Significance
The last steps of energy conversion in aerobic eukaryotes take place in cellular organelles called mitochondria. The process involves a number of protein complexes, referred to as the respiratory chain, that reside in the inner mitochondrial membrane. This chain shuttles electrons, which originate from degradation of foodstuff, to the acceptor dioxygen. The process is regulated to optimize the energy supply, depending on requirements and external factors. We found that two small proteins that have been found to associate with the respiratory–enzyme complexes regulate the electron flux at the last two components of the chain. Furthermore, we identified the regulatory mechanism at the molecular level.
Keywords: cytochrome c oxidase, electron transfer, membrane protein, cytochrome aa3, cytochrome bc1
Abstract
The respiratory supercomplex factors (Rcf) 1 and 2 mediate supramolecular interactions between mitochondrial complexes III (ubiquinol-cytochrome c reductase; cyt. bc1) and IV (cytochrome c oxidase; CytcO). In addition, removal of these polypeptides results in decreased activity of CytcO, but not of cyt. bc1. In the present study, we have investigated the kinetics of ligand binding, the single-turnover reaction of CytcO with O2, and the linked cyt. bc1-CytcO quinol oxidation-oxygen-reduction activities in mitochondria in which Rcf1 or Rcf2 were removed genetically (strains rcf1Δ and rcf2Δ, respectively). The data show that in the rcf1Δ and rcf2Δ strains, in a significant fraction of the population, ligand binding occurs over a time scale that is ∼100-fold faster (τ ≅ 100 μs) than observed with the wild-type mitochondria (τ ≅ 10 ms), indicating structural changes. This effect is specific to removal of Rcf and not dissociation of the cyt. bc1–CytcO supercomplex. Furthermore, in the rcf1Δ and rcf2Δ strains, the single-turnover reaction of CytcO with O2 was incomplete. This observation indicates that the lower activity of CytcO is caused by a fraction of inactive CytcO rather than decreased CytcO activity of the entire population. Furthermore, the data suggest that the Rcf1 polypeptide mediates formation of an electron-transfer bridge from cyt. bc1 to CytcO via a tightly bound cyt. c. We discuss the significance of the proposed regulatory mechanism of Rcf1 and Rcf2 in the context of supramolecular interactions between cyt. bc1 and CytcO.
The respiratory chain in mitochondria drives formation and maintenance of a proton electrochemical gradient across the inner membrane. The free energy stored in this gradient is used, for example, to generate ATP from ADP in the F1Fo-ATP-synthase or to drive transmembrane transport. In mammalian mitochondria, the first component of the respiratory chain is the integral membrane-bound complex I (or type I NADH dehydrogenase), which receives electrons from NADH. In Saccharomyces cerevisiae, complex I is replaced by type II NADH dehydrogenases, which are peripheral dimeric membrane proteins (1, 2). In both cases, the electrons are used for reduction of quinone to quinol, which diffuses within the membrane to donate electrons to complex III [ubiquinol-cytochrome c reductase (cyt. bc1)]. Cyt. bc1 reduces water-soluble cyt. c, which is the electron donor to complex IV [cytochrome c oxidase (CytcO)]. CytcO catalyzes oxidation of four molecules of cyt. c, which is linked to reduction of molecular oxygen to water and proton pumping across the membrane (e.g., reviewed in ref. 3).
The catalytically active core of CytcO is composed of the mitochondrially encoded subunits I–III, which are largely conserved across the family of homologous prokaryotic and eukaryotic oxidases. The mitochondrial CytcOs are typically composed of a number of additional, smaller subunits that are encoded in the nucleus in S. cerevisiae (eight additional subunits; Fig. 1) and Bos taurus (10 additional subunits). In S. cerevisiae, CytcO associates with cyt. bc1, forming supercomplexes (4–12) composed of a cyt. bc1 dimer bound to either one or two CytcOs. These supercomplexes are stabilized by cardiolipin (13–16). In recent years, two additional polypeptides, the respiratory supercomplex factors 1 and 2 (Rcf1 and Rcf2, respectively), were shown to associate with CytcO and cyt. bc1 (8, 17–20), thereby stabilizing the cyt. bc1–CytcO interactions.
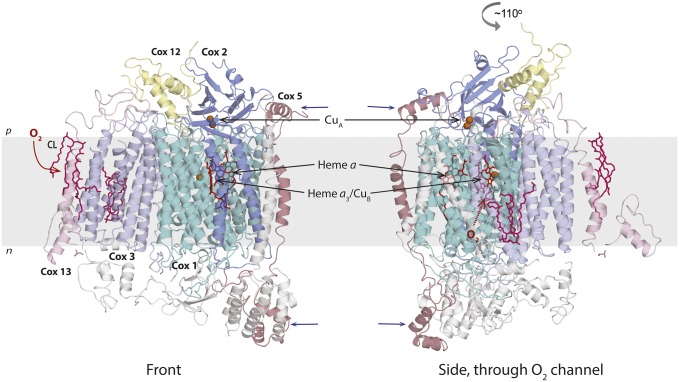
Fig. 1.
Structural model of the S. cerevisiae CytcO. The core subunits I–III (Cox1–Cox3) (in cyan, marine, and light blue, respectively) are shown. The accessory subunits are shown in gray, except for Cox12 (VIb, yellow), Cox13 (VIa, pink), and Cox5 (IV, light red) (the roman numerals refer to the numbering of the CytcO from bovine heart). Cardiolipin (CL) is shown in dark pink. Cox2 holds CuA, the entry point for electrons from cyt. c. From CuA, electrons are transferred via heme a to the catalytic site composed of heme a3 and CuB in Cox1. Results from functional studies indicate that Cox12 and Cox13 (as well as Cox3) interact with Rcf1 and that Rcf1 mediates the cyt. bc1–CytcO interactions (main text). However, the latter is not consistent with structural models indicating that the cyt. bc1–CytcO interactions are seen near Cox5 (blue arrows; Discussion). The O2 entry channel is indicated (Left) and is seen as a cavity (Right). The Protein Data Base structural model is from Maréchal et al. (22). The figure was prepared using the program Pymol (57).
Although the Rcf2 polypeptide is conserved only among fungi, Rcf1 orthologs are conserved in prokaryotes and eukaryotes. The role of the Rcf polypeptides in supercomplex formation at a molecular level is not yet fully understood. Results from recent studies indicate that in the absence of Rcf1, interactions required for stability of the CytcO–cyt. bc1 supercomplex are disrupted (17–19). In S. cerevisiae, the Rcf1 polypeptide is not required for correct assembly of the core subunits of CytcO itself (or the cyt. bc1 complex), although effects on assembly of the CytcO subunits Cox12 and Cox13 (Fig. 1) were noted, which may be linked to a diminished stability of the cyt. bc1–CytcO supercomplex (18) [but in Podaspora anserina, Rcf1 appears to be important for CytcO assembly (20)]. Effects of Rcf2 deficiency are less defined. The polypeptide also has a role in stabilizing the supramolecular interactions between CytcO and cyt. bc1, but the effects of removal of Rcf2 are less pronounced than the effects of removing Rcf1. However, when both Rcf1 and Rcf2 were deleted, the fraction of CytcO and cyt. bc1 in supercomplexes was significantly smaller than seen upon removal of Rcf1 alone (18). In Fig. 1, we have indicated the approximate, putative interaction surface between CytcO and cyt. bc1, which is based on structural modeling (7, 9).
On a functional level, removal of Rcf1 alone resulted in reduced activity of CytcO (17–19), which could be fully restored upon expression of His-tagged Rcf1 in a S. cerevisiae strain lacking Rcf1 (18). Interestingly, deletion of Rcf2 alone had essentially no effect on CytcO turnover (17, 18), even though it resulted in a significant increase in levels of incompletely reduced dioxygen (17). Furthermore, removal of both Rcf1 and Rcf2 resulted in a larger decrease of CytcO activity than removal of Rcf1 alone (18), indicating overlapping functions of the two Rcf polypeptides.
Crystal structures of the mitochondrial CytcO from B. taurus as well as from several bacteria have been determined at atomic resolution (reviewed in ref. 21), and a homology model of the S. cerevisiae CytcO has also been published (22). Early spectroscopic studies and the more recent CytcO structures show that all these enzymes harbor the same four redox-active metal sites. The water-soluble cyt. c docks near CuA, which is bound in subunit II. After reduction of CuA, the electron is transferred consecutively to the intermediate electron acceptor, heme a, and the catalytic site composed of heme a3 and CuB, all bound in subunit I (the structure and function of CytcOs are reviewed in refs. 21 and 23–29).
Rcf1 is very similar to the protein products of members of the hypoxia-induced gene 1 (Hig1) family. Because the Hig1 polypeptides are expressed at low oxygen tensions, members of this protein family may modulate oxygen binding to the CytcO catalytic site. Such a regulatory role is supported by the observation that the Rcf1 polypeptide interacts with Cox3, Cox12, and Cox13 (17–19) (i.e., near the O2 channel in CytcO). However, these three subunits are located at a distance from the cyt. bc1–CytcO interaction surface in the structural models of the supercomplex (7, 9) (Fig. 1), which is not compatible with the role of Rcf1 in stabilizing the supramolecular interactions. To address the functional role of the Rcf polypeptides in S. cerevisiae, we investigated the quinol oxidation/O2-reduction activity of the cyt. bc1 and CytcO complexes, as well as the kinetics of ligand binding and the single-turnover reaction of CytcO with O2. All measurements were performed with inner mitochondrial membranes, either wild type or mutants in which Rcf1 (rcf1Δ), Rcf2 (rcf2Δ), or the cyt. bc1 complex (qcr8Δ) was removed genetically. The data suggest a mechanism by which the Rcf polypeptides regulate the activity of the respiratory chain in S. cerevisiae.
Results
Optical Absorbance Spectra.
