously described (44), with modifications. A detailed description of affinity purification and in vivo cross-linking procedures are provided in SI Materials and Methods.
In Vivo DSP Cross-Linking of [35S]-Pulse–Labeled Cells.
Strains MC4100 containing pET9aFLAG3-BamA and pLptD/LptD4213/LptD61-Myc3 were used in in vivo DSP cross-linking of [35S]-pulse–labeled cells. A 25-mL culture was grown to OD600 ∼0.6 in M63/glucose minimal media supplemented with 18 amino acids (minus methionine and cysteine) at 37 °C. The culture was pulse-labeled with [35S]-methionine (200 μCi/mL final concentration; American Radiochemicals) for 2 min and then chased with cold methionine (5 mM) at 37 °C. At the indicated time points during the chase, a 5-mL culture aliquot was pelleted by centrifuging at 14,000 × g for 1 min. The cell pellet was resuspended in 300 μL of 150 mM NaCl and 10 mM NaH2PO4, pH 7.2, pelleted again at 18,000 × g for 30 s, and resuspended in 300 μL of the same buffer. Three microliters of DMSO containing 75 μg DSP was added to the resuspended cells and incubated at 37 °C for 30 min. The cross-linking reaction was quenched by addition of 1 M Tris⋅HCl (pH 8.0) to a final concentration of 20 mM followed by addition of 50 μL of trichloroacetic acid (TCA, 70% (wt/vol) in water) and incubated on ice for 20 min. Precipitated proteins were pelleted at 18,000 × g for 10 min at 4 °C, washed with 1 mL of ice-cold acetone, and then solubilized in 150 μL of 100 mM Tris⋅HCl, pH 8.0, containing 1% (wt/vol) SDS. The sample was sonicated for 30 s to aid solubilization. The cross-linked products were collected from the samples by immunoprecipitation as described previously (24), with modifications. After immunoprecipitation, 15 μL of eluted sample was applied to SDS/PAGE directly; 4–20% Tris⋅HCl polyacrylamide gels were used (running conditions: 150 V for 90 min). The gel was then dried and exposed to phosphor storage screens for autoradiography. Further details are provided in SI Materials and Methods.
Site-Specific in Vivo Photo–Cross-Linking.
Photo–cross-linking experiments are based on techniques as previously described (21), with modifications. A detailed description of affinity purification and in vivo photo–cross-linking procedures are provided in SI Materials and Methods.
SI Materials and Methods
Bacterial Strains and Growth Conditions.
The WT strain used is MC4100 [F− araD139 Δ(argF-lac) U169 rpsL150 relA1 flbB5301 ptsF25 deoC1 ptsF25 thi]. The lptDΔ330–352 (lptD4213) mutant strain used is NR698 (27). The lptDΔ330–352, N274I (lptD4213sup) mutant strain used is NR731 (27). To limit levels of lptE expression, plasmid pBBR1MCSlptE was constructed by subcloning the XbaI-XhoI lptE fragment from pET23/42lptE (22) into the same sites of plasmid pBBR1MCS. The resultant plasmid was transformed into a strain [MC4100 ara+ ΔlptE ∆(λatt-lom)::bla PBAD-lptE] where expression of chromosomal lptE is arabinose-dependent to yield strain MG1559. When grown in the absence of arabinose, MG1559 remains viable but produces limiting levels of LptE. Lysogeny (LB) broth and M63/glucose minimal broth and agar were prepared as described previously (45). Growth of strains was carried out at 37 °C unless explicitly indicated. When appropriate, carbenicillin (50 μg/mL), chloramphenicol (30 μg/mL), kanamycin (50 μg/mL), and spectinomycin (50 μg/mL) were added. Amino acids were added at 50 μg/mL where indicated. pBPA (Bachem Americas) was used at 0.9 mM. The suppressor plasmid pSup-BpaRS-6TRN was a generous gift from Peter G. Schultz, Scripps Institute, San Diego, CA.
Plasmid Construction.
To construct pET9aHis-BamA, a cassette containing the coding sequence of the His-BamA was inserted into pET9a. Briefly, the pET23/42His-BamA template was amplified by PCR (using primers 5′- AAT TGG ATC CAT GGC GAT GAA AAA GTT GCT C -3′ and 5′- ATA TGC TCA GCT TAC CAG GTT TTA CCG ATG TTA AAC -3′) and the resulting PCR product was digested with BamHI and BlpI for >3 h at 37 °C. The digested PCR product was ligated to a pET9a vector that had been digested with BamHI and BlpI for >3 h at 37 °C. NovaBlue (Novagen) cells were transformed with 1 μL of the ligation product and plated onto LB plates containing 50 μg/mL kanamycin. Proper plasmid construction was confirmed by DNA sequencing.
