Abstract
Urine is a highly desirable biospecimen for biomarker analysis because it can be collected recurrently by non-invasive techniques, in relatively large volumes. Urine contains cellular elements, biochemicals, and proteins derived from glomerular filtration of plasma, renal tubule excretion, and urogenital tract secretions that reflect, at a given time point, an individual's metabolic and pathophysiologic state. High-resolution mass spectrometry, coupled with state of the art fractionation systems are revealing the plethora of diagnostic/prognostic proteomic information existing within urinary exosomes, glycoproteins, and proteins. Affinity capture pre-processing techniques such as combinatorial peptide ligand libraries and biomarker harvesting hydrogel nanoparticles are enabling measurement/identification of previously undetectable urinary proteins. Future challenges in the urinary proteomics field include a) defining either single or multiple, universally applicable data normalization methods for comparing results within and between individual patients/data sets, and b) defining expected urinary protein levels in healthy individuals.
Keywords: biomarker, combinatorial peptide ligand libraries, diabetes, glycoprotein, hydrogel nanoparticles, Lyme Disease, mass spectrometry, protein, proteomics, selected reaction monitoring, urine
1. Introduction
“The composition of the blood is determined not by what the mouth ingests but what the kidney keeps.” [1] This simple quote underscores the complex process of urine formation via plasma filtration by renal glomeruli, selective reabsorption/secretion of water, glucose, and ions from the renal tubules, and shedding of proteins from the kidney and urinary tract [1-3]. 30% of the urinary proteome is derived from glomerular plasma filtration and 70% of the urinary proteome originates from the urogenital tract; therefore the urine proteome is expected to contain a variety of potential biomarkers related to overall health as well as specific urogenital organ pathophysiology [1, 2].
Urine biochemicals such as glucose, ketones, and total protein have been routinely measureable for many years, however the unique protein/peptide composition of urine has been under-explored. Even though urine is generally available in milliliter quantities and can be collected frequently and non-invasively, urine proteomic analysis has lagged behind that of serum/plasma, despite blood's more complex protein repertoire [4]. For example, manual annotation of the literature by the UniProt consortium has revealed between 13,841 and 17,132 human protein-coding genes [5]. Yet, by 2015, only 4,430 urinary proteins had been identified, a large number of which were detected due to recent improvements in pre-analytical fractionation/concentration techniques [6, 7].
Physiologically, the urine proteome is less complex compared to the plasma proteome but this fact alone does not fully explain the smaller number of identified proteins. The dearth of information about the urine proteome/peptidome has been due to historically labor-intensive methods such as 2-D gel electrophoresis that fail to distinguish protein isoforms [8], poor precision of enzyme linked immunosorbent assays (ELISA) with urine [9], the variable urine matrix/proteome between and within individuals [10-14], the presence of high abundance proteins (e.g. uromodulin, albumin, alpha-1-antitrypsin, transferrin, immunoglobulin, alpha-1-microglobulin and complement) that mask low abundance proteins [15, 16], endogenous proteases [17], and a lack of technologies for rapid profiling of the urine proteome, with or without quantitative analyses (Table 1) [16].
Table 1.
Advantages and disadvantages of urine specimens for protein biomarker discovery [1,2,28,30,31,44,89,155].
