Abstract
Many of the continuing scientific advances achieved through computational biology are predicated on the availability of ongoing increases in computational power required for detailed simulation and analysis of cellular processes on biologically-relevant timescales. A critical challenge facing the development of future exascale supercomputer systems is the development of new computing hardware and associated scientific applications that dramatically improve upon the energy efficiency of existing solutions, while providing increased simulation, analysis, and visualization performance. Mobile computing platforms have recently become powerful enough to support interactive molecular visualization tasks that were previously only possible on laptops and workstations, creating future opportunities for their convenient use for meetings, remote collaboration, and as head mounted displays for immersive stereoscopic viewing. We describe early experiences adapting several biomolecular simulation and analysis applications for emerging heterogeneous computing platforms that combine power-efficient system-on-chip multi-core CPUs with high-performance massively parallel GPUs. We present low-cost power monitoring instrumentation that provides sufficient temporal resolution to evaluate the power consumption of individual CPU algorithms and GPU kernels. We compare the performance and energy efficiency of scientific applications running on emerging platforms with results obtained on traditional platforms, identify hardware and algorithmic performance bottlenecks that affect the usability of these platforms, and describe avenues for improving both the hardware and applications in pursuit of the needs of molecular modeling tasks on mobile devices and future exascale computers.
Keywords: Molecular modeling, Heterogeneous architectures, GPU computing, High-performance computing, Mobile computing, Energy efficiency
I. Introduction
Computer simulations provide a powerful tool for probing the dynamics of cellular processes at temporal and spatial resolutions that are not accessible to experimental methods alone. These studies require tremendous computational capabilities for both the simulations and the analysis and visualization of the results, made possible through the availability of clusters and supercomputers and parallel simulation and analysis software that makes efficient use of them. Petascale supercomputers are beginning to stretch the practical limits to space, power consumption, and cooling, with leading systems such as ORNL Titan and NCSA Blue Waters requiring on the order of 10 MW of electrical power. It is expected that many exascale systems may be constrained to power levels on the order of 20 MW, and a critical challenge facing the development of these systems is the development of new computing hardware and associated scientific applications that dramatically improve upon the energy efficiency of existing solutions, while providing increased simulation, analysis, and visualization performance. Exascale computing will require reduction of data movement and awareness of data locality within operating systems, applications, and algorithms [1].
Graphics processing units (GPUs) have evolved from their origins as fixed-function hardware accelerators solely intended for computer graphics and image processing workloads into much more flexible massively parallel computing devices that can be programmed for use by general purpose scientific applications [2], [3], and for molecular modeling in particular [4], [5]. Due to their massively parallel hardware architecture, GPUs are capable of substantially outperforming conventional multi-core CPUs on applications that contain large amounts of fine-grain parallelism, and in many cases they also significantly improve upon the energy efficiency achieved by conventional hardware platforms [6]. GPUs have become an effective tool for acceleration of a wide variety of computationally demanding molecular modeling applications [4], [7], [8], [9], [3], [10], [11], [12], [13], [5], [14], and these successes have contributed to the use of GPUs in the top performing supercomputers in the world.
Contemporary GPUs are not entirely self-sufficient; they depend on a host computer to run operating system software, and for execution of low-parallelism or latency-sensitive code that is poorly suited to execution on massively parallel hardware. The evolution of mobile phones into ubiquitous browsing, gaming, and communication devices has created tremendous demand for power-efficient system-on-chip (SoC) processor designs that integrate GPU hardware on-chip, or that are tightly coupled with external GPUs. ARM® processors are currently one of the leading CPU architectures used in mobile phones and tablet computers, with billions of ARM processors currently in the field.
The performance of SoCs and mobile computing platforms has steadily increased in recent years. In combination with integrated or discrete GPUs, these systems are now capable of performing molecular visualization and analysis tasks that have previously required conventional laptop or desktop computers. The small physical size and portability of these hardware platforms presents unique opportunities for their use as convenient multi-modal displays, as client systems for remote visualization of molecular structures, and as self-contained systems driving head mounted displays (HMDs) for immersive stereoscopic viewing of molecular structures.
We describe early experiences adapting several biomolecular simulation and analysis applications for heterogeneous computing platforms based on the combination of multi-core ARM SoCs with GPUs. We specifically evaluate emerging heterogeneous computing platforms that combine ARM multicore CPUs with both mobile GPUs and state-of-the-art discrete GPUs, with the goal of identifying performance and energy efficiency bottlenecks in hardware and software on these platforms, and their applicability to key computational biology applications.
Lattice Microbes (LM) is a suite of programs for performing stochastic biological simulations. Designed from the outset to leverage heterogeneous computing systems that combine traditional multi-core CPUs and GPUs, LM simulates well-stirred reactions and spatially-resolved reaction-diffusion processes with a focus on complex, realistic biological environments [15], [16] and supports single and multi-GPU hardware platforms [17]. Due to its focus on heterogeneous computing platforms, LM is an excellent candidate application to use in testing emerging energy-efficient computing platforms that combine multi-core ARM CPUs with high performance GPU accelerators.
NAMD is a GPU-accelerated parallel molecular dynamics simulation package that specializes in simulating large biomolecular complexes [18]. NAMD is based on the Charm++ runtime system, and is designed for execution on workstations, clusters, and petascale computers [4], [11]. We evaluate performance, energy efficiency, and GPU acceleration factors for NAMD on the CARMA and KAYLA platforms, and on a conventional Intel x86 platform.
VMD is a widely used tool for preparation, visualization, and analysis of biomolecular and cellular simulations. VMD was one of the very first molecular modeling applications to employ GPUs for general purpose scientific computations [4], and it incorporates a broad range of GPU-accelerated algorithms. We explore the performance scaling and energy efficiency of three of these algorithms and identify bottlenecks that hinder performance and limit the potential energy efficiency gains on the CARMA and KAYLA test platforms, and we present kernel-level performance results for one of the algorithms on all test platforms, including two Intel x86 system configurations.
We compare the performance and energy efficiency of these scientific applications running on emerging platforms with results obtained on traditional platforms, identify hardware and algorithmic performance bottlenecks that affect the usability of these platforms, and describe avenues for improving both the hardware and applications in pursuit of the needs of future exascale systems.
II. Hardware and Operating System Overview
We evaluate five heterogeneous computing platforms that combine multi-core CPUs based on the ARM architecture with on-chip, on-board, and add-in-board GPUs made by NVIDIA. We performed rudimentary evaluation of the NVIDIA Jetson TK1 and Jetson TX1, and the AppliedMicro X-Gene. The Jetson TK1 uses an NVIDIA Tegra K1 (32-bit) with an on-chip integrated GPU based on the Kepler GPU architecture, the first Tegra that is capable of running CUDA kernels natively in its on-chip GPU. The Jetson TK1 ran Ubuntu 14.04 with GCC 4.8.2 and CUDA 6.5. The Jetson TX1 is similar to the TK1, but is based on a newer Tegra X1 (64-bit) SoC capable of higher clock rates. The Jetson TX1 ran developmental operating system software limited to 32-bit addressing, based on Ubuntu 14.04 with GCC 4.8.4 and CUDA 7.0. The AppliedMicro X-Gene is also one of the first 64-bit ARM CPUs. The X-Gene host system ran an early developmental operating system with 64-bit addressing, but which had a number of software limitations relative to the other platforms. The X-Gene was paired with an NVIDIA Tesla K20 discrete add-in-board GPU for all tests, and used GCC 4.8.1 and the ARM64 version of CUDA 6.5.
We evaluated two of the energy-efficient heterogeneous computing platforms in much greater detail, both based on the NVIDIA Tegra 3 system-on-chip (SoC), which utilize 32-bit ARM Cortex-A9 CPU cores and an NVIDIA GPU accelerator. Both are single-board computers produced by SECO: CARMA, a heterogeneous computing development board with an on-board NVIDIA Quadro 1000M GPU, and KAYLA, a development board with one PCIe ×16 expansion slot supporting a discrete add-in-board GPU accelerator. The NVIDIA Tegra 3 SoC includes four high performance CPU cores and a fifth low speed energy efficient CPU core for use during idle periods. Tegra 3 supports ARM NEON 4-way vector instructions, accessible through GCC compiler intrinsics. Both development boards include SATA, ethernet, on-board video, and serial I/O. Both development systems were installed with Ubuntu 12.04 running a kernel supporting the ARM hardware floating point ABI, and each of the applications described were compiled using an ARM-native compiler toolchain based on GCC 4.6.3, and the CUDA 5.5 toolkit for the ARM platform.
