Significance
Legionella pneumophila, the bacterium causing Legionnaires’ disease, invades lung cells by creating intracellular vacuoles where it is protected and where it can multiply. For expansion, the vacuoles must hijack intracellular vesicles while avoiding destruction by lysosomes. To achieve these goals, Legionella exports effector proteins into the cytoplasm and into the membrane of the vacuole. Here we report that certain Legionella effectors mimic intracellular fusion proteins of the SNARE family, allowing them to selectively fuse with specific intracellular transport vesicles, thus providing more space for the dividing bacteria. In contrast to SNAREs, however, the Legionella proteins can be used only once, thus allowing the bacteria to control the amount of hijacked vesicles.
Keywords: Legionella pneumophila, SNAREs, NSF, membrane fusion
Abstract
Legionella pneumophila, the Gram-negative pathogen causing Legionnaires’ disease, infects host cells by hijacking endocytic pathways and forming a Legionella-containing vacuole (LCV) in which the bacteria replicate. To promote LCV expansion and prevent lysosomal targeting, effector proteins are translocated into the host cell where they alter membrane traffic. Here we show that three of these effectors [LegC2 (Legionella eukaryotic-like gene C2)/YlfB (yeast lethal factor B), LegC3, and LegC7/YlfA] functionally mimic glutamine (Q)-SNARE proteins. In infected cells, the three proteins selectively form complexes with the endosomal arginine (R)-SNARE vesicle-associated membrane protein 4 (VAMP4). When reconstituted in proteoliposomes, these proteins avidly fuse with liposomes containing VAMP4, resulting in a stable complex with properties resembling canonical SNARE complexes. Intriguingly, however, the LegC/SNARE hybrid complex cannot be disassembled by N-ethylmaleimide-sensitive factor. We conclude that LegCs use SNARE mimicry to divert VAMP4-containing vesicles for fusion with the LCV, thus promoting its expansion. In addition, the LegC/VAMP4 complex avoids the host’s disassembly machinery, thus effectively trapping VAMP4 in an inactive state.
Legionnaires’ disease in humans is caused by Legionella pneumophila (1), which enters human monocytes and alveolar macrophages by macropinocytosis. After endocytotic uptake, Legionella prevents fusion with lysosomes to escape host degradation and establishes a replication niche, called the Legionella-containing vacuole (LCV) (2). To achieve these goals, Legionella translocates effector proteins through a type IVB (Icm/Dot) secretion system into the host cytoplasm or into the LCV membrane (2–5). Around 300 Legionella effectors were identified by genetic or bioinformatic approaches (6–8). Whereas growth and survival of Legionella depends on these effector proteins, they appear to be highly redundant because as many as 71 effector-encoding genes can be deleted in a single strain that retains the ability to grow in macrophages (9).
For membrane expansion, LCVs recruit trafficking vesicles from the host cell. Mainly, these vesicles originate from trafficking vesicles shuttling between the endoplasmic reticulum (ER) and the cis-face of the Golgi apparatus (10), although other sources of membrane, e.g., of endosomal sources, cannot be excluded (11). Thus, Legionella is capable of redirecting trafficking vesicles to fuse with the LCV, but the mechanisms by which this is achieved are only slowly emerging. In eukaryotes, the specificity of membrane traffic is governed by sets of regulatory proteins, which ultimately converge to regulate vesicle fusion carried out by SNARE [soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein receptor] proteins. The regulatory proteins include small GTPases of the Rab and Arf families. These proteins operate as molecular switches that, once activated, recruit effector proteins from the cytoplasm to ensure fusion with the correct target by controlling SNAREs. SNAREs comprise a family of small and mostly membrane-anchored proteins (12). They are characterized by coiled-coil (CC)–forming SNARE motifs that assemble between the membranes and thus initiate fusion. SNARE motifs are classified into four subfamilies termed Qa-, Qb-, Qc-, and arginine (R)-SNAREs, with one of each required for assembly of a fusion-competent SNARE complex (13). Whereas each intracellular fusion step appears to involve its own specific set of SNARE proteins, SNAREs on their own are rather promiscuous, with members of the same subfamily being capable of substituting for each other in cells, and even more so in vitro (12).
Although the function of most Legionella effectors is still unknown, several of them were shown to target trafficking protein, including small GTPases such as Rab1 and Arfs, or to interfere with the formation of autophagosomes (14). Moreover, LCV formation is associated with the formation of noncognate SNARE complexes between an R-SNARE functioning in trafficking between the ER and the Golgi apparatus (mSec22b) and glutamine (Q)-SNAREs normally operating at the plasma membrane [syntaxins 2, 3, 4, and synapotosomal-associated protein (SNAP)-23] (15). Complex formation appears to be enhanced by DrrA, a Legionella effector that binds to the SNARE Syntaxin 3, a reaction that appears to be regulated by the small GTPase Rab1 (16).
Intriguingly, some effectors bear superficial similarity to SNAREs and thus may interfere with SNARE function. For example, the IncA effector of Chlamydia, was shown to interfere with SNARE assembly (17, 18). More recently, another putative Legionella SNARE paralog was identified by bioinformatic searches (LseA) and shown to interact with host cell SNAREs (19).
In this study, we have investigated whether three structurally related Legionella effectors (LegC2/YlfB, LegC3, and LegC7/YlfA) (20) interact with mammalian SNAREs, and if so, whether this interaction affects SNARE function. These LegC-proteins comprise a group of transmembrane proteins that possess coiled-coil motifs reminiscent of SNARE proteins, raising the possibility that they may form hybrid complexes with endogenous SNARE proteins. Indeed, LegC3 was shown previously to inhibit SNARE-mediated homotypic fusion of yeast vacuoles in vitro, but a direct interaction between LegC3 and SNAREs was not observed (21). We found that all three LegC proteins selectively interact with the R-SNARE vesicle-associated membrane protein 4 (VAMP4), and furthermore, that the availability of VAMP4 is rate limiting for the intracellular proliferation of Legionella. Using in vitro fusion assays, we also report that the three LegC proteins together are capable of substituting for the endogenous Q-SNARE partners of VAMP4, resulting in the formation of hybrid LegC/SNARE complexes and fusion with an efficiency comparable to that observed with the endogenous SNARE partners of VAMP4. Intriguingly, the hybrid LegC/VAMP4 complex formed during fusion cannot be dissociated by the SNARE disassembly enzyme NSF.
Results
LegC2, LegC3, and LegC7 Resemble Mammalian Q-SNAREs and Interact with Host VAMP4.
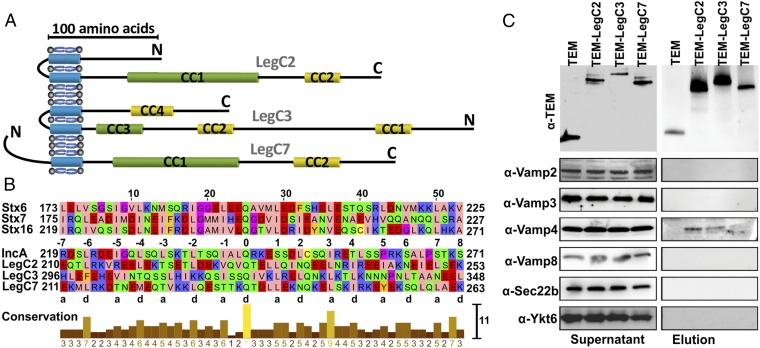
LegC proteins contain one (LegC7) or two (LegC2 and LegC3) predicted transmembrane domains (TMDs) and two predicted coiled-coil motifs of which one is reminiscent of SNARE motifs (Fig. 1A). Alignment with SNARE motifs from mammalian Q-SNAREs revealed the typical heptad repeat structure, with hydrophobic residues in the “a” and “d” positions, and a conserved glutamine (Q) residue in the center of the motifs, just as in the “0” layer of Q-SNAREs (13) (Fig. 1B). The similarities appear to be higher than in IncA, which contains helix-breaking proline residues in the C-terminal part (Fig. 1B). Furthermore LegC2 and LegC3 with two consecutive TMDs resemble the unique hairpin-type autophagosomal Qa-SNARE–Stx17 (22). Hence, we investigated whether the Legionella effector proteins LegC2/YlfB, LegC3, and LegC7/YlfA interact with host cell SNAREs to modulate membrane fusion. To examine whether LegC proteins interact with host cell R-SNAREs, we infected phorbol 12-myristate 13-acetate (PMA)-differentiated THP-1 macrophage-like cells with Legionella strains overexpressing either β-lactamase (TEM) or N-terminally TEM-tagged versions of one of each of the effector proteins LegC2, LegC3, or LegC7. Six hours after infection, LegC proteins were immunoprecipitated from cell lysates using anti-TEM antibodies. Immunoblotting for TEM revealed bands of the expected size in all three cases both in the lysates and immunoprecipitates [TEM (30 kDa), TEM-LegC2 (76 kDa), TEM-LegC3 (93 kDa), and TEM-LegC7 (78 kDa)]. We then examined the precipitated material for the presence of mammalian R-SNAREs including VAMP2/Synaptobrevin and VAMP3/Cellubrevin (exocytosis) (23), VAMP4 (trans-Golgi network) (24), VAMP8 (late endosome/lysosome), as well as Sec22b and Ykt6 (ER–Golgi transport) (25). Whereas all six R-SNARE proteins were clearly detectable in the lysate, VAMP4 was the only R-SNARE that selectively coprecipitated with each of the tagged LegC proteins (Fig. 1C). Conversely, immunoprecipitations using anti-VAMP4 antibodies resulted in the coprecipitation of TEM-fused LegC2, LegC3, and LegC7, confirming that the interaction is specific (Fig. S1).