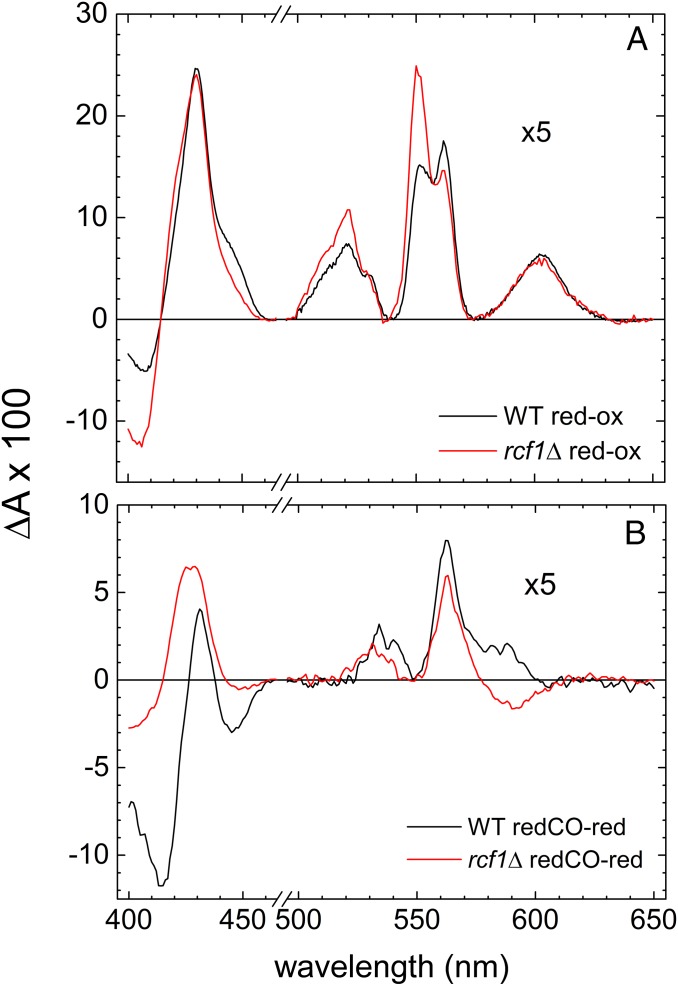
As seen in Fig. 2A, the absorbance difference spectrum (reduced minus oxidized) of wild-type mitochondria (with the outer membrane removed; Materials and Methods) shows the characteristics of CytcO and cyt. bc1: a peak at 605 nm (mainly heme a); a shoulder at 445 nm (about equal contributions from hemes a and a3); peaks at ∼560 nm and 430 nm (hemes b); and a peak at 550 nm, a shoulder at ∼420 nm, and a trough at ∼410 nm (hemes c). For the rcf1Δ mitochondria, the peaks originating from hemes b and c were larger, indicating larger amounts of cyt. bc1 relative to CytcO in this sample (the spectra were normalized to the heme a peak at 605 nm). In particular, we noted that the relative amount of cyt. c was significantly increased in the rcf1Δ sample (Fig. 2A). In the Soret region, the shoulder at 445 nm is smaller in rcf1Δ mitochondria but larger at 420 nm, which indicates that a fraction of heme a3 remains oxidized after addition of the reductant. After reduction of the mitochondria, the reduced form of CytcO binds CO to heme a3 (the “as-isolated” oxidized form does not bind CO). The reduced CO-bound minus reduced difference spectrum is shown in Fig. 2B. Here, the spectrum of the wild-type mitochondria shows the characteristic features of CO binding to reduced heme a3 in CytcO: peaks at 430 nm and 590 nm and a trough at 445 nm. In the difference spectrum of rcf1Δ mitochondria, the “430-nm peak” is larger and slightly blue-shifted. The trough at 445 nm (arising from subtraction of the peak at 445 nm in the reduced minus oxidized spectrum) is very small in rcf1Δ, consistent with the lack of a shoulder at 445 nm in Fig. 2A. Furthermore, with the rcf1Δ mitochondria, at 590 nm, the expected peak is replaced by a trough, which is explained by further reduction of the cyt. bc1 complex upon addition of CO.
Fig. 2.
Optical absorbance difference spectra. (A) Reduced (red) minus oxidized (ox) difference spectra of wild-type and rcf1Δ mitochondria, where the two spectra have been normalized to the absorbance at 605 nm (the rcf1Δ data were multiplied by a factor of 2). (B) Difference spectra of reduced CO-bound minus reduced mitochondria. In both panels, the data in the range 515–650 nm have been multiplied by a factor of 5 for clarity. The samples were prepared in a buffer of 20 mM Hepes (pH 7.4) and 60 mM sorbitol, reduced with 100 μM sodium dithionite (final concentration). The CO pressure in B was ∼130 kPa (∼1.3 mM).
Ligand Binding.
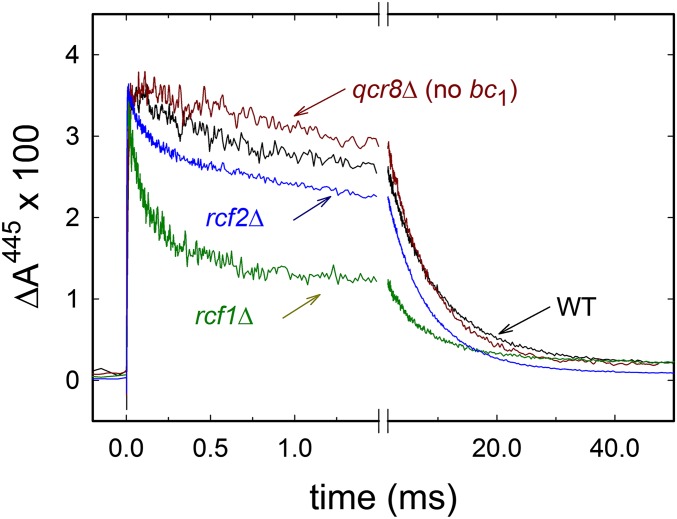
Mitochondria with added excess ascorbate and the electron mediator TMPD were incubated under an atmosphere of pure CO (∼1.3 mM), which binds to the reduced heme a3 at the catalytic site of CytcO, seen as an absorbance decrease at 445 nm (Fig. 2B) (in principle, CO may also bind to other heme proteins; Discussion). Upon illumination of the sample with a short laser flash, the ligand dissociates and rebinds, which results in absorbance changes that were measured as a function of time (Fig. 3). The increase in absorbance at zero time at 445 nm is the result of CO dissociation, presumably from heme a3, whereas the following decrease in absorbance is associated with rebinding of the CO ligand. With the wild-type mitochondria, we observed a main component with a time constant of 8.9 ± 0.9 ms (SD, n = 6), which is similar to the time constant measured with detergent-solubilized CytcOs (30, 31) and whole cells (32). In addition, we observed a small component with a time constant of 270 ± 130 μs (n = 6; the large error is a consequence of the very small amplitude) and amplitude of 15 ± 5% (n = 6) of the total absorbance change at 445 nm (Fig. 3). The time constants are summarized in Table 1.
Fig. 3.
Absorbance changes associated with flash-induced CO dissociation and recombination. The sample was illuminated by a laser flash at t = 0, which results in an increase in absorbance associated with CO dissociation. The following decrease is associated with CO recombination. With the wild-type and qcr8Δ strains, mainly one component was observed with a time constant of ∼8.9 ms (in addition, there was a small component with a time constant of ∼270 μs; main text). With the rcf1Δ and rcf2Δ mitochondria, the slow component displayed a time constant of ∼6.5 ms, whereas the significantly larger rapid component displayed a time constant of ∼110 μs. All data were normalized to yield the same CO-dissociation absorbance change at t = 0. A laser artifact at t = 0 has been truncated. Experimental conditions: The samples were prepared in a buffer of 20 mM Hepes (pH 7.4) and 60 mM sorbitol and reduced with 1 μM phenylmethylsulfonyl (PMS) and 4 mM ascorbate (final concentrations) in an atmosphere of ∼130 kPa CO (∼1.3 mM).
Table 1.
Summary of the activities as well as time and rate constants of the reactions studied in this work
| Mitochondria | CO ligand-binding time constant, ms (fraction, %) | CytcO activity, e−⋅s−1 | Cyt. bc1 activity, e−⋅s−1 | Coupled cyt. bc1-CytcO activity,* e−⋅s−1 | ||||
| Rapid | Slow | S.c. cyt. c | Horse cyt. c | Horse cyt. c (+KCN) | S.c. cyt. c | Horse cyt. c | ||
| Wild type | 0.27 (15) | 8.9 | 230 | 34 | 50 | 70 | 130 | 170 |
| rcf1Δ | 0.11 (55) | 6.5 | 70 | 34 | 53 | 65 | 52 | 102 |
| rcf2Δ | 0.11 (28) | 6.5 | 184 | n.d. | n.d. | n.d. | n.d. | n.d. |
Errors are given in the main text. n.d., not determined; S.c.; S. cerevisiae.
Coupled activity refers to measurements of the O2-consumption rate upon addition of excess reduced DQH2.
The signal obtained with mitochondria from rcf1Δ cells showed an increase in the relative amplitude of the rapid component to 55 ± 2% (n = 4) of the total absorbance change at 445 nm, but the time constants of the two components were similar to the time constants measured with the wild-type mitochondria [110 ± 20 μs and 6.5 ± 0.8 ms (n = 4), respectively]. In the absence of Rcf2, the fraction of rapid component was larger (∼28%) than with the wild-type mitochondria, but smaller than in the rcf1Δ mitochondria (Fig. 3).
As noted above, results from earlier studies suggest that removal of the Rcf1 polypeptide may destabilize the CytcO–cyt. bc1 supercomplex (17–19). To investigate if the rapid component is a result of removal of the Rcf1 polypeptide or an indirect effect of dissociation of the supercomplex, we also measured the CO ligand binding to mitochondria extracted from a S. cerevisiae strain in which the cyt. bc1 complex was removed genetically (qcr8Δ). The variant displayed similar behavior to the wild-type S. cerevisiae mitochondria (Fig. 3), which indicates that the effects seen in strains rcf1Δ and rcf2Δ are specific to interactions of Rcf with the CytcO and not disruption of the CytcO–cyt. bc1 supercomplex.