| Parameter | Advantages | Disadvantages |
|---|---|---|
| Specimen volume | Large quantities available | |
| Ease of collection | Non-invasive collection | |
| Urine production | Urine is produced continuously throughout the day and can be collected to match normal diurnal variation. First morning urine samples are generally more concentrated than randomly collected specimens. |
Some urinary proteins and chemical constituents are affected by diet, hydration, exercise, metabolism, circadian rhythms and hormones. |
| Protein quantity | Abundant source of soluble proteins/peptides that is less complex than blood. | |
| Protein stability | Urine proteins remain relatively stable at room temperature and 4°C for up to 6 hours. | |
| Pathophysiology | Urine reflects the plasma proteome as well as kidney proteome due to glomerular filtration, tubular reabsorption, and secretion. | A wide spectrum of renal pathologies produces similar urinary proteomic markers, e.g. albuminuria. |
| Constituents | Multiple chemical, cellular, and proteomic constituents are present in urine, many of which are well-defined in normal and pathologic states. Multiple analytes can be selected for data comparison between and within individuals/data sets. | Inter and intra-individual variability requires the use of multiple parameters for comparing urinary proteome data. Thoroughly annotated clinical information and/or urine biochemical and microscopic analyses may be needed to accurately compare urine proteomic data. |
| Analysis | Low molecular weight soluble proteins can be readily quantified by mass spectrometry. Urine proteins can be quantified by multiple methods. Urine biochemical analysis can be easily performed with urine test strips (dipstick). Cellular elements can be identified with bright field microscopy. Strategies for detecting low abundance urinary proteins are well documented and commercially available. |
High abundance proteins mask the presence of low abundance proteins, thus requiring depletion and/or enrichment strategies for detecting low abundance proteins. |
However, in the past decade, analytical advances in quantitative mass spectrometry and fractionation techniques [18, 19], introduction of capillary electrophoresis mass spectrometry (CE-MS) [20, 21], and novel pre-analytical sample processing methods are advancing the field of urinary proteomics [22-25]. Several database repositories and urine proteomic resources are now available for locating information regarding urinary protein names, pathophysiology, literature citations, and consensus documents for analytical methods (Table 2). Many of the issues encountered in proteomic profiling and biomarker discovery, including sample preparation and pre-analytical variables [2, 26-31], interfering substances such as bacteria, antibiotics, and x-ray contrast media [32-34], the need for standardized procedures [35-39] and inter-laboratory comparisons [40], have been debated and extensively reviewed elsewhere [36, 41-45]. This review presents a snapshot of the current state of urinary protein biomarker discovery, with emphasis on pre-analytical processing and fractionation methodologies for improving the detection of low abundance proteins and/or protein classes, such as glycoproteins.
Table 2.
Human kidney/bladder/urine proteome/peptidome resources for best practice guidelines, data repositories, and integrative knowledge databases.
| Resource | Acronym | URL | Procedures/ Standards |
Human Protein Data | Proteomic Methodologies |
Reference |
|---|---|---|---|---|---|---|
| Chorus Project | https://chorusproject.org | n/a | User/experiment specific Cloud based platform for storage, analysis, and sharing of mass spectrometry data |
Mass Spectrometry | [106] | |
| Human Kidney & Urine Proteome Project | HKUPP | www.hkupp.org alos accessible from www.hupo.org | Urine exosome collection and processing | Glomerulus Renal medulla Urine-normal Urine-exosome |
2-D Gel Electrophoresis CE-MS LC-MS/MS SELDI-TOF MS |
[69,100] |
| Human Proteinpedia and Human Protein Reference Database | HPRD |
http://www.humanproteinpedia.org/ http://www.hprd.org/ |
n/a | Portal for sharing/integrating proteomic data | Mass Spectrometry Immunohistochemistry Immunofluorescence Western blotting Co-Immunoprecipitation Protein/Peptide microarray Yeast two hybrid screens |
[70] |
| Human Proteome Organization | HUPO | www.hupo.org | Data interpretation guidelines | Links to multiple proteomic initiatives | Mass Spectrometry, antibodies, and the Knowledge Base | [159] |
| Human Protein Atlas | HPA | http://www.proteinatlas.org/ | n/a | High-resolution images showing the spatial distribution of proteins in tissues | Immunohistochemistry Immunofluorescence RNASeq |
[5,93,156,160,164] |
| Human Urine Database v3.0 | http://mosaiques-diagnostics.de/diapatpcms/mosaiquescms/front_content.php?idcat=257 | n/a | 25,000 peptides from independent samples for urine specimens from normal and diseased patients | CE-MS | [162] | |