Fig. 1.
LegC2, LegC3, and LegC7 resemble mammalian Q-SNAREs and interact with host VAMP4. (A) Predicted TMDs and the CC motifs in LegC proteins. CC motifs with probability scores <99% are shown in yellow and those with probability scores >99% are shown in green. (B) Multiple sequence alignments of the high-score CC motifs (green) of LegC effectors with the SNARE motifs of the endosomal Q-SNAREs Stx6 (173–225), Stx7 (175–227), and Stx16 (219–271). The hydrophobic residues in positions “a” and “d” of the heptad-repeats are indicated. LegC2, LegC3, and LegC7 also contain the conserved Q residue as in the central 0 layer of Q-SNARE. In comparison, the predicted CC motif of IncA (219–271) shows lower similarities in the C-terminal region. (C) VAMP4 but no other R-SNARE coprecipitates with LegC2, LegC3, and LegC7, respectively. Differentiated THP-1 cells were infected with Legionella strains either overexpressing TEM or individual N-terminal TEM-fused effector proteins for 6 h. Cells were lysed and supernatants were precleared with protein A/G agarose, followed by incubation with protein A/G agarose containing purified anti-TEM antibodies. The beads were washed and eluted at low pH buffer. The precleared supernatants and elutions from coimmunoprecipitation experiments were analyzed by Western blotting using anti-TEM, anti-VAMP2, anti-VAMP3, anti-VAMP4, anti-VAMP8, anti-Sec22b, or anti-Ykt6 antibodies, respectively. Of the six targeted R-SNAREs, only VAMP4 coprecipitates with the LegC proteins.
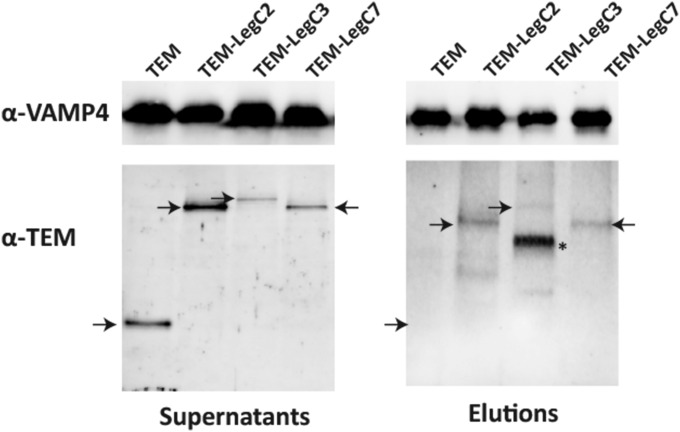
Fig. S1.
LegC effectors interact with VAMP4. Immunoprecipitation of VAMP4 reveals coprecipitation of TEM-tagged LegC2, LegC3 (rather weak only), and LegC7. Both the starting supernatants and the eluted antigen complexes were analyzed for VAMP4 and TEM by immunoblotting, the position of the LegC proteins is marked with arrows. The band marked with an asterisk was not identified; it may represent a breakdown product.
VAMP4 Knockdown in THP-1 Cells Reduces Intracellular Growth of Legionella.
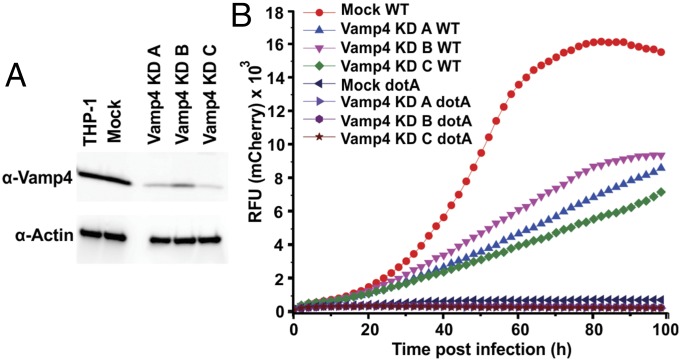
Next we asked whether VAMP4 is required for infection and/or intracellular proliferation of Legionella. We generated three different THP-1 cell lines in which VAMP4 expression is stably knocked down (Fig. 2A), whereas cell viability remains unaffected (Fig. S2). Intracellular growth of Legionella was monitored by fluorescence increase after infection (26) with a mCherry-expressing wild-type strain (27). Growth was significantly reduced in all three knockdown cell lines (Fig. 2B). As a control, we infected the cells with a Legionella dotA mutant that fails to grow intracellularly (2) and found negligible growth for both mock and knockdown constructs (Fig. 2B). In contrast, no differences were observed in pathogen uptake (Fig. S3), showing that VAMP4 is not rate limiting for phagocytosis, but rather for intracellular growth.
Fig. 2.
Decreased expression of VAMP4 in THP-1 cells reduces intracellular growth of Legionella. (A) Immunoblot of lysates from THP-1 macrophage cell lines showing significant reduction in levels of VAMP4 with the three different knockdown constructs KD_A, KD_B, and KD_C in comparison with mock and negative control. (B) Intracellular growth of Legionella is reduced in cells in which VAMP4 expression is knocked down. For monitoring growth, a wild-type Legionella strain expressing mCherry as fluorescent marker was used, allowing for quantification of Legionella using fluorescence intensity. As controls, cells were also infected with a dotA mutant of Legionella, which fails to grow in both mock and knockdown constructs. The Legionella growth in VAMP4 KD cell lines were repeated in at least three independent experiments with similar results. The graph shows the result of one representative independent experiment. Each data point represents the average of technical triplicates. The nonparametric t test was performed between the indicated KD cell lines with the mock cell line infected with wild-type Legionella. The following P values were obtained comparing the mock-KD cells to the VAMP4 KD cell lines: VAMP4_KD_A (P = 0.0007); VAMP4_KD_B (P = 0.0043); and VAMP4_KD_C (P < 0.0001). All of the P values in these t tests are <0.01. Thus, the Legionella growth in Vamp4_KD A, B, and C are statistically different from wild-type Legionella growth in mock cell line at P value of 0.01.
Fig. S2.
Knockdown of VAMP4 does not cause cytotoxicity. (A–C) Cell viability, assayed by the MTT reaction, upon infection with increasing concentrations of Legionella shows no significant differences between the mock-transfected and the VAMP4 knockdown cell lines.
Fig. S3.
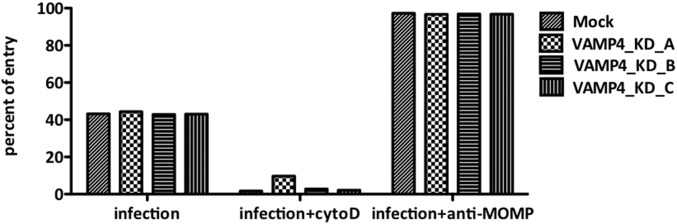
VAMP4 knockdown does not affect Legionella uptake. Percent of entry of the Legionella showing no significant difference between VAMP4 knockdown and mock constructs. Percentage of entry for negative control with Cytochalasin D and positive control with anti-MOMP antibody as comparison.
LegCs Mediate Membrane Fusion Along with Host R-SNAREs and Form SNARE-Like, SDS-Resistant Complexes.