One of the key power efficiency features of the Tegra 3, Tegra K1, Tegra X1, and many similar SoCs is the inclusion of (one or multiple) CPU cores that are specialized for low-speed, high-efficiency execution during idle periods. When the system reaches a low utilization state, the hardware and operating system cooperate to migrate processes and threads off of the four high-performance CPU cores, and move them to the energy-efficient CPU core(s). Once processes have been migrated, the four main CPU cores are shut off, essentially eliminating associated power consumption and leakage current. This energy efficiency feature is particularly valuable for mobile computing scenarios. One unusual implementation detail that arises is that the operating system reports varying numbers of both available and active CPUs through standard POSIX operating system APIs. This behaviour is unusual as compared to traditional HPC hardware platforms, and it can confound existing scientific applications and numerical libraries which have been written to spawn a number of threads, processes, or MPI ranks equal to the number of available CPUs as reported by the operating system during the program’s initialization. We have made appropriate modifications to each of the applications we discuss below, thereby enabling them to utilize all of the CPU cores, regardless of the system’s power management state at the time the application is launched. It seems likely that, as energy efficiency becomes an increasing concern, future desktop computers and server platforms will also employ mechanisms to shut down entire CPU cores as a means of optimizing energy consumption.
Since the CARMA and KAYLA systems use the same Tegra 3 SoC, but have different GPUs, they have very different performance and power consumption characteristics, as shown in Table I. Several performance tests were used to establish basic performance and power consumption characteristics for the two test platforms. Idle power consumption was measured for both systems after booting into the Linux OS and allowing them to become fully quiescent. Interactive VMD sessions were run with and without CUDA GPU acceleration support, and idle power consumption was measured. In all three idle power consumption tests, the Linux OS shutdown the high-performance CPU cores, migrating all threads to the low-power fifth core. The idle power consumption shown for both the “Linux idle” test and the non-GPU-accelerated VMD are nearly identical, and these results match the minimum power consumption observed from system power-on, prior to booting Linux.
TABLE I.
Basic hardware attributes and power consumption.
| Test Case | CARMA, Quadro 1000M | KAYLA, GeForce GTX Titan | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Perf. | Watts | Energy Eff. | Perf. | Watts | Energy Eff. | |
|
| ||||||
| Linux idle | - | 11W | - | - | 29W | - |
| VMD text-mode, idle | - | 11W | - | - | 30W | - |
| VMD-CUDA text-mode, idle | - | 18W | - | - | 80W | - |
| STREAMBW-copy | 1,090 MB/s | 13W | 84MB/J | 1,102 MB/s | 32W | 34 MB/J |
| CUDA-PCIEBW-pinned | 449 MB/s | 20W | 22MB/J | 451 MB/s | 97W | 4.6 MB/J |
| CUDA-MEMBW | 24,500 MB/s | 34W | 720 MB/J | 241,000 MB/s | 201W | 1,199 MB/J |
| CUDA-MADD | 175 GFLOP/s | 35W | 5.0 GFLOP/J | 2,923 GFLOP/s | 202W | 14.4 GFLOP/J |
In contrast to the CPU-only idle power results above, a very pronounced increase in idle power consumption was observed for the GPU-accelerated VMD-CUDA test case, with the idle power increasing by factors of 1.6× (CARMA) and 2.76× (KAYLA) over the “Linux idle” and non-GPU-accelerated VMD idle tests. We examined this in detail and determined that the significant increase in idle power consumption is associated with the creation of a CUDA “context”, and that the idle power consumption remains high even if the associated process is stopped in the scheduler or otherwise blocked in kernel wait. We found that idle power consumption remains increased for as long as any process holds a CUDA context, regardless of whether that process is considered “runnable” by the scheduler or not. This behaviour is the result of the GPU device driver initializing the GPU and raising it out of the idle power state, presumably as a means of improving responsiveness and overall performance in the common scenario where there are a series of CUDA kernel launches or host– device memory copies interspersed among very brief periods of GPU inactivity. The CUDA runtime system currently lacks a mechanism to allow an application to indicate that it is entering an idle phase where the GPU could be placed into a minimum-power idle state. If such a mechanism were added to CUDA, the GPU driver would have the opportunity to significantly improve the idle power consumption of GPU-accelerated applications that are interactive and must sometimes wait for user input, and for applications that only utilize the GPU for a subset of algorithms or workloads.
The remaining tests in Table I measured both performance and power consumption associated with several CPU and GPU microbenchmarks that measured peak CPU and GPU memory bandwidths (STREAMBW-copy, and CUDA-MEMBW), host–GPU PCIe transfer bandwidths (CUDA-PCIEBW-pinned), and peak GPU single-precision multiplyadd arithmetic throughput (CUDA-MADD). Each of the microbenchmarks used are implemented as built-in system benchmarking routines in VMD. Power consumption measurements were made for short-running tests, e.g. CUDA-MADD, by running the tests continuously in a loop. For each performance test, power consumption is reported in Watts, and energy efficiency in terms of work units per Joule.
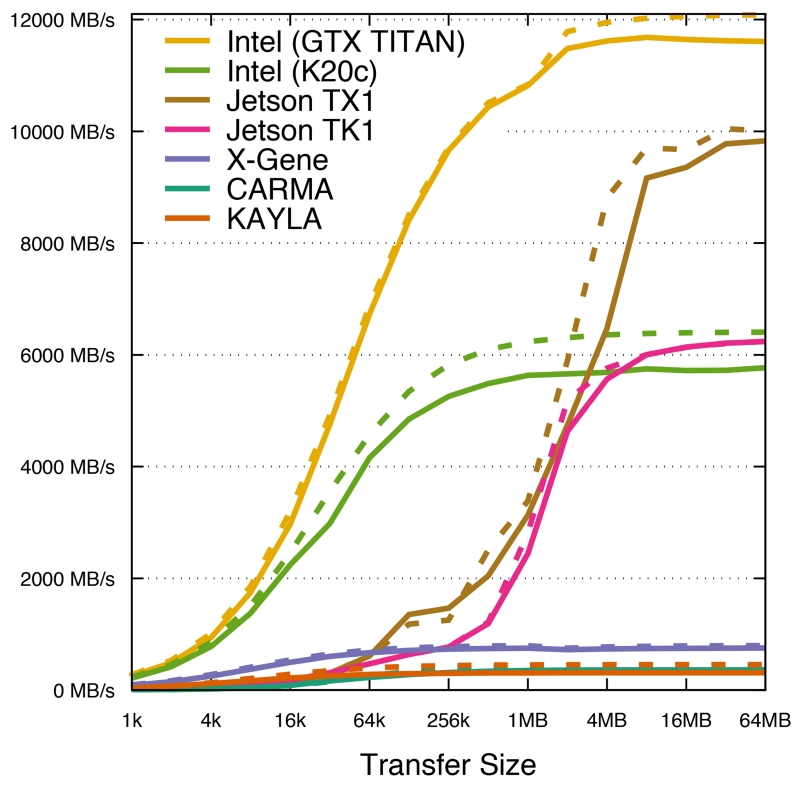
To better understand the PCIe performance of these systems, Fig. 2 shows the achieved transfer bandwidth across a range of payload sizes. An Intel i7-3960X system was used to evaluate performance for GPUs connected to a host with PCIe 2.0 or PCIe 3.0 link rates. All of the ARM-based platforms incur substantially higher latencies for initiating small-size GPU data transfers than the Intel i7-3960X platform. CARMA, KAYLA, and the X-Gene are an order of magnitude less performant for memory transfers than the Intel i7-3960X host. The Jetson TK1 and TX1 systems perform considerably better, likely due to their use of a unified memory architecture for their CPUs and GPUs. The Jetson TK1 and TX1 are able to achieve full bandwidth beginning with transfer sizes just beyond 4 MB.