To construct pET9aBamD-His, a cassette containing the coding sequence of the BamD-His was inserted into pET9a. Briefly, pET23/42BamD-His was digested with XbaI and BlpI for >3 h at 37 °C. The digested product was gel-purified and the coding sequence of the BamD-His was ligated to a pET9a vector that had been digested with XbaI and BlpI for >3 h at 37 °C. NovaBlue (Novagen) cells were transformed with 1 μL of the ligation product and plated onto LB plates containing 50 μg/mL kanamycin. Proper plasmid construction was confirmed by DNA sequencing.
To construct pET9aFLAG3-BamA, a cassette containing the coding sequence of the FLAG3 tag was inserted into pET9aHis-BamA to replace the original His6 tag. Briefly, the entire pET9aHis-BamA template was amplified by PCR (using primers 5′-A GAT CAT GAT ATC GAC TAT AAA GAC GAT GAT GAC AAA GAA GGG TTC GTA GTG AAA GAT-3′ and 5′-TCG ATA TCA TGA TCT TTG TAG TCG CCG TCG TGA TCT TTA TAA TCA GCA CCG TAT ACG GTG-3′) and the resulting PCR product mixture digested with DpnI for >1 h at 37 °C. NovaBlue (Novagen) cells were transformed with 1 μL of digested PCR product and plated onto LB plates containing 50 μg/mL kanamycin. Proper plasmid construction was confirmed by DNA sequencing.
Isolation of OM for Analysis of LptD Oxidation States.
OM analyses were performed as previously described (24). Briefly, cells were pelleted by centrifugation at 5,000 × g for 20 min and then resuspended in 5 mL Tris-B buffer [10 mM Tris⋅HCl (pH 8.0)] containing 20% (wt/wt) sucrose, 1 mM phenylmethylsulfonyl fluoride (PMSF, Sigma), 50 µg/mL DNase I (Sigma), and 50 mM iodoacetamide (IAM, Sigma). Cells were lysed by a single passage through a French Press (Thermo Electron) at 8,000 psi. Approximately 8 mL of cell lysate was layered onto a two-step sucrose gradient [top: 4 mL Tris-B buffer containing 40% (wt/wt) sucrose; bottom: 1 mL Tris-B buffer containing 65% (wt/wt) sucrose] and centrifuged at 39,000 rpm for 16 h in a Beckman SW41 rotor in an ultracentrifuge (Model XL-90, Beckman). OM fragments (∼0.5 mL) were isolated from the 40%/65% interface by puncturing the side of the tube with a syringe. Next, 1 mL of 20 mM Tris⋅HCl (pH 8.0) was added to the OM fragments to lower the sucrose concentration to below 20% (wt/wt). The OM fragments were then pelleted in a microcentrifuge at 18,000 × g for 30 min and then resuspended in 200–250 µL TBS containing 5 mM IAM. Protein concentrations of these OM preparations were determined using the Bio-Rad DC protein assay after precipitating in 10% (vol/vol) TCA and resolubilizing in TBS containing 2% (wt/vol) SDS. The same amount of OM (based on protein content) for each strain was analyzed by nonreducing SDS/PAGE and immunoblotted using antibodies directed against LptD and LptE.
In Vivo DSP Cross-Linking.
Strains MC4100, NR698, NR731, and MC4100 ara+ lptE− λatt(PBAD-lptE) containing either pET9aHis-BamA or pET9aBamD-His were used in in vivo DSP cross-linking experiments. Cells were grown in 500 mL of LB to OD600 ∼0.6 unless explicitly indicated. Cells were pelleted by centrifugation at 5,000 × g for 15 min and the cell pellets were washed twice with 25 mL of 150 mM NaCl and 10 mM NaH2PO4, pH 7.2, and resuspended in the same buffer. DSP dissolved in DMSO was added to the cell suspension at a final concentration of 200 μg/mL and the cells were incubated for 30 min at 37 °C. The cross-linking reaction was quenched by addition of 1 M Tris⋅HCl (pH 8.0) to a final concentration of 20 mM, and cells were harvested by centrifugation at 5,000 × g for 10 min.
