Significance
Disorders affecting neuromuscular tissues are among the most devastating human diseases, characterized by progressive loss of voluntary movement, muscle wasting, and premature death. Neurodegenerative disorders are genetically heterogeneous, with most disease-causing mutations still unidentified. Here we show that mice lacking zinc finger protein 106 (ZFP106) develop muscle and motor nerve atrophy and degeneration and die prematurely. We found that ZFP106 is a skeletal muscle-enriched RNA-binding protein that directly interacts with the core splicing factor RNA binding motif protein 39 in nuclear speckles. Analysis of Zfp106 knockout tissues revealed gene expression and alternative splicing changes consistent with disease-causing abnormalities found in human neurodegenerative diseases. These findings identify ZFP106 as a candidate gene for autosomal-recessive neurodegenerative disorders in humans.
Keywords: ZNF106, amyotrophic lateral sclerosis (ALS), neuromuscular junction (NMJ), motor neuron disease (MND), pre-mRNA splicing
Abstract
Innervation of skeletal muscle by motor neurons occurs through the neuromuscular junction, a cholinergic synapse essential for normal muscle growth and function. Defects in nerve–muscle signaling cause a variety of neuromuscular disorders with features of ataxia, paralysis, skeletal muscle wasting, and degeneration. Here we show that the nuclear zinc finger protein ZFP106 is highly enriched in skeletal muscle and is required for postnatal maintenance of myofiber innervation by motor neurons. Genetic disruption of Zfp106 in mice results in progressive ataxia and hindlimb paralysis associated with motor neuron degeneration, severe muscle wasting, and premature death by 6 mo of age. We show that ZFP106 is an RNA-binding protein that associates with the core splicing factor RNA binding motif protein 39 (RBM39) and localizes to nuclear speckles adjacent to spliceosomes. Upon inhibition of pre-mRNA synthesis, ZFP106 translocates with other splicing factors to the nucleolus. Muscle and spinal cord of Zfp106 knockout mice displayed a gene expression signature of neuromuscular degeneration. Strikingly, altered splicing of the Nogo (Rtn4) gene locus in skeletal muscle of Zfp106 knockout mice resulted in ectopic expression of NOGO-A, the neurite outgrowth factor that inhibits nerve regeneration and destabilizes neuromuscular junctions. These findings reveal a central role for Zfp106 in the maintenance of nerve–muscle signaling, and highlight the involvement of aberrant RNA processing in neuromuscular disease pathogenesis.
Signaling between the presynaptic terminus of a motor neuron and the postsynaptic membrane of a skeletal myofiber occurs at the neuromuscular junction (NMJ), the site of communication that allows for neural control of myofiber identity, growth, and contractility (1). Motor neuron damage or perturbation of nerve–muscle signaling through disruption of the NMJ causes a variety of neurodegenerative disorders, including amyotrophic lateral sclerosis (ALS), Charcot-Marie-Tooth disease (CMT), spinal muscular atrophy (SMA), and hereditary spastic paraplegia (HSP) (2–6). These neurodegenerative diseases are genetically heterogeneous, with mutations in proteins associated with diverse cellular and molecular functions (7–10). Although the precise disease mechanisms underlying the pathogenesis of these disorders remain unclear, abnormalities in apoptotic signaling, oxidative stress, protein folding, and RNA processing in motor neurons and skeletal muscle have been implicated in these diseases (11–15).
In a bioinformatics screen for nuclear proteins with enriched expression in skeletal muscle, we identified ZFP106, a zinc finger protein first characterized as an immunodominant cytotoxic determinant of the mouse H3 minor histocompatibility complex (16, 17). Zinc fingers (ZnFs) mediate DNA, RNA, and protein interactions and are commonly found in regulatory proteins that govern gene transcription, translation, RNA trafficking, and splicing (18). ZFP106 is annotated as a multizinc finger and WD40 repeat-containing protein that localizes to the nucleus. A number of cellular functions have been proposed for ZFP106, such as signaling from the insulin receptor, regulation of rRNA transcription, nucleosome assembly through interaction with the testis-specific nucleolar protein TSG118 (Knop1) and the Y-encoded-like protein (TSPYL), and testis development (17, 19, 20). The necessity of ZFP106 for motor and sensory neuronal maintenance and survival has recently been described (21); however, the molecular role of ZFP106 required for maintaining normal nerve–muscle signaling remains unclear.
Here we show that mice lacking ZFP106 develop postnatal motor neuron atrophy and loss accompanied by progressive hindlimb ataxia, paralysis, and severe muscle wasting and degeneration. We found that ZFP106 shares homology with a subunit of the U2 small nuclear ribonucleoprotein (snRNP) complex and directly interacts with the U2 snRNP auxiliary factor-related protein RBM39 (RNA binding motif protein 39), suggesting that ZFP106 functions in pre-mRNA processing. Consistent with this function, biochemical and cell-based assays showed that ZFP106 is an RNA-binding protein that localizes to nuclear speckles in close proximity to the spliceosome. Differential gene expression and alternative splicing defects in skeletal muscle and spinal cord of Zfp106 knockout mice were consistent with disease-causing defects found in human and mouse models of neurodegenerative disorders. Together, these findings identify a previously unrecognized role for ZFP106 as a component of the RNA processing machinery whose function is required for the maintenance of nerve–muscle signaling and health.
Results
Skeletal Muscle-Enriched Expression of Zfp106.
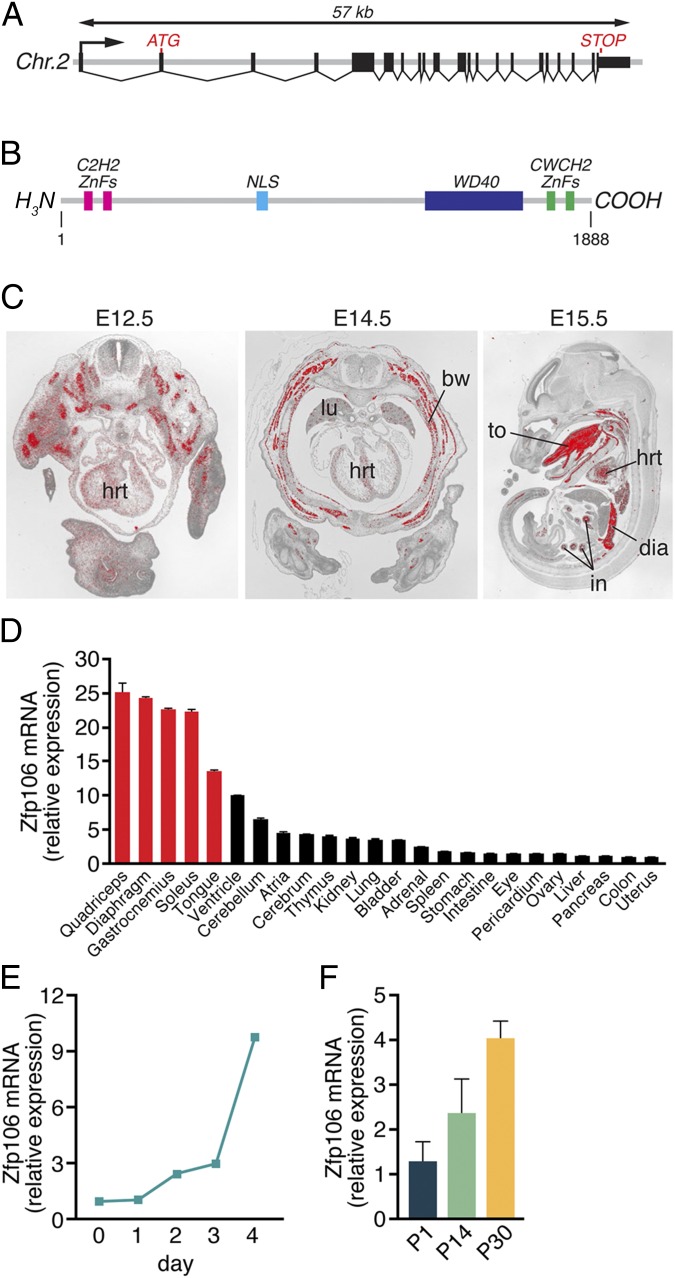
In a bioinformatics screen for uncharacterized nuclear proteins enriched in skeletal muscle, we identified ZFP106, a multizinc finger and WD40-containing protein of unknown function. The mouse Zfp106 gene spans ∼57 kb on chromosome 2 and contains 21 exons (Fig. 1A). The Zfp106 transcript encodes a protein of 1,888 amino acids that contains two C2H2 class ZnFs at its N terminus, two CWCH2 ZnFs at its C terminus, and a C-terminal WD40 domain composed of six WD repeats (Fig. 1B). Zfp106 orthologs are found in diverse vertebrate species, including mammals, birds, reptiles, amphibians, and fish (Fig. S1A) (22).
Fig. 1.
Zfp106 gene structure and expression. (A) The mouse Zfp106 gene is located on chromosome 2 and is composed of 21 exons, spanning ∼57 kb. (B) The mouse ZFP106 protein is 1,888 amino acids long and contains a bipartite nuclear localization signal (NLS), four zinc fingers, and a WD40 domain. (C) Section in situ hybridization using a probe antisense to Zfp106 showed strong expression in skeletal muscle, heart, intestine, and lungs during embryogenesis. bw, body wall muscle; dia, diaphragm; hrt, heart; in, intestine; lu, lung; to, tongue. (D) The expression of Zfp106 in adult mouse tissues was measured using qRT-PCR. Zfp106 transcript expression was highest in fast- and slow-type skeletal muscles (red bars), heart, and brain. Detectable levels of Zfp106 transcripts were broadly expressed in the adult tissues examined. (E and F) Zfp106 expression, quantified using qRT-PCR, increased during C2C12 muscle cell differentiation in vitro (E) and in gastrocnemius–plantaris muscles isolated from P1, P14, and P30 mice (F). Mean ± SD.