Fig. 2.
PCI-Express transfer rates for host to device copies (solid lines), and device to host copies (dashed lines) for the ARM SoC platforms tested. Also shown is the transfer rate for a conventional Intel i7-3960X system with a Tesla K20c GPU, demonstrating PCIe 2.0 ×16 transfer rates of approximately 6 000 MB/s, and an Intel i7-3960X system with a GeForce GTX TITAN, demonstrating PCIe 3.0 ×16 rates of 12 000 MB/s. CARMA and KAYLA obtain a top speed of 450 MB/s, and X-Gene transfers at a maximum rate of 800 MB/s. The Jetson TK1 and TX1 systems have unified memory systems that support both the CPU and GPU, so memory copies don’t pass through a PCIe bus. The Jetson TK1 is able to perform at speeds equivalent to PCIe 2.0 ×16; the TX1 falls short of full PCIe 3.0 ×16 speed, but is the best performer of all of the ARM SoC systems. For small data transfers, performance is latency bound for the ARM systems. CARMA, KAYLA, TK1, and TX1 all take over 180 μs to perform a 1 kB copy. The X-Gene latency improves upon these results dramatically completing the transfer in 12 μs. The Intel i7-3960X has the lowest PCIe overhead, completing the transfer in as little as 4 μs.
III. Power Monitoring Approach
We have taken two approaches for measuring power consumption through the use of commercial AC power monitoring devices, and with custom-made DC power monitoring circuits constructed from low-cost components and commercial analog-to-digital conversion interfaces (A/D) or digital storage oscilloscopes (DSOs).
The CARMA board contains an NVIDIA Tegra 3 SoC, on-chip and on-board I/O peripherals, and an NVIDIA Quadro 1000M GPU, all powered by main board circuit traces. The CARMA board is powered by an external Mean Well switching power supply that provides 19.0 V DC at up to 4.74 A (90 W). Due to the lack of circuit diagrams, bus extenders, or test probe cards for the custom daughter cards containing the Tegra 3 and Quadro 1000M, power measurements monitored system level power at the board-level DC input, and at the AC input to the power supply. The CARMA board and associated power monitoring instrumentation are shown in the Fig.1 photograph. In the future, we plan to use the same power monitoring approach taken for the CARMA board for detailed application power profiling on the Jetson TK1, Jetson TX1, and similar single-board systems that require only low-voltage DC input power.
Fig. 1.
Photograph of power monitoring instrumentation attached to the CARMA development board, while running VMD molecular orbital benchmark tests described in Sec. IV-C, and Table V.
The KAYLA development board contains the NVIDIA Tegra 3 SoC, on-chip and on-board I/O peripherals, and it provides a single PCIe 2.0 ×16 (physical) slot for discrete GPUs with PCIe interfaces. The KAYLA main board obtains power from an external ATX switching power supply through a standard ATX motherboard power connector. The discrete PCIe GPU installed in the KAYLA board obtains its power from both the PCIe bus itself and additional PCIe GPU power cables attached to the ATX power supply. In the present work we have obtained measurements of the KAYLA AC power input only. Accurate DC power monitoring for KAYLA, conventional Intel x86 systems, the AppliedMicro X-Gene system, and similar platforms would require multiple current sensors, multi-channel A/D, and post-processing and calibration of per-channel measurements, which we leave as future work.
Commercially available power monitoring devices provide inexpensive means to measure overall system power consumption at AC power supply inputs. AC power consumption measurements were performed using a commercially made Kill A Watt® model p4400 meter, produced by P3 International. The Kill A Watt meter provides readings that are rated to 0.2% accuracy and AC measurements reported were obtained by running workloads that yielded long-term constant average power consumption readings enabling simple single-value measurements to be made. Since hardware configurations vary between the systems under test, e.g. different models of SSDs, hard drives, and memory capacity, the Kill A Watt provides more than sufficient accuracy to support the low-temporal-resolution parts of the power efficiency evaluations and conclusions presented in this work. Many alternative commercial AC power monitoring solutions exist, but the Kill A Watt devices are inexpensive and widely available, and we have experience using them in previous power monitoring of molecular modeling workloads on GPU clusters [6]. We note that these approaches provided only coarse temporal resolution on the order of one sample per-second, due to limitations in the underlying commercial power monitoring instrumentation itself.
We sought to be able to monitor power consumption with very fine temporal resolution sufficient to resolve the power consumption of individual GPU kernel functions. Ge et al. constructed sophisticated power monitoring infrastructure using commercially available power monitoring components capable of monitoring individual subsystems and components within cluster nodes [19]. We note that in the case of platforms based on SoC designs such as CARMA and KAYLA, it is difficult or impossible to directly measure power consumption of on-chip circuitry with external instrumentation, and that it may only be practical to instrument outboard components such as disks, and external GPUs.
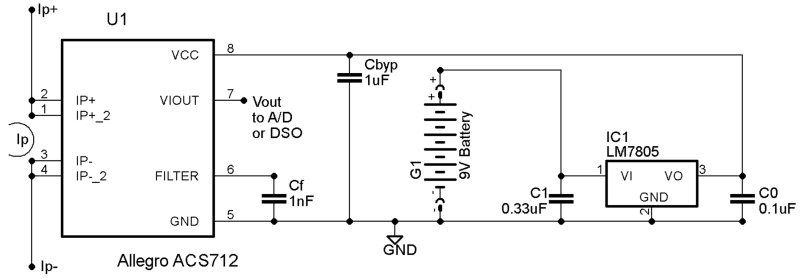
To overcome the limitations of existing monitoring devices, we built our own DC power monitoring instrumentation combined with commercial A/D instruments and DSOs. Our DC power monitoring hardware is composed of an Allegro Microsystems ACS 712 Hall effect current sensor, powered by either an LM 7805 voltage regulator driven by a common 9 V battery, or a sufficiently low-noise 5 V power supply, as shown in Figs. 3, and 4. The reported results were obtained using an Allegro ACS 712 rated for currents of up to 5 A and total error below 1.5%. The 5 A model yields the best measurement precision for the input voltages and currents required by the SECO CARMA board under test. The 5 A ACS 712 produces an output voltage signal of 2.500 V plus 185 mV/A of measured current, which is measured by external A/D instrumentation or a DSO. The peak frequency response or temporal resolution provided by the ACS 712 is roughly 80 kHz, and it can be set to a lower frequency by increasing filtering capacitor Cf in Fig. 3. The output voltage of the ACS 712 is proportional to its 5 V supply, so low-noise measurements require a low-noise power supply to the ACS 712. Our measurements were conducted using a simple battery-driven LM 7805 voltage regulator to minimize noise and to avoid ground loops or other undesirable coupling to the attached DSO or A/D instrumentation, but good results can be achieved using any sufficiently low-noise power supply.
Fig. 3.
Schematic of low-voltage DC power monitoring circuit based on the Allegro Microsystems ACS 712 Hall-effect current sensor, and common voltage regulator parts.
Fig. 4.
Photograph of our power monitoring circuit, connected to a networked Labjack UE9 high speed analog-to-digital conversion interface.
The high sample rate power profiles presented in this paper were obtained using a Labjack UE9 analog A/D interface and a Rigol DS2102 100 MHz digital storage oscilloscope. The UE9 provides multi-channel A/D conversion and sample rates up to 57,000 samples/s, depending on the number of channels sampled and the required precision. We operate the UE9 in a streaming mode, continuously sending A/D samples over the network, allowing high-frequency power measurements to be recorded for long running applications. The Rigol DS2102 DSO provides two input channels, and when set for maximum recording time, can record at a rate of 1M samples/s for up to 1.4 s. The extremely high sample rate of the DS2102 exceeds the frequency response of the ACS 712 sensor by over a factor of ten, but multiple samples can be averaged to reduce noise. The DSO was particularly useful for identifying and eliminating power supplies that generated excessive ripple or noise on their DC output.