| Kidney & Urinary Pathway Knowledge Base | KUPKB | http://www.kupkb.org/#tab0 | n/a | Information extracted from scientific publications and other related renal databases using Semantic Web search tools | Multiple proteomics data from published literature | [157] (note new URL after publication) |
| Max-Planck Unified Proteome Database | MAPU 2.0 | http://www.mapuproteome.com/urine/ | n/a | Protein sequences, function, motifs, references, & links to tissue sources and peptides | LTQ-FT and LTQ-Orbitrap mass spectrometer data | [67] |
| PeptideAtlas | www.peptideatlas.org/hupo/hkup | n/a | Data repository for annotation of eukaryotic genomes via validation of expressed proteins | Mass Spectrometry | [42,71] | |
| ProteomeXchange | http://www.proteomexchange.org/ | n/a | Provide a harmonized submission of MS proteomics data to the main existing repositories, and encourage data distribution | Shotgun mass spectrometry data sets | [163] | |
| Sys-Body Fluid Database | http://www.biosino.org/bodyfluid/ | n/a | 11 body fluid databases compiled from scientific literature | Multiple data types | [158] | |
| Urinary Exosomes Protein Database | https://hpcwebapps.cit.nih.gov/ESBL/Database/Exosome/ | n/a | Urinary exosomes protein database with GO annotations | Mass Spectrometry | [68,69] | |
| UniProt | UniProt | http://www.uniprot.org/ | n/a | Unified view of protein sequence and functional information | Multiple data types | [165] |
| Urine Proteomics | Urineproteomics.org | n/a | Links to methods, databases, companies and organizations | Mass Spectrometry | [42,114] | |
| Yale Protein Expression Database | YPED | http://yped.med.yale.edu/repository/Welcome.do;jsessionid=ECB816BF54EE909E9FFDC1F7A70F2551 | n/a | Public access data repository | LCMS, MudPIT, ICAT, iTRAQ, SILAC, 2D Gel and DIGE, Progenesis and Skyline Label Free Quantitation, MRM analysis and SWATH | [89,161] |
2. Summary of urinary proteomic biomarker discovery to date
Technological advances in proteomics and genomics are rapidly advancing our understanding of the molecular basis of disease. Translational research embodies these new diagnosis and treatment paradigms and is globally referred to as precision medicine, molecular medicine, or personalized medicine. The common goal, regardless of the name, is to design therapies based on a patient's phenotype and their genomic and/or proteomic disease profile, coupled to our knowledge of biomarkers and prognostic factors [46, 47]. Urinary proteomics is a perfect example of the embodiment of precision medicine in which research is enhancing our assessment of disease risk, elucidating disease mechanisms, and predicting optimal therapy [48]. Extensive reviews of the translational research progress in the urinary proteomic field have been published [2, 41, 49-51]. Potential urinary biomarkers are being discovered in a variety of non-kidney associated diseases including acute appendicitis [52, 53], infectious diseases such as Tuberculosis [54], Chagas Disease and Lyme Disease [55-57], cancer [49, 50, 58-61], cardiovascular disease [62-64], and aging [65, 66]. Selected examples of the progress in urinary protein identification are described below, highlighting a) improvements in mass spectrometry-based urinary proteomic discovery, b) the number of identifiable proteins, and c) pathophysiologic associations over the last decade.
In 2001, Sphar et. al. established the efficacy of urine protein identification [16]. 124 proteins/expressed sequence tags were identified from unfractionated, pooled normal male urine by applying liquid chromatography coupled mass spectrometry (LC-MS) with iterative peptide ion analysis on a hybrid quadrupole-time-of-flight (Q-TOF) instrument. The combined abundance of 115 proteins represented less than 10% of the sample by mass, thus highlighting the magnitude of low abundance proteins in urine [16]. By 2006, linear ion trap-Fourier transform (LTQ-FT) and linear ion trap-orbitrap (LTQ-Orbitrap) mass spectrometers were being utilized for urine protein discovery due to the instruments' high resolution, mass accuracy, wide dynamic range, and fast cycle times. Adachi et. al. identified 1543 urinary proteins in 10 specimens from healthy individuals (one individual and a pool of 9 specimens) [67]. This study identified plasma membrane and lysosomal proteins in the urine, some of which are known to be associated with urinary exosomes, thus underscoring the complex source of urine proteins/peptides [68, 69].
Using high resolution Fourier transform mass spectrometry, Marimuthu et. al. discovered 1823 urine proteins, 671 of which were previously unreported in the urine, from a urine pool specimen comprising 24 healthy individuals [70]. A unique aspect of this study was lectin affinity chromatography with concanavalin A, wheat germ agglutinin and jacalin, to enrich glycoprotein constituents.