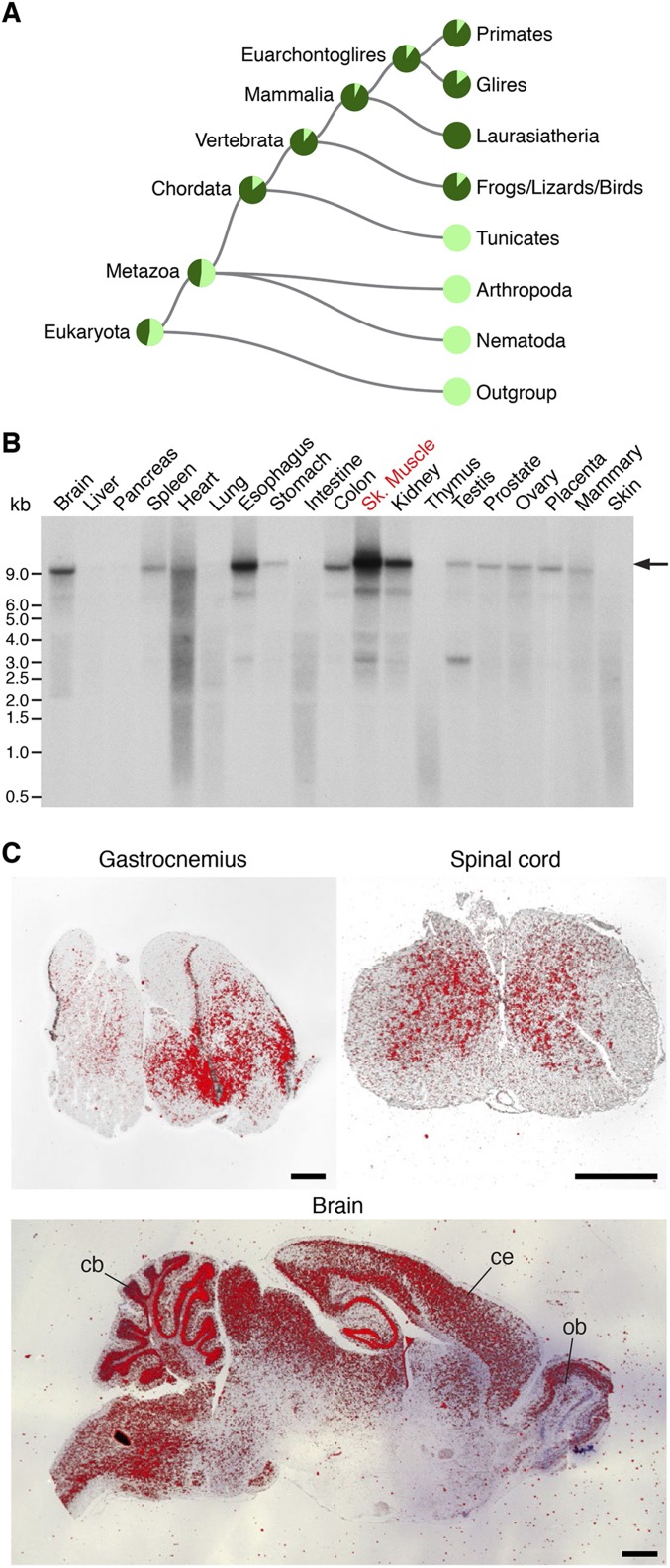
Fig. S1.
Conservation and expression of Zfp106. (A) Phylogram generated by Treefam (22) showing that Zfp106 orthologs are found in diverse vertebrate species, including mammals, birds, reptiles, amphibians, and fish. (B) Northern blot analysis of adult mouse tissues using a probe specific to Zfp106 showed enrichment in skeletal muscle, esophagus, kidney, colon, and brain. The arrow marks the predicted transcript size of the full-length Zfp106 transcript encoding the 1,888-aa ZFP106 protein. (C) Section in situ hybridization on adult mouse tissues using an antisense probe to Zfp106 showing expression in gastrocnemius, spinal cord, and brain. cb, cerebellum; ce, cerebral cortex; ob, olfactory bulb. (Scale bars, 1 mm.)
In situ hybridization using an antisense radiolabeled probe specific to Zfp106 revealed strong enrichment in skeletal muscle and cardiac tissues in the developing mouse between embryonic day (E)12.5 and E15.5 (Fig. 1C). In adulthood, Zfp106 is broadly expressed in multiple tissues but remains highest in skeletal muscle, heart, and brain, as detected by quantitative real-time PCR (qRT-PCR), in situ hybridization, and Northern blot (Fig. 1D and Fig. S1 B and C). Zfp106 is expressed in both fast- and slow-type skeletal muscles (quadriceps and soleus, respectively) and is significantly up-regulated during C2C12 myoblast differentiation in vitro and postnatal skeletal muscle maturation in vivo (Fig. 1 E and F).
Neurological Dysfunction and Premature Lethality of Zfp106 KO Mice.
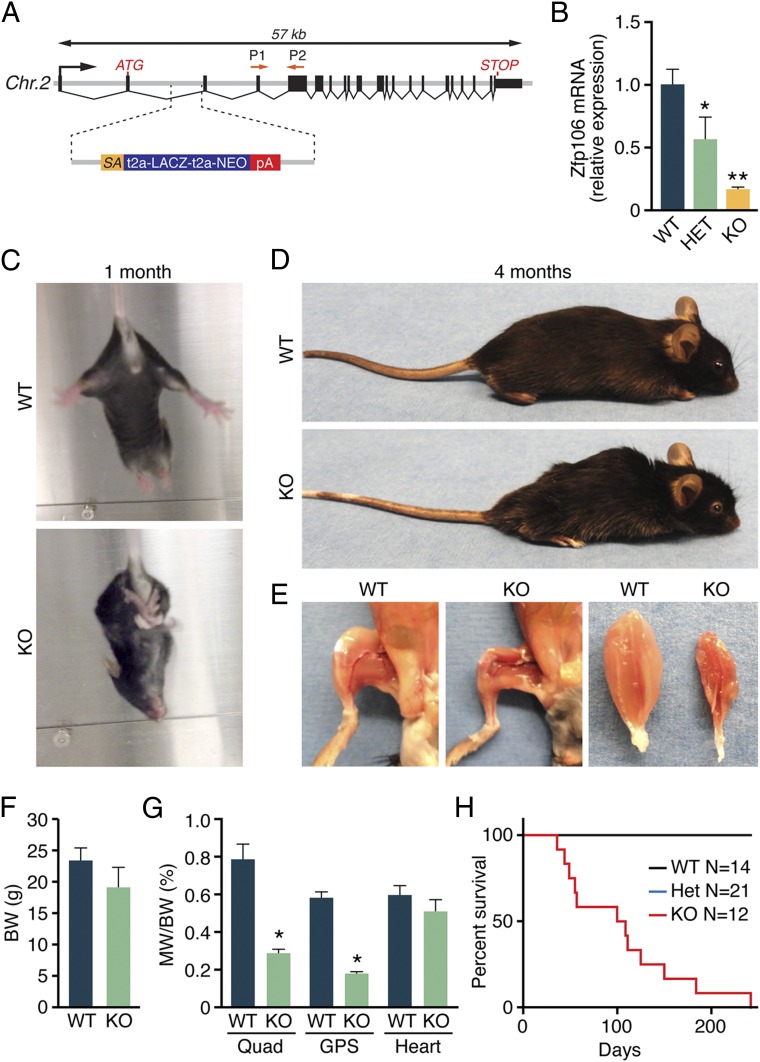
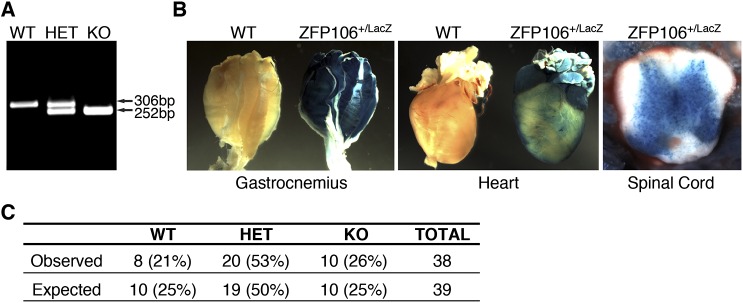
To investigate the function of Zfp106 in vivo, we used in vitro fertilization to rederive Zfp106 knockout (KO) mice using sperm obtained from the University of California, Davis Knockout Mouse Project (KOMP) Repository. The targeted Zfp106 KO allele contains a β-galactosidase (LacZ) reporter and polyadenylation cassette inserted downstream of the first coding exon of the Zfp106 locus, which we genotyped using a PCR-based strategy (Fig. 2A and Fig. S2A). In mice heterozygous (HET) for the knock-in allele, LacZ expression was detected in skeletal muscle, heart, and spinal cord, consistent with the endogenous expression pattern of Zfp106 in adult mice (Fig. S2B).
Fig. 2.