Both the UE9 and DS2102 can be triggered by external trigger signals or by software, but in the current work we used software-based triggering. We developed C++ libraries to drive both the Labjack UE9 and the Rigol DS2102, with the aim of linking the software into existing CPU and GPU performance profiling tools developed by others. To the best of our knowledge, our DC power monitoring instrumentation achieves a sampling rate higher than has been reported by recent efforts. In many cases our sample rate is two orders of magnitude higher, based on the limitations of the hardware instrumentation described or specific sample rates reported in each case [20], [19], [21], [22], [23], [24]. Our approach is more limited in scope compared to more general power monitoring frameworks due to the minimalistic nature of the SoC-based CARMA and KAYLA hardware platforms we study, and the complexities involved in monitoring outboard PCIe GPU power consumption [23], [24].
IV. Application Test Cases and Key Algorithms
Test cases were selected for evaluation of the performance and energy efficiency of the CARMA and KAYLA platforms as compared with conventional workstation and cluster node architectures based on 64-bit Intel and AMD processors. The test cases were selected to determine whether particular features of the hardware architecture or algorithms used in the science applications exposed strengths or weaknesses in relation to performance and energy efficiency.
A. Lattice Microbes
Several Lattice Microbes (LM) [15], [16], [17] test cases were selected to exercise CPU and GPU implementations of its stochastic solvers, providing direct comparison of energy efficiency on the test architectures.
A serial process at the heart of the CME algorithm consumes a profusion of GPU-generated random numbers to determine if a reaction occurs, with a probability determined by the “propensity matrix”, R. While algorithm execution is synchronous, random number generation and data output occur concurrently.
On the other hand, the lattice-based RDME algorithm enjoys fine-grained parallelism as described by:
| (1) |
The CME is solved independently on each lattice site (v ∈ V), assuming that only the contained particles (α) can interact. Inter-site () diffusion of particles–which occurs with probability D–enjoys considerable spatial locality.
A bimolecular reaction, where two particles combine or separate at rates k1 and k2, is a quintessential test case with regular, predictable serial execution. To test ranges of biologically relevant particle counts, simulations are run varying the number of A and B particles at t0 from 1K–250K. Lattice sizes for the bimolecular reactions are varied from 323 to 2563 (memory use between 256 kB and 512 MB). Each lattice site is 8 bytes in size and can store one particle per byte. The first set of simulations begin with 1,000 A and B particles over all lattice sizes. A second set of simulations scale the number of A and B with the lattice size.
For simulating RDME processes, we employ our multi-particle diffusion algorithm (MPD-RDME [15], [16]). The algorithm is composed of four CUDA kernels on the GPU: Three kernels perform x-, y-, and z-axis particle diffusion and one kernel to perform reactions. The host then checks for lattice site overflow events: The GPU overflow buffer must be copied to host memory, and then if necessary, the host corrects lattice overflows before beginning the next timestep. Beyond overflow handling, the CPU does little more than schedule GPU operations.
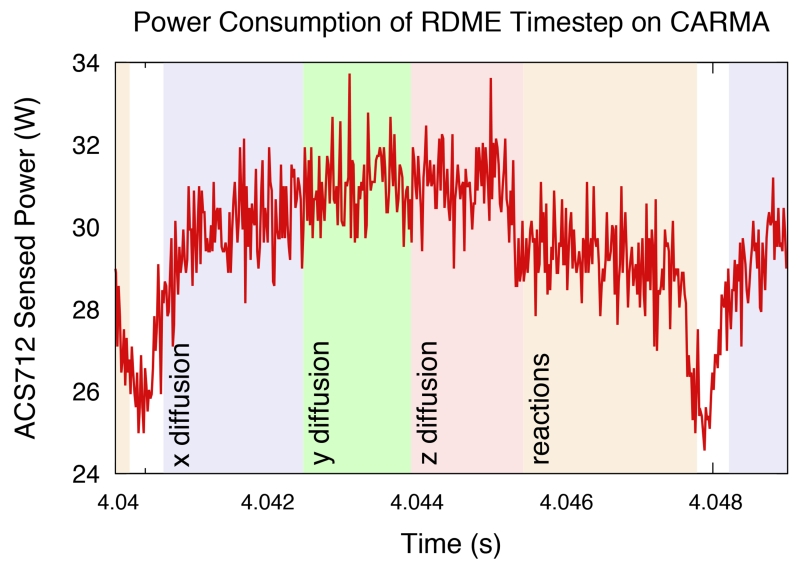
Figure 5 shows a high-resolution power profile for an RDME timestep annotated with the GPU kernel that is being executed. It is interesting to learn that the GPU utilization and efficiency of the kernel are evident from examination of power usage. The y- and z-axis kernels draw more power than the other two, however their execution time is shorter. This is consistent with the predicted GPU occupancy for each kernel. The y- and z-axis kernels have an occupancy of 66% whereas the occupancy of the x-axis and reaction kernels is 33%. Since they are less able to fully occupy the GPU, the power draw is below peak. One can also determine certain execution characteristics, such as the timestep length, solely from high-resolution power measurements. This opens the door for “black-box” instrumentation where execution profiling can be performed without any source-level or runtime modifications.
Fig. 5.
High-resolution power trace showing the CARMA platform running Lattice Microbes RDME timestep on the 643 lattice. The y- and z-axis diffusion kernels exhibit shorter run times and higher power usage than x-axis diffusion and reaction kernels due to higher GPU occupancy.
Table II shows RDME simulation rates, power usage, and resulting efficiency for CARMA and KAYLA. Table III compares a computer with an Intel i7-3960X processor and a Tesla K20c against the Applied Micro X-Gene system with the same GPU. Each successive lattice size requires significantly more work; as the lattice edge length doubles, the lattice volume increases by a factor of eight. MPD-RDME workload scales linearly with the number of lattice sites and with the number of particles and reactions present in the simulation [17]. KAYLA, with the discrete GPU, outperforms CARMA and is more energy efficient in all but the 323 lattice models. Lattice Microbes performance is predominantly dependent on the GPU, and not the CPU. This is demonstrated clearly with the comparison between the Intel-based system and the X-Gene system where the GPU is identical. The X-Gene system draws considerably less power than the Intel system, but the application performance is similar. An approximate 25% increase in energy efficiency is gained from running on the ARM-based X-Gene platform.
TABLE II.
Lattice Microbes RDME performance and power efficiency for the CARMA and KAYLA ARM development boards. Simulation rate values are in steps per second, power in watts, and efficiency values are in timesteps per Joule of energy expended.
| Lattice Size | Particles | CARMA, Quadro 1000M | KAYLA, GeForce GTX Titan | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Sim. Rate | Power | Efficiency | Sim. Rate | Power | Efficiency | ||
|
| |||||||
| 323 | 2, 000 | 726 steps/s | 31W | 23.4 steps/J | 1304 steps/s | 137W | 9.5 steps/J |
| 323 | 64, 000 | 169 steps/s | 30W | 5.6 steps/J | 226 steps/s | 143W | 1.6 steps/J |
| 643 | 2, 000 | 184 steps/s | 34W | 5.4 steps/J | 1000 steps/s | 158W | 6.3 steps/J |
| 643 | 128, 000 | 130 steps/s | 34W | 3.8 steps/J | 863 steps/s | 175W | 4.9 steps/J |
| 1283 | 2, 000 | 27.9 steps/s | 34W | 0.82 steps/J | 301 steps/s | 193W | 1.6 steps/J |
| 1283 | 256, 000 | 21.7 steps/s | 34W | 0.64 steps/J | 253 steps/s | 203W | 1.2 steps/J |
| 2563 | 2, 000 | 3.50 steps/s | 34W | 0.10 steps/J | 45.2 steps/s | 205W | 0.22 steps/J |
| 2563 | 512, 000 | 3.05 steps/s | 35W | 0.09 steps/J | 40.4 steps/s | 212W | 0.19 steps/J |
TABLE III.