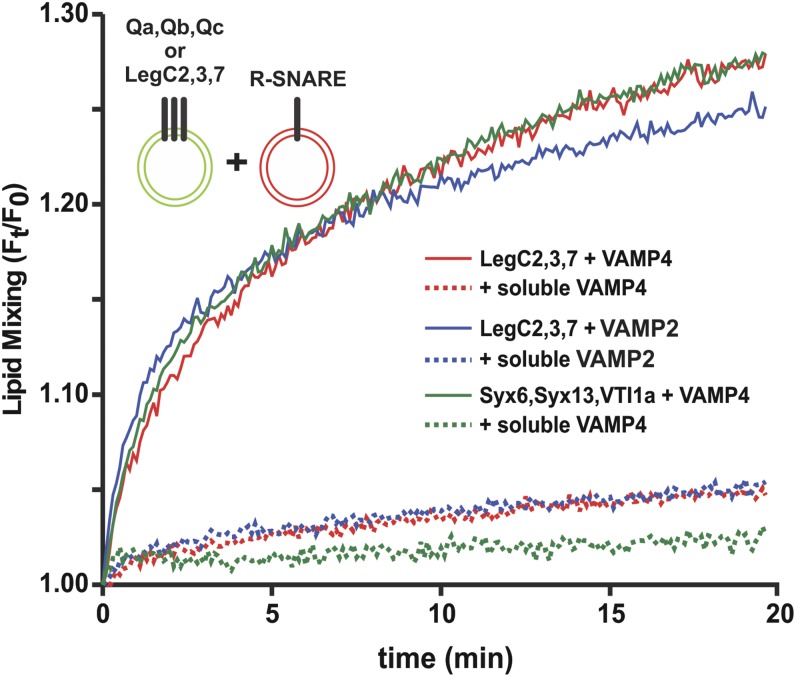
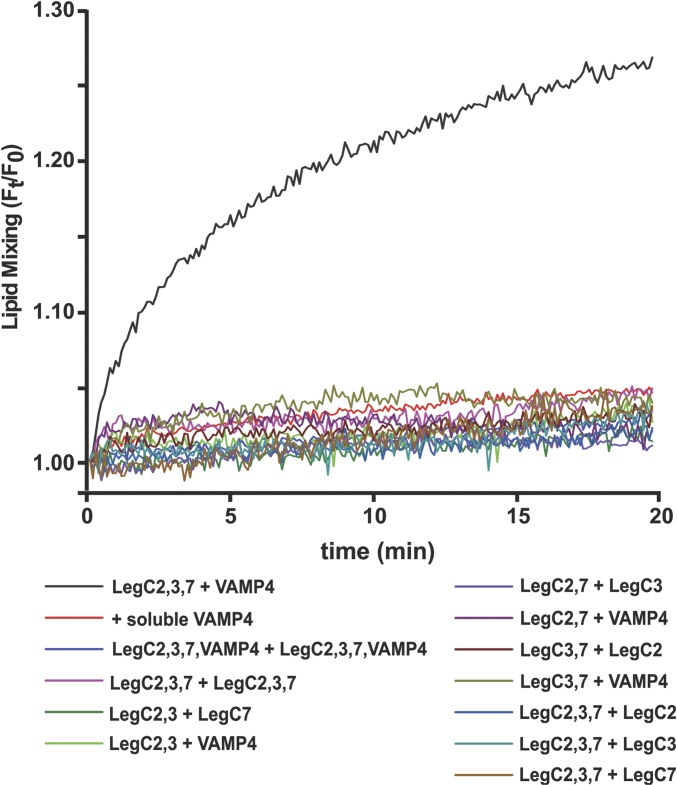
We then asked whether the LegC proteins can functionally substitute for Q-SNAREs by mediating fusion with membranes containing VAMP4. To this end, we reconstituted purified VAMP4 and the three LegC proteins in small unilamellar vesicles (SUVs), respectively. Using a standard FRET assay (28, 29) robust fusion, comparable to that of vesicles containing the canonical Q-SNAREs Syntaxin 13, Syntaxin 6, and VTI1a, was observed (Fig. 3). Because SNAREs show promiscuity (12) both in vivo (30–34) and in vitro (35, 36), we replaced the R-SNARE VAMP4 with VAMP2 and still observed fusion (Fig. 3). We have observed SNARE promiscuity in fusion assays earlier as well (37). The fusions could be completely blocked by competitive inhibition (29) using the cytoplasmic domains of either VAMP4 or VAMP2 (Fig. 3). To test whether LegC effectors can mediate membrane fusion in other topologies, we reconstituted them on liposomes together with VAMP4 in 12 different combinations. Fusion was monitored by the same FRET-based assay (Fig. S4). Fusion was observed only when proteoliposomes reconstituted with LegC2, LegC3, and LegC7 were mixed with VAMP4 reconstituted proteoliposomes. Hence, no fusion was observed if one of the LegC proteins was omitted or if various combinations of LegC proteins were used in the absence of the R-SNARE (Fig. S4).
Fig. 3.
Reconstitution of fusion by LegCs and host R-SNAREs. SUVs reconstituted with LegC2, LegC3, and LegC7 fuse with SUVs reconstituted with either VAMP4 (red) or VAMP2 (blue). The fusion rate was comparable to that observed with the endogenous endosomal Qa, Qb, and Qc SNAREs (green). Fusion was monitored as FRET based on lipid-mixing assay and all fusion reactions were inhibited by the soluble domains (20) of the corresponding R-SNAREs (dotted traces).
Fig. S4.
LegC-reconstituted liposomes fuse with VAMP4 liposomes in only one topology. Twelve different pairs of liposomes were prepared containing LegC2, LegC3, LegC7, and VAMP4, in various combinations as indicated. Each pair was then tested for fusion as in Fig. 3. LegC effectors were found to drive in vitro membrane fusion together with VAMP4 only in the topology LegC2:LegC3:LegC7 + VAMP4.
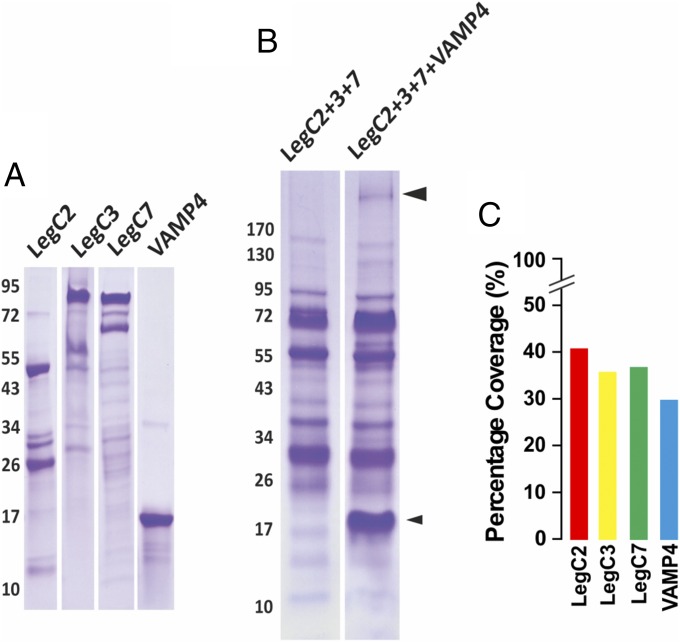
To test whether the LegC proteins form hybrid complexes with VAMP4, mixtures of purified LegC2, LegC3, and LegC7 were incubated overnight at 4 °C in the presence or absence of purified VAMP4 (Fig. 4A) and then resolved by SDS/PAGE without heat denaturation of the samples, followed by Coomassie staining (Fig. 4B). This procedure has shown previously to preserve, at least in part, SNARE complexes that then migrate with higher Mr during SDS/PAGE (38). A band of lower mobility was visible only when VAMP4 was present during the incubation (Fig. 4B). This band was cut out and analyzed by mass spectrometry. The resulting peptide hits were searched against a local database comprising the proteins LegC2, LegC3, LegC7, and VAMP4 using Mascot (Matrix Science). This led to unequivocal identification of all four proteins as indicated (Fig. 4C) by the percentage coverage (which is the percentage of all of the amino acids for a given protein that were detected in the sample). Together, these data show that LegC2, LegC3, and LegC7 form a SDS-resistant complex with VAMP4.
Fig. 4.
LegC2, LegC3, LegC7, and VAMP4 form SDS-resistant complexes. (A) Relative purity of the recombinant, purified LegC2, LegC3, LegC7, and VAMP4 proteins used in this study, illustrated as Coomassie-stained bands upon SDS/PAGE. Note that the strongly stained upper bands correspond to the expected Mr of the respective proteins. (B) Purified LegC2, LegC3, and LegC7 were incubated with or without VAMP4, followed by SDS/PAGE and Coomassie staining. The lower (smaller) arrowhead indicates VAMP4; the upper (bigger) arrowhead indicates the LegC–VAMP4 hybrid complex (band of slower mobility). (C) Percentage coverage of all four proteins derived from proteomic analysis of the latter band.
Unlike SNARE Complexes, the LegC–R-SNARE Complex Is Resistant to NSF/α-SNAP–Mediated Disassembly.