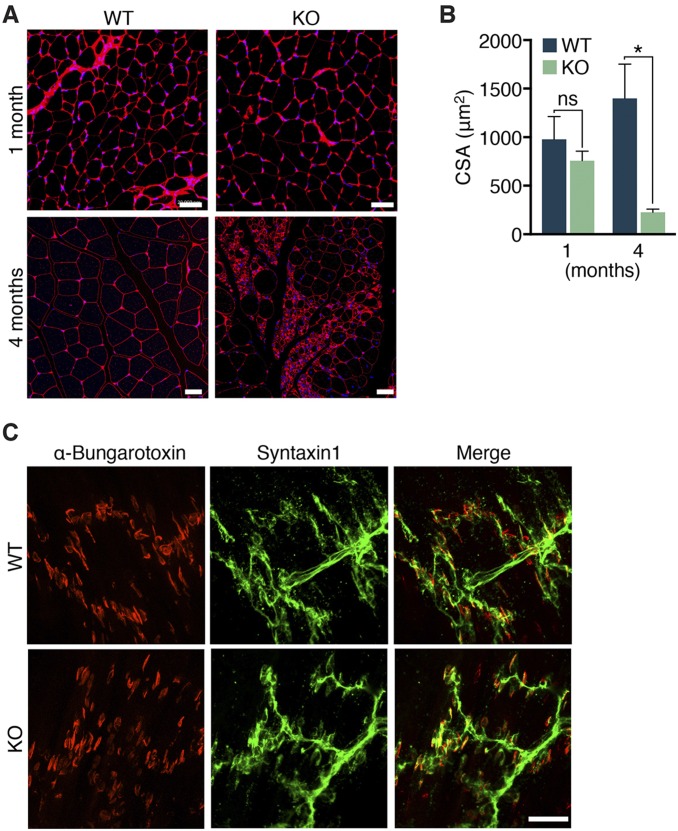
Postnatal muscle wasting in Zfp106 KO mice. (A) Diagram showing the Zfp106 LacZ knock-in allele obtained from the KOMP Repository. P1 and P2 indicate the locations of Zfp106 qRT-PCR primers. (B) Zfp106 transcript levels were reduced by 85% in mice homozygous for the LacZ allele (KO), measured by qRT-PCR in gastrocnemius–plantaris muscles. (C) At ∼1 mo of age, Zfp106 KO mice display abnormal hindlimb clasping. (D) At 4 mo of age, Zfp106 KO mice are approximately the same size as their WT littermates but show kyphotic curvature of the spine. (E) Gross examination of hindlimb skeletal muscles of 4-mo-old Zfp106 KO mice showed severe muscle wasting. (F and G) At 4 mo of age, no differences were detected in body weight or heart weight of Zfp106 KO and WT mice. Skeletal muscle weight was significantly reduced in Zfp106 KO mice. BW, body weight; GPS, gastrocnemius–plantaris–soleus muscle group; MW, muscle weight; Quad, quadriceps muscle. (H) Kaplan–Meier survival plot showing premature lethality of Zfp106 KO mice. *P < 0.01; **P < 0.0001; Mean ± SD.
Fig. S2.
Characterization of Zfp106 KO mice. (A) Duplex PCR-based genotyping strategy used to identify wild-type (WT), heterozygous (HET), and knockout (KO) mice for the Zfp106 LacZ knock-in allele. (B) Staining of adult tissues heterozygous for the Zfp106 LacZ knock-in allele showed β-galactosidase staining in skeletal muscles, heart, and spinal cord. (C) Number of mice born versus expected from heterozygous intercrosses of Zfp106 LacZ knock-in mice.
Mice heterozygous for the LacZ knock-in allele appeared grossly normal and were fertile and indistinguishable from WT littermates. At birth, Zfp106 homozygous KO mice generated from heterozygous intercrosses also showed no overt signs of pathology and were born in normal Mendelian ratios, suggesting that Zfp106 is not essential for normal embryonic development (Fig. S2C). Zfp106 transcripts, quantified by qRT-PCR using primers downstream of the LacZ knock-in allele, were reduced by 85% in skeletal muscles isolated from Zfp106 KO mice (Fig. 2B).
By 1 mo of age, Zfp106 KO mice displayed evidence of neurological dysfunction, characterized by hindlimb clasping when suspended by the tail (Fig. 2C). This phenotype became progressively pronounced and, by 4 mo of age, Zfp106 KO animals displayed severe ataxia, partial hindlimb paralysis, and kyphosis (Fig. 2D). Although total body weight was not significantly decreased in Zfp106 KO mice, gross dissection revealed visible signs of muscle wasting and significant reduction in hindlimb skeletal muscle weight compared with WT littermates (Fig. 2 E–G). No significant differences in heart weight were observed in Zfp106 KO animals in comparison with WT littermates (Fig. 2G). Zfp106 KO mice died prematurely, and only a few survived beyond 6 mo of age (Fig. 2H).
Skeletal Muscle Atrophy and Denervation of Zfp106 KO Mice.
Histological analyses of skeletal muscles isolated from 1-mo-old Zfp106 KO mice revealed no significant difference in myofiber cross-sectional area or level of endomysial fibrosis compared with WT littermates (Fig. 3A and Fig. S3 A and B). In contrast, by 4 mo of age, hindlimb skeletal muscles of Zfp106 KO mice showed significant myofiber atrophy, measured by an 80% decrease in average myofiber cross-sectional area (Fig. S3 A and B). In addition to muscle wasting, the occurrence of increased fibrosis, as well as pseudohypertrophied and regenerating myofibers marked by centralized nuclei, was widespread in Zfp106 KO muscle (Fig. 3A). Further examination using transmission electron microscopy revealed highly disorganized sarcomere structures in Zfp106 KO muscles isolated from 4-mo-old animals (Fig. 3A).
Fig. 3.
Severe wasting and denervation in Zfp106 KO muscle. (A) Transverse sections of gastrocnemius muscles from 1- and 4-mo-old WT and Zfp106 KO mice were stained with Masson’s trichrome. Electron micrographs (EM) of WT and KO gastrocnemius revealed disorganized sarcomere structures in 4-mo-old muscles from Zfp106 KO muscle. (Scale bars, 1 μm.) (B) Gastrocnemius muscle from 1- and 4-mo-old WT and Zfp106 KO mice were stained with anti-Syntaxin 1 (green) and Texas red-conjugated α-bungarotoxin (red) to assess neuromuscular junctions. White arrowheads indicate end plates that lack nerve terminal juxtaposition in Zfp106 KO gastrocnemius muscles. (Scale bars, for 1-mo WT and KO mice, 20 μm; for 4-mo WT and KO mice, 50 μm.)
Fig. S3.
Progressive skeletal muscle atrophy in Zfp106 KO mice. (A and B) Wheat germ agglutinin (red) and DAPI (blue) staining of transverse myofibers of 1- and 4-mo-old Zfp106 KO mice showed an 80% decrease in myofiber cross-sectional area (CSA) in Zfp106 KO gastrocnemius muscle. *P < 0.01. ns, not significant. Mean ± SD. (Scale bars, 20 μm.) (C) Motor axons and nerve terminals were stained using an antibody against Syntaxin 1 (green) and Texas red-conjugated α-bungarotoxin to label the motor end plates at the neuromuscular junctions (NMJs). At postnatal day 1 (P1), no differences in NMJ structure, number, or innervation by motor nerves were detected in gastrocnemius muscles of Zfp106 KO compared with WT littermates. (Scale bar, 50 μm.)
The concurrence of ataxia and muscle wasting in Zfp106 KO mice suggested a neurological contribution to the phenotype. Therefore, we characterized NMJ structure and skeletal muscle innervation by peripheral motor neurons using α-bungarotoxin to identify postsynaptic acetylcholine receptors (AChRs) at the motor end plates and an antibody against Syntaxin 1 to label presynaptic nerves and nerve terminals. At postnatal day 1 (P1), we did not detect any overt differences in NMJ structure, number, or innervation in gastrocnemius muscles of Zfp106 KO mice compared with WT littermates (Fig. S3C). Gastrocnemius muscles isolated from 1- and 4-mo-old KO animals retained normal NMJ structure and number; however, they exhibited progressive denervation, characterized by end plates that lacked nerve terminal juxtaposition (Fig. 3B). Additionally, the morphology of motor nerves in Zfp106 KO mice showed increased branching and defasciculation of the nerve bundles. These data suggest that a loss of nerve–muscle signaling due to myofiber denervation contributes to the observed ataxia and muscle wasting present in Zfp106 KO mice.
Motor Neuron Atrophy and Degeneration in Zfp106 KO Mice.
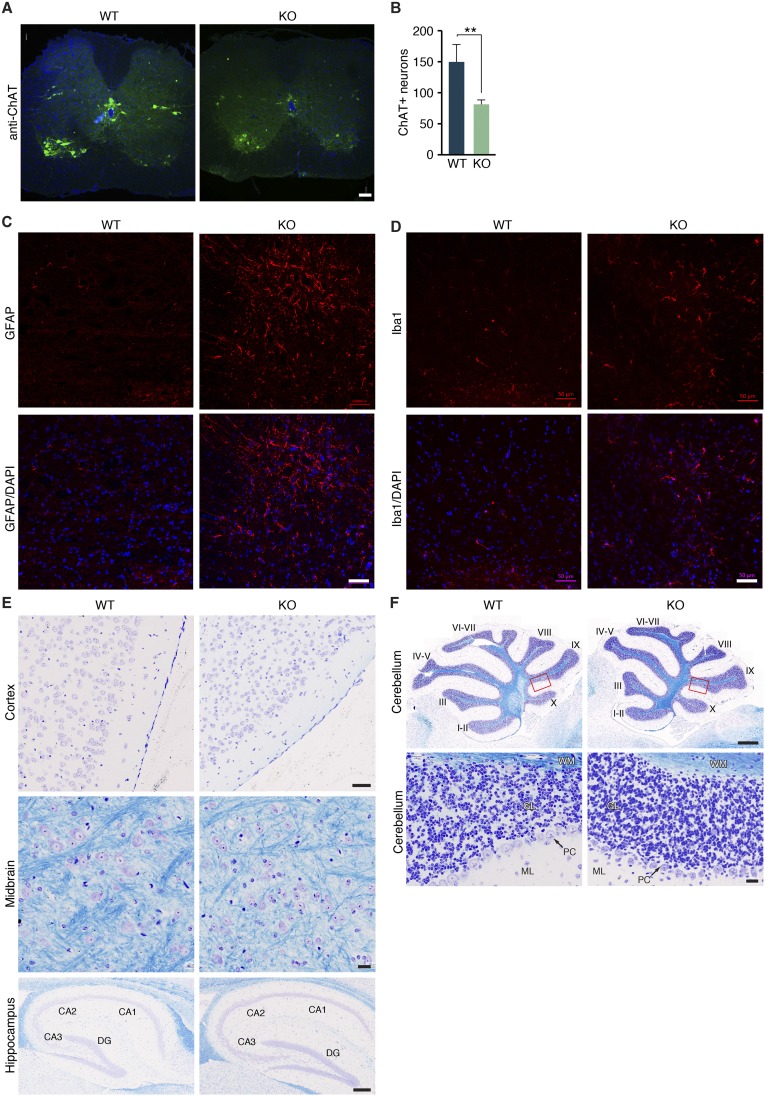
Due to the nerve fasciculation and innervation defects observed in Zfp106 KO mice, we further examined motor neuron morphology in sciatic nerves by light and electron microscopy. At 2 wk of age, histological analysis of transverse sections of sciatic nerves stained with toluidine blue showed a slight reduction in overall bundle diameter; however, myelination and motor nerve axon diameters appeared grossly normal (Fig. 4 A–C). By 4 mo of age, degenerating motor neurons were more prevalent and axon diameters were greatly reduced in sciatic nerves of Zfp106 KO animals compared with WT littermate controls (Fig. 4 A–C). Consistent with axonal degeneration, we observed a decrease in the number of motor neuron cell bodies in the ventral horns of the spinal cord of Zfp106 KO animals, measured by a 43% decrease in the number of choline acetyltransferase-positive (ChAT+) neurons in lumbar spinal segments (L3–L5) (Fig. S4 A and B). Concurrent with the decrease in lumbar neurons, we observed a significant increase in the number of microglia and active astrocytes present in the lumbar spinal cord of Zfp106 KO animals, indicating the presence of microgliosis and astrogliosis, respectively, found in other models of motor neuron degeneration (Fig. S4 C and D) (23, 24). Interestingly, we did not detect any overt differences in the number, morphology, or myelination of neurons in the motor cortex or cerebellum at 3 mo of age, suggesting that the observed pathology is restricted to peripheral motor neurons at this time point (Fig. S4 E and F).