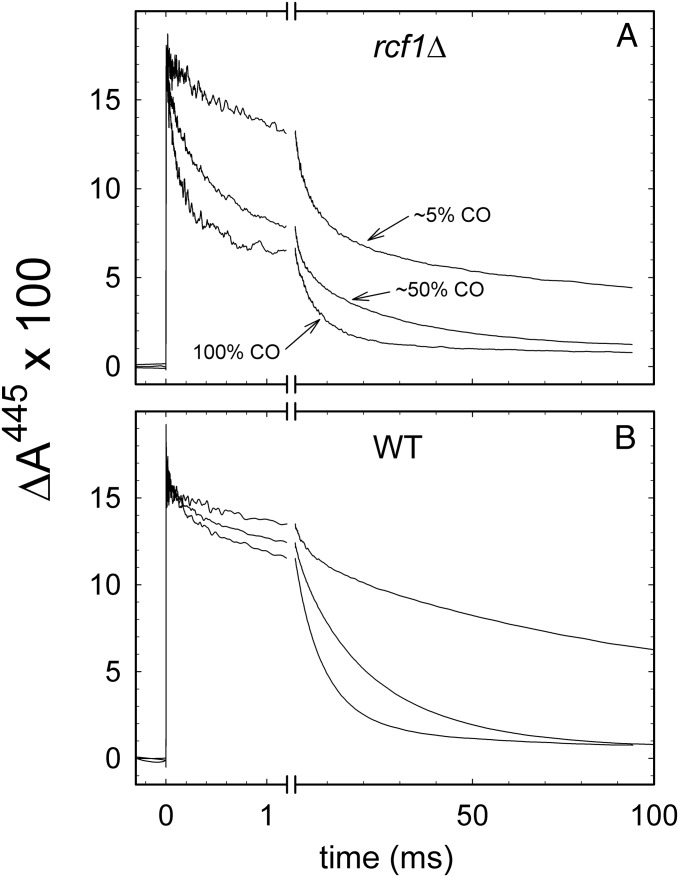
The faster component (τ ≅ 110 μs for rcf1Δ) could, in principle, be associated with events other than CO recombination [e.g., a structural relaxation as a consequence of illumination (33)]. To test whether or not both components are associated with CO recombination, we repeated the measurements with different concentrations of CO. If, after flash-induced dissociation, CO leaves the enzyme and equilibrates with the surrounding bulk solution before rebinding, this rebinding is expected to be dependent on the bulk CO concentration. As seen in Fig. 4, both kinetic components displayed a CO-concentration dependence, which indicates that they are associated with rebinding of CO from the bulk solution after flash-induced dissociation.
Fig. 4.
Kinetics of CO recombination at lower CO concentrations. (A) The 100% plot is the same as shown for rcf1Δ in Fig. 3 (but normalized to the other traces). The CO partial pressure was lowered to ∼5% and ∼50%, where the remaining part of the gas phase was replaced by N2. (B) Data with the wild-type mitochondria for approximately the same CO concentrations as in A. The CO concentration was only approximately determined by changing the partial pressures of CO and N2, respectively, and was estimated from the time constant of the slow component. The data show that both the rapid and slow components are dependent on the CO concentration in bulk water. Experimental conditions were the same as in Fig. 3, except for the different CO concentrations.
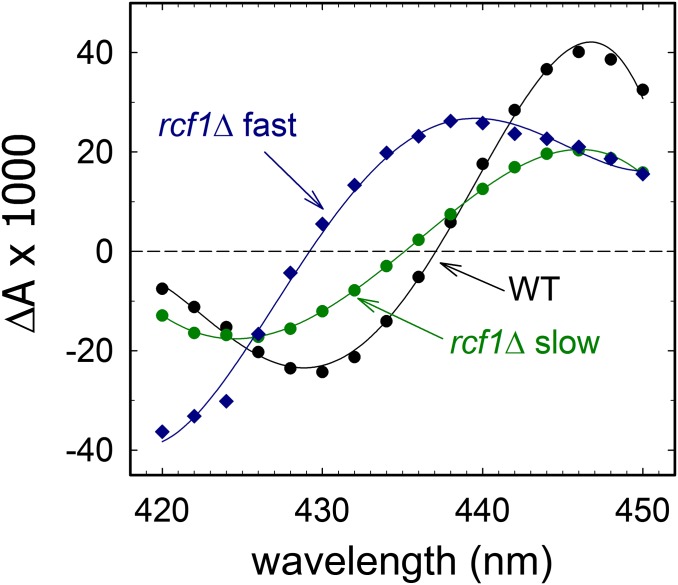
Next, we repeated the measurements at a number of wavelengths in the range of 420–450 nm (the signal-to-noise ratio with mitochondria was too small to perform measurements in the alpha region of the spectrum). Fig. 5 shows the relative amplitudes of the slow component for the wild-type mitochondria and both components for rcf1Δ mitochondria as a function of wavelength (i.e., a kinetic difference spectrum). The kinetic difference spectra of the slow components with the wild-type and rcf1Δ mitochondria were similar. As noted above, the time constant of the slower component in the CO recombination was similar to the time constant observed with pure CytcOs, and its difference spectrum agrees well with CO recombination to heme a3 (e.g., ref. 32). However, a comparison of the difference spectra of the slow (τ ≅ 6.5 ms) and fast (τ ≅ 110 μs) components from the rcf1Δ mitochondria shows that they were not the same. In other words, the fast component reflects either CO binding to CytcO in a different structural state or CO binding to a site other than heme a3.
Fig. 5.
Kinetic difference spectra. Each point in the difference spectra is the total absorbance change at a specific wavelength for each of the two components (only the main, slow component for the wild-type mitochondria is shown) in the CO recombination (compare data at 445 nm in Fig. 3). The solid lines are added as a guide for the eye. Conditions were the same as in Fig. 3.
Activity of CytcO.
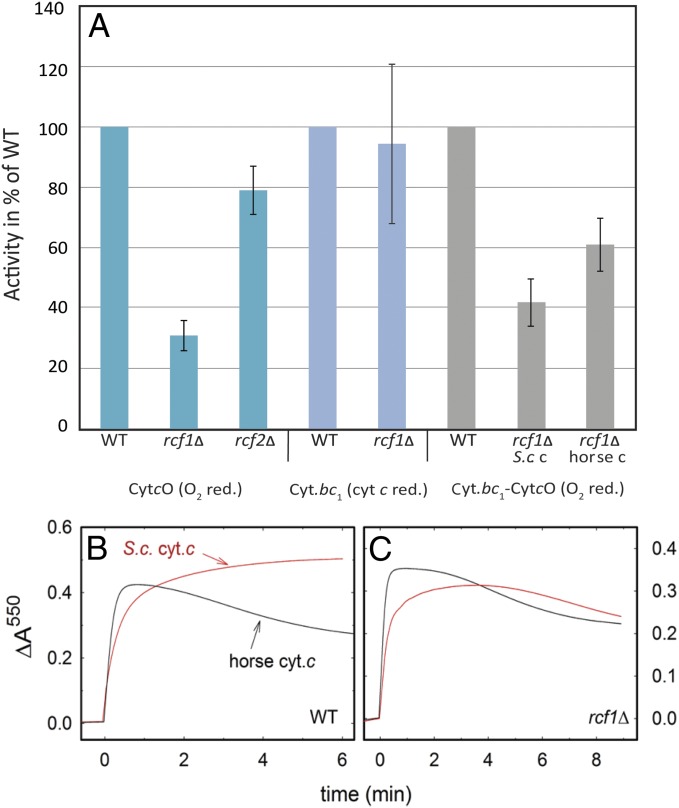
Using ascorbate, TMPD, and cyt. c as electron donors, we measured the O2-reduction activity, which reflects the CytcO activity. As seen in Fig. 6A (also Table 1), these activities for the rcf1Δ and rcf2Δ mitochondria were 31 ± 5% and 80 ± 8% (SD, n = 3), respectively, of the activities measured with wild-type mitochondria, consistent with data obtained previously (17–19). Using horse heart cyt. c as an electron donor/mediator, a typical preparation of wild-type mitochondria displayed an activity of 230 ± 20 e−⋅s−1 (SD, n = 3), normalized to the total CytcO concentration (legend for Fig. 6A). We also investigated inhibition of the wild-type and rcf1Δ CytcO by potassium cyanide (KCN) and did not observe any significant differences in the remaining residual activities after addition of KCN (∼10 s−1, measured using an O2 electrode).
Fig. 6.
Steady-state activities with whole mitochondria. (A) Activity of CytcO: CytcO activity was measured upon addition of ascorbate (5 mM), TMPD (0.5 mM), and horse-heart cyt. c (20 μM) and then by following in time O2 consumption (starting concentration ∼270 μM O2) using a Clark-type oxygen electrode in 20 mM Hepes (pH 7.4), 60 mM sorbitol, and 0.1 mM EDTA at room temperature. The activity was normalized to the CytcO concentration (Materials and Methods), typically in the range of 2–20 nM. Activity of cyt. bc1: Activity of the cyt. bc1 complex was measured by following in time absorbance changes at 550 nm (reduction of cyt. c) upon addition of DQH2 (as shown in B and C). The activities were determined from the initial slopes of the 550-nm absorbance changes. The rate of uncatalyzed reduction of cyt. c by DQH2 was typically 6–7% of the cyt. bc1-catalyzed rate (this value was subtracted). The same cyt. bc1 activities were obtained in the presence of KCN, which blocks CytcO. The activities were normalized to the concentration of cyt. bc1 in each sample, which was determined as described in Materials and Methods (typically ∼100 nM). Conditions were as follows: 40 μM cyt. c, 100 μM DQH2, 1 mM KCN (when present), and 20 mM Hepes (pH 7.4) at room temperature. Coupled cyt. bc1-CytcO activity: Coupled activity was measured after addition of 100 μM DQH2 and cyt. c from either horse heart or S. cerevisiae in the presence of O2 (∼270 μM). The O2 reduction was followed in time using a Clark-type oxygen electrode and normalized to the total concentration of CytcO. Conditions were as follows: 20 μM cyt. c, 20 mM Hepes (pH 7.4), 60 mM sorbitol, and 0.1 mM EDTA at room temperature. The reduction levels of cyt. c from horse heart (B) and S. cerevisiae (C), measured at 550 nm upon addition of DQH2 to wild-type and rcf1Δ mitochondria in the presence of O2, are shown. The increase in absorbance at 550 nm is associated with reduction of cyt. c by the cyt. bc1 complex, whereas a decrease is associated with oxidation by CytcO. The concentrations of cyt. bc1 were 16 nM and 15 nM for rcf1Δ and wild type, respectively, and the CytcO concentrations were ∼7.5 nM in both strains. Conditions were as follows: 40 μM cyt. c, 100 μM DQH2, 20 mM Hepes (pH 7.4), and 60 mM sorbitol.