Cell pellets were resuspended in 5 mL of 20 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, and 1% (wt/vol) Anzergent 3-14 containing lysozyme (50 μg/mL), DNase I (50 μg/mL), and RNase I (50 μg/mL), and were lysed by sonication. To remove cell debris after lysis, the mixture was centrifuged at 10,000 × g for 10 min. The cleared lysate was supplemented with 20 mM imidazole (pH 8.0), and incubated with 200 μL of pre-equilibrated Ni-NTA resin for 30 min at 4 °C. The column was then washed four times with 10 mL of 20 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, 40 mM imidazole, 0.1% Triton X-100, and 0.1% SDS. The column was eluted with 4 mL of 20 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, and 200 mM imidazole. The eluate was concentrated via ultrafiltration at 5,000 × g for 20 min. The concentrated sample was used for SDS/PAGE and Western blot analysis.
In Vivo EGS Cross-Linking.
Strains MC4100 and NR698 containing pET9aHis-BamA were used in in vivo EGS cross-linking experiments. The protocol is largely similar to that of in vivo DSP cross-linking with modifications. Cells were grown in 500 mL of LB to OD600 ∼0.6, pelleted by centrifugation at 5,000 × g for 15 min. The cell pellets were washed twice with 25 mL of 150 mM NaCl and 10 mM NaH2PO4 (pH 7.2) and resuspended in the same buffer. EGS dissolved in DMSO was added to the cell suspension at a final concentration of 200 μg/mL, and the cells were incubated for 30 min at 37 °C. The cross-linking reaction was quenched by addition 1 M Tris⋅HCl (pH 7.4) to a final concentration of 20 mM, and cells were harvested by centrifugation at 5,000 × g for 10 min. Cell pellets were resuspended in 5 mL of 20 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, and 1% Anzergent 3-14 (wt/vol) containing lysozyme (50 μg/mL), DNase I (50 μg/mL), and RNase I (50 μg/mL), and were lysed by sonication. To remove cell debris after lysis, the mixture was then centrifuged at 10,000 × g for 10 min. The cleared lysate was supplemented with 20 mM imidazole (pH 8.0), and incubated with 200 μL of pre-equilibrated Ni-NTA resin for 30 min at 4 °C. The column was then washed four times with 10 mL of 20 mM Tris buffer (pH 8.0), with 300 mM NaCl, 40 mM imidazole, 0.1% Triton X-100 (vol/vol), and 0.1% SDS (wt/vol). The column was eluted with 4 mL of 20 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, and 200 mM imidazole. The eluate was concentrated via ultrafiltration at 5,000 × g for 20 min (∼30 μL) and added to 470 μL of freshly made 1 M hydroxylamine-HCl (pH 8.5), 0.02% Anzergent 3-14 (wt/vol), and incubated at 37 °C for 4 h, followed by dialysis against 1 L of 20 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 0.02% (wt/vol) Anzergent 3-14 overnight. The resulting solution was concentrated via ultrafiltration at 18,000 × g for 20 min. The concentrated sample was used for SDS/PAGE and Western blot analysis.
Immunoprecipitation After in Vivo DSP Cross-Linking of [35S]-Pulse–Labeled Cells.
After the chase and TCA precipitation, 1.4 mL of ice-cold IP buffer [50 mM Tris⋅HCl (pH 8.0) containing 150 mM NaCl, 2% (vol/vol) Triton X-100, 1 mM EDTA] was added to each sample aliquot and was centrifuged at 18,000 × g for 10 min at 4 °C. Next, 1.4 mL of the supernatant was transferred to another tube containing 25 μL of anti-FLAG M2 magnetic beads (Sigma). The beads were washed and pre-equilibrated with 3 × 1 mL IP buffer before use. The mixture was incubated on a rotary shaker for 3∼4 h at 4 °C, and the beads were washed with 4 × 1 mL of ice-cold high-salt buffer [50 mM Tris⋅HCl (pH 8.0) containing 1 M NaCl, 1% (vol/vol) Triton X-100, 1 mM EDTA) and 1 × 1 mL ice-cold 10 mM Tris⋅HCl (pH 8.0) using a magnetic separation rack (New England Biolabs). Next, 160 μL 100 mM Tris⋅HCl (pH 8.0) containing 1% (wt/vol) SDS was then added to the beads and the mixture heated for 10 min at 100 °C to elute the bound proteins. Ten microliters of the eluate was set aside and used for SDS/PAGE analysis.
Next, 1.4 mL of ice-cold IP buffer was added to the remaining 150 μL of the eluate and the sample was centrifuged at 18,000 × g for 10 min at 4 °C; 1.4 mL of the supernatant was transferred to another tube containing 120 μL of anti-Myc agarose beads (Sigma). The beads were washed and pre-equilibrated with 3 × 1 mL IP buffer before use. The mixture was incubated on a rotary shaker for 3∼4 h at 4 °C, and the beads were washed with 4 × 1 mL of ice-cold high salt buffer. Samples were then eluted with 1 mL ice-cold 10 mM Tris⋅HCl (pH 8.0). Then 80 μL 2× SDS nonreducing sample buffer was added to the beads and the mixture heated for 10 min at 100 °C to elute the bound proteins.