Fig. 4.
Motor axon atrophy and degeneration in Zfp106 KO mice. (A) Light microscopy of toluidine blue-stained transverse sections of sciatic nerves isolated from WT and Zfp106 KO mice at 2 wk and 4 mo of age. (Scale bar, 100 µm.) (B) Axon morphology in sciatic nerves shown in A. The yellow arrowhead points to a degenerating axon in a Zfp106 KO sciatic nerve section. (Scale bar, 20 µm.) (C) Electron microscopy analysis showed degenerating motor axons present in the sciatic nerve of Zfp106 KO mice. Yellow arrowheads point to degenerating motor axons. SC, Schwann cell nucleus. (Scale bar, 2 µm.)
Fig. S4.
Peripheral neurodegeneration in Zfp106 KO mice. (A and B) Transverse sections through the lumbar spinal cord of WT and Zfp106 KO mice stained with an antibody against choline acetyltransferase (ChAT) showed a 43% decrease in the number of ChAT+ motor neurons at 2 mo of age. **P < 0.01. Mean ± SD. (Scale bar, 100 μm.) (C and D) Immunohistochemistry using antibodies specific to GFAP and Iba1 in the lumbar spinal cord revealed an increase in the number of active astrocytes and microglia cells in Zfp106 KO mice compared with WT littermates, respectively. (Scale bars, 50 μm.) (E and F) Nissl/Luxol fast blue staining of brain tissues revealed no overt differences in the morphology, myelination, or number of neurons in 3-mo-old Zfp106 KO mice compared with WT littermates. Red box shows the magnified area below. [Scale bars, 50 μm (cortex), 20 μm (midbrain), 200 μm (hippocampus), 500 μm (cerebellum; Top), and 20 μm (cerebellum; Bottom).] DG, dentate gyrus; GL, granular layer; ML, molecular layer; PC, Purkinje cells; WM, white matter.
ZFP106 Is an RNA-Binding Protein That Interacts with the Splicing Factor RBM39.
ZFP106 contains well-conserved ZnFs and WD40 repeats, suggesting that these domains play an important role in mediating the function of ZFP106 through binding to nucleic acids or proteins. A search of the Conserved Domain Database identified sequence homology between an N-terminal region of ZFP106 containing a C2H2 ZnF (amino acids 44–111) and a homolog of splicing factor 3A2 (SF3A2/PRP11) (amino acids 55–122) (Fig. S5A) (25). SF3A2 is a zinc finger protein that forms a subunit of the U2 snRNP complex that binds directly to pre-mRNAs and is required for splicing, suggesting that ZFP106 may play a similar role in pre-mRNA processing. Consistent with this function, ZFP106 was recently identified by mass spectrometry in a screen for polyA mRNA-binding proteins in mammalian cells (26).
Fig. S5.
Sequence homology between ZFP106 and RNA-binding ZnF proteins. (A) A search of the Conserved Domain Database using the amino acid sequence of ZFP106 identified homology to the zinc finger splicing factor PRP11, a homolog of mammalian SF3A2. Protein alignment shows that the region encompasses an N-terminal ZnF of ZFP106 and ZnF of PRP11. (B) The N-terminal ZnFs of ZFP106 share significant homology with the ZnFs of zinc finger RNA-binding protein (ZFR). (C) Alignment of the ZnFs of ZFP106 and other RNA-binding ZnF proteins that contain the conserved C2H2 ZnF motif C-X2-C-X12-H-X5-H. Cysteine and histidine residues comprising the C-X2-C-X12-H-X5-H motif are highlighted in red. “*” denotes identical, “:” denotes strongly similar and “.” denotes weakly similar residues.
A bioinformatics search using BLAST further revealed that the N-terminal ZnFs of ZFP106 share significant homology with the ZnFs of zinc finger RNA-binding protein (ZFR), a component of the spliceosome that associates with pre-mRNA processing complexes (Fig. S5B) (27). Interestingly, ZFP106, SF3A2, and ZFR all contain a specific C2H2 ZnF motif (C-X2-C-X12-H-X5-H) that was previously identified to be conserved in Matrin 3 (MATR3) and several other RNA-binding proteins, which differs from the typical DNA-binding C2H2 ZnF motif (Fig. S5C) (28, 29).
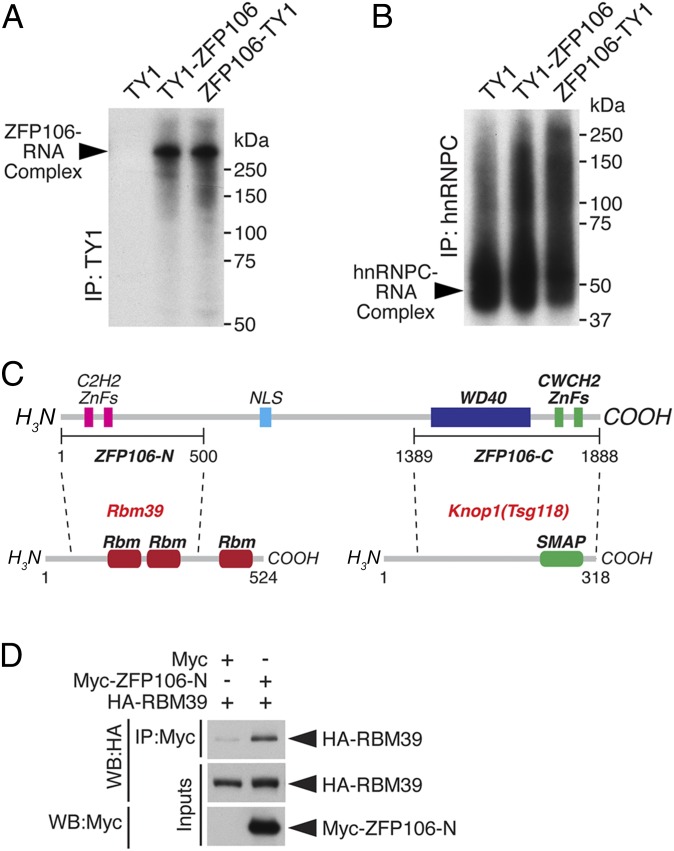
To determine whether ZFP106 is capable of binding directly to RNA, we performed an iCLIP assay using full-length ZFP106 as a fusion to either an N- or C-terminal TY1-epitope tag in the mouse cell line Neuro2a (Fig. 5A) (30). To detect ZFP106–RNA complexes, cells transfected with ZFP106 fusions were cross-linked with UV light and immunoprecipitated using an anti-TY1 antibody. Radiolabeling of RNA and SDS/PAGE resolved a strong band corresponding to ZFP106–RNA complexes at the molecular mass of ZFP106 but not from cells transfected with a vector encoding the TY1 epitope alone (Fig. 5A). As a control for RNA integrity, protein–RNA complexes from each sample were immunoprecipitated using a primary antibody specific for the heterogeneous nuclear ribonucleoprotein C (hnRNPC), which bound similar levels of RNA from each sample (Fig. 5B). These data show that ZFP106 is an RNA-binding protein that shares homology with RNA-binding zinc finger proteins involved in pre-mRNA processing.
Fig. 5.
ZFP106 is an RNA-binding protein that interacts with RBM39. (A) Autoradiograph resolving the ZFP106–RNA complex after cross-linking and immunoprecipitation (IP) of TY1 epitope-tagged ZFP106 in Neuro2a cells. (B) Immunoprecipitation using a primary antibody against hnRNPC was used as a control for RNA integrity. (C) Diagram showing the ZFP106 fragments used for a yeast two-hybrid screen of a mouse normalized cDNA library. The 500-aa N-terminal fragment (ZFP106-N) and the 500-aa C-terminal fragment (ZFP106-C) identified RBM39 and KNOP1 as ZFP106-interacting proteins, respectively. SMAP, small acidic protein. (D) Western blot (WB) analysis showing that HA epitope-tagged RBM39 coprecipitated with a Myc epitope-tagged fragment of the N-terminal ZFP106 fragment (Myc–ZFP106-N) in COS7 cells.
To identify proteins that directly interact with ZFP106, we performed a yeast two-hybrid screen of a mouse normalized cDNA library using the N- or C-terminal 500 amino acids of ZFP106. The N-terminal fragment contains the two C2H2 ZnFs, and the C-terminal fragment contains both the WD40 domain and the two CWCH2 ZnFs (Zfp106-N and -C, respectively) (Fig. 5C). Using the N-terminal fragment of ZFP106, we identified the U2 snRNP auxiliary factor-related protein RBM39 as a direct ZFP106-interacting protein. Mutations in RBM39 are associated with skeletal muscle myopathies and RBM39 is predicted to be a candidate ALS disease-causing gene (31, 32). Coimmunoprecipitation experiments using the Myc epitope-tagged N-terminal fragment of ZFP106 and an HA epitope-tagged RBM39 fusion protein confirmed the ZFP106–RBM39 interaction in mammalian cells (Fig. 5D). Consistent with previous reports, we also identified the uncharacterized nucleolar protein TSG118 (KNOP1) as a binding partner using the C-terminal fragment of ZFP106 (ZFP106-C) (Fig. 5C) (19, 20). Thus, the N-terminal region of ZFP106 is both homologous to and interacts with subunits of the U2 snRNP pre-mRNA splicing complex.