Activity of the Cyt. bc1 Complex.
The Rcf1 polypeptide was shown earlier to associate not only with CytcO but also, to a minor extent, with the cyt. bc1 complex (17–19). Consequently, we also measured the cyt. bc1 activities in the wild-type and rcf1Δ mitochondria by following in time absorbance changes at 550 nm (the initial slope), associated with reduction of horse heart cyt. c upon addition of the electron donor decylubiquinol (DQH2) (Fig. 6). When normalized to the cyt. bc1 concentration, the activities were 50 ± 14 e−⋅s−1 and 53 ± 13 e−⋅s−1 (SD, n = 6) for the wild-type and rcf1Δ mitochondria, respectively. In other words, these activities were about the same, which is also consistent with the data obtained previously (17–19). We noted that for both wild-type and rcf1Δ mitochondria, the cyt. bc1 activities were lower when measured with S. cerevisiae cyt. c (68 ± 14% and 66 ± 12% of the cyt. bc1 activities obtained with horse heart cyt. c, respectively). The activities obtained in the presence of KCN, which blocks CytcO, were ∼70 e−⋅s− and 65 e−⋅s− for the wild-type and rcf1Δ mitochondria, respectively (i.e., similar to those activities obtained in the absence of KCN).
The Coupled Quinol Oxidation/O2-Reduction Activity by Cyt. bc1-CytcO.
Next, we combined the two experiments above and followed in time reduction of O2 upon addition of DQH2, which reflects the coupled activity of cyt. bc1 and CytcO. In this reaction, electrons are transferred from DQH2 via cyt. bc1 to cyt. c and then from cyt. c via CytcO to O2. Compared with the artificial electron donors (ascorbate and TMPD), the rates in the cyt. bc1-coupled system were slightly lower (∼75%), indicating that the electron transfer within or from cyt. bc1 to cyt. c was at least partly rate-limiting. As seen in Fig. 6A, for the rcf1Δ mitochondria, the activities were ∼60% (horse heart cyt. c) and ∼40% (S. cerevisiae cyt. c) of the activities obtained with wild-type mitochondria, which reflects the lower activity of CytcO in rcf1Δ mitochondria. However, the observed effect of Rcf1 removal in the coupled assay was smaller than when measuring only the CytcO activity, presumably because the rate-limiting step(s) is not only associated with CytcO itself. In addition, the rcf1Δ mitochondria contain more cyt. bc1 than CytcO (Fig. 2A), which would increase the electron flux via cyt. c into the CytcO in rcf1Δ mitochondria.
For the wild-type mitochondria, the coupled cyt. bc1-CytcO activities were 170 ± 24 e−⋅s− (n = 4) and 130 ± 11 e−⋅s− (n = 3) for cyt. c from horse heart and S. cerevisiae cyt. c, respectively (normalized to the CytcO concentration in the membrane). We note that these coupled cyt. bc1-CytcO activities were higher than the activities of the cyt. bc1 complex alone, which presumably is due to the higher concentration of cyt. bc1 than CytcO in the mitochondrial membrane. In other words, more than one cyt. bc1 complex per CytcO contributes to the overall reduction rate of cyt. c.
Next, we measured absorbance changes at 550 nm upon addition of DQH2 to a solution of wild-type and rcf1Δ mitochondria in the presence of O2, with cyt. c from either S. cerevisiae (red traces) or horse heart (black traces) as an electron mediator (Fig. 6 B and C). The initial increase in absorbance is associated with reduction of cyt. c by the cyt. bc1 complex, whereas the following decrease in absorbance is associated with reoxidation of cyt. c by CytcO. As indicated above, this increase, when normalized to the total amount of cyt. bc1 complex, displayed about the same slopes with the rcf1Δ as with the wild-type strains, reflecting similar activities of the cyt. bc1 complex in the two strains. The most significant differences were observed upon comparison of the oxidation kinetics of cyt. c by CytcO. Fig. 6B shows data obtained with the wild-type mitochondria. Although horse heart cyt. c was reoxidized by CytcO after the initial rapid reduction by cyt. bc1, S. cerevisiae cyt. c was first rapidly reduced, but the reduction level then continued to increase slowly over the time scale of measurement. Fig. 6C shows data from the same experiment obtained with the rcf1Δ mitochondria. Here, the behavior observed for the two cyt. c species was similar; the absorbance first increased and then slowly decreased again. We note in particular the significant difference in the traces obtained with the S. cerevisiae cyt. c for the wild-type and rcf1Δ mitochondria; although the activity of the CytcO in wild-type mitochondria was larger than in the rcf1Δ mitochondria, the cyt. c reoxidation rate was larger in the latter. Such a behavior can be explained by binding of at least one S. cerevisiae cyt. c (but not of horse heart cyt. c) at a site where it can mediate direct electron transfer from cyt. bc1 to CytcO in the wild-type mitochondria, but not in the rcf1Δ mitochondria (Discussion).
Time-Resolved Single-Turnover Reaction of Reduced CytcO with O2.
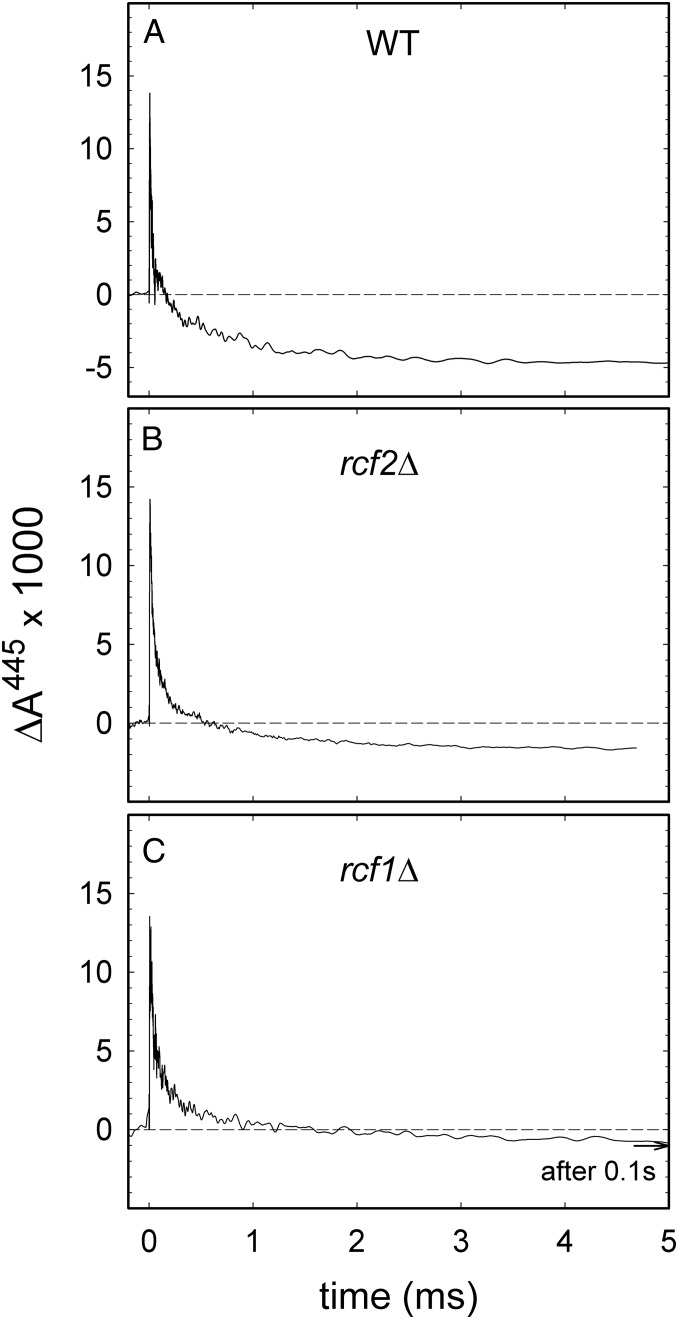
The above-discussed data indicate that the overall activity of CytcO is significantly lower in rcf1Δ than in the wild-type mitochondria. However, results from measurements of the turnover activity cannot discriminate between a situation where the entire CytcO population displays a lower activity and a situation where a fraction of CytcO is inactive (or significantly less active), whereas a fraction is fully active. In an attempt to resolve this issue, we studied the single-turnover reaction of CytcO with O2 using the so-called flow-flash technique. In the past, this approach has been used to investigate the kinetics of specific steps of the reaction of reduced CytcO with O2 (reviewed in refs. 34 and 35). When using this technique, the CytcO is fully reduced, after which the sample is incubated under an atmosphere of CO. This CytcO–CO complex is then rapidly mixed with an O2-saturated solution, after which the CO ligand is dissociated by a short laser flash (∼10 ns) that displaces the CO ligand and allows O2 to bind at the catalytic site of CytcO. The reaction of the reduced CytcO with O2 is then followed in time with microsecond time resolution by monitoring absorbance changes at wavelengths that are specific (e.g., to redox changes of the hemes). In earlier studies, primarily with the detergent-solubilized enzyme, the reaction of reduced CytcO with O2 was investigated in detail using several spectroscopic techniques, which offered detailed information on the sequence of electron- and proton-transfer reactions (reviewed in refs. 34 and 35). With the detergent-solubilized S. cerevisiae CytcO (36), at 445 nm, we observed three kinetic components with time constants of 23 μs (binding of O2 and electron transfer from heme a to the catalytic site, forming a state that is called “peroxy” and denoted as PR), 0.4 ms, and 6 ms, where the two latter time constants are associated with electron transfer from CuA/heme a to the catalytic site, forming the fully oxidized state, denoted O. Formation of the so-called oxoferryl state, denoted F, with a time constant of ∼90 μs, is not seen at 445 nm (36). Here, with the wild-type mitochondria, we observed about the same absorbance changes at 445 nm as observed previously with detergent-solubilized S. cerevisiae CytcO (36) (Fig. 7). Because the slowest component in this reaction has a time constant of ∼1 ms, the CytcO is ∼90% oxidized after a few milliseconds (Fig. 7A). As seen in Fig. 7 B and C, absorbance changes with similar time constants were observed with the rcf1Δ and rcf2Δ mitochondria, but the reaction was incomplete with the mutants and stopped (over a time scale of ∼0.1 s) at a level where a fraction of CytcO was incompletely oxidized. As discussed elsewhere in this report (Discussion), these data indicate that in the rcf1Δ and rcf2Δ mitochondria, a fraction of CytcO is fully active, whereas another fraction is inactive.