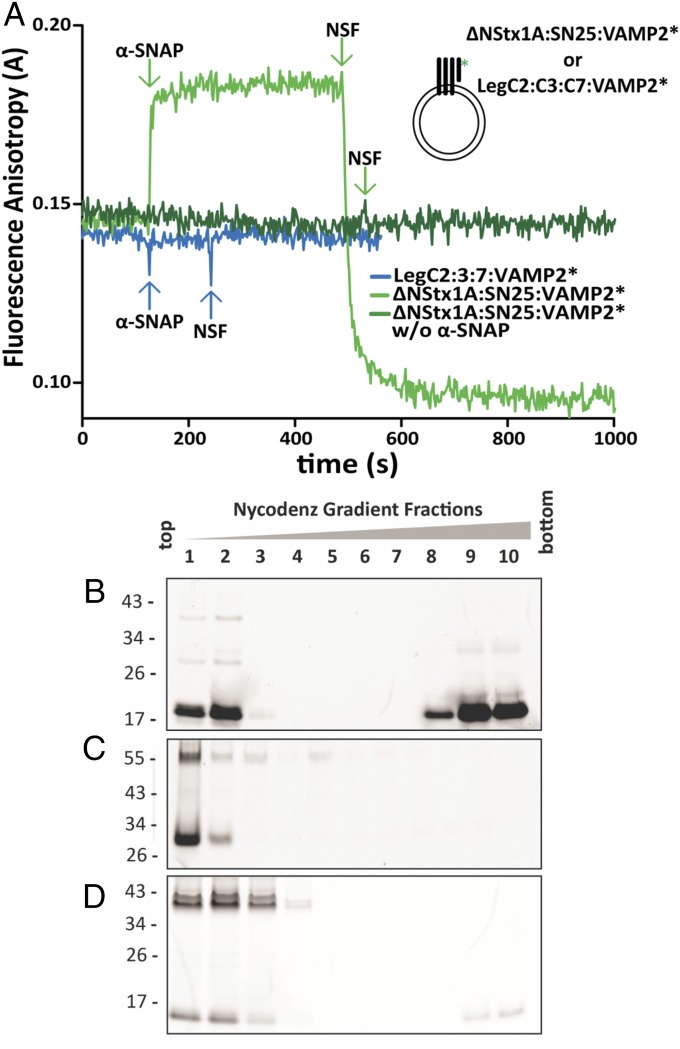
Endogenous SNARE complexes are disassembled by the AAA+ ATPase NSF, which regenerates fusion-competent SNAREs (12). For disassembly, the cofactor α-SNAP first binds to SNARE complexes, followed by the recruitment of NSF (39). To test whether the LegC–R-SNARE hybrids can be disassembled, we reconstituted a complex comprising LegC2, LegC3, LegC7, and Oregon Green-labeled cytoplasmic domain of Synaptobrevin (VAMP2*) (40) on SUVs and measured mobility changes associated with protein binding and complex disassembly by fluorescence anisotropy. A truncated neuronal SNARE ternary complex (41) comprising the same VAMP2*, Syntaxin 1 (183–288), and SNAP-25 (1–206) was reconstituted on SUVs (42), as positive control. Addition of α-SNAP caused increase in anisotropy due to its binding to the SNARE complex, followed by a decrease in anisotropy upon addition of NSF due to the dissociation of the VAMP2* (Fig. 5A). In contrast, adding α-SNAP and NSF sequentially to the LegC–VAMP2* hybrid complex did not change anisotropy, suggesting that the complex was not recognized by α-SNAP. As an additional control, the proteoliposomes were incubated with α-SNAP and NSF in the presence of Mg+2 and ATP for 10 min followed by flotation gradients to separate liposomes and soluble proteins (Fig. 5B). In the case of proteoliposomes reconstituted with the neuronal SNARE complex, the majority of the VAMP2* was detected in the soluble fraction, whereas in the case of the LegC–VAMP2* proteoliposomes, the VAMP2* remained in the liposomal fraction, confirming that NSF is unable to disassemble the LegC–R-SNARE hybrid complexes.
Fig. 5.
LegC–R-SNARE complexes are resistant to NSF-mediated disassembly. (A) Binding of α-SNAP and disassembly by NSF monitored by fluorescence anisotropy of labeled, cytoplasmic Syb (VAMP2*) reconstituted on SUVs with neuronal Q-SNAREs or LegCs. As positive control, sequential addition of α-SNAP (500 nM) and NSF (60 nM) to the truncated, ternary neuronal SNARE complex, led to expected rise and fall in anisotropy, respectively (light green trace). In the absence of α-SNAP, the same complex showed no change in anisotropy upon NSF addition (dark green trace). There was no change in anisotropy upon addition of α-SNAP and NSF to the LegC–VAMP2* hybrid complex, indicating that α-SNAP does not bind the complex, preventing binding of NSF (blue trace). For all SUVs, the protein:lipid (molar ratio) was 1:1,000. For purity of the recombinant proteins see Fig. 4A. (B–D) Proteoliposomes (as in A) reconstituted with truncated ternary neuronal SNARE complexes or LegC–VAMP2* hybrid complexes were incubated with α-SNAP, NSF, Mg+2, and ATP for 10 min followed by loading the samples on the bottom of a Nycodenz density gradient. After ultracentrifugation, the liposomes are enriched in the Top fractions of the gradient, whereas proteins not associated with membranes remain at the Bottom. The fractions were resolved by SDS/PAGE and the Oregon-Green–labeled, cytoplasmic Syb (VAMP2*) was detected in gel by a fluorescence imager. (B) In proteoliposomes containing the neuronal SNARE complexes, the majority of the VAMP2* was released and remained at the bottom of the gradient. Part of the unassembled VAMP2* seen in the Upper fractions illustrates the successful reconstitution of the complex on liposomes. (C) Identical to B, but ATP was replaced with its analog ATPγS. No dissociation of VAMP2* was detectable. Furthermore an SDS-resistant band of approximately 55 kDa is visible as expected for a truncated ternary neuronal SNARE complex. (D) For proteoliposomes containing the LegC–VAMP2 hybrid complexes, VAMP2* was not released (note that the weak signal on the bottom of the gradient represent the excess VAMP2* that did not form the complex). In addition, prominent SDS-resistant bands of higher Mr are detectable, representing nondissociated complexes.
Discussion
In this study, we have shown that the three Legionella effectors LegC2, LegC3, and LegC7 together form fully functional SNARE acceptor complexes that mediate membrane fusion by forming SNARE-like hybrid complexes with R-SNAREs. We have also shown that, although promiscuous in vitro, the LegC proteins selectively bind to the R-SNARE VAMP4, and that VAMP4 is required for supporting the growth of Legionella in infected cells.
Our findings provide a fascinating example of molecular mimicry by convergent evolution. Despite the similarities between LegC coiled-coil domains and SNARE motifs (Fig. 1), none of the LegC domains is significantly homologous (43) to SNAREs, indicating that they are not evolutionarily related. Due to this lack of homology, SNARE mimicry is difficult to distinguish from functionally unrelated coiled-coil motifs using bioinformatics approaches.
The capability of fully substituting for the endogenous SNAREs (at least in vitro) is unexpected and highly surprising. Whereas there is some variability in SNARE structure, the TMDs of SNAREs are typically at the C-terminal end. More importantly, they are separated from the interacting SNARE motifs only by few amino acids, with the distance between membrane anchor and SNARE motifs thought to be critical for their ability to transmit energy from the zippering to the fusion reaction (44). Whereas the overall domain structure of the LegC proteins resembles many SNAREs (TMD connected by a linker to the coiled-coil motif), linker length in all cases is substantially higher than in SNAREs. Even more surprisingly, in both LegC2 and LegC7, the orientation of the coiled-coil motifs is inverted, which may put constraints on the alignment with SNAREs (parallel or antiparallel). Presently we do not know yet in which way the proteins interact which each other, which of the coiled-coils are participating, and to what extent the hybrid complexes are structurally similar to the highly conserved SNARE four-helix bundles. However, it appears that the structural tolerance of the SNARE-based zipper mechanism for membrane fusion is much higher than anticipated (45), which is also supported by the surprising structural diversity of artificial SNARE mimetics capable of inducing fusion in vitro by some kind of zippering mechanism (46, 47). Further structural investigation of the LegC/SNARE hybrid complex can thus be expected to yield novel insights not only into the degree of structural conservation but also into the mechanisms involved in SNARE-mediated membrane fusion.
The ability of Legionella to use the R-SNARE VAMP4 as a “one-shot” device, enabling fusion with vesicles needed for expansion of the LCV, but then preventing the reactivation of the LegC/SNARE hybrid complex by blocking its disassembly, leads to an irreversible hijacking of the SNARE machinery by the pathogen. This is a fascinating novel example of the emerging arsenal used by intracellular pathogens for manipulating host cells. At present the contribution of this pathway to the overall growth of the LCV in comparison with the pathway involving ER-derived trafficking vesicles is difficult to evaluate (Fig. 6). It is also possible that other effectors are functionally equivalent to the LegC proteins studied here. Taken together, the ability of pathogen effectors to form coiled-coil complexes with host SNAREs that not only inhibit but also functionally substitute for endogenous SNAREs constitutes yet another mechanism by which intracellular pathogens manipulate the membrane traffic of host cells for survival and growth.
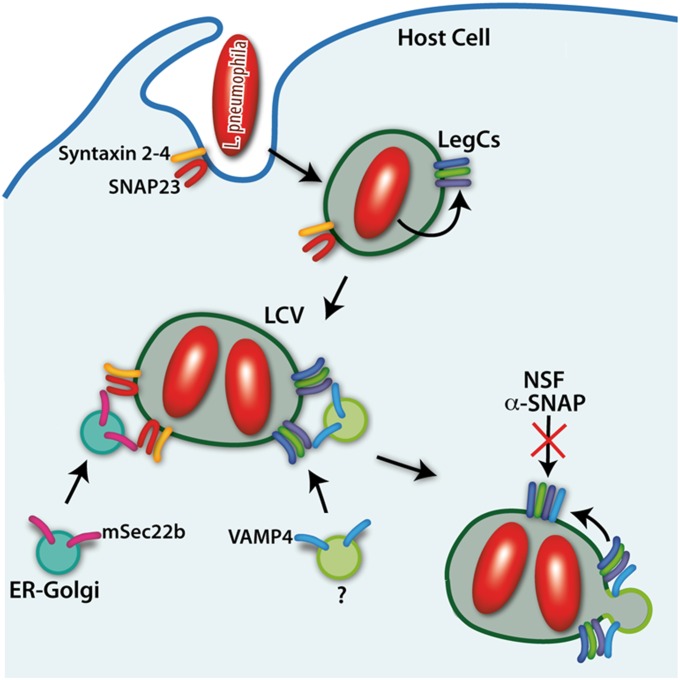
Fig. 6.