Lattice Microbes RDME performance and power efficiency on Intel and X-Gene ARM systems, both with a single Tesla K20c GPU. Comparable performance is found on both platforms, but the X-Gene is 25% more efficient for many of the test systems.
| Lattice Size | Particles | Intel i7-3960X, Tesla K20c | X-Gene, Tesla K20c | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Sim. Rate | Power | Efficiency | Sim. Rate | Power | Efficiency | ||
|
| |||||||
| 323 | 2, 000 | 5463 steps/s | 226W | 24.2 steps/J | 4638 steps/s | 142W | 32.6 steps/J |
| 323 | 64, 000 | 1718 steps/s | 229W | 7.5 steps/J | 744 steps/s | 139W | 5.4 steps/J |
| 643 | 2, 000 | 1991 steps/s | 247W | 8.1 steps/J | 1844 steps/s | 166W | 11.1 steps/J |
| 643 | 128, 000 | 1343 steps/s | 252W | 5.2 steps/J | 1265 steps/s | 170W | 7.4 steps/J |
| 1283 | 2, 000 | 417 steps/s | 264W | 1.6 steps/J | 395 steps/s | 186W | 2.1 steps/J |
| 1283 | 256, 000 | 305 steps/s | 266W | 1.2 steps/J | 300 steps/s | 189W | 1.6 steps/J |
| 2563 | 2, 000 | 58.9 steps/s | 268W | 0.22 steps/J | 54.1 steps/s | 192W | 0.28 steps/J |
| 2563 | 512, 000 | 48.3 steps/s | 270W | 0.18 steps/J | 47.7 steps/s | 195W | 0.24 steps/J |
For smaller simulations, especially on the CARMA and KAYLA platforms, the PCIe bus plays a factor in performance. The overflow buffer, which is 4 kB in size, is examined at the end of every timestep to check for any particles that need to be re-inserted into the simulation. Figure 2, shows that the performance of the ARM-based systems lags considerably behind the Intel i7-3960X for 4 kB transfers, with the X-Gene performing the best among the ARM systems for transfers below 64 kB. On CARMA and KAYLA, this copy takes approximately 0.2 ms. On the Intel system, it takes 0.03 ms for the copy, and 0.05 ms on the X-Gene. That is significant overhead from PCIe latency for CARMA and KAYLA, especially for the small lattice sizes where the GPU kernel runtimes are very short. On KAYLA, 323 lattice with 2, 000 particles runs at 1, 304 steps per second; 23% of that runtime is copying the overflow buffer. If ARM platforms supported host-mapped memory, this copy could be entirely avoided.
Only the 323 lattice with 64, 000 particles simulation exhibits overflows. Copying the lattice to host memory and back to the device for overflow correction is costly when PCIe bandwidth is low. On both CARMA and KAYLA, an average of 3.5ms is spent on correcting overflows, accounting for 65% of the runtime on CARMA and 87% on KAYLA, where the GPU is not in use. This inefficiency is reflected in the relatively poor performance reported for the 323/64, 000 simulation. The dependence on CPU performance for overflow handling is also apparent in comparing the Intel system to X-Gene for this simulation. The Intel system is 2.3 times faster, and is the only test model where the Intel system is superior in terms of energy efficiency.
CME is primarily implemented on the CPU, and is accelerated by the GPU by offloading random number generation. This partitioning keeps a CPU core busy but does not keep the GPU consistently busy; as a result multiple independent CME replicas can be simultaneously run on multiple CPUs while sharing a single GPU. We initially observed very poor performance when multiple replicas shared the GPU on both the CARMA and KAYLA platforms. Upon inspection, the majority of the time spent the GPU spent copying the random numbers to the host due to the low PCIe bandwidth. Delayed completion of memory copies due to bus contention would occasionally make the CME threads stall until more random numbers could be produced, even though the actual generation is very quick. We reduced the amount of data that needed to be transferred by reducing the batch size for the random number generation, which reduced contention. This requires the kernel to be called more often, however these calls are fully overlapped with the CPU computation.
The CARMA platform executes the random number kernel in 1.7 ms, and the KAYLA platform with the GTX Titan in 252 μs for a batch of 256K double-precision numbers. The random number generation kernel produces data at rates of 1.2 GB/s and 7.9 GB/s respectively, while the observed bandwidth for device-to-host memory transfers is roughly 450 MB/s, further demonstrating the imbalance between computation and communication efficiency on these hardware platforms.
B. NAMD
Porting NAMD and the Charm++ runtime system to the ARM architecture initially took roughly two days of effort. One limitation of existing ARM platforms with respect to support for the NVIDIA CUDA toolkit and driver is the lack of support for host-mapped memory, which is normally used to implement a low-latency mechanism to inform the Charm++ runtime system running on the host CPUs when GPU work units have completed. By having GPU kernels writing work completion status to host-mapped memory, it becomes possible for CPU-side operations on GPU output to begin sooner than if they had to wait for enqueued GPU kernels to complete. The lack of support for host-mapped memory on the ARM platform is the result of limitations in the implementation of PCIe transfers and interaction with the ARM CPU caches, and is therefore unlikely to change in the near future.
NAMD decomposes its simulation work across the available CPUs and GPU accelerators, moving the most demanding non-bonded force calculations onto the GPU, and performing comparatively low-cost bond, angle, dihedral, and improper force terms on the host CPU(s). As a result of the decomposition of force computations across both the CPU and GPU hardware, NAMD must perform frequent host-device memory transfers to exchange atom positions, forces, and energies between the GPU and the host. One of the hardware limitations of the CARMA and KAYLA platforms that has a strong impact on the performance of these transfers is the PCIe host-device transfer bandwidth, as reported in the CUDA-PCIEBW-pinned tests shown in Table I, and plotted in Fig. 2.
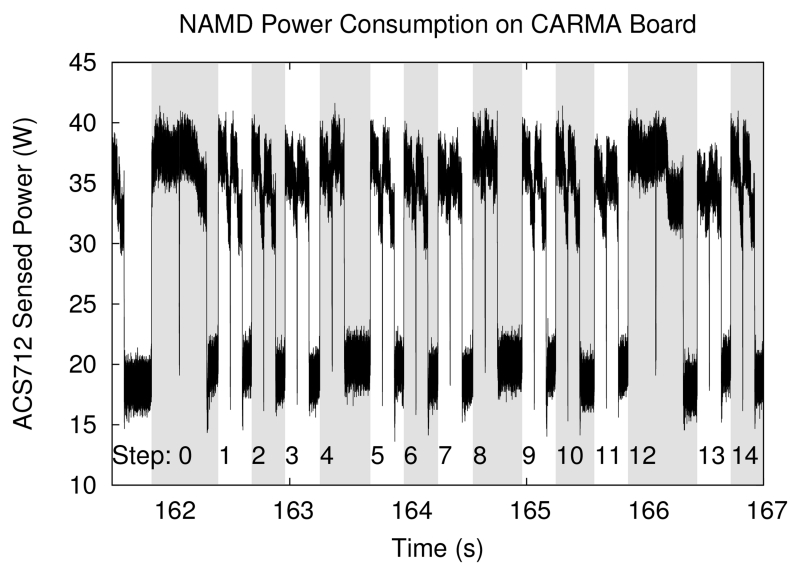
We chose to test NAMD on a moderately sized ApoA 1 system because it represents a typical per-node workload for GPU-accelerated systems, and it was used as a test case in our previous work [11]. Power and performance results are shown in Figs. 6 and 7, and in Table IV. For NAMD, driving the GPU with a high-performance CPU maximizes power efficiency as well as performance.
Fig. 6.
Measured power consumption of NAMD running ApoA 1 benchmark. Timestep boundaries are marked by shaded bands. Steps correspond to Fig. 7. Step 0 is preceded by atom migration; steps 0, 4, 8, and 12 include PME long-range electrostatics; steps 0 and 12 also include pairlist regeneration; all other steps calculate short-range forces only. The GPU calculation for each step is divided into two grids, with the boundaries between grids visible as brief drops in power usage.
Fig. 7.
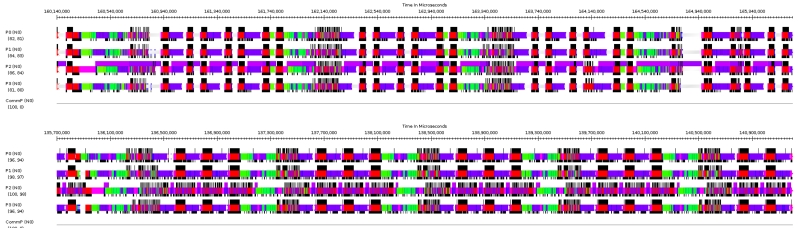
Execution of NAMD running ApoA 1 benchmark. Top: CARMA (Quadro 1000M). Bottom: KAYLA (GeForce GTX Titan). Both figures span 5.5 seconds. CARMA corresponds to Fig. 6; note PME work indicated in green. GPU execution is indicated by violet bands above P2 traces. Note that GPU is mostly busy on CARMA (top), mostly idle on KAYLA (bottom). Integration (red blocks) is split into two blocks per timestep on CARMA (top), corresponding to separate GPU grid completions, while on KAYLA (bottom) these blocks appear merged because GPU results are available before CPU is idle. Traces generated with Projections tool for Charm++ parallel runtime system.