A comprehensive kidney, urine, and plasma proteome comparison was conducted by Farrah et. al. in 2013 [42]. By combining kidney, urine, and plasma datasets collected from different laboratories, with an estimate of relative protein abundance based on spectral counting and a normalization strategy to compare the proteomes, they were able to identify 2491 non-redundant proteins in the PeptideAtlas (www.peptideatlas.org/hupo/hkup) (Table 2) [71].
3. Advancements in mass spectrometry based urinary proteomics
The ever-increasing depth and breadth of the urinary proteome is attributable to the versatility of mass spectrometry for protein discovery, identification, relative and absolute quantification. Top-down protein profiling begins with a statistically significant number of complex biological samples which are separated by chromatography or 2-D gel electrophoresis. Specific fractions or gel spots containing proteins are analyzed in a mass spectrometer (e.g. MALDI-TOF, SELDI TOF, or electrospray) to identify potential biomarkers. Top-down proteomics provides protein identity, relative quantity, and mass information for naturally occurring small peptides, post-translational modifications, and protein cleavages.
Bottom-up proteomic studies start with a few enzymatically digested protein samples to create a complex peptide mixture. The peptide mixture can be analyzed by liquid chromatography tandem mass spectrometry (e.g. LC-MS/MS or LC-MALDI MS/MS), providing high-resolution identity and relative quantity of many peptide sequences. Bottom-up approaches are low-throughput methods to analyze a few specimens (10 or less) from simple model systems such as treated/untreated cultured cell lines [18, 19, 72]. Specific details of mass spectrometry for proteomics are thoroughly described in the updated reviews by Bantscheff et.al. [18, 19]
Several different groups have developed tactics for enhancing identification and quantification of urinary proteins. A simple approach for improving urine protein detection was developed by combining silver staining with classic cellulose acetate membrane electrophoresis, prior to mass spectrometric analysis. Cellulose acetate membrane electrophoresis (CAME) is the classical method for identifying free light chain immunoglobulins (Bence-Jones proteins) in urine, based on relative mobility of the immunoglobulin fractions [73]. CAME allows discrimination of well-separated proteins on the cellulose acetate membrane, as well as the overlapping proteins. Nakayama et. al. have improved on the technique by incorporating a silver staining step immediately following electrophoresis to better fractionate the CAME protein patterns [74]. Well-separated protein bands are observed with <800mg/l total protein loaded per lane. Proteins are extracted over 8 hours in Laemmli sample buffer. The protein bands are excised from the cellulose acetate membrane and extracted for analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and identified via mass spectrometry. This method successfully demonstrated the ability to identify 31 proteins from the urine samples of two tubulointerstitial nephritis patients. Of the 31 proteins identified, only 11 had been previously identified by immunofixation [74].
Protein post-translational modifications such as phosphorylation can indicate the activation state of cell signaling pathways [75]. Phosphoprotein enrichment is often performed using titanium dioxide chromatography during specimen processing [76]. Li et. al. developed two methods, Integrated Multidimensional Liquid Chromatography (IMDL) and Yin-yang multidimensional liquid chromatography (MDLC) tandem mass spectrometry, to identify urine proteins/phosphoproteins without using a titanium dioxide pre-fractionation step [77]. Although their method did not identify a significant number of previously unidentified urinary proteins, they were able to identify 1310 non-redundant proteins including 59 phosphorylation sites [77].
Following identification of potential proteomic biomarkers using top-down/bottom-up mass spectrometry, validation and absolute quantification of urine biomarkers is best achieved by Selected Reaction Monitoring (SRM) [35, 78, 79]. SRM, also known as Multiple Reaction Monitoring (MRM), allows previously defined peptides and fragment ions, known as signature peptides, to be quantified by monitoring multiple fragment ions produced during collision-induced dissociation [18]. For robust SRM analysis with strong signal intensity, the peptides for SRM analysis must be unique to the protein of interest, should not contain post-translational modifications, and should not have genetically encoded variations [78]. Absolute quantification can be performed with stable isotope-labeled peptides corresponding to the signature peptides. The stable isotope-labeled peptide differs from the signature peptide only by mass, thus acting as an internal standard for comparison to the peptides of interest [79]. PeptideAtlas and PRIDE databases provide publically available lists of peptides suitable for SRM [18, 42, 71] (Table 2). Liquid chromatography SRM has been shown to be a robust method across laboratories, with a coefficient of variation (CV) <30% between 4 laboratories, when combined with pre-analytical enrichment strategies in serum, urine, and seminal fluid specimens [80].