Model showing manipulation of membrane traffic by Legionella LegC proteins using SNARE mimicry. LegC proteins on the LCV membrane recruit VAMP4 vesicles (among others) to acquire membranes for vacuolar growth to facilitate proliferation of the pathogen.
Materials and Methods
Detailed protocols for all sections are described in SI Materials and Methods.
Bacterial Strains, Plasmids, and Oligonucleotides.
Legionella strains are all derivatives of L. pneumophila, Philadelphia-1, and their construction is described in detail in SI Materials and Methods. Oligonucleotide sequences used to amplify relevant Legionella genes are listed in SI Materials and Methods. The resulting plasmid constructs are also described in SI Materials and Methods.
Cell Culture.
THP-1 cells were obtained from American Type Culture Collection and grown in Advanced RPMI 1640 (Invitrogen) supplemented with 10% (vol/vol) FBS and 2 mM glutamine at 37 °C in a CO2 incubator. THP-1 cells were differentiated into macrophage-like cells by resuspending them into RPMI + 2 mM glutamine + 10% (vol/vol) FBS + 30 μM PMA. Following 72 h of treatment with PMA, the differentiated THP-1 cells were washed and resuspended in RPMI + 2 mM glutamine + 10% (vol/vol) FBS for infection (20).
Infection of THP-1 Cells by Legionella and Coimmunoprecipitation of LegC Proteins and VAMP4.
Coimmunoprecipitation experiments were carried following the manual of the Pierce Crosslink IP Kit (Thermo Fisher Scientific); additional details are described in SI Materials and Methods.
VAMP4 Knockdown.
Expression of VAMP4 in THP-1 cells was decreased by creating stable cell lines that express a shRNA complementary to the VAMP4 transcript carried on a lentivirus vector as detailed in SI Materials and Methods.
Protein Purification.
Proteins were overexpressed as N-terminal 6×-His tagged recombinant polypeptides using pET15b vector (Novagen) in the Escherichia coli strain BL21 (DE3) and affinity purified using Ni2+-nitrilotriacetic acid (NTA) agarose (Qiagen) resin followed by ion-exchange chromatography using the ÄKTA system (GE Healthcare) as described earlier (37, 39). The truncated, ternary neuronal SNARE complex was formed by mixing Syntaxin 1A (183–288), SNAP-25 (1–206), and Oregon-Green–labeled (at position S28C) cytoplasmic domain (1–96) of Syb (VAMP2*), in the molar ratio = 1:1:1.5 and purified as described previously (41); additional details are described in SI Materials and Methods.
Proteoliposome Preparation and Fusion Assay.
Proteoliposomes were prepared by reconstituting the proteins and lipids (both dissolved in detergents) into SUVs by detergent removal through a Sephadex G-50 column. Fusion of liposomes was monitored by lipid mixing assay based on FRET using a spectrofluorimeter (FluoroMax-2; Jobin Yvon) and NSF-mediated disassembly was monitored by fluorescence anisotropy using a FluoroLog 3 spectrometer in a T configuration equipped for polarizers (model FL322; Jobin Yvon); additional details are described in SI Materials and Methods.
SI Materials and Methods
In Silico Analysis of the LegC Effectors.
Coiled-coil (CC) motifs were predicted using the protein sequences of the Legionella effectors LegC2 (YP_095901.1), LegC3 (AAU27781.1), and LegC7 (AAU28360.1) using COILS (48) (window = 28 and cutoff = 0.9). Sequences of the LegC effectors and mammalian Qa SNAREs like Stx6, Stx7, and Stx16 were aligned using MUSCLE (49) and conservation scores were calculated using Jalview (50) based on the AMAS method (51). Details about the Zappo color scheme can be found at: www.jalview.org/help/html/colourSchemes/clustal.html. Transmembrane helices (TMHs) were predicted by the SMART 7 (52). CC motifs and TMDs shown in Fig. 1B are as follows: LegC2, TMD1 (79–101), TMD2 (105–127), CC1 (169–289), and CC2 (330–361); LegC3, CC1 (59–96), CC2 (232–259), CC3 (310–357), TMD1 (373–395), TMD2 (402–424), and CC4 (472–508); and LegC7, TMD (97–119), CC1 (155–272), and CC2 (325–371).
Legionella Strains and Plasmids.
The desired genes were amplified by PCR reactions using chromosomal DNA from Legionella strain KS79 with the following combinations of primers. Gene legC2 was amplified using primers: ATCGCATATGACAGACACTCCAAAAGC and ATCGCTCGAGCTAACCTGTGAGAGTTTG. Gene legC3 was amplified using primers: ATCGCATATGATTATGTTTTTGGCCAACTGC and ATCGCTCGAGTTACGCTATCTCATTAACTG. Gene legC7 was amplified with primers: ATCGCATATGGCTACTAATGAAACAGAGC and ATCGCTCGAGTTAATTGACTAAAGCAATAG. The PCR products were individually cloned into NdeI and XhoI sites of pET15b and correct clones were confirmed by sequencing. These plasmids were used to express 6×-His–LegC2, 6×-His–LegC3, and 6×-His–LegC7. All of the experiment were performed with L. pneumophila JR32 (53) a Philadelphia-1–derived strain, with dotA (a JR32 dotA::Tn903dIIlacZ strain LELA3118) or with KS79 (20) a hypercompetent comR mutant of JR32. Legionella strains carrying a plasmid expressing β-lactamase effector protein fusions were constructed as previously described (20). Legionella strains harboring a plasmid pXDC50 to express mCherry protein for Legionella growth curve were constructed in a previous study (27).
Cell Culture.
THP-1 cells were obtained from ATCC and grown in Advanced RPMI 1640 (Invitrogen) supplemented with 10% (vol/vol) FBS and 2 mM glutamine at 37 °C in a CO2 incubator. THP-1 cells were differentiated into macrophage-like cells by resuspending them into RPMI + 2 mM glutamine + 10% (vol/vol) FBS + 30 μM PMA. Following 72 h of treatment with PMA, the differentiated THP-1 cells were washed and resuspended in RPMI + 2 mM glutamine + 10% (vol/vol) FBS for infection (20).
Infection of THP-1 Cells by Legionella and Coimmunoprecipitation of LegC Proteins and VAMP4.
Coimmunoprecipitation experiments were carried out following the manual of the Pierce Crosslink IP Kit (Thermo Fisher Scientific). Legionella strains overexpressing either TEM or N-terminal TEM-tagged versions of the effector proteins LegC2, LegC3, or LegC7, were used to infect differentiated THP-1 macrophages. After 6 h of infection, 4 × 107 infected THP-1 cells were harvested and lysed on ice with lysis buffer [25 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 5% (vol/vol) glycerol, pH 7.2] in the presence of protease inhibitors (Halt Protease Inhibitor Mixture; Thermo Fisher Scientific). The supernatant was collected and precleared with 50% (wt/vol) Protein A/G agarose. The precleared supernatant was incubated overnight with Protein A/G agarose charged with 500 µg affinity-purified, polyclonal, rabbit anti-TEM antibodies (prepared at Pocono Farms). The beads were washed three times and eluted with elution buffer at pH 2. The precleared supernatants and elutions were analyzed by immunoblot analysis with anti-TEM antibody and different human R-SNARE antibodies (Fig. 1C and Fig. S1). TEM (30 kDa), TEM-LegC2 (76 kDa), TEM-LegC3 (93 kDa), and TEM-LegC7 (78 kDa) are present in the precleared supernatant as well as in eluted samples at the expected sizes.
VAMP4 Knockdown.