TABLE IV.
NAMD power efficiency across platforms.
| Platform | Time/step | Power | Steps/kJ | Speedup |
|---|---|---|---|---|
|
| ||||
| CARMA (Q 1000M) | 0.350 s | 34W | 84 | 4.3X |
| KAYLA (GT 640) | 0.267 s | 45W | 83 | 6.2X |
| KAYLA (GTX Titan) | 0.283 s | 93W | 38 | 5.9X |
| i7-3960X (GTX Titan) | 0.0185 s | 444W | 122 | 5.8X |
C. VMD
VMD is a GPU-accelerated tool for preparing, analyzing, and visualizing molecular dynamics simulations [25], [4]. VMD has recently been extended with features for visualization and analysis of cellular simulations [16], and has been adapted for parallel analysis and visualization of large scale simulation trajectories on petascale computers [26], [27], [28], [29], [30], [31].
The main limitation of the CARMA and KAYLA hardware that impacts the performance and energy efficiency of the VMD algorithms we evaluate is the PCIe host-GPU transfer bandwidth, as reported in the CUDA-PCIEBW-pinned tests shown in Table I, and plotted in Fig. 2. Below, we find that this limitation, as with the NAMD benchmarks, has a profound impact on the performance scaling of the GPU algorithms in VMD. This is due in part to the fact that our algorithm designs assumed high-bandwidth PCIe connectivity, and long-term storage of visualization data structures in comparatively large size host memory, thereby enabling GPU memory to be used for detailed renderings of petascale trajectories [27]. Surveying the attributes of other recently announced and currently available ARM SoCs, we find that all of the existing platforms have limitations on PCIe bandwidth, with none providing more than 8-lane PCIe 2.0 bandwidth to our knowledge. This observation leads us to conclude that in order to obtain best performance from similar platforms, we must begin to adapt our existing visualization and analysis algorithms to reduce dependencies on high-bandwidth host-GPU transfers. Below we demonstrate the potential performance gains that can be achieved by redesigning our molecular orbital visualization algorithm to reduce host-GPU transfers.
We evaluated the performance of an electrostatics algorithm [32] that has been evaluated previously on conventional GPU-accelerated platforms [6], and GPU-accelerated algorithms for visualization of molecular orbitals [33], [34] and molecular surfaces [35], [27]. Table V lists the performance results and energy efficiency for each of the tests measured on the CARMA and KAYLA boards. In all three of the VMD test cases, performance results were adversely affected by the very limited PCIe bandwidth.
TABLE V.
VMD performance power consumption measurements.
| Test Case | CARMA, Quadro 1000M | KAYLA, GeForce GTX Titan | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Perf. | Watts | Energy Eff. | Perf. | Watts | Energy Eff. | |
|
| ||||||
| C60 orbital calc+viz (orig. alg.) | 1.43 frames/s | 26W | 55 frames/kJ | 2.12 frames/s | 88W | 24 frames/kJ |
| C60 orbital calc+viz (new alg.) | 2.62 frames/s | 26W | 100 frames/kJ | 3.89 frames/s | 89W | 49 frames/kJ |
| Ankryin surface calc+viz | 0.49 frames/s | 22W | 22 frames/kJ | 0.61 frames/s | 86W | 7.1 frames/kJ |
| STMV electrostatic potential map | 13.41 s/frame | 35W | 2.12 frames/kJ | 12.05 s/frame | 102W | 0.81 frames/kJ |
The “STMV electrostatic potential” test case computed a 3-D electrostatic potential map for satellite tobacco mosaic virus, using the multilevel summation method [32]. Performance of the electrostatic potential test case was limited by significant 3-D potential interpolation work that is not currently GPU-accelerated. The interpolation work was previously an inconsequential component of overall runtime, but the high speed of the CARMA and KAYLA GPUs relative to Tegra 3 SoC makes this algorithm step a significant performance bottleneck.
The C60 molecular orbital visualization test case stresses both the CUDA molecular orbital kernels, and the subsequent marching cubes stage that extracts the orbital surface and creates the triangle mesh for rendering via OpenGL. Two molecular orbital algorithms were tested with the C60 case. The C60 “orig. alg.” case presents performance and efficiency data for our original GPU-accelerated molecular orbital algorithm [33], [34]. The C60 “new alg.” case presents results for a revised algorithm, designed to overcome the extremely limited bandwidth of host-GPU PCIe transfers, by moving the marching cubes algorithm from the CPU (per the original algorithm), entirely onto the GPU, resulting in a 1.8× increase in performance and energy efficiency for the C60 test case on both test platforms.
The “Ankyrin molecular surface” test case computes and visualizes molecular surfaces for an Ankyrin unfolding simulation. The VMD molecular surface algorithm used in the performance tests already implements the same optimization described for the C60 molecular orbital test case above, however the final stages of the Ankyrin molecular surface test case generate a much larger triangle mesh, demonstrating that even a highly streamlined GPU algorithm can succumb to performance bottlenecks caused by low host-GPU transfer bandwidth. The final host-GPU transfer could be eliminated from the molecular surface algorithm assuming that the GPU had sufficient memory capacity for both intermediate data structures and the resulting triangle mesh, but all current GPU hardware falls short in this respect.
An exciting development in the field of molecular visualization is the recent availability of commodity head mounted displays (HMDs) which are an ideal platform for immersive stereoscopic visualization of large biomolecular complexes [36], [37], [38]. The GPU workloads associated with rasterization or ray tracing of very large biomolecular complexes are currently beyond the capabilities of the platforms discussed in this paper, but they appear to be capable of supporting remote visualization schemes where remote supercomputers render omnidirectional stereoscopic projections at very high resolution and stream the results to remote clients, which perform local view-dependent reprojection and display for the user’s HMD orientation [31], [38]. Rudimentary performance tests were performed on the CARMA and Jetson TK1 SoC platforms, which are physically small and have a low enough power requirement that they could potentially be self-contained to drive commodity HMDs. The performance results for these tests indicate that the CARMA and Jetson TK1 can easily perform the required omnidirectional image reprojection tasks using OpenGL. The test platforms were able to perform omnidirectional texture mapping reprojection and HMD lens distortion corrections at frame rates exceeding 150 Hz when driving a display at 1920 × 1080 resolution, with sufficient performance headroom that we believe that the rasterization part of the workload will not impede the use of these platforms for driving HMDs for remote rendering. The remaining components of the workload include H.264 video stream decoding, which could in principle be handled by on-chip video decoding hardware present on the NVIDIA Tegra 3 and Tegra K1 SoCs. We leave the evaluation of H.264 video stream decode performance as future work.
V. Results Discussion and Hardware Outlook
The CARMA and KAYLA test platforms that we have evaluated in detail above combine low-power NVIDIA Tegra 3 ARM SoCs which are commonly incorporated into mobile phones and tablet computing devices with GPUs that achieve substantially higher aggregate performance, albeit with increased power consumption. The net energy efficiency benefits obtained from the use of such platforms depends critically on how effectively the high-performance and high-power consumption parts of the system are utilized.
In the case of the platforms we evaluated, we have shown that it is critical for the GPU hardware to be fully utilized in order to achieve best energy efficiency. One of the limitations we observed in the performance of the CARMA and KAYLA development boards is that the Tegra 3 ARM SoCs on these systems lack full PCIe 2.0 ×16 bus bandwidth, creating a performance bottleneck for some algorithms, particularly those that regularly exchange large datasets between the CPU and the GPU during the course of execution. In light of these observations, we revised the GPU-accelerated molecular orbital algorithm in VMD to avoid host-GPU transfers between algorithm stages, thereby achieving a 1.8× performance and energy efficiency improvement for the C60 test case on both the CARMA and KAYLA platforms.