Fig. 7.
Reaction of the reduced CytcO with O2. A sample containing mitochondria was reduced and then incubated under an atmosphere of CO. The sample was transferred to a stopped-flow device, where it was mixed with an O2-saturated solution. About 200 ms after mixing, the mixture was illuminated by a short laser flash (at t = 0), which resulted in a rapid increase in absorbance, associated with dissociation of the CO ligand and binding of O2 (decrease in absorbance after the rapid increase). The following absorbance changes were associated with stepwise oxidation of the CytcO. Data are shown with wild-type (A), rcf2Δ (B), and rcf1Δ (C) mitochondria. Experimental conditions were as follows (before mixing): 1 μM PMS, 4 mM sodium ascorbate, ∼1.3 mM CO, 20 mM Hepes (pH 7.4), and 60 mM sorbitol. The sample containing mitochondria was mixed 1:1 with an O2-containing (∼1.2 mM O2) buffer composed of 20 mM Hepes (pH 7.4) and 60 mM sorbitol. A laser artifact at t = 0 has been truncated.
Discussion
Ligand Binding.
We first discuss the kinetics of CO ligand binding after light-induced photolysis. The ligand binds mainly to the iron of heme a3 in the catalytic site of CytcO, and the kinetics of CO binding reveal structural changes at this site. Results from earlier studies with oxidases from several different species have shown that after light-induced dissociation from heme a3, the CO ligand equilibrates with CuB, after which it dissociates into solution (31) (Scheme 1)
The dissociation rate constant from heme a3 in the dark is very small (e.g., with the bovine mitochondrial CytcO, it is ∼0.03 s−1); upon pulsed illumination (hν), the ligand moves to CuB in <<10 ns. With the bovine heart CytcO, the dissociation rate constant from CuB is ∼7⋅105 s−1 (31). The recombination of the CO ligand also occurs via CuB, where binding of CO to CuB is a second-order process (1⋅108 M−1⋅s−1). The internal transfer of CO from CuB to heme a3 displays a rate constant of 1⋅103 s−1. With the rate constants in Scheme 1, the observed CO-recombination rate is approximately given by the fraction of CO bound to CuB (middle state) multiplied by the CO-transfer rate from CuB to heme a3 [i.e., 0.13 × 1,000 s−1 = 130 s−1 (τ ≅ 8 ms) at 1 mM CO].
The rcf1Δ and rcf2Δ mitochondria displayed clearly biphasic CO-recombination kinetics (Fig. 3); that is, there were two components, one of which displayed a rate constant similar to the rate constant observed with the wild-type mitochondria (110 s−1, τ ≅ 9 ms, main, ∼85% component) and one that was about 100-fold faster (9⋅103 s−1, τ ≅ 110 μs). We note that the kinetic difference spectrum of the slow component agrees well with a CO-reduced minus reduced static difference spectrum (32) (Fig. 5). Consequently, we assume that the fraction of CytcOs corresponding to the slow CO-recombination component in the rcf1Δ/rcf2Δ strains is in the same state as the major population of the wild-type CytcO. Then, we address the origin of the fast component (τ ≅ 110 μs) while focusing on rcf1Δ, because the effect was more pronounced with rcf1Δ than with rcf2Δ. The kinetic difference spectrum of this fast component differs from the kinetic difference spectrum of the slower component. Assuming the model in Scheme 1 for the yeast CytcO, the observed CO-recombination rate cannot be accelerated beyond the value of 103 s−1 (i.e., the rate constant for CO transfer from CuB to heme a3), unless there are structural changes or loss of CuB at the catalytic site. In this context, we note that the fraction of rapid component at 445 nm presumably does not quantitatively reflect the fraction of the CytcO population with altered CO-binding kinetics because the absorption coefficient associated with the rapid process is not known.
Because the Rcf proteins were shown to interact with both cyt. bc1 and CytcO, we also considered the possibility that the rapid CO-recombination component is associated with binding of the ligand to another heme [e.g., the high-potential heme bH (heme bL is not reduced by ascorbate), the heme c of the cyt. bc1 complex, or heme a in CytcO]. We exclude water-soluble heme proteins, other than cyt. c, because they were presumably washed away during preparation of the samples. Because hemes b and c normally do not bind CO, a scenario where any of these hemes would bind CO would also require structural changes caused by removal of the Rcf1 or Rcf2 polypeptide. It is not straightforward to identify the CO-binding species from the kinetic difference spectrum of the rapid component (Fig. 5) because CO binding would be caused by weakening or dissociation of one of the two axial amino acid ligands of the six-coordinated heme, resulting in unknown spectral changes. However, we note that for CO binding to heme c of the cbb3 oxidase (37), genetically modified S. cerevisiae cyt. c (38), or carboxymethylated cyt. c (39), the difference spectra for CO dissociation are more blue-shifted than the ∼6-nm shift of the rapid component observed here. Consequently, CO binding to heme c is less likely. Furthermore, we note that dissociation of an axial ligand would also lower the midpoint potential of the heme (40, 41), which would alter the activity of the enzyme containing the altered heme. No such changes were observed for cyt. bc1 in the rcf1Δ variant. Collectively, these data suggest that the rapid CO-recombination component is associated with a fraction of structurally modified CytcO. However, in future studies, the possibility of CO binding to hemes other than CytcO hemes should be kept in mind.
As indicated above, there is a possibility that removal of the Rcf1 polypeptide results in a weaker interaction between one of the heme a axial ligands and the iron, allowing CO to bind. We discuss this scenario because, in a recent study, Hayashi et al. (42) showed that the Higd1A polypeptide, which belongs to the same protein family as Rcf1, associates with CytcO from B. taurus heart, resulting in increased CytcO turnover activity and a structural change around heme a. The effect on the activity of Higd1A binding to the B. taurus CytcO is similar to the effect observed for Rcf1 binding to the S. cerevisiae CytcO. However, the observed structural changes at heme a upon Higd1A binding yielded a fraction of high-spin heme a, which cannot explain the data with rcf1Δ in the S. cerevisiae mitochondria. At present, the interaction surface of the Rcf1 protein with the surface of CytcO is not known. Even though the structure of two Hig1 proteins has been determined using NMR (43), the structure of the Rcf1 protein is not known because this protein has an additional stretch of ∼65 amino acid residues, which could participate in the Rcf1–CytcO interactions.
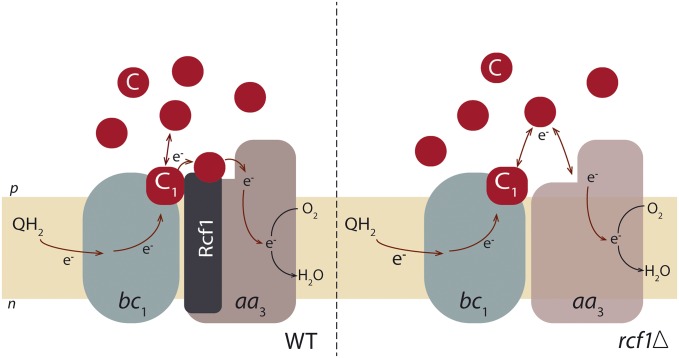
Collectively, the data described above indicate that removal of the Rcf1 or Rcf2 polypeptides results in structural changes, presumably in CytcO. Because these changes are specific to removal of Rcf1 or Rcf2, but not to dissociation of the supercomplex (Fig. 3), these protein components appear to have an influence on CytcO function (Fig. 8).
Fig. 8.
Schematic illustration of the data interpretation. The picture illustrates cyt. bc1 and CytcO in the preparation of mitochondria with the outer membrane removed, as studied in this work, and with added cyt. c. DQH2 delivers electrons to cyt. bc1, which reduces the cyt. c pool. Electrons from the cyt. c pool are transferred to CytcO, which reduces O2. With the wild-type mitochondria, the CytcO is fully active, and in the presence of added S. cerevisiae cyt. c, electrons are transferred from cyt. c1 of the cyt. bc1 complex via a cyt. c that is attached at CytcO or in the interface between cyt. bc1 and CytcO, possibly at Rcf1 (Left). Under steady-state conditions, the direct electron transfer, via the bound cyt. c, is faster than the equilibration of electrons with the cyt. c pool (during this direct electron transfer, the cyt. c pool is slowly reduced because the pool is in equilibrium with the bound cyt. c). This direct electron transfer does not take place in the absence of Rcf1 [i.e., electrons are only transferred via the cyt. c pool: first to the pool and then from the pool (Right)]. When using horse heart cyt. c, there is no prebound cyt. c and electron transfer between cyt. bc1 and fully active CytcO occurs only via the cyt. c pool (i.e., as on the Right, but with fully active CytcO).