Escherichia coli strains carrying pCMV or pMD helper plasmids were grown in LB medium with 100 μg/mL carbenicillin. E. coli strains carrying plasmids pGIPZ (vector control) or pGIPZ shRNA for knockdown of VAMP4 (V3LHS_364980, V3LHS_364977, and V3LHS_364975), obtained from Thermo Fisher Scientific and were grown in low-salt LB medium with 100 μg/mL ampicillin and 25 μg/mL zeocin. pCMV or pMD helper plasmids were prepared with Qiagen Plasmid Maxi Kit and pGIPZ shRNA plasmids were extracted with Qiagen Plasmid Midi Kit following supplier’s protocols. Phoenix cells (a gift from Jose Silva, Columbia University, New York) were cultivated in DMEM with 10% (vol/vol) FBS and transfected with jetPEI transfection reagent (Polyplus) following supplier’s protocols. Briefly, 2 × 105 cells were seeded in a six-well plate 24 h before transfection and media were replaced by fresh DMEM with 10% (vol/vol) FBS 1 h before transfection. One microgram of pMD, 1 μg of pCMV, and 2 μg of pGIPZ shRNA or pGIPZ plasmid alone were diluted into 150 mM NaCl to a final volume of 100 μL, and 8 μL of jetPEI transfection reagent was diluted with 150 mM NaCl to a final volume of 100 μL. The plasmid solution was mixed with jetPEI transfection reagent solution briefly and further incubated at room temperature for 20 min. The mixture was added to Phoenix cells and incubated at 37 °C in a CO2 incubator. After 20 h of transfection, the medium was replaced with 2 mL fresh DMEM + 10% (vol/vol) FBS. After 48 h of transfection, 2 mL DMEM supernatant containing viral particles was collected and filtered with Millex-HA filter. To construct stable VAMP4 knockdown cell lines in THP-1 cells, 1 × 106 THP-1 cells in 1.25 mL Advanced RPMI + 2 mM glutamine + 10% (vol/vol) FBS, were seeded in a six-well plate shortly before infection. A mixture of 0.75 mL viral suspension produced in Phoenix cells as described above and 8 μg Polybrene were added to the THP-1 cells. These THP-1 cells were centrifuged at 1,000 × g for 1 h at room temperature and further incubated at 37 °C in a CO2 incubator. After 4 h of infection, THP-1 cells were spun down, washed once with Advanced RPMI + 2 mM glutamine + 10% FBS, resuspended into 3 mL Advanced RPMI + 2 mM glutamine + 10% (vol/vol) FBS, and returned to the CO2 incubator at 37 °C. After 48–60 h of infection, infected THP-1 cells were sorted using a MoFlo-HTS cell sorter (Dako Cytomation) and GFP+ THP-1 cells were collected. The GFP+ cells were grown for several generations before further experiments.
Protein Purification.
N-terminal 6× His-tagged, full-length versions of LegC2, LegC3, and LegC7 were cloned into pET15b (Novagen) vector for overexpression in the E. coli strain BL21 (DE3) and affinity purified using Ni2+-nitrilotriacetic acid (NTA) agarose (Qiagen) resin followed by thrombin cleavage to remove the 6×-His tags. The proteins were further purified by ion-exchange chromatography using the ÄKTA system (GE Healthcare) as described earlier for purification of SNAREs (54). Purity of the proteins was analyzed by SDS/PAGE and Coomassie blue staining (Fig. S4A). N-terminal 6× His-tagged, full-length VAMP4 (1–220) and α-SNAP (1–298) were overexpressed and purified as described earlier (37, 39). Recombinant NSF from Chinese hamsters was expressed and purified as described earlier (39). A cytoplasmic fragment of a cysteine mutant (S28C) of VAMP2 (Syb2, 1–96) was expressed using a pET28a vector and purified as described earlier (40) and then labeled with Oregon-Green 488 Iodoacetamide (Molecular Probes) as per manufacturer’s instructions. Soluble fragments of R-SNAREs were expressed and purified as described earlier (55). As positive control for disassembly, a truncated, ternary neuronal SNARE complex was formed by mixing Syntaxin 1A (183–288), SNAP-25 (1–206), and Oregon-Green–labeled (at position S28C) cytoplasmic domain (1–96) of Syb (VAMP2*), in the molar ratio = 1:1:1.5 and purified as described earlier (41). Purified proteins were snap frozen with liquid nitrogen and stored at −80 °C until use.
Characterization of LegC/SNARE Hybrid Complexes.
For the formation of SDS-resistant complexes, purified, full-length versions of LegC2, LegC3, and LegC7 were mixed in equal stoichiometric ratio in the presence or absence of VAMP4 and incubated overnight at 4 °C on a rotary mixer. Both samples were resolved by SDS/PAGE without prior heat denaturation, followed by Coomassie blue staining (Fig. 4B). See Fig. 4A for the individual components used in the experiment. The band of lower mobility (larger arrowhead), visible only in the presence of VAMP-4, was cut out and analyzed by nano LC/MS-MS as described earlier (56). In short, proteins were separated by 1D SDS/PAGE [4–12% (wt/vol) NuPAGE Bis⋅Tris gel; Invitrogen] followed by Coomassie staining. The desired band was sliced out as a gel piece, reduced with 10 mM DTT for 55 min at 56 °C, alkylated with 55 mM IAA (iodoacetaide) for 20 min at 26 °C, and digested with modified trypsin (Serva) overnight at 37 °C. Tryptic peptides were injected into a C18 precolumn [2.5 cm, 360 μm o.d. (outer diameter), 150 μm i.d. (inner diameter), Reprosil-Pur 120 Å, 5 μm, C18-AQ; Dr. Maisch] at a flow rate of 10 μL/min. Bound peptides were eluted and separated on a C18 capillary column (15 cm, 360 μm o.d., 75 μm i.d., Reprosil-Pur 120 Å, 3 μm, C18-AQ; Dr. Maisch) at a flow rate of 300 nL/min, with a gradient from 7.5% to 37.5% (vol/vol) ACN (acetonitrile) in 0.1% formic acid for 50 min using an Agilent 1100 nano-flow LC system (Agilent Technologies) coupled to an LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Electron). The mass spectrometer was operated in the data-dependent mode to automatically switch between MS and MS/MS acquisition. Survey MS spectra were acquired in the Orbitrap (m/z 350–1,600) with the resolution set to 30,000 at m/z 400 and automatic gain control target at 5 × 105. The eight most intense ions were sequentially isolated for CID MS/MS fragmentation and detection in the linear ion trap. Ions with single and unrecognized charge states were excluded. The peptide hits generated were searched against a local database comprising the proteins LegC2, LegC3, LegC7, and VAMP4 using Mascot (Matrix Science) and results were viewed using the Scaffold Viewer (Proteome Software).
Proteoliposome Preparation and Fusion Assay.
For the fusion assays, SUVs (average diameter of 50 nm) were prepared by detergent removal as described earlier (57). In brief, purified, full-length LegC2, LegC3, and LegC7 in 1% CHAPS were mixed (molar ratio 1:1:1) overnight at 4 °C and then added to the lipid mixture (DOPC:DOPE:DOPS:cholesterol:Oregon-Green-DHPE (1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine) mol% ratio 55:22:11:11:1 in 5% cholate), with final protein:lipid molar ratio of 1:1,000. The mixture was passed through a Sephadex G-50 column using the HP150 (20 mM Hepes, 150 mM KCl, pH 7.4) buffer to form small unilamellar proteoliposomes by detergent removal. All lipids were bought from Avanti Polar Lipids. Using the same procedure, proteoliposomes containing purified, full-length VAMP4 or VAMP2 (with 1 mol% Texas-Red-DHPE label) were prepared (final protein:lipid molar ratio 1:1,000). Liposome concentrations were quantified as total lipid content based on phospholipid concentrations measured by Fiske’s reagent (58, 59) to ensure using them in equal stoichiometric ratios for the fusion assay. Proteoliposome fusion was monitored by a lipid mixing assay based on FRET using a spectrofluorimeter (FluoroMax-2; Jobin Yvon) as described earlier (29). In short, 25 μL of Oregon-Green–labeled LegC SUVs in 1.2 mL HP150 buffer (20 mM Hepes pH 7.4, 150 mM KCl), were excited at 496 nm in a quartz cuvette; the emission was monitored at 515 nm. The emission was allowed to stabilize upon the initial drop. To start the fusion reaction, equal amounts of Texas-Red–labeled R-SNARE SUVs were added and fusion was monitored via FRET, λex = 496 nm and λem = 605 nm. Acceptor fluorescence (Ft) was normalized to the initial value (F0) (at the instant (t0) of addition) as Ft/F0 and plotted with respect to time (t). For competitive inhibition, the LegC SUVs were incubated with excess soluble SNARE domains of the R-SNAREs for 30 min, before being used for the assay. Raw data were analyzed and plotted using Origin Pro-9.0 (OriginLabs).
NSF-Mediated Disassembly of SNARE Complexes Monitored by Fluorescence Anisotropy.
Because both VAMP4 and VAMP2 proteoliposomes were able to fuse with LegC proteoliposomes (Fig. 3A) due to in vitro “promiscuity” (typical for many SNAREs), we decided to use an Oregon-Green–labeled cytoplasmic fragment of Synaptobrevin (VAMP2*) as the R-SNARE partner with neuronal Q-SNAREs or LegCs to monitor NSF-mediated disassembly of such complexes using fluorescence anisotropy. To check whether the soluble VAMP2 formed complexes with the neuronal Q-SNAREs and LegC effectors to be reconstituted on the respective SUVs, the proteoliposomes were subjected to coflotation assay by density gradient centrifugation as described earlier (60) followed by detection of the fluorescently labeled VAMP2*. Full-length LegC2, LegC3, LegC7, and Oregon-Green–labeled (at residue 28) cytoplasmic domain of Synaptobrevin (VAMP2*) were mixed overnight at 4 °C to generate LegC–VAMP2* hybrid complex. The complex was reconstituted into SUVs by detergent removal (57) using a protein:lipid ratio of 1:1,000. As control, truncated, ternary neuronal SNARE complexes (with the same VAMP2* fragment) were also reconstituted into SUVs with a protein:lipid ratio of 1:1,000. For the anisotropy experiments, we used 10 μL of proteoliposomes in 0.6 mL “disassembly buffer” (20 mM Hepes-KOH pH = 7.4, 20 mM KCH3COO, 120 mM KGlu) (30). Anisotropy of VAMP2* was measured using a FluoroLog 3 spectrometer in T configuration equipped for polarizers (model FL322; Jobin Yvon). All experiments were done at 37 °C. Disassembly was performed adding 60 nM NSF, 500 nM α-SNAP, 2 mM ATP, and 5 mM Mg+2 (39). The G factor was calculated according to G = IHV/IHH, where I is the fluorescence intensity, the first subscript letter indicates the direction of the exciting light, and the second subscript letter the direction of emitted light. The intensity of the vertically (V) and horizontally (H) polarized emission light after excitation by vertically polarized light was measured. The anisotropy (r) was determined according to r = (IVV − G × IVH)/(IVV + 2G × IVH).