While the CARMA and KAYLA platforms we study in detail provide relatively low host-GPU transfer rates, limitations on PCIe bandwidth are also common among several more recently announced ARM platforms, including server-oriented processors such as the AppliedMicro X-Gene, and the AMD A1100. One potential resolution to PCIe bandwidth limitations would be the incorporation of support for GPU I/O buses such NVIDIA’s NVLink into ARM SoCs, thereby enabling an alternative data path for host-GPU transfers, but it is unclear if this is economically viable in low-cost mass market SoCs. New on-chip and die-stacked memory systems have begun to debut in discrete GPUs from AMD and have been announced by NVIDIA, increasing GPU global memory bandwidths by up to 4× relative to the GeForce GTX Titan used in the KAYLA and Intel i7-3960X benchmarks reported here.
We expect that upcoming 64-bit ARM SoCs will begin to address some of performance bottlenecks reported here, making them interesting targets for future evaluation. We have recently begun porting and testing efforts on the NVIDIA Jetson TK1 (32-bit Tegra K1 SoC), Jetson TX1 (64-bit Tegra X1 running 32-bit operating system software), and AppliedMicro X-Gene (64-bit). While the ARM SoCs share many of the same design attributes and performance issues we have already described based on our testing of the CARMA and KAYLA development boards, they have improved CPU clock rates, increased PCIe bandwidths, and they support newer versions of the CUDA compiler toolchain and drivers. The two models of Jetson boards will provide a new opportunity to evaluate heterogeneous platforms that provide a unified memory system that is shared by both the CPU and GPU, thereby potentially eliminating the need for explicit data transfers between host memory and GPU memory in many cases.
We have performed additional early testing on NVIDIA Jetson TK1 and TX1, and AppliedMicro X-Gene test platforms, albeit limited to tests that could be performed on the X-Gene system which has no display output and comparatively minimalistic operating system software. The results in Table VI (photo of systems shown in Fig. 8) compare performance for just the molecular orbital computation phase of the C60 molecular orbital visualizations described previously. The best performing ARM platform is the X-Gene system, which is limited to PCIe 2.0 ×8 transfer rates, but achieves the best combination of GPU kernel performance and low latency for small size CPU-GPU data transfers. We compared the performance of the ARM platforms against the Intel i7-3960X CPU, which operates at a much higher clock rate than the ARM systems and also benefits from full-bandwidth PCIe 3.0 ×16 GPU connectivity and substantially lower latency for small CPU-GPU data transfers.
TABLE VI.
Comparison of VMD molecular orbital kernel performance.
| Hardware platform, CPU, and GPU | VMD molecular orbital kernel runtime for C60 |
|---|---|
|
| |
| CARMA Tegra 3, Quadro 1000M | 2.170 s |
| Jetson TK1 Tegra K1 | 2.020 s |
| Jetson TX1 Tegra X1 (beta software) | 1.210 s |
| KAYLA Tegra 3, GeForce 640 | 0.989 s |
| KAYLA Tegra 3, GeForce Titan | 0.396 s |
| Applied Micro X-Gene, Tesla K20c | 0.243 s |
| Intel Core i7-3960X, Tesla K20c | 0.208 s |
| Intel Core i7-3960X, GeForce Titan | 0.157 s |
Fig. 8.
Photograph of NVIDIA/SECO CARMA and KAYLA, NVIDIA Jet-son TK1, and AppliedMicro X-Gene test platforms used for the performance comparisons in Table VI. The CARMA board is shown in the top center, KAYLA and GeForce 640 in lower left, two Jetson TK1s are shown at bottom, the AppliedMicro X-Gene system is shown at right, with a Jetson TX1 sitting on top of it.
The results in Table VI show performance that strongly correlates with the speed of the attached GPU and the peak host-GPU transfer bandwidth provided by each platform, shown in Fig. 2. The Intel i7-3960X system paired with the GeForce GTX Titan gains a significant additional performance boost due to its much higher PCIe 3.0 ×16 host-GPU transfer bandwidth, combined with low latency for small transfers. We note that the Jetson TK1 achieved performance comparable to that of CARMA, and the Jetson TX1 was close behind the KAYLA platform paired with the GeForce 640, both using only the SoCs on-chip integrated GPUs in each case. We expect that future tests with detailed power monitoring of the Jetson TK1 and Jetson TX1 may demonstrate significant increases in energy efficiency compared with CARMA and KAYLA. The AppliedMicro X-Gene performs competitively with the Intel i7-3960X given the substantial host-GPU PCIe transfer bandwidth advantage enjoyed by the i7-3960X. At the time of writing, the X-Gene platform and operating system don’t support a windowing system or graphics output, but we expect that if it did, that the overall C60 molecular orbital visualization performance would likely track the performance ratio we observe for the molecular orbital kernel since the visualization performance is also impacted affected by PCIe bandwidth and transfer latency. We look forward to repeating tests on the AppliedMicro X-Gene and Jetson TX1 with future operating system software that is more mature and robust. It is possible that the relatively large latencies that we observed for small-size CPU-GPU transfers will be addressed with improved operating system software and CUDA GPU kernel drivers.
VI. Conclusions
We have evaluated the performance of key computational biology applications on several emerging heterogeneous computing platforms that combine energy-efficient multi-core ARM CPUs with massively parallel GPU accelerators. The 25% energy efficiency increase obtained for the Lattice Microbes application running on the AppliedMicro X-Gene and NVIDIA Tesla K20c GPU was the most successful among our tests in terms of demonstrating the potential for ARM-based heterogeneous computing platforms to achieve significant energy efficiency gains for molecular and cellular simulation workloads. Our performance tests and analysis have shown areas where existing energy-efficient SoCs have architectural limitations that impact performance of molecular modeling applications, and we have demonstrated that careful adaptation or redesign of GPU-accelerated application kernels can help reduce the impact of such limitations on performance. We have presented power monitoring instrumentation that allows highly detailed snapshots of heterogeneous computing kernels and their power consumption behavior, correlated with individual GPU kernels. We have provided early results from benchmarks of molecular orbital kernels on two of the latest ARM SoCs, giving us a perspective of how future systems may evolve and the level of performance that we can hope to achieve as the platforms mature.
Acknowledgment
The authors wish to acknowledge support of the CUDA Center of Excellence at the University of Illinois, and NIH funding through grants 9P41GM104601, 5R01GM098243-02 and PHS 5 T32 GM008276.