Subcellular Localization and Translocation of ZFP106 in Mammalian Cells.
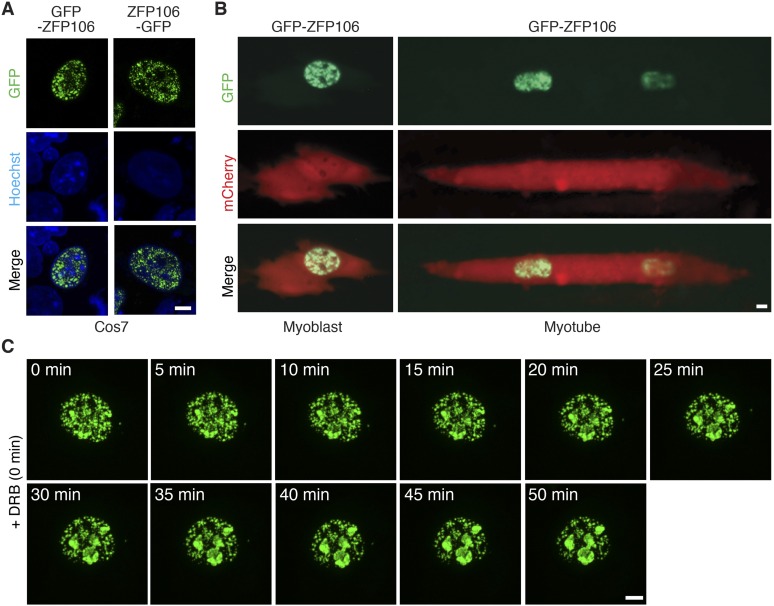
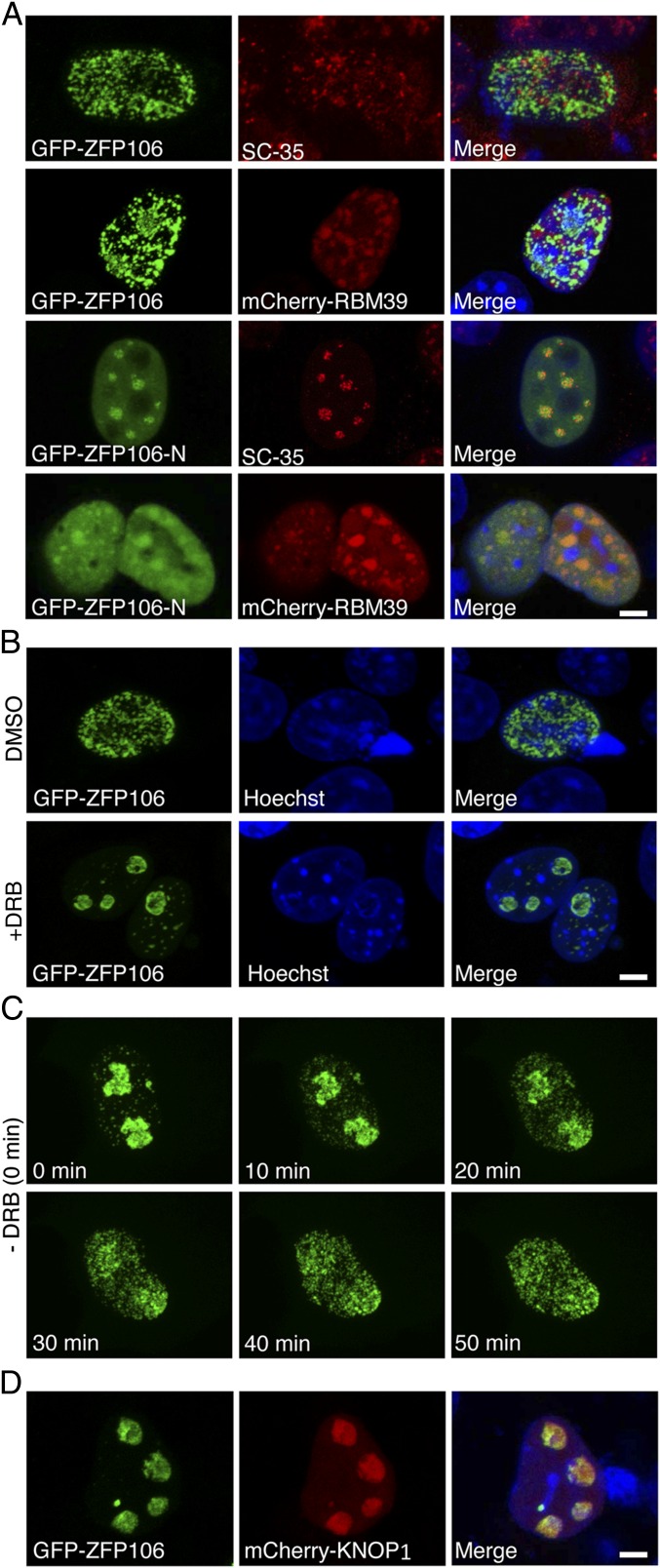
To further investigate the molecular function of ZFP106, we expressed the full-length (1,888-aa) protein as an N- or C-terminal fusion to GFP in COS7 or C2C12 muscle cells (Fig. S6 A and B). Both GFP-ZFP106 and ZFP106-GFP fusion proteins localized to nuclear speckles, a heterogeneous subnuclear domain that functions as a reservoir for small nuclear RNAs (snRNAs) and proteins involved in pre-mRNA splicing (33, 34). The ZFP106 speckle domain was in close proximity to splicing speckles labeled with a primary antibody specific to the splicing factor SC-35, but did not show appreciable colocalization (Fig. 6A). ZFP106 showed a greater degree of overlap with an mCherry fusion to RBM39 (Fig. 6A). A GFP fusion with the N-terminal fragment of ZFP106 (GFP–ZFP106-N), which we used to identify the RBM39 interaction, localized to the nucleus in a pattern that overlapped with SC-35 splicing speckles and the expression of an mCherry-RBM39 fusion protein (Fig. 6A). These findings support an interaction between ZFP106 and components of the splicing machinery, but suggest that additional molecular interactions contribute to the normal subcellular localization of full-length ZFP106.
Fig. S6.
Subcellular localization of ZFP106 in COS7 and C2C12 muscle cells. (A) Confocal microscopy of transiently transfected COS7 cells showed GFP-ZFP106 and ZFP106-GFP localized to nuclear speckles. (B) The nuclear speckle pattern of GFP-ZFP106 was also found in both C2C12 myoblasts and myotubes. Coexpression of an unlocalized mCherry fluorescent protein was used to denote cell shape. (Scale bars, 5 µm.) (C) Inhibition of RNA synthesis by DRB treatment of COS7 cells transfected with GFP-ZFP106. A panel of fluorescence micrographs acquired by live-cell imaging every 5 min over the course of 50 min showed GFP-ZFP106 progressively localized to nucleoli.
Fig. 6.
Nuclear localization and translocation of ZFP106. (A) Confocal microscopy of COS7 cells showing coexpression and localization of GFP-ZFP106 with SC-35 or mCherry-RBM39. The N-terminal fragment of ZFP106 (ZFP106-N) localized to a nuclear speckle pattern that overlapped with SC-35 and an mCherry-RBM39 fusion protein. (B) Inhibition of total cellular RNA synthesis by DRB treatment induced the translocation of GFP-ZFP106 from nuclear speckles to the nucleolus. (C) After removal of DRB, GFP-ZFP106 regained a nuclear speckle pattern over the course of 50 min. (D) Coexpression of GFP-ZFP106 and mCherry-KNOP1 induced the translocation of ZFP106 from nuclear speckles to the nucleolus. (Scale bars, 5 µm.)
Many RNA processing proteins that localize to nuclear speckles translocate to the nucleolus upon drug-inhibition of RNA synthesis (35, 36). To determine whether ZFP106 behaved in a similar fashion, we inhibited RNA polymerase II elongation in COS7 cells by treating with the adenosine analog 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) and visualized the localization of GFP-ZFP106 over time. Strikingly, treatment with DRB induced an almost complete translocation of GFP-ZFP106 from nuclear speckles to the nucleolus within 50 min (Fig. 6B and Fig. S6C). Upon recovery of RNA synthesis by removal of DRB, GFP-ZFP106 reacquired a nuclear speckle pattern within a similar time frame (Fig. 6C). This finding suggests that the localization of ZFP106 to nuclear speckles is dynamic and dependent upon active pre-mRNA transcription.
Additionally, we observed that overexpression of the nucleolar protein KNOP1 (mCherry-KNOP1), which we identified as a direct ZFP106-interacting partner, similarly induced the translocation of ZFP106 to the nucleolus, where the two proteins completely overlapped (Fig. 6D). Thus, the translocation of ZFP106 from nuclear speckles to the nucleolus may be mediated through its affinity for KNOP1 at its C terminus.
Gene Expression and Alternative Splicing Changes in Zfp106 KO Tissues.