Reaction with O2.
The conclusion above is also supported by the data from the flow-flash experiments. In these experiments, we followed in time absorbance changes associated with the single-turnover reaction of the four-electron reduced CytcO with O2 with microsecond time resolution, which allowed observation of all kinetic steps associated with progressive oxidation of CytcO. First, these data show that in the absence of Rcf1 or Rcf2, oxidation was incomplete in a fraction of the CytcO population over a time scale of ∼0.1 s because the final absorbance level was found to be above the absorbance level observed with the wild-type mitochondria. Furthermore, we note that the fraction of CytcO that did become oxidized with rcf1Δ/rcf2Δ reacted over about the same time scale (a few milliseconds) as CytcO in the wild-type mitochondria. In addition, the kinetic components observed with the wild-type mitochondria (formation of states PR and O with time constants of 23 μs and 0.4/6 ms, respectively; Results) were also represented in the rcf1Δ/rcf2Δ mitochondria, but with smaller relative amplitudes. These observations indicate that with the rcf1Δ/rcf2Δ mitochondria, the same sequence of reactions takes place as in the wild-type mitochondria, but in a smaller population of the CytcO. Consequently, the data suggest that in the rcf1Δ mitochondria, there is a fraction of CytcO that is fully active and a fraction that is inactive (compare a situation in which the entire CytcO population is 1/3 active with a situation in which 1/3 of the population is fully active and the remaining part (2/3) is inactive; compare Fig. 6). It is possible that there is an equilibrium between the two forms of the oxidase and this equilibrium is altered toward the inactive form by removal of either of the two Rcf polypeptides, but with a more significant effect for Rcf1 than for Rcf2 (Fig. 8). Even though the data discussed here are independent of the presence or absence of supercomplexes, we note that dynamic equilibrium of supercomplexes and free forms of cyt. bc1 and CytcO was noted by Cui et al. (44).
A lower activity of CytcO, without a decrease in the electron flux into the respiratory chain, would result in accumulation of reducing equivalents (e.g., at the quinol pool). These reducing equivalents could react with O2, resulting in an increase in the amount of nonenzymatically (partly) reduced O2, collectively referred to by the unspecific term “reactive oxygen species” (ROS). Effects of ROS formation in the rcf1Δ and rcf2Δ mutants have been discussed (17, 19).
Steady-State Activity.
Measurements of the levels of reduced cyt. c upon addition of DQH2 in the presence of O2 (Fig. 6 B and C) showed that when using cyt. c from horse heart, the cyt. c was first reduced (by cyt. bc1) and then reoxidized (slower reoxidation by CytcO) with both wild-type and rcf1Δ mitochondria. The oxidation rate was slower with rcf1Δ than with the wild-type mitochondria, reflecting the lower activity of the CytcO in the former. However, when using S. cerevisiae cyt. c, the data displayed a very different behavior. The reductive phase (increase in absorbance) was similar to the reductive phase seen with the horse heart cyt. c, but oxidation of cyt. c by CytcO displayed a reversed behavior (i.e., the apparent rate of cyt. c oxidation was faster with rcf1Δ than with the wild-type mitochondria).
This behavior (Fig. 6 B and C) can be explained as follows. We do not expect the horse heart cyt. c to form a 1:1 complex with the S. cerevisiae cyt. bc1 or CytcO. Consequently, when using horse heart cyt. c, the entire cyt. c population would be present in the bulk water solution, in equilibrium with the membrane-bound cyt. bc1-CytcO. Because the observed absorbance changes at 550 nm (Fig. 6 B and C) reflect this bulk population (cyt. c is added in large excess to cyt. bc1-CytcO), the absorbance first increases, followed in time by a decrease. After this decrease, the reduced cyt. c level would reach a steady-state level, which is dependent on the relative concentration of DQH2 and O2, as well as the turnover rate of cyt. bc1-CytcO.
To explain the behavior with the S. cerevisiae cyt. c, we assume formation of a tight complex between cyt. bc1-CytcO and at least one cyt. c molecule, as reported previously (4, 32). Assuming this scenario, in the wild-type mitochondria, the bound cyt. c would bridge electron transfer between the two respiratory–enzyme complexes (also refs. 12 and 45). Upon addition of DQH2, the cyt. c pool would be rapidly reduced by cyt. bc1. This cyt. c pool would then remain largely reduced, and electron transfer to CytcO would take place primarily via the tightly bound cyt. c. Consequently, no decrease in absorbance, due to oxidation by CytcO, is observed with the S. cerevisiae cyt. c (Fig. 6B) because the direct electron transfer from cyt. bc1 to CytcO is presumably faster than via bulk solution. In this context, we note that the O2-reduction turnover activity of wild-type mitochondria upon addition of DQH2 was about the same when using cyt. c from horse heart or S. cerevisiae (Fig. 6A). In other words, even though the reduction levels of the cyt. c pools differ significantly for the two types of cyt. c (Fig. 6B), the electron flux from DQH2 to O2, via cyt. bc1 and CytcO, is not significantly different, suggesting that the behavior observed upon addition of S. cerevisiae cyt. c to the wild-type mitochondria is not due to an altered steady-state activity. The scenario outlined above is also consistent with the findings of Trouillard et al. (32) (i.e., that the prebound cyt. c would be in redox equilibrium with the cyt. c pool).
In conclusion, these data indicate that in the S. cerevisiae mitochondria, there is at least one cyt. c bound to the cyt. bc1–CytcO supercomplex, that this cyt. c mediates electron transfer between the two complexes, and that the binding of cyt. c to the supercomplex is dependent on the presence of the Rcf1 protein (Fig. 8). The involvement of the Rcf1 protein in coordinating cyt. c is further supported by the observation that the amount of expressed cyt. c was elevated in rcf1Δ cells (Fig. 2). In other words, under conditions when direct electron transfer, presumably via a prebound cyt. c, is lost, the amount of bulk cyt. c is increased by the cell.
Structural and Functional Considerations and Conclusions.
The detailed molecular interactions between Rcf1 and CytcO are not known, but combined data from several studies point to possible interactions with subunits Cox3, Cox12, and Cox13 (17–19). It is not straightforward to discuss these findings in the framework of the structural models of the S. cerevisiae cyt. bc1–CytcO supercomplex (7, 9) because these models indicate that the cyt. bc1–CytcO interaction surface is located away from Cox12 or Cox13. In other words, there is presently disagreement between the data from the functional and structural studies published earlier because the former point to interactions of Rcf1 with Cox3, Cox12, and Cox13, whereas the latter point to interactions near Cox5. The functional model in Fig. 8 is based on the structural models of the cyt. bc1–CytcO supercomplex (7, 9), suggesting interactions as indicated by the blue arrow near Cox5.
Even though our data suggest interactions of Rcf1 near Cox5, we discuss possible interactions of Rcf1 with Cox13, which would have to occur independently. Subunit VIa (B. taurus mitochondrial CytcO), which is a homolog of Cox13 in the S. cerevisiae CytcO, houses an allosteric nucleotide binding site close to the N terminus on its matrix side (46). In a recent study, Maréchal et al. (22) noted that in the model of the S. cerevisiae CytcO, the residues presumably involved in nucleotide binding are not conserved. Instead, in the S. cerevisiae CytcO, there is a stretch of 37 amino acid residues, which together form an extension at the N terminus (not present in the B. taurus CytcO). This domain was suggested to be a possible regulatory site of the S. cerevisiae CytcO (22), where Rcf1 binding may play a role independent of the supramolecular interactions discussed above.
The data from the present study indicate that Rcf1 directly regulates the activity of CytcO, but this regulatory role is independent of the role of Rcf1 in stabilizing the cyt. bc1–CytcO supramolecular interactions (the function of CytcO, as assessed, e.g., in the CO-binding experiments, is independent of removal of the cyt. bc1 complex). An obvious question is whether or not there is a functional relation between a decrease in CytcO activity and dissociation of the cyt. bc1–CytcO supercomplex. A possible answer to this question is offered by the data in Fig. 6, discussed in detail above. If the Rcf1 polypeptide does coordinate cyt. c that mediates direct electron transfer from cyt. bc1 to CytcO, an increased amount of Rcf1 would stimulate CytcO activity, induce formation of the cyt. bc1–CytcO supercomplex, and recruit a tightly bound cyt. c that would bridge the distance between cyt. bc1 and CytcO (9). This role would be equivalent to the role of the membrane-anchored cyt. cγ in Rhodobacter sphaeroides or Rhodobacter capsulatus (47, 48).
In future studies, it will be important to address structural aspects of the Rcf1–CytcO interactions because this information is key to understanding the effects observed here at the molecular level (and the currently available data are not consistent). It would also be interesting to address the role of the Rcf proteins in controlling the ratio of O2 reduction over ATP formation (the P/O ratio) because changes in structure, such as those changes presumably induced by removal of Rcf1, may interfere with proton pumping (49–52).
Materials and Methods
Materials.
All chemicals were purchased from Sigma–Aldrich, except Zymolyase-20T (from Arthrobacter luteus), which was purchased from Nacalai Tesque. Yeast strains were purchased from the European Saccharomyces Cerevisiae Archive for Functional Analysis (EUROSCARF, University of Frankfurt). Decylubiquinone (DQ) was reduced as follows: A few grains of sodium borohydrate were added to 20 mM DQ dissolved in ultrapure ethanol and left for a few minutes on ice. When the solution was clear, a few drops of HCl were added, followed by centrifugation to remove excess borohydrate. The reduced DQH2 was kept at −20 °C before use.
Strains and Growth Conditions.
All strains used were S. cerevisiae BY4741 carrying the following deletions: wild type (Mat a; his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), rcf1Δ (Mat a; his3Δ1, leu2Δ0, met15Δ0, ura3Δ0; YML030w::kanMX4), rcf2Δ (Mat a; his3Δ1, leu2Δ0, met15Δ0, ura3Δ0; YNR018W::kanMX4), and qcr8Δ (Mat a; his3Δ1, leu2Δ0, met15Δ0, ura3Δ0; YJL166W::kanMX4). All deletions were confirmed using PCR. The S. cerevisiae cells were grown at 30 °C under vigorous shaking in standard YP medium (pH 5.4) supplemented with either 2% (vol/vol) glycerol (wild type) or 2% (wt/vol) galactose (deletion strains).