Flotation Assay to Separate Soluble VAMP2* Released from NSF-Mediated Disassembly of Complexes.
The same proteoliposomes as above were incubated with α-SNAP (500 nM), NSF (60 nM), Mg+2 (5 mM), and ATP (2 mM) for 10 min at room temperature in disassembly buffer (20 mM Hepes-KOH pH = 7.4, 20 mM KCH3COO, 120 mM KGluconate). To distinguish proteins attached to liposomes from those solubilized upon disassembly, samples were subjected to flotation assay based on density gradient centrifugation using a Nycodenz gradient as described earlier (60). Fractions from the gradient were resolved by SDS/PAGE and distribution of the fluorescent VAMP2* among the fractions (in the gel) was detected (Fig. 5 B–D) using the FLA-7000 Imager (Fujifilm).
Immunoprecipitation of VAMP4 Confirms Interaction with LegC2, LegC3, and LegC7 Using Anti-VAMP4 Antibody.
Similar to the experiment shown in Fig. 1C, we used lysates of THP-1 cells that were infected with Legionella strains overexpressing either TEM or TEM-tagged versions of LegC2, LegC3, or LegC7, respectively. Precleared supernatants of the corresponding cell lysates were then incubated with anti-VAMP4 antibody immobilized on Protein A/G agarose, followed by washing and elution of the bound antigen complex as described in Materials and Methods. Starting supernatants and eluted antigen complexes were analyzed by immunoblotting using anti-VAMP4 and anti-TEM antibodies, respectively (Fig. S1). For details see Materials and Methods above and the legend to Fig. 1C.
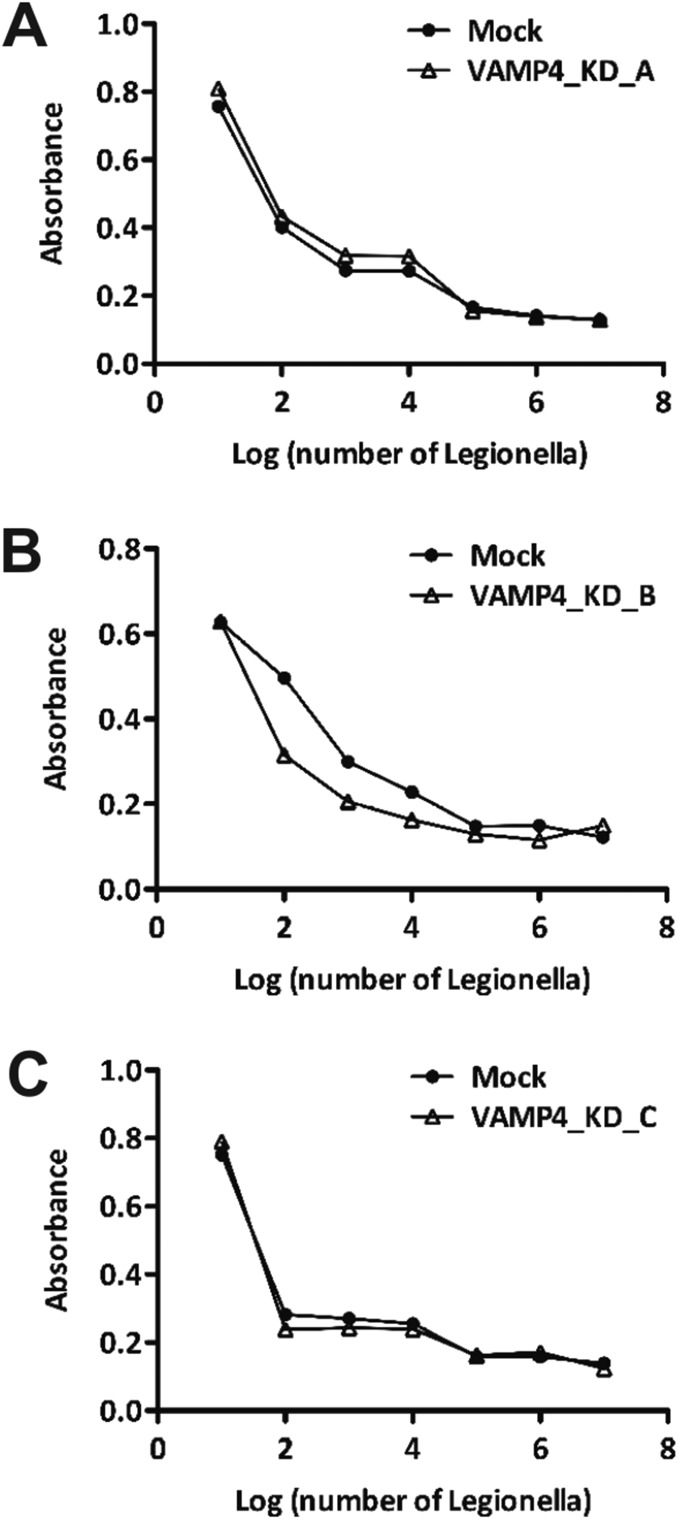
Measurement of Cell Viability of Infected Cells After Knockdown of VAMP4.
To test whether VAMP4 knockdown has a negative impact on cell viability after infection with Legionella, we infected control cells and three VAMP4-knockdown cell lines (Materials and Methods) with wild-type Legionella and measured cell viability using MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium) as reporter (61). In brief, 1 × 105 cells were plated in 96-well plates, followed by infection with 10-fold serial dilutions from 1 × 107 bacteria per well to 10 bacteria per well of L. pneumophila as described in Materials and Methods. After 120 h of infection, 20 μL of 5 mg/mL MTT was added to each well and the plate was further incubated for 4 h at 37 °C in a CO2 incubator. Afterward, the supernatant was removed by aspiration and 100 μL of isopropanol containing 40 mM HCl was added to each well to solubilize the Formazan dye. A total of 20 μL of 10% (wt/vol) SDS was added and the samples were mixed by incubating 10 min at room temperature on a shaker. A570 was measured using a plate reader (TECAN Infinite 200 PRO). The results (Fig. S2) showed no significant differences between the mock transfected and the three VAMP4 knockdown cell lines (KD_A, KD_B, and KD_C). Hence cytotoxicity can be excluded as a cause for the retarded intracellular growth of Legionella in the THP-1 macrophage cell lines with VAMP4 knockdown.
Assay of Legionella Uptake.
Legionella wild-type or dotA mutant strains overexpressing mCherry (27) were used to infect THP-1 cell lines transfected with VAMP4 knockdown or mock constructs. As a negative control, cells were treated with Cytochalasin D (an inhibitor for actin elongation, which inhibits the entry of Legionella) 30 min before infection. As a positive control, cells were incubated with anti-MOMP (major outer membrane protein) antibodies 30 min before infection. After 40 min of infection, cells were fixed and further stained with rabbit anti-MOMP antibodies and anti-rabbit Alexa Fluor 647 for staining external bacteria. The percent of entry (number of intracellular bacteria/total bacteria) was calculated and plotted (Fig. S3). There was no significant difference in the levels of uptake of Legionella into the cell lines, between the VAMP4 knockdown and mock constructs.
Acknowledgments
We thank Christine Labno and Dr. Vytas Bindokas (University of Chicago Light Microscopy Core Facility) for help with confocal imaging; Ursel Ries for technical assistance; Tobias Klöpper and Nickias Kienle for assistance with in silico analysis; Henning Urlaub and Monika Raabe for carrying out proteomic analysis of hybrid SNARE complexes; and Hartmut Sebesse for help with Fig. 6. This work was supported in part by Grant SFB 803 from the Deutsche Forschungsgemeinschaft (to R.J.) and NIH Grant AI23549 (to H.A.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608755113/-/DCSupplemental.