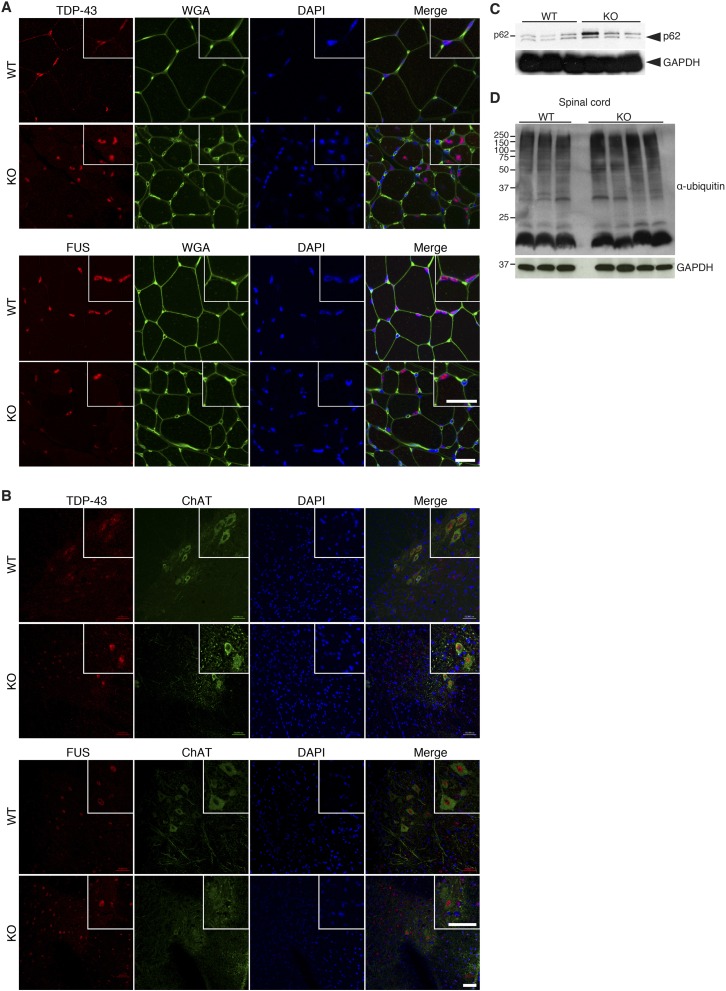
Defects in diverse molecular pathways in motor neurons and neighboring tissues drive neurodegenerative disease phenotypes. To investigate the underlying molecular basis for the observed pathology found in Zfp106 KO mice, we performed microarray analysis and qRT-PCR on mRNA isolated from hindlimb skeletal muscle and lumbar spinal cord at 1 mo of age, before overt disease onset (Fig. 7 A and B and Dataset S1). Gene expression changes in skeletal muscle of Zfp106 KO mice were characteristic of a denervated muscle phenotype, with robust up-regulation of the alpha and embryonic isoforms of acetylcholine receptors (Chrna1 and Chrng, respectively). The most up-regulated gene in Zfp106 KO skeletal muscle was aldehyde dehydrogenase 3 family member A1 (Aldh3a1), an enzyme that protects against oxidative stress by metabolizing cytotoxic aldehydes produced by reactive oxygen species-dependent pathways (37). Interestingly, many of the significant gene expression changes (>1.5-fold) observed in skeletal muscle were also observed in spinal cord, suggesting that a common molecular pathway could underlie the pathology of these different tissues (Fig. 7A). Among the genes dysregulated in both tissues, many are also aberrantly expressed in mouse models of ALS (Lgals1, Lgals3, Gpnmb, and Hspb1) or associated with CMT (Mpz, Pmp22, Hspb1, and Prx) (Fig. 7 A and B) (38, 39). However, in skeletal muscle and spinal cord tissues of Zfp106 KO mice, we did not observe several common pathological features associated with motor neuron diseases, including the mislocalization of the DNA/RNA-binding proteins TDP-43 and FUS or increased markers of protein ubiquitination and autophagy (Fig. S7). This suggests that the disease mechanisms underlying the phenotype of Zfp106 KO mice are specific to the tissues and molecular pathways dependent on ZFP106 expression and function, respectively.
Fig. 7.
Gene profiling and alternative splicing changes in Zfp106 KO tissues. (A) Genes up-regulated and down-regulated in both skeletal muscle and spinal cord of 1-mo-old Zfp106 KO mice. (B) Quantitative real-time PCR for a subset of genes dysregulated in Zfp106 KO tissues. *P < 0.05. ND, not detected. (C) Quantitative real-time PCR showing expression of Nogo and Nogo-A transcript levels in WT and Zfp106 KO gastrocnemius–plantaris muscles at 1 mo of age. *P < 0.01. (D) Robust up-regulation of NOGO-A protein was observed in gastrocnemius–plantaris muscle of 1-mo-old Zfp106 KO mice, detected using an antibody specific to the A isoform of Nogo. Western blot analysis for GAPDH was used as a loading control. Mean ± SD.
Fig. S7.
Protein processing and degradation in Zfp106 KO mice. (A and B) Immunohistochemistry using antibodies specific to TDP-43 and FUS revealed normal subcellular localization in skeletal muscle (A) and spinal cord (B) of 3-mo-old Zfp106 KO mice compared with WT littermates. WGA, wheat germ agglutinin. Insets are magnification showing stained cell nuclei. (Scale bars, A, 50 μm; B, 25 μm.) (C) Western blot analysis revealed similar levels of p62 protein in spinal cord from Zfp106 KO and WT littermate mice. (D) Normal levels of polyubiquitinated proteins were detected in Zfp106 KO spinal cord lysates using an antibody against ubiquitin.
Skeletal muscle-specific signals are essential for neuromuscular synapse formation and regeneration (40–43). Defects in these signals contribute to myofiber denervation by promoting the release of presynaptic nerve terminals and inhibiting compensatory regeneration of neuromuscular synapses. Notably, ectopic expression of the “A” isoform of neurite outgrowth inhibitor Nogo (Rtn4) in skeletal muscle can lead to myofiber denervation, and has recently been shown to correlate with disease severity in ALS patients and in mouse models of ALS (44, 45). To determine whether altered levels of NOGO-A might contribute to the Zfp106 disease phenotype, we quantified the relative expression of Nogo transcripts using qRT-PCR primers to an exon common to all Nogo isoforms or specific to Nogo-A. Although the relative abundance of all Nogo transcripts was not significantly changed, we observed robust up-regulation of Nogo-A transcripts and protein in skeletal muscle of Zfp106 KO mice compared with WT littermates (Fig. 7 C and D). Together, these findings identify gene expression and alternative splicing changes in Zfp106 KO mice that likely contribute to a disease pathology characteristic of neurodegenerative disorders such as ALS.
Discussion
Here we identify ZFP106 as a muscle-enriched RNA-binding protein that localizes to nuclear speckles and interacts with the core splicing factor RBM39. Disruption of Zfp106 in mice resulted in a postnatal, progressive neuromuscular disease phenotype characterized by atrophy and degeneration of muscle and peripheral motor neurons. The progressive neuromuscular phenotype observed in Zfp106 KO mice shares many phenotypic abnormalities observed in ALS and CMT patients, including myofiber denervation, kyphosis, ataxia, and premature death. Gene expression and alternative splicing changes present in Zfp106 KO mice are characteristic of those found in human neurodegenerative disorders, suggesting that a common disease mechanism may underlie these similar pathologies.
The recent identification of ZFP106 in a screen for mammalian polyA mRNA-interacting proteins suggests it functions as a regulator of pre-mRNA processing (26). The trafficking, splicing, and stability of eukaryotic pre-mRNAs require a combination of snRNAs and a number of RNA-binding proteins, many of which contain ZnF motifs (46). ZFP106 shares a specific C2H2 ZnF motif that is conserved among other RNA-binding proteins, notably SF3A2, ZFR, and MATR3. Additionally, we found that ZFP106 directly interacts with RBM39, a subunit of the U2 snRNP complex that is required for normal assembly of the mammalian spliceosome by binding to the pre-mRNA branch site within introns. Together, these data suggest that ZFP106 functions as a previously unidentified component of the RNA processing machinery and that aberrant RNA processing underlies the disease phenotype of Zfp106 KO mice.
For most neurodegenerative disorders, it remains a mystery as to how mutations in broadly expressed proteins result in specific muscle and motor neuron pathologies. ZFP106 is most highly expressed in skeletal muscle both during development and in adulthood, but is broadly expressed at low levels in most tissues throughout the body. The severe degeneration of skeletal muscle observed in Zfp106 KO mice exceeds typical denervation abnormalities and suggests an additional primary function for ZFP106 in skeletal muscle, in addition to its role in motor neurons. Primary defects in skeletal muscle play an important role in the induction and progression of neurodegenerative diseases, including ALS (47, 48). The robust up-regulation of NOGO-A in skeletal muscle of Zfp106 KO mice, as well as in human ALS patients, highlights the role of skeletal muscle in the pathogenesis of neuromuscular disease phenotypes. Notably, skeletal muscle-specific overexpression of NOGO-A in WT mice has been demonstrated to be sufficient to induce myofiber denervation, whereas genetic deletion of NOGO-A decreases ALS disease pathogenesis. Future studies that modulate the expression of NOGO-A levels in Zfp106 KO mice will be important for understanding its contribution to disease pathogenesis and whether it is a direct target of ZFP106 activity in skeletal muscle or other tissues.
Because the Zfp106 KO allele that we have characterized disrupts Zfp106 expression in all tissues examined, it remains undetermined whether primary defects originating in skeletal muscle or other tissues give rise to the disease phenotype in Zfp106 KO mice. Conditional disruption of Zfp106 in specific tissues or cell types will be helpful for understanding the etiology of the muscle and motor neuron pathologies that result from loss of function of ZFP106 or other RNA processing components. We further note residual transcript expression in both skeletal muscle and spinal cord tissues of Zfp106 KO mice, which may reflect incomplete transcript termination from the knock-in polyadenylation cassette or alternative promoter use downstream of the knock-in site (49). Alternative approaches to disrupt Zfp106 expression in different tissues could perhaps enhance disease severity or decrease age of onset.
Neurodegenerative disorders are genetically heterogeneous, with mutations identified in many functionally diverse proteins associated with disease pathology. Defects in RNA processing stemming from mutations in DNA/RNA-binding proteins are now appreciated as a major cause of neurodegenerative disease in humans. Notably, causative mutations in the nuclear DNA/RNA-binding proteins FUS and TDP-43 give rise to ∼8% of familial ALS (50–53), and mutations in the RNA-binding proteins SMN1 and SMN2 give rise to spinal muscular atrophy (54). Interestingly, mutations in ZnF proteins related to ZFP106 are associated with human neurodegenerative diseases. Causative mutations in MATRIN 3, a DNA/RNA-binding protein that shares a similar C2H2 ZnF motif with ZFP106 and interacts with TDP-43, give rise to a rare form of familial ALS (ALS21) and distal myopathies (55, 56). Similarly, mutations in the DNA/RNA-binding protein ZFR cause hereditary spastic paraplegia 71 (SPG71), a motor neuron disease that shares many phenotypic characteristics with ALS (57).