Site-Specific in Vivo Photo–Cross-Linking.
For affinity pulldowns with pBPA-containing LptE-His MC4100 or NR698, strains harboring pSup-BpaRS-6TRN and pET23/42lptE-His containing the TAG stop codon at the indicated positions were grown overnight, diluted 1:100 into 100 mL of the same media, and grown to midlog phase. After normalization by optical density, each culture was split in half and used directly for irradiation with UV light at 365 nm for 10 min. All samples were subsequently kept at 4 °C. Samples were resuspended in 5 mL ice-cold TBS containing 1% (wt/vol) Anzergent 3-14 (Anatrace), 100 μg/mL lysozyme, 1 mM PMSF, and 50 μg/mL DNase I, lysed by sonication, and centrifuged at 15,000 × g in a table-top centrifuge for 30 min. The supernatant was then passaged three times over Ni-NTA beads and washed twice with 4 mL ice-cold TBS containing 0.02% Anzergent 3-14, and 20 mM imidazole. Samples were eluted with ice-cold TBS containing 0.02% Anzergent 3-14 and 200 mM imidazole. Eluates were supplemented with 10% TCA by volume (100 μL) and incubated on ice for 30 min. Precipitated proteins were pelleted at 18,000 × g for 10 min at 4 °C and washed twice with 1 mL of ice-cold acetone. All samples were finally resuspended in 125 μL of SDS/PAGE buffer and boiled for 5 min. Eight microliters of each sample were analyzed by SDS/PAGE and immunoblotting.
Antibodies.
Monoclonal α-His conjugated to horseradish peroxidase was purchased from Qiagen. Monoclonal α-FLAG conjugated to horseradish peroxidase was purchased from Sigma. α-LptD (46) and α-LptE (22) antiserum were obtained as already described.
Acknowledgments
Special thanks to the T.J.S. and D.E.K. laboratories for their helpful discussions. This work was supported by funds from the National Institutes of Health Awards F31GM116210 (to J.L.), F32GM108258 (to J.S.W.), GM34821 (to T.J.S.), GM100951 (to N.R.), and AI081059 (to D.E.K.); and from National Science Foundation Graduate Research Fellowship Program Award DGE1148900 (to H.A.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604100113/-/DCSupplemental.
References
- 1.Paschen SA, et al. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature. 2003;426(6968):862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- 2.Reumann S, Davila-Aponte J, Keegstra K. The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: Identification of a cyanobacterial homolog. Proc Natl Acad Sci USA. 1999;96(2):784–789. doi: 10.1073/pnas.96.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiedemann N, et al. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424(6948):565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- 4.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299(5604):262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 5.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121(2):235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Eggert US, et al. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science. 2001;294(5541):361–364. doi: 10.1126/science.1063611. [DOI] [PubMed] [Google Scholar]
- 7.Malinverni JC, et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61(1):151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 8.Sklar JG, et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104(15):6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagan CL, Kim S, Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328(5980):890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagan CL, Kahne D. The reconstituted Escherichia coli Bam complex catalyzes multiple rounds of β-barrel assembly. Biochemistry. 2011;50(35):7444–7446. doi: 10.1021/bi2010784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gessmann D, et al. Outer membrane β-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc Natl Acad Sci USA. 2014;111(16):5878–5883. doi: 10.1073/pnas.1322473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kutik S, et al. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell. 2008;132(6):1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 13.Ieva R, Tian P, Peterson JH, Bernstein HD. Sequential and spatially restricted interactions of assembly factors with an autotransporter beta domain. Proc Natl Acad Sci USA. 2011;108(31):E383–E391. doi: 10.1073/pnas.1103827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleinschmidt JH, den Blaauwen T, Driessen AJ, Tamm LK. Outer membrane protein A of Escherichia coli inserts and folds into lipid bilayers by a concerted mechanism. Biochemistry. 1999;38(16):5006–5016. doi: 10.1021/bi982465w. [DOI] [PubMed] [Google Scholar]
- 15.Burgess NK, Dao TP, Stanley AM, Fleming KG. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J Biol Chem. 2008;283(39):26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol. 2002;45(5):1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 17.Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci USA. 2004;101(25):9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sampson BA, Misra R, Benson SA. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122(3):491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature. 2014;511(7507):108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- 20.Dong H, et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature. 2014;511(7507):52–56. doi: 10.1038/nature13464. [DOI] [PubMed] [Google Scholar]