References
- 1.McDade JE, et al. Legionnaires’ disease: Isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med. 1977;297(22):1197–1203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- 2.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 3.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7(1):7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 4.Segal G, Purcell M, Shuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci USA. 1998;95(4):1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279(5352):873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 6.Isaac DT, Isberg R. Master manipulators: An update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future Microbiol. 2014;9(3):343–359. doi: 10.2217/fmb.13.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franco IS, Shuman HA, Charpentier X. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol. 2009;11(10):1435–1443. doi: 10.1111/j.1462-5822.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 8.Ensminger AW. Legionella pneumophila, armed to the hilt: Justifying the largest arsenal of effectors in the bacterial world. Curr Opin Microbiol. 2016;29:74–80. doi: 10.1016/j.mib.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 9.O’Connor TJ, Adepoju Y, Boyd D, Isberg RR. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci USA. 2011;108(36):14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorer MS, Kirton D, Bader JS, Isberg RR. RNA interference analysis of Legionella in Drosophila cells: Exploitation of early secretory apparatus dynamics. PLoS Pathog. 2006;2(4):e34. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Personnic N, Bärlocher K, Finsel I, Hilbi H. Subversion of retrograde trafficking by translocated pathogen effectors. Trends Microbiol. 2016;24(6):450–462. doi: 10.1016/j.tim.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Jahn R, Scheller RH. SNAREs: Engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7(9):631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 13.Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95(26):15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choy A, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338(6110):1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arasaki K, Roy CR. Legionella pneumophila promotes functional interactions between plasma membrane syntaxins and Sec22b. Traffic. 2010;11(5):587–600. doi: 10.1111/j.1600-0854.2010.01050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arasaki K, Toomre DK, Roy CR. The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE-dependent membrane fusion. Cell Host Microbe. 2012;11(1):46–57. doi: 10.1016/j.chom.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delevoye C, et al. SNARE protein mimicry by an intracellular bacterium. PLoS Pathog. 2008;4(3):e1000022. doi: 10.1371/journal.ppat.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paumet F, et al. Intracellular bacteria encode inhibitory SNARE-like proteins. PLoS One. 2009;4(10):e7375. doi: 10.1371/journal.pone.0007375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King NP, et al. Soluble NSF attachment protein receptor molecular mimicry by a Legionella pneumophila Dot/Icm effector. Cell Microbiol. 2015;17(6):767–784. doi: 10.1111/cmi.12405. [DOI] [PubMed] [Google Scholar]
- 20.de Felipe KS, et al. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 2008;4(8):e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett TL, et al. LegC3, an effector protein from Legionella pneumophila, inhibits homotypic yeast vacuole fusion in vivo and in vitro. PLoS One. 2013;8(2):e56798. doi: 10.1371/journal.pone.0056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151(6):1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Calakos N, Bennett MK, Peterson KE, Scheller RH. Protein-protein interactions contributing to the specificity of intracellular vesicular trafficking. Science. 1994;263(5150):1146–1149. doi: 10.1126/science.8108733. [DOI] [PubMed] [Google Scholar]
- 24.Steegmaier M, Klumperman J, Foletti DL, Yoo JS, Scheller RH. Vesicle-associated membrane protein 4 is implicated in trans-Golgi network vesicle trafficking. Mol Biol Cell. 1999;10(6):1957–1972. doi: 10.1091/mbc.10.6.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang T, Hong W. Ykt6 forms a SNARE complex with syntaxin 5, GS28, and Bet1 and participates in a late stage in endoplasmic reticulum-Golgi transport. J Biol Chem. 2001;276(29):27480–27487. doi: 10.1074/jbc.M102786200. [DOI] [PubMed] [Google Scholar]
- 26.Levi A, Folcher M, Jenal U, Shuman HA. Cyclic diguanylate signaling proteins control intracellular growth of Legionella pneumophila. MBio. 2011;2(1):e00316–e10. doi: 10.1128/mBio.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charpentier X, et al. Chemical genetics reveals bacterial and host cell functions critical for type IV effector translocation by Legionella pneumophila. PLoS Pathog. 2009;5(7):e1000501. doi: 10.1371/journal.ppat.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Struck DK, Hoekstra D, Pagano RE. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20(14):4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 29.van den Bogaart G, et al. One SNARE complex is sufficient for membrane fusion. Nat Struct Mol Biol. 2010;17(3):358–364. doi: 10.1038/nsmb.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsui MM, Banfield DK. Yeast Golgi SNARE interactions are promiscuous. J Cell Sci. 2000;113(Pt 1):145–152. doi: 10.1242/jcs.113.1.145. [DOI] [PubMed] [Google Scholar]
- 31.Wendler F, Tooze S. Syntaxin 6: The promiscuous behaviour of a SNARE protein. Traffic. 2001;2(9):606–611. doi: 10.1034/j.1600-0854.2001.20903.x. [DOI] [PubMed] [Google Scholar]
- 32.Hohenstein AC, Roche PA. SNAP-29 is a promiscuous syntaxin-binding SNARE. Biochem Biophys Res Commun. 2001;285(2):167–171. doi: 10.1006/bbrc.2001.5141. [DOI] [PubMed] [Google Scholar]
- 33.Kweon Y, Rothe A, Conibear E, Stevens TH. Ykt6p is a multifunctional yeast R-SNARE that is required for multiple membrane transport pathways to the vacuole. Mol Biol Cell. 2003;14(5):1868–1881. doi: 10.1091/mbc.E02-10-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bethani I, et al. The specificity of SNARE pairing in biological membranes is mediated by both proof-reading and spatial segregation. EMBO J. 2007;26(17):3981–3992. doi: 10.1038/sj.emboj.7601820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang B, et al. SNARE interactions are not selective. Implications for membrane fusion specificity. J Biol Chem. 1999;274(9):5649–5653. doi: 10.1074/jbc.274.9.5649. [DOI] [PubMed] [Google Scholar]
- 36.Fasshauer D, Antonin W, Margittai M, Pabst S, Jahn R. Mixed and non-cognate SNARE complexes. Characterization of assembly and biophysical properties. J Biol Chem. 1999;274(22):15440–15446. doi: 10.1074/jbc.274.22.15440. [DOI] [PubMed] [Google Scholar]
- 37.Brandhorst D, et al. Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity. Proc Natl Acad Sci USA. 2006;103(8):2701–2706. doi: 10.1073/pnas.0511138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi T, et al. Synaptic vesicle membrane fusion complex: Action of clostridial neurotoxins on assembly. EMBO J. 1994;13(21):5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winter U, Chen X, Fasshauer D. A conserved membrane attachment site in alpha-SNAP facilitates N-ethylmaleimide-sensitive factor (NSF)-driven SNARE complex disassembly. J Biol Chem. 2009;284(46):31817–31826. doi: 10.1074/jbc.M109.045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fasshauer D, Eliason WK, Brünger AT, Jahn R. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry. 1998;37(29):10354–10362. doi: 10.1021/bi980542h. [DOI] [PubMed] [Google Scholar]
- 41.Pobbati AV, Stein A, Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313(5787):673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 42.Schuette CG, et al. Determinants of liposome fusion mediated by synaptic SNARE proteins. Proc Natl Acad Sci USA. 2004;101(9):2858–2863. doi: 10.1073/pnas.0400044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kloepper TH, Kienle CN, Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol Biol Cell. 2007;18(9):3463–3471. doi: 10.1091/mbc.E07-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chernomordik LV, Zimmerberg J, Kozlov MM. Membranes of the world unite! J Cell Biol. 2006;175(2):201–207. doi: 10.1083/jcb.200607083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490(7419):201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan YH, van Lengerich B, Boxer SG. Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides. Proc Natl Acad Sci USA. 2009;106(4):979–984. doi: 10.1073/pnas.0812356106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lygina AS, Meyenberg K, Jahn R, Diederichsen U. Transmembrane domain peptide/peptide nucleic acid hybrid as a model of a SNARE protein in vesicle fusion. Angew Chem Int Ed Engl. 2011;50(37):8597–8601. doi: 10.1002/anie.201101951. [DOI] [PubMed] [Google Scholar]
- 48.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252(5009):1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 49.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2: A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livingstone CD, Barton GJ. Protein sequence alignments: A strategy for the hierarchical analysis of residue conservation. Comput Appl Biosci. 1993;9(6):745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 52.Letunic I, Doerks T, Bork P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40(Database issue):D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadosky AB, Wiater LA, Shuman HA. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun. 1993;61(12):5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460(7254):525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Antonin W, Fasshauer D, Becker S, Jahn R, Schneider TR. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat Struct Biol. 2002;9(2):107–111. doi: 10.1038/nsb746. [DOI] [PubMed] [Google Scholar]
- 56.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 57.Hernandez JM, Kreutzberger AJ, Kiessling V, Tamm LK, Jahn R. Variable cooperativity in SNARE-mediated membrane fusion. Proc Natl Acad Sci USA. 2014;111(33):12037–12042. doi: 10.1073/pnas.1407435111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fiske CH, Subbarow Y. The colorimetric determination of phosphorus. J Biol Chem. 1925;66:375–400. [Google Scholar]
- 59.Böttcher CJF, Pries C, van Gent CM. A rapid and sensitive colorimetric microdetermination of free and bound choline. Recl Trav Chim. 1961;80:1169–1178. [Google Scholar]
- 60.Hernandez JM, et al. Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science. 2012;336(6088):1581–1584. doi: 10.1126/science.1221976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marra A, Horwitz MA, Shuman HA. The HL-60 model for the interaction of human macrophages with the Legionnaires’ disease bacterium. J Immunol. 1990;144(7):2738–2744. [PubMed] [Google Scholar]