For ALS, causative mutations can be inherited in an autosomal-dominant, autosomal-recessive, or X-linked manner (58). However, only 5–10% of cases are inherited, whereas a majority (90–95%) of disease-causing mutations are sporadic and show no clear genetic linkage. It has been suggested that many sporadic cases of ALS may arise through an autosomal-recessive pattern of inheritance, in which a disease-causing mutation has not been identified due to asymptomatic heterozygous individuals. The autosomal-recessive inheritance of the Zfp106 KO phenotype and asymptomatic condition of the heterozygotes make Zfp106 a strong candidate for such cases of inherited human neurodegenerative disorders that lack an identified disease-causing mutation.
Although human disease-causing mutations have not yet been attributed to mutations in Zfp106 (ZNF106 in humans), it is notable that the human ZNF106 locus resides within a region of chromosome 15 that is associated with both a juvenile recessive form of amyotrophic lateral sclerosis (ALS5) and axonal autosomal-recessive Charcot-Marie Tooth disease (CMT2X) (59, 60). Mutations in Spatacsin (SPG11), a gene first associated with spastic paraplegia, have been associated with patients with ALS5 or CMT2X (61). However, for reasons currently unknown, the same mutations identified in SPG11 can manifest with symptoms of ALS or SPG, which suggests that mutations in ZNF106, located ∼2 Mb away from SPG11 on human chromosome 15, may contribute to disease pathology in these patients. Future studies of disease-associated mutations in ZFP106 will undoubtedly provide useful insights into the mechanisms driving human neurodegenerative disorders.
Materials and Methods
Mice and Genotyping.
All experiments involving animals were approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center. The Zfp106 knockout mouse strain was rederived by in vitro fertilization using germ plasm obtained from the KOMP Repository (https://www.komp.org; Project ID CSD26348). Please see Dataset S2 for genotyping primer sequences.
Radioisotopic in Situ Hybridization.
In situ hybridizations were performed as previously described (62). Primer sequences used to clone the Zfp106 cDNA template are listed in Dataset S2.
Northern Blot Analysis.
A multitissue Northern blot of mouse tissues (Zyagen; MN-MT-1) was hybridized with a [32P]dCTP-labeled DNA probe specific to a fragment of Zfp106, generated using a RadPrime Kit (Life Technologies). The Zfp106 cDNA template used for Northern blot analysis was identical to that used for radioisotopic in situ hybridization. Please see Dataset S2 for primer sequences.
β-Galactosidase Staining of Adult Tissues.
Adult mouse tissues isolated from mice heterozygous for the Zfp106 LacZ knock-in allele were stained as previously described (63).
UV Cross-Linking, Immunoprecipitation, and Labeling of ZFP106-Associated RNA.
Neuro2a neuroblastoma cells were transiently transfected using Lipofectamine 2000 (Life Technologies) with constructs encoding N- or C-terminal TY1 epitope-tagged ZFP106 fusion proteins. Forty-eight hours after transfection, cells were washed twice with PBS and irradiated on ice with UV light at 254 nm (150 mJ/cm2). Immediately after irradiation, cells were lysed in three cell-pellet volumes of lysis buffer (50 mM Tris, pH 7.4, 1% Nonidet P-40, 100 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, EDTA-free protease inhibitor mixture). Lysates were incubated with a low concentration of RNase I (Ambion) for 3 min at 37 °C. Lysates were subsequently cleared by centrifugation at 16,000 × g for 20 min at 4 °C. TY1 fusion proteins and hnRNPC proteins were captured by incubation with a TY1 antibody (Diagenode) or hnRNPC antibody (Abcam) and 100 µL Dynabeads overnight at 4 °C. The following day, the Dynabeads were washed two times with high-salt buffer (1 M NaCl, 50 mM Tris, pH 7.5, 1 mM EDTA, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, EDTA-free protease inhibitor mixture) and two times with wash buffer (20 mM Tris, pH 7.4, 10 mM MgCl2, 0.2% Tween 20). The beads were then incubated with 1 U/μL T4 polynucleotide kinase (NEB) and 0.1 μCi/μL [γ-32P]ATP (Fisher) for 5 min at 37 °C to label the 5′ ends of cross-linked RNAs. The Dynabeads were then incubated in Laemmli loading buffer for 10 min at 70 °C and immunoprecipitates were resolved on a 4–12% NuPAGE Bis-Tris gel (Life Technologies) and blotted on nitrocellulose membranes. Cross-linked RNA–protein complexes were visualized by autoradiography.
Transfection and Immunofluorescence Confocal Microscopy.
COS7 cells were transiently transfected using FuGENE 6 (Promega). Following 24 h, cells were washed in PBS and fixed at room temperature in 4% (vol/vol) paraformaldehyde in PBS for 10 min. For immunostaining, cells were permeabilized with 1% Triton X-100 in PBS for 15 min at room temperature. Immunofluorescence staining for SC-35 (Abcam; ab11826), GFAP (Abcam; ab7260), Iba1 (Abcam; ab107159), TDP-43 (Proteintech; 10782-1-AP), FUS (Abcam; ab70381), p62 (SQSTM1; Santa Cruz; sc-25575), and ubiquitin (Santa Cruz; sc-8017) was performed using standard techniques. Fluorescence microscopy of fixed cells was carried out using a 63× N.A. 1.4 Plan-Apochromat objective on an LSM 780 multiphoton Zeiss microscope. Images represent projections of the entire nuclear fluorescence stack.
Inhibition of RNA Synthesis in Mammalian Cells.
Cells were incubated with 100 μM DRB (Sigma) for 1 h at 37 °C and subsequently fixed in 4% paraformaldehyde and directly imaged.
Tissue Histology and Staining.
For skeletal muscle histology, whole muscles were dissected and then fixed in 4% (vol/vol) paraformaldehyde in PBS overnight at room temperature. Muscles were dehydrated and embedded in paraffin for routine histological analyses and staining for hematoxylin and eosin and Masson’s trichrome as previously described (62). For staining of nerve terminals and neuromuscular junctions, whole muscles were fixed in 2% formaldehyde in 0.1 M phosphate buffer (pH 7.3) overnight at 4 °C. Muscle samples were incubated with 2 nM Texas red-conjugated α-bungarotoxin (Life Technologies) for 30 min and then with an antibody against Syntaxin 1 overnight at 4 °C. Muscle samples were subsequently incubated with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG overnight at 4 °C. Muscle samples were washed with PBS and mounted in Vectashield mounting medium.
For spinal cord and CNS histology, anesthetized animals were fixed first by transcardial perfusion with 4% paraformaldehyde. Brains were dissected and further fixed by immersion for 48 h in 4% paraformaldehyde before paraffin embedding and staining according to standard procedures. Staining for Luxol fast blue and cresyl echt violet (LFB-CEV) was carried out manually, subjecting the slides to 0.1% LFB overnight at 60 °C and then destaining and sequential differentiation in 0.05% lithium carbonate and 70% ethanol before final staining for 10 min in 0.1% CEV. Slides were rinsed in water, dehydrated, cleared, and coverslipped with synthetic mounting medium.
Electron Microscopy.
Skeletal muscle tissues were harvested from mice perfused with a solution of 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) containing 50 mM CaCl2. Tissues were subsequently postfixed in buffered 2% osmium tetroxide containing 0.8% potassium ferricyanide for 3 h. Tissues were rinsed with dH2O, en bloc stained in 4% uranyl acetate in 50% ethanol, dehydrated with a graded series of ethanol, and embedded in EMbed 812 resin (Electron Microscopy Sciences). Thin sections were cut on a Leica Ultracut UCT ultramicrotome and then stained with 2% uranyl acetate and lead citrate. Images were acquired on an FEI Tecnai G2 Spirit electron microscope equipped with a LaB6 source Gatan CCD camera and operated at 120 kV.
Supplementary Material
Acknowledgments
We thank Jose Cabrera for assistance with graphic images. We thank Abhijit Bugde and Dr. Kate Luby-Phelps for help with confocal imaging at the Live Cell Imaging Core Facility at UT Southwestern, John Shelton for help with histology, in situ hybridizations, and imaging, and Dr. Karen Rothberg for assistance and imaging at the Electron Microscopy Core Facility at UT Southwestern. This work was supported in part by grants from the National Institutes of Health (HL-077439, DK-099653, HL-130253, NS-055028, HD-087351, and AR-067294), a Robert A. Welch Foundation Grant (1-0025; to E.N.O.), a Muscular Dystrophy Association development Grant (MDA377340; to D.M.A.), an American Heart Association Predoctoral Fellowship (14PRE19830031; to K.M.A.), and a fellowship from University of Pavia (DR 21/11/2014, 1671; to J.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608423113/-/DCSupplemental.