References
- [1].Shalf J, Dosanjh SS, Morrison J. Exascale computing technology challenges. In: Palma JMLM, Dayd MJ, Marques O, Lopes JC, editors. VECPAR. Vol. 6449. Springer; 2010. pp. 1–25. ser. Lect. Notes in Comp. Sci. [Google Scholar]
- [2].Nickolls J, Buck I, Garland M, Skadron K. Scalable parallel programming with CUDA. ACM Queue. 2008;6(2):40–53. [Google Scholar]
- [3].Garland M, Le Grand S, Nickolls J, Anderson J, Hardwick J, Morton S, Phillips E, Zhang Y, Volkov V. Parallel computing experiences with CUDA. IEEE Micro. 2008;28(4):13–27. [Google Scholar]
- [4].Stone JE, Phillips JC, Freddolino PL, Hardy DJ, Trabuco LG, Schulten K. Accelerating molecular modeling applications with graphics processors. J. Comp. Chem. 2007;28:2618–2640. doi: 10.1002/jcc.20829. [DOI] [PubMed] [Google Scholar]
- [5].Stone JE, Hardy DJ, Ufimtsev IS, Schulten K. GPU-accelerated molecular modeling coming of age. J. Mol. Graph. Model. 2010;29:116–125. doi: 10.1016/j.jmgm.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Enos J, Steffen C, Fullop J, Showerman M, Shi G, Esler K, Kindratenko V, Stone JE, Phillips JC. Quantifying the impact of GPUs on performance and energy efficiency in HPC clusters. International Conference on Green Computing. 2010:317–324. [Google Scholar]
- [7].Owens JD, Houston M, Luebke D, Green S, Stone JE, Phillips JC. GPU computing. Proc. IEEE. 2008;96:879–899. [Google Scholar]
- [8].Anderson JA, Lorenz CD, Travesset A. General purpose molecular dynamics simulations fully implemented on graphics processing units. J. Chem. Phys. 2008;227(10):5342–5359. [Google Scholar]
- [9].Ufimtsev IS, Martinez TJ. Quantum chemistry on graphical processing units. 1 Strategies for two-electron integral evaluation. J. Chem. Theor. Comp. 2008;4(2):222–231. doi: 10.1021/ct700268q. [DOI] [PubMed] [Google Scholar]
- [10].Lindholm E, Nickolls J, Oberman S, Montrym J. NVIDIA Tesla: A unified graphics and computing architecture. IEEE Micro. 2008;28(2):39–55. [Google Scholar]
- [11].Phillips JC, Stone JE, Schulten K. SC ’08: Proceedings of the 2008 ACM/IEEE Conference on Supercomputing. IEEE Press; Piscataway, NJ, USA: 2008. Adapting a message-driven parallel application to GPU-accelerated clusters; p. 9. [Google Scholar]
- [12].Giupponi G, Harvey M, Fabritiis GD. The impact of accelerator processors for high-throughput molecular modeling and simulation. Drug Discovery Today. 2008;13(23-24):1052–1058. doi: 10.1016/j.drudis.2008.08.001. [DOI] [PubMed] [Google Scholar]
- [13].Friedrichs MS, Eastman P, Vaidyanathan V, Houston M, Legrand S, Beberg AL, Ensign DL, Bruns CM, Pande VS. Accelerating molecular dynamic simulation on graphics processing units. J. Comp. Chem. 2009;30(6):864–872. doi: 10.1002/jcc.21209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stone JE, Gohara D, Shi G. OpenCL: A parallel programming standard for heterogeneous computing systems. Comput. in Sci. and Eng. 2010;12:66–73. doi: 10.1109/MCSE.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Roberts E, Stone JE, Sepulveda L, Hwu WW, Luthey-Schulten Z. Long time-scale simulations of in vivo diffusion using GPU hardware. Proceedings of the IEEE International Parallel & Distributed Processing Symposium. 2009:1–8. [Google Scholar]
- [16].Roberts E, Stone JE, Luthey-Schulten Z. Lattice microbes: High-performance stochastic simulation method for the reaction-diffusion master equation. J. Comp. Chem. 2013;34:245–255. doi: 10.1002/jcc.23130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hallock MJ, Stone JE, Fry ERC, Luthey-Schulten Z. Simulation of reaction diffusion processes over biologically relevant size and time scales using multi-GPU workstations. J. Paral. Comp. 2014;40:86–99. doi: 10.1016/j.parco.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ge R, Feng X, Song S, Chang H-C, Li D, Cameron KW. Powerpack: Energy profiling and analysis of high-performance systems and applications. IEEE Trans. Parallel Distrib. Syst. 2010;21(5):658–671. [Google Scholar]
- [20].Feng X, Ge R, Cameron K. Power and energy profiling of scientific applications on distributed systems. Parallel and Distributed Processing Symposium, 2005. Proceedings. 19 th IEEE International. 2005 Apr;:34–34. [Google Scholar]
- [21].Esmaeilzadeh H, Cao T, Xi Y, Blackburn SM, McKinley KS. Looking back on the language and hardware revolutions: Measured power, performance, and scaling. SIGARCH Comput. Archit. News. 2011 Mar.39(1):319–332. [Google Scholar]
- [22].Lively CW, Wu X, Taylor VE, Moore S, Chang H-C, Cameron KW. Energy and performance characteristics of different parallel implementations of scientific applications on multicore systems. IJHPCA. 2011;25(3):342–350. [Google Scholar]
- [23].Johnsson B, Ganestam P, Doggett M, Akenine-Möller T. Proceedings of the Fourth ACM SIGGRAPH / Eurographics Conference on High-Performance Graphics. Eurographics Association; Aire-la-Ville, Switzerland, Switzerland: 2012. Power efficiency for software algorithms running on graphics processors; pp. 67–75. ser. EGGH-HPG’12. [Google Scholar]
- [24].Laros JH, III, Pokorny P, DeBonis D. The Third International Workshop on Power Measurement and Profiling in conjunction with IEEE IGCC 2013. Arlington, Va: 2013. Powerinsight - a commodity power measurement capability. [Google Scholar]
- [25].Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- [26].Stone JE, Isralewitz B, Schulten K. Early experiences scaling VMD molecular visualization and analysis jobs on Blue Waters. Extreme Scaling Workshop (XSW), 2013. 2013 Aug.:43–50. [Google Scholar]
- [27].Stone JE, Vandivort KL, Schulten K. Proceedings of the 8th International Workshop on Ultrascale Visualization. ACM; New York, NY, USA: 2013. GPU-accelerated molecular visualization on petascale supercomputing platforms; pp. 6:1–6:8. ser. UltraVis ’13. [Google Scholar]
- [28].Phillips JC, Stone JE, Vandivort KL, Armstrong TG, Wozniak JM, Wilde M, Schulten K. SC’14 workshop on High Performance Technical Computing in Dynamic Languages. IEEE Press; 2014. Petascale Tcl with NAMD, VMD, and Swift/T; pp. 6–17. ser. SC ’14. [Google Scholar]
- [29].Stone JE, McGreevy R, Isralewitz B, Schulten K. GPU-accelerated analysis and visualization of large structures solved by molecular dynamics flexible fitting. Faraday Discuss. 2014;169:265–283. doi: 10.1039/c4fd00005f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Klein MD, Stone JE. Cray User Group Conf. Cray; May, 2014. Unlocking the full potential of the Cray XK7 accelerator. [Google Scholar]
- [31].Stone JE, Sener M, Vandivort KL, Barragan A, Singharoy A, Teo I, Ribeiro JV, Isralewitz B, Liu B, Goh BC, Phillips JC, MacGregor-Chatwin C, Johnson MP, Kourkoutis LF, Hunter CN, Schulten K. Atomic detail visualization of photoynthetic membranes with GPU-accelerated ray tracing. Parallel Computing. 2016 doi: 10.1016/j.parco.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hardy DJ, Stone JE, Schulten K. Multilevel summation of electrostatic potentials using graphics processing units. J. Paral. Comp. 2009;35:164–177. doi: 10.1016/j.parco.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stone JE, Saam J, Hardy DJ, Vandivort KL, Hwu WW, Schulten K. Proceedings of the 2nd Workshop on General-Purpose Processing on Graphics Processing Units, ACM International Conference Proceeding Series. Vol. 383. ACM; New York, NY, USA: 2009. High performance computation and interactive display of molecular orbitals on GPUs and multi-core CPUs; pp. 9–18. [Google Scholar]
- [34].Stone JE, Hardy DJ, Saam J, Vandivort KL, Schulten K. GPU-accelerated computation and interactive display of molecular orbitals. In: Hwu W, editor. GPU Computing Gems. Morgan Kaufmann Publishers; 2011. pp. 5–18. ch. 1. [Google Scholar]
- [35].Krone M, Stone JE, Ertl T, Schulten K. Fast visualization of Gaussian density surfaces for molecular dynamics and particle system trajectories. EuroVis - Short Papers 2012. 2012:67–71. [Google Scholar]
- [36].Stone JE, Kohlmeyer A, Vandivort KL, Schulten K. Immersive molecular visualization and interactive modeling with commodity hardware. Lect. Notes in Comp. Sci. 2010;6454:382–393. [Google Scholar]
- [37].Stone JE, Vandivort KL, Schulten K. Immersive out-of-core visualization of large-size and long-timescale molecular dynamics trajectories. Lect. Notes in Comp. Sci. 2011;6939:1–12. [Google Scholar]
- [38].Stone JE, Sherman WR, Schulten K. Parallel and Distributed Processing Symposium Workshop (IPDPSW), 2016 IEEE International. IEEE; May, 2016. Immersive molecular visualization with omnidirectional stereoscopic ray tracing and remote rendering. [DOI] [PMC free article] [PubMed] [Google Scholar]