Preparation of Mitochondria.
Cells were harvested by centrifugation at 3,000 × g, and mitochondria were prepared essentially according to the protocol of Meisinger et al. (53) with some adjustments. Briefly, after harvest, the cells were incubated for 10 min in 100 mM Tris⋅base at pH ∼10, followed by washing in 1.2 M sorbitol, and they were then incubated in a zymolyase buffer [1.2 M sorbitol, 20 mM KPi (pH 7.4), zymolyase (3 mg/g wet weight)] for ∼60 min at 30 °C while shaking. Spheroplasts were centrifuged at 3,000 × g and resuspended in 6.5 mL/g wet weight of homogenization buffer [10 mM Tris⋅HCl (pH 7.4), 1 mM EDTA, 1 mM PMSF, 0.6 M sorbitol] and broken using a glass-Teflon homogenizer (10 times). The homogenized spheroplasts were spun (3,000 × g, 5 min, 4 °C), and the supernatant was kept while the pellet was homogenized once more. After centrifugation, the supernatants were pooled and spun (17,000 × g, 12 min, 4 °C), and the pellet was resuspended in a buffer composed of 20 mM Hepes (pH 7.4) containing 0.6 M sorbitol, frozen in liquid nitrogen, and stored at −80 °C until use.
Preparation of Fully Reduced and CO-Bound Mitochondria.
Mitochondria were centrifuged (10,000 × g, 12 min, 4 °C) and resuspended in a hypotonic buffer [20 mM Hepes (pH 7.4) containing 60 mM sorbitol] to remove the outer mitochondrial membrane. Mitochondrial inner membranes were then washed three times in the same buffer and resuspended in a volume that was adjusted to yield 0.2–4 μM CytcO (concentration determination is discussed below). The redox mediator phenylmethylsulfonyl was added (1 μM final concentration) to the samples before they were transferred to an anaerobic, locally modified Thunberg cuvette (total sample volume of ∼1.5 mL, path length of 1.00 cm) prepared with ascorbate in the side bulb to yield a final concentration of ∼4 mM after mixing. The gas in the cuvette was exchanged for N2 on a vacuum line before ascorbate was mixed in. Samples were allowed to reduce (for ∼10 min) before N2 was replaced with CO (∼130 kPa). The effects of the gas exchange, reduction, and CO binding were monitored spectrophotometrically (Cary 4000 UV-Vis; Agilent Technologies).
Flash-Photolysis Kinetics.
The Thunberg cuvette was placed in a flash photolysis/flow-flash setup from Applied Photophysics. The absorbance of the reduced sample under a CO atmosphere was monitored at a specific wavelength that could be adjusted to any value in the visible range. The CO ligand was dissociated by a short laser flash (8 ns at 532 nm, Brilliant B; Quantel) that illuminated the sample at a 90° angle. The flash-induced absorbance changes, associated with CO dissociation and recombination, were monitored with a time resolution of ∼100 ns at a specific chosen wavelength using a photomultiplier tube.
For the kinetic difference spectra, absorbance changes were determined every 2 nm in the wavelength interval of 420–450 nm. The amplitudes of the kinetic components were determined from a fit of the data with a sum of exponential functions. The amplitudes of these components were plotted at each wavelength. To investigate the CO-concentration dependence, the CO-recombination rates were first monitored at 445 nm with a CO-saturated sample, after which a fraction of CO was removed on the vacuum line and replaced with N2.
Flow-Flash Kinetics.
The flow-flash setup was purchased from Applied Photophysics. It is composed of a stopped-flow apparatus, connected to a laser (8 ns at 532 nm, Brilliant B) that is used to illuminate the cuvette (1.00-cm path length) at a specified time after mixing. The samples were prepared as described above for the flash-photolysis studies. In a flow-flash experiment, the anaerobic CO-bound sample was mixed in ∼10 ms at a ratio of 1:1 with a buffer (same composition as the mitochondrial sample) that was saturated with pure O2 (∼1.2 mM). About 200 ms after mixing, CO was removed by the laser flash, which allowed O2 to bind to the CytcO catalytic site to initiate the reaction (under the conditions used, O2 binding and trapping is a factor of ∼103 faster than CO recombination). Absorbance changes were recorded with a time resolution of ∼100 ns. The rate constants of the absorbance changes at 445 nm (contributions of both hemes a and a3) were determined using the software Pro-K (Applied Photophysics).
Determination of CytcO and Cyt. bc1 Concentrations.
Concentration determination of CytcO.
Mitochondria in suspension are highly light scattering. To obtain a sufficiently transparent sample, the samples were diluted with appropriate volumes of buffer by eye. The CytcO concentration was determined either from the dithionite reduced-oxidized absorbance difference spectrum at 605 nm (absorption coefficient of 24 mM−1⋅cm−1) or from the laser flash-induced absorbance change at 445 nm using the absorption coefficient of 67 mM−1⋅cm−1. The CytcO concentration was typically in the range 0.2–4 μM.
Determination of the cyt. bc1 concentration.
To determine the concentration of the cyt. bc1 complex, the samples were reduced with dithionite (both hemes b are reduced). The equation used for determining the concentration (Cbc1) was as follows (54, 55):
where ΔAx−y is the difference in absorbance at wavelength x minus the absorbance at wavelength y.
The concentration of cyt. c was determined using the absorption coefficient 21.1 mM−1⋅cm−1 for the reduced minus oxidized spectrum at 550 nm (56).
Steady-State Activity Measurements.
Oxygen consumption.
Multiple turnover activity was measured either for CytcO alone, by reduction with ascorbate/TMPD and cyt. c, or as the coupled activity of cyt. bc1 and CytcO upon addition of DQH2 as an electron donor to cyt. bc1 using cyt. c as an electron mediator. The oxygen reduction was monitored using a Clark-type oxygen electrode (Hansatech) with a chamber volume of 1 mL. When measuring the CytcO activity alone, a baseline was recorded first with measurement buffer [20 mM Hepes (pH 7.4), 60 mM sorbitol, 0.1 mM EDTA] containing 5 mM ascorbate and 0.5 mM TMPD before addition of inner mitochondrial membranes. The final concentration of CytcO varied between preparations and was adjusted in the range of 2–20 nM. The ratio of substrate to mitochondria was adjusted to make sure that electron input was not rate-limiting. After recording for a few minutes, KCN was added to block the CytcO, verifying that oxygen consumption was mediated by the oxidase. In the case of coupled cyt. bc1-CytcO activity, a baseline in the measurement buffer containing 0.1 mM DQH2 and 20 μM cyt. c was recorded before inner mitochondrial membranes were added. As above, the activity was blocked with KCN at the end of the experiment.
Cyt. bc1 and CytcO activities as measured by absorbance changes of cyt. c.
The steady-state activity of cyt. bc1 was measured by following in time absorbance changes at 550 nm (reduction of cyt. c) upon addition of DQH2 in the presence or absence of KCN to block cyt. c oxidation by CytcO. First, oxidized cyt. c (from horse heart or S. cerevisiae at 20 μM) and mitochondria were resuspended in measurement buffer, and a baseline was recorded. The reaction was initiated by addition of DQH2 (100 μM). Similarly, if KCN was omitted, the coupled activity (reduction of cyt. c by cyt. bc1, followed by its reoxidation by CytcO) was measured following absorbance changes at 550 nm. Direct reduction of cyt. c by DQH2 was measured independently, and the rate was found to be negligible compared with the rate of the enzyme-catalyzed reaction.