- 21.Freinkman E, Chng S-S, Kahne D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc Natl Acad Sci USA. 2011;108(6):2486–2491. doi: 10.1073/pnas.1015617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chng S-S, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci USA. 2010;107(12):5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz N, Chng S-S, Hiniker A, Kahne D, Silhavy TJ. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc Natl Acad Sci USA. 2010;107(27):12245–12250. doi: 10.1073/pnas.1007319107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chng S-S, et al. Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science. 2012;337(6102):1665–1668. doi: 10.1126/science.1227215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J Bacteriol. 2007;189(2):446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen C, Heutink M, Tommassen J, de Cock H. The assembly pathway of outer membrane protein PhoE of Escherichia coli. Eur J Biochem. 2000;267(12):3792–3800. doi: 10.1046/j.1432-1327.2000.01417.x. [DOI] [PubMed] [Google Scholar]
- 27.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: A genetic strategy to probe organelle assembly. Cell. 2005;121(2):307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 28.Wu T, et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103(31):11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chimalakonda G, et al. Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2011;108(6):2492–2497. doi: 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK. Lateral opening and exit pore formation are required for BamA function. Structure. 2014;22(7):1055–1062. doi: 10.1016/j.str.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noinaj N, et al. Structural insight into the biogenesis of β-barrel membrane proteins. Nature. 2013;501(7467):385–390. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S, et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317(5840):961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 33.Hagan CL, Silhavy TJ, Kahne D. β-Barrel membrane protein assembly by the Bam complex. Annu Rev Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 34.Hagan CL, Westwood DB, Kahne D. Bam lipoproteins assemble BamA in vitro. Biochemistry. 2013;52(35):6108–6113. doi: 10.1021/bi400865z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagan CL, Wzorek JS, Kahne D. Inhibition of the β-barrel assembly machine by a peptide that binds BamD. Proc Natl Acad Sci USA. 2015;112(7):2011–2016. doi: 10.1073/pnas.1415955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2002;99(17):11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fairman JW, Noinaj N, Buchanan SK. The structural biology of β-barrel membrane proteins: A summary of recent reports. Curr Opin Struct Biol. 2011;21(4):523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konovalova A, Perlman DH, Cowles CE, Silhavy TJ. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proc Natl Acad Sci USA. 2014;111(41):E4350–E4358. doi: 10.1073/pnas.1417138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho S-H, et al. Detecting envelope stress by monitoring β-barrel assembly. Cell. 2014;159(7):1652–1664. doi: 10.1016/j.cell.2014.11.045. [DOI] [PubMed] [Google Scholar]
- 40.Kleinschmidt JH, Bulieris PV, Qu J, Dogterom M, den Blaauwen T. Association of neighboring β-strands of outer membrane protein A in lipid bilayers revealed by site-directed fluorescence quenching. J Mol Biol. 2011;407(2):316–332. doi: 10.1016/j.jmb.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 41.Kleinschmidt JH. Folding kinetics of the outer membrane proteins OmpA and FomA into phospholipid bilayers. Chem Phys Lipids. 2006;141(1-2):30–47. doi: 10.1016/j.chemphyslip.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Huysmans GHM, Baldwin SA, Brockwell DJ, Radford SE. The transition state for folding of an outer membrane protein. Proc Natl Acad Sci USA. 2010;107(9):4099–4104. doi: 10.1073/pnas.0911904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleinschmidt JH. Folding of β-barrel membrane proteins in lipid bilayers—Unassisted and assisted folding and insertion. Biochim Biophys Acta. 2015;1848(9):1927–1943. doi: 10.1016/j.bbamem.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Thanabalu T, Koronakis E, Hughes C, Koronakis V. Substrate-induced assembly of a contiguous channel for protein export from E. coli: Reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 1998;17(22):6487–6496. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silhavy TJ, Berman ML, Enquist LW. Experiments with Gene Fusions. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1984. [Google Scholar]
- 46.Narita S, Masui C, Suzuki T, Dohmae N, Akiyama Y. Protease homolog BepA (YfgC) promotes assembly and degradation of β-barrel membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2013;110(38):E3612–E3621. doi: 10.1073/pnas.1312012110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casadaban MJ. Regulation of the regulatory gene for the arabinose pathway, araC. J Mol Biol. 1976;104(3):557–566. doi: 10.1016/0022-2836(76)90120-0. [DOI] [PubMed] [Google Scholar]
- 48.Kovach ME, Phillips RW, Elzer PH, Roop RM, 2nd, Peterson KM. pBBR1MCS: A broad-host-range cloning vector. Biotechniques. 1994;16(5):800–802. [PubMed] [Google Scholar]
- 49.Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat Methods. 2006;3(4):263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]