References
- 1.Hirsch NP. Neuromuscular junction in health and disease. Br J Anaesth. 2007;99(1):132–138. doi: 10.1093/bja/aem144. [DOI] [PubMed] [Google Scholar]
- 2.Goulet BB, Kothary R, Parks RJ. At the “junction” of spinal muscular atrophy pathogenesis: The role of neuromuscular junction dysfunction in SMA disease progression. Curr Mol Med. 2013;13(7):1160–1174. doi: 10.2174/15665240113139990044. [DOI] [PubMed] [Google Scholar]
- 3.Moloney EB, de Winter F, Verhaagen J. ALS as a distal axonopathy: Molecular mechanisms affecting neuromuscular junction stability in the presymptomatic stages of the disease. Front Neurosci. 2014;8:252. doi: 10.3389/fnins.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollari E, Goldsteins G, Bart G, Koistinaho J, Giniatullin R. The role of oxidative stress in degeneration of the neuromuscular junction in amyotrophic lateral sclerosis. Front Cell Neurosci. 2014;8:131. doi: 10.3389/fncel.2014.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tintignac LA, Brenner HR, Rüegg MA. Mechanisms regulating neuromuscular junction development and function and causes of muscle wasting. Physiol Rev. 2015;95(3):809–852. doi: 10.1152/physrev.00033.2014. [DOI] [PubMed] [Google Scholar]
- 6.Blackstone C, O’Kane CJ, Reid E. Hereditary spastic paraplegias: Membrane traffic and the motor pathway. Nat Rev Neurosci. 2011;12(1):31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoyle JC, Isfort MC, Roggenbuck J, Arnold WD. The genetics of Charcot-Marie-Tooth disease: Current trends and future implications for diagnosis and management. Appl Clin Genet. 2015;8:235–243. doi: 10.2147/TACG.S69969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tisdale S, Pellizzoni L. Disease mechanisms and therapeutic approaches in spinal muscular atrophy. J Neurosci. 2015;35(23):8691–8700. doi: 10.1523/JNEUROSCI.0417-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters OM, Ghasemi M, Brown RH., Jr Emerging mechanisms of molecular pathology in ALS. J Clin Invest. 2015;125(5):1767–1779. doi: 10.1172/JCI71601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A. Hereditary spastic paraplegia: Clinical-genetic characteristics and evolving molecular mechanisms. Exp Neurol. 2014;261:518–539. doi: 10.1016/j.expneurol.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Ferraiuolo L, Kirby J, Grierson AJ, Sendtner M, Shaw PJ. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7(11):616–630. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- 12.Tapia R. Cellular and molecular mechanisms of motor neuron death in amyotrophic lateral sclerosis: A perspective. Front Cell Neurosci. 2014;8:241. doi: 10.3389/fncel.2014.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barber SC, Shaw PJ. Oxidative stress in ALS: Key role in motor neuron injury and therapeutic target. Free Radic Biol Med. 2010;48(5):629–641. doi: 10.1016/j.freeradbiomed.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 14.Bäumer D, Ansorge O, Almeida M, Talbot K. The role of RNA processing in the pathogenesis of motor neuron degeneration. Expert Rev Mol Med. 2010;12:e21. doi: 10.1017/S1462399410001523. [DOI] [PubMed] [Google Scholar]
- 15.Sreedharan J, Brown RH., Jr Amyotrophic lateral sclerosis: Problems and prospects. Ann Neurol. 2013;74(3):309–316. doi: 10.1002/ana.24012. [DOI] [PubMed] [Google Scholar]
- 16.Zuberi AR, Christianson GJ, Mendoza LM, Shastri N, Roopenian DC. Positional cloning and molecular characterization of an immunodominant cytotoxic determinant of the mouse H3 minor histocompatibility complex. Immunity. 1998;9(5):687–698. doi: 10.1016/s1074-7613(00)80666-4. [DOI] [PubMed] [Google Scholar]
- 17.Salvatore P, Hanash CR, Kido Y, Imai Y, Accili D. Identification of sirm, a novel insulin-regulated SH3 binding protein that associates with Grb-2 and FYN. J Biol Chem. 1998;273(12):6989–6997. doi: 10.1074/jbc.273.12.6989. [DOI] [PubMed] [Google Scholar]
- 18.Laity JH, Lee BM, Wright PE. Zinc finger proteins: New insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11(1):39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 19.Grasberger H, Bell GI. Subcellular recruitment by TSG118 and TSPYL implicates a role for zinc finger protein 106 in a novel developmental pathway. Int J Biochem Cell Biol. 2005;37(7):1421–1437. doi: 10.1016/j.biocel.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Ide S, Dejardin J. End-targeting proteomics of isolated chromatin segments of a mammalian ribosomal RNA gene promoter. Nat Commun. 2015;6:6674. doi: 10.1038/ncomms7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joyce PI, et al. Deficiency of the zinc finger protein ZFP106 causes motor and sensory neurodegeneration. Hum Mol Genet. 2016;25(2):291–307. doi: 10.1093/hmg/ddv471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruan J, et al. TreeFam: 2008 update. Nucleic Acids Res. 2008;36(Database issue):D735–D740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philips T, Robberecht W. Neuroinflammation in amyotrophic lateral sclerosis: Role of glial activation in motor neuron disease. Lancet Neurol. 2011;10(3):253–263. doi: 10.1016/S1474-4422(11)70015-1. [DOI] [PubMed] [Google Scholar]
- 24.Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32(12):638–647. doi: 10.1016/j.tins.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchler-Bauer A, et al. CDD: NCBI’s Conserved Domain Database. Nucleic Acids Res. 2015;43(Database issue):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149(6):1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 27.Chen YI, et al. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 2007;35(12):3928–3944. doi: 10.1093/nar/gkm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsushima Y, Matsumura K, Kitagawa Y. Zinc finger-like motif conserved in a family of RNA binding proteins. Biosci Biotechnol Biochem. 1997;61(5):905–906. doi: 10.1271/bbb.61.905. [DOI] [PubMed] [Google Scholar]
- 29.Rossi F, et al. Involvement of U1 small nuclear ribonucleoproteins (snRNP) in 5′ splice site-U1 snRNP interaction. J Biol Chem. 1996;271(39):23985–23991. doi: 10.1074/jbc.271.39.23985. [DOI] [PubMed] [Google Scholar]
- 30.Konig J, et al. iCLIP—Transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J Vis Exp. 2011;(50):e2638. doi: 10.3791/2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Couthouis J, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci USA. 2011;108(52):20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordin A, Larsson E, Holmberg M. The defective splicing caused by the ISCU intron mutation in patients with myopathy with lactic acidosis is repressed by PTBP1 but can be derepressed by IGF2BP1. Hum Mutat. 2012;33(3):467–470. doi: 10.1002/humu.22002. [DOI] [PubMed] [Google Scholar]
- 33.Lamond AI, Spector DL. Nuclear speckles: A model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4(8):605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 34.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3(2):a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16(11):5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumaran RI, Muralikrishna B, Parnaik VK. Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J Cell Biol. 2002;159(5):783–793. doi: 10.1083/jcb.200204149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lassen N, et al. Multiple and additive functions of ALDH3A1 and ALDH1A1: Cataract phenotype and ocular oxidative damage in Aldh3a1(-/-)/Aldh1a1(-/-) knock-out mice. J Biol Chem. 2007;282(35):25668–25676. doi: 10.1074/jbc.M702076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez de Aguilar JL, et al. Gene profiling of skeletal muscle in an amyotrophic lateral sclerosis mouse model. Physiol Genomics. 2008;32(2):207–218. doi: 10.1152/physiolgenomics.00017.2007. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka H, et al. The potential of GPNMB as novel neuroprotective factor in amyotrophic lateral sclerosis. Sci Rep. 2012;2:573. doi: 10.1038/srep00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burden SJ, Yumoto N, Zhang W. The role of MuSK in synapse formation and neuromuscular disease. Cold Spring Harb Perspect Biol. 2013;5(5):a009167. doi: 10.1101/cshperspect.a009167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pérez-García MJ, Burden SJ. Increasing MuSK activity delays denervation and improves motor function in ALS mice. Cell Reports. 2012;2(3):497–502. doi: 10.1016/j.celrep.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yumoto N, Kim N, Burden SJ. Lrp4 is a retrograde signal for presynaptic differentiation at neuromuscular synapses. Nature. 2012;489(7416):438–442. doi: 10.1038/nature11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams AH, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326(5959):1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jokic N, et al. The neurite outgrowth inhibitor Nogo-A promotes denervation in an amyotrophic lateral sclerosis model. EMBO Rep. 2006;7(11):1162–1167. doi: 10.1038/sj.embor.7400826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jokic N, et al. Nogo expression in muscle correlates with amyotrophic lateral sclerosis severity. Ann Neurol. 2005;57(4):553–556. doi: 10.1002/ana.20420. [DOI] [PubMed] [Google Scholar]
- 46.Nagai K, et al. Structure and assembly of the spliceosomal snRNPs. Novartis Medal Lecture. Biochem Soc Trans. 2001;29(Pt 2):15–26. doi: 10.1042/0300-5127:0290015. [DOI] [PubMed] [Google Scholar]
- 47.Dobrowolny G, et al. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab. 2008;8(5):425–436. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Wong M, Martin LJ. Skeletal muscle-restricted expression of human SOD1 causes motor neuron degeneration in transgenic mice. Hum Mol Genet. 2010;19(11):2284–2302. doi: 10.1093/hmg/ddq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grasberger H, Ye H, Mashima H, Bell GI. Dual promoter structure of ZFP106: Regulation by myogenin and nuclear respiratory factor-1. Gene. 2005;344:143–159. doi: 10.1016/j.gene.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 50.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lagier-Tourenne C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15(11):1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 53.Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9(10):995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 54.Tizzano E, Baiget M. [Molecular basis of spinal muscular atrophy: The SMN gene] Neurologia. 2000;15(9):393–400. Spanish. [PubMed] [Google Scholar]
- 55.Leblond CS, et al. 2016. Replication study of MATR3 in familial and sporadic amyotrophic lateral sclerosis. Neurobiol Aging 37:209.e17–209.e21.
- 56.Johnson JO, et al. ITALSGEN Consortium Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17(5):664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Novarino G, et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343(6170):506–511. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen S, Sayana P, Zhang X, Le W. Genetics of amyotrophic lateral sclerosis: An update. Mol Neurodegener. 2013;8:28. doi: 10.1186/1750-1326-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hentati A, et al. Linkage of a commoner form of recessive amyotrophic lateral sclerosis to chromosome 15q15-q22 markers. Neurogenetics. 1998;2(1):55–60. doi: 10.1007/s100480050052. [DOI] [PubMed] [Google Scholar]
- 60.Montecchiani C, et al. ALS5/SPG11/KIAA1840 mutations cause autosomal recessive axonal Charcot-Marie-Tooth disease. Brain. 2016;139(Pt 1):73–85. doi: 10.1093/brain/awv320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orlacchio A, et al. SPATACSIN mutations cause autosomal recessive juvenile amyotrophic lateral sclerosis. Brain. 2010;133(Pt 2):591–598. doi: 10.1093/brain/awp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shelton JM, Lee MH, Richardson JA, Patel SB. Microsomal triglyceride transfer protein expression during mouse development. J Lipid Res. 2000;41(4):532–537. [PubMed] [Google Scholar]
- 63.Anderson DM, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.