Acknowledgments
We thank Hannah Dawitz for sequencing the mutant strains and Prof. Fevzi Daldal for valuable discussions. These studies were supported by grants from the Knut and Alice Wallenberg Foundation and the Swedish Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Luttik MAH, et al. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J Biol Chem. 1998;273(38):24529–24534. doi: 10.1074/jbc.273.38.24529. [DOI] [PubMed] [Google Scholar]
- 2.Iwata M, et al. The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates. Proc Natl Acad Sci USA. 2012;109(38):15247–15252. doi: 10.1073/pnas.1210059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rich PR, Maréchal A. The mitochondrial respiratory chain. Essays Biochem. 2010;47:1–23. doi: 10.1042/bse0470001. [DOI] [PubMed] [Google Scholar]
- 4.Schägger H. Respiratory chain supercomplexes. IUBMB Life. 2001;52(3-5):119–128. doi: 10.1080/15216540152845911. [DOI] [PubMed] [Google Scholar]
- 5.Stuart RA. Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J Bioenerg Biomembr. 2008;40(5):411–417. doi: 10.1007/s10863-008-9168-4. [DOI] [PubMed] [Google Scholar]
- 6.Chaban Y, Boekema EJ, Dudkina NV. Structures of mitochondrial oxidative phosphorylation supercomplexes and mechanisms for their stabilisation. Biochim Biophys Acta. 2014;1837(4):418–426. doi: 10.1016/j.bbabio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Heinemeyer J, Braun HP, Boekema EJ, Kouřil R. A structural model of the cytochrome C reductase/oxidase supercomplex from yeast mitochondria. J Biol Chem. 2007;282(16):12240–12248. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- 8.Winge DR. Sealing the mitochondrial respirasome. Mol Cell Biol. 2012;32(14):2647–2652. doi: 10.1128/MCB.00573-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mileykovskaya E, et al. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J Biol Chem. 2012;287(27):23095–23103. doi: 10.1074/jbc.M112.367888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genova ML, Lenaz G. Functional role of mitochondrial respiratory supercomplexes. Biochim Biophys Acta. 2014;1837(4):427–443. doi: 10.1016/j.bbabio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Acin-Perez R, Enriquez JA. The function of the respiratory supercomplexes: The plasticity model. Biochim Biophys Acta. 2014;1837(4):444–450. doi: 10.1016/j.bbabio.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Boumans H, Grivell LA, Berden JA. The respiratory chain in yeast behaves as a single functional unit. J Biol Chem. 1998;273(9):4872–4877. doi: 10.1074/jbc.273.9.4872. [DOI] [PubMed] [Google Scholar]
- 13.Pfeiffer K, et al. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278(52):52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 14.Bazán S, et al. Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. J Biol Chem. 2013;288(1):401–411. doi: 10.1074/jbc.M112.425876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277(46):43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M, Mileykovskaya E, Dowhan W. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J Biol Chem. 2005;280(33):29403–29408. doi: 10.1074/jbc.M504955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vukotic M, et al. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15(3):336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Strogolova V, Furness A, Robb-McGrath M, Garlich J, Stuart RA. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol Cell Biol. 2012;32(8):1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YC, et al. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15(3):348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer F, Filippis C, Osiewacz HD. RCF1-dependent respiratory supercomplexes are integral for lifespan-maintenance in a fungal ageing model. Sci Rep. 2015;5:12697. doi: 10.1038/srep12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshikawa S, Shimada A. Reaction mechanism of cytochrome c oxidase. Chem Rev. 2015;115(4):1936–1989. doi: 10.1021/cr500266a. [DOI] [PubMed] [Google Scholar]
- 22.Maréchal A, Meunier B, Lee D, Orengo C, Rich PR. Yeast cytochrome c oxidase: A model system to study mitochondrial forms of the haem-copper oxidase superfamily. Biochim Biophys Acta. 2012;1817(4):620–628. doi: 10.1016/j.bbabio.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosler JP, Ferguson-Miller S, Mills DA. Energy transduction: Proton transfer through the respiratory complexes. Annu Rev Biochem. 2006;75:165–187. doi: 10.1146/annurev.biochem.75.062003.101730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Namslauer A, Brzezinski P. Structural elements involved in electron-coupled proton transfer in cytochrome c oxidase. FEBS Lett. 2004;567(1):103–110. doi: 10.1016/j.febslet.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 25.Brzezinski P, Ädelroth P. Design principles of proton-pumping haem-copper oxidases. Curr Opin Struct Biol. 2006;16(4):465–472. doi: 10.1016/j.sbi.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Richter OMH, Ludwig B. Electron transfer and energy transduction in the terminal part of the respiratory chain - lessons from bacterial model systems. Biochim Biophys Acta. 2009;1787(6):626–634. doi: 10.1016/j.bbabio.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Ferguson-Miller S, Hiser C, Liu J. Gating and regulation of the cytochrome c oxidase proton pump. Biochim Biophys Acta. 2012;1817(4):489–494. doi: 10.1016/j.bbabio.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rich PR, Maréchal A. Functions of the hydrophilic channels in protonmotive cytochrome c oxidase. J R Soc Interface. 2013;10(86):20130183. doi: 10.1098/rsif.2013.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaila VRI, Verkhovsky MI, Wikström M. Proton-coupled electron transfer in cytochrome oxidase. Chem Rev. 2010;110(12):7062–7081. doi: 10.1021/cr1002003. [DOI] [PubMed] [Google Scholar]
- 30.Salomonsson L, Lee A, Gennis RB, Brzezinski P. A single-amino-acid lid renders a gas-tight compartment within a membrane-bound transporter. Proc Natl Acad Sci USA. 2004;101(32):11617–11621. doi: 10.1073/pnas.0402242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Einarsdóttir O, et al. Photodissociation and recombination of carbonmonoxy cytochrome oxidase: Dynamics from picoseconds to kiloseconds. Biochemistry. 1993;32(45):12013–12024. doi: 10.1021/bi00096a011. [DOI] [PubMed] [Google Scholar]
- 32.Trouillard M, Meunier B, Rappaport F. Questioning the functional relevance of mitochondrial supercomplexes by time-resolved analysis of the respiratory chain. Proc Natl Acad Sci USA. 2011;108(45):E1027–E1034. doi: 10.1073/pnas.1109510108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Namslauer A, Brändén M, Brzezinski P. The rate of internal heme-heme electron transfer in cytochrome C oxidase. Biochemistry. 2002;41(33):10369–10374. doi: 10.1021/bi025976y. [DOI] [PubMed] [Google Scholar]
- 34.Brzezinski P, Gennis RB. Cytochrome c oxidase: Exciting progress and remaining mysteries. J Bioenerg Biomembr. 2008;40(5):521–531. doi: 10.1007/s10863-008-9181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brzezinski P, Johansson AL. Variable proton-pumping stoichiometry in structural variants of cytochrome c oxidase. Biochim Biophys Acta. 2010;1797(6-7):710–723. doi: 10.1016/j.bbabio.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 36.Näsvik Öjemyr L, et al. Reaction of wild-type and Glu243Asp variant yeast cytochrome c oxidase with O2. Biochim Biophys Acta. 2014;1837(7):1012–1018. doi: 10.1016/j.bbabio.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 37.Huang Y, Reimann J, Singh LMR, Ädelroth P. Substrate binding and the catalytic reactions in cbb3-type oxidases: The lipid membrane modulates ligand binding. Biochim Biophys Acta. 2010;1797(6-7):724–731. doi: 10.1016/j.bbabio.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 38.Silkstone G, Stanway G, Brzezinski P, Wilson MT. Production and characterisation of Met80X mutants of yeast iso-1-cytochrome c: Spectral, photochemical and binding studies on the ferrous derivatives. Biophys Chem. 2002;98(1-2):65–77. doi: 10.1016/s0301-4622(02)00085-6. [DOI] [PubMed] [Google Scholar]
- 39.Brzezinski P, Wilson MT. Photochemical electron injection into redox-active proteins. Proc Natl Acad Sci USA. 1997;94(12):6176–6179. doi: 10.1073/pnas.94.12.6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darrouzet E, et al. Substitution of the sixth axial ligand of Rhodobacter capsulatus cytochrome c1 heme yields novel cytochrome c1 variants with unusual properties. Biochemistry. 1999;38(25):7908–7917. doi: 10.1021/bi990211k. [DOI] [PubMed] [Google Scholar]
- 41.Mao J, Hauser K, Gunner MR. How cytochromes with different folds control heme redox potentials. Biochemistry. 2003;42(33):9829–9840. doi: 10.1021/bi027288k. [DOI] [PubMed] [Google Scholar]
- 42.Hayashi T, et al. Higd1a is a positive regulator of cytochrome c oxidase. Proc Natl Acad Sci USA. 2015;112(5):1553–1558. doi: 10.1073/pnas.1419767112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klammt C, et al. Facile backbone structure determination of human membrane proteins by NMR spectroscopy. Nat Methods. 2012;9(8):834–839. doi: 10.1038/nmeth.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui TZ, Conte A, Fox JL, Zara V, Winge DR. Modulation of the respiratory supercomplexes in yeast: Enhanced formation of cytochrome oxidase increases the stability and abundance of respiratory supercomplexes. J Biol Chem. 2014;289(9):6133–6141. doi: 10.1074/jbc.M113.523688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Genova ML, Lenaz G. A critical appraisal of the role of respiratory supercomplexes in mitochondria. Biol Chem. 2013;394(5):631–639. doi: 10.1515/hsz-2012-0317. [DOI] [PubMed] [Google Scholar]
- 46.Anthony G, Reimann A, Kadenbach B. Tissue-specific regulation of bovine heart cytochrome-c oxidase activity by ADP via interaction with subunit VIa. Proc Natl Acad Sci USA. 1993;90(5):1652–1656. doi: 10.1073/pnas.90.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myllykallio H, Drepper F, Mathis P, Daldal F. Membrane-anchored cytochrome cy mediated microsecond time range electron transfer from the cytochrome bc1 complex to the reaction center in Rhodobacter capsulatus. Biochemistry. 1998;37(16):5501–5510. doi: 10.1021/bi973123d. [DOI] [PubMed] [Google Scholar]
- 48.Daldal F, et al. Mobile cytochrome c2 and membrane-anchored cytochrome cy are both efficient electron donors to the cbb3- and aa3-type cytochrome c oxidases during respiratory growth of Rhodobacter sphaeroides. J Bacteriol. 2001;183(6):2013–2024. doi: 10.1128/JB.183.6.2013-2024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nilsson T, et al. Lipid-mediated protein-protein interactions modulate respiration-driven ATP synthesis. Sci Rep. 2016;6:24113. doi: 10.1038/srep24113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Ballmoos C, Biner O, Nilsson T, Brzezinski P. Mimicking respiratory phosphorylation using purified enzymes. Biochim Biophys Acta. 2016;1857(4):321–331. doi: 10.1016/j.bbabio.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 51.Vilhjálmsdóttir J, Johansson A-L, Brzezinski P. Structural changes and proton transfer in cytochrome c oxidase. Sci Rep. 2015;5:12047. doi: 10.1038/srep12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Ballmoos C, et al. Mutation of a single residue in the ba3 oxidase specifically impairs protonation of the pump site. Proc Natl Acad Sci USA. 2015;112(11):3397–3402. doi: 10.1073/pnas.1422434112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- 54.Vanneste WH. Molecular proportion of the fixed cytochrome components of the respiratory chain of Keilin-Hartree particles and beef heart mitochondria. Biochim Biophys Acta. 1966;113(1):175–178. doi: 10.1016/s0926-6593(66)80132-7. [DOI] [PubMed] [Google Scholar]
- 55.Baymann F, Robertson DE, Dutton PL, Mäntele W. Electrochemical and spectroscopic investigations of the cytochrome bc1 complex from Rhodobacter capsulatus. Biochemistry. 1999;38(40):13188–13199. doi: 10.1021/bi990565b. [DOI] [PubMed] [Google Scholar]
- 56.van Gelder B, Slater EC. The extinction coefficient of cytochrome c. Biochim Biophys Acta. 1962;58(3):593–595. doi: 10.1016/0006-3002(62)90073-2. [DOI] [PubMed] [Google Scholar]
- 57.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA: 2002. [Google Scholar]