SRM can be performed for specific peptide isoforms and post-translational modifications; however, the methods may require optimization due to potentially weak SRM signal intensity [18, 78]. Using uromodulin as a model protein, Fu et. al. provided details for an experimental workflow to identify trypsin-digested signature peptide fragments for use in SRM quantification assays [78]. Uromodulin (Tamm-Horsfall glycoprotein, 68 kDa), the most abundant urine protein, is produced by the ascending loop of Henle in the kidney [1, 81]. Uromodulin function is not completely understood, but it has been associated with renal calculi formation [82], hypertension [83, 84], and kidney disease [85, 86]. Furthermore, uromodulin peptide abundance has been shown by CE-MS to be increased in patients with pre-eclampsia [87] and rheumatoid arthritis [88], as well as with age [66]. However, uromodulin forms large aggregates, impeding typical mass spectrometry analysis. Using pooled healthy urine specimens and specimens from participants in the Atherosclerosis Risk in Communities study of 1987-1989, Fu et. al. identified 4 signature uromodulin peptides for SRM analysis that exhibited linearity and correlation to uromodulin protein abundance over 3 orders of magnitude [78]. Their process required extensive optimization due to complexities linked to composition of the urine sample matrix, biological characteristics of the target protein, and chromatography and spectroscopy incompatibility with data-dependent MS testing. Their workflow included: a) Identifying potential peptides within the detectable mass/charge (m/z) range, b) Refining candidate peptides based on a peptide length of 6-21 amino acids, c) Eliminating readily oxidizable peptides and peptides prone to post-translational modifications, d) Eliminating peptides only found in specific isoforms, unless the specific isoform was the intended target for quantification, e) Creating a correlation matrix to identify peptides that did not appear in chromatograms in a predictable fashion, and f) Selecting signature peptides that correlated well with each other, under a multitude of experimental conditions [78]. Signature peptides of uromodulin were identified that illustrated strong correlation of abundance (r2 > 0.90) with each other and with the target protein within a complex milieu of pooled urine samples. The results were comparable to those derived by ELISA and demonstrated significant statistical superiority to data-dependent MS screening methods and unfocused SRM quantification techniques [78].
Cantley et. al. developed a workflow, termed the Targeted Urine Proteome Assay (TUPA), for evaluating a large number of urinary protein biomarkers [89]. To develop the assay, Cantley et. al. performed literature searches plus multiple urine proteomic assays from four patients with immediate or delayed graft function following kidney transplantation. 416 potential biomarkers of kidney disease were identified [89]. The multiple proteomic assays included difference gel electrophoresis, iTRAQ, and label free quantitation in undepleted, high abundance protein depleted, and moderate abundance protein depleted samples. High abundance plasma proteins were depleted using a chicken polyclonal antibody column (Seppro® IgY14 LC10), with subsequent removal of moderately abundant plasma proteins on a Seppro® Supermix LC2 column [89]. The IgY antibodies in the Supermix LC2 column are directed against plasma proteins present in the flow-through of the LC10 columns [89]. Trypsin-digested peptides accounting for 200 of the proteins were identified, and synthetic, isotope labeled standards were assayed to eliminate peptides that exhibited deviation due to urine interference in MRM assays. The resulting TUPA assay featured 224 peptides corresponding to 167 proteins. However, a caveat of the TUPA assay is that the avian antibody depletion strategy removes plasma proteins, but not necessarily the highly abundant urinary protein, uromodulin, which may mask low abundance proteins. This plasma protein depletion strategy may explain their findings that 97 proteins from the high+moderate abundance depleted fractions did not show significant fold differences as they did in the non-depleted specimen [89].
Further methods are being developed to provide reproducible, high throughput, quantitative urinary proteomic data. In the classical data-dependent acquisition (DDA) method, LC-MS peak intensities of peptide precursor ions are subjected to fragmentation (sequencing) as they are eluted from the liquid chromatography system, without defining the peptides beforehand [18]. DDA is a sequential operation, analyzing one peptide at a time. However, under-sampling is a problem due to the molecular complexity and dynamic expression range of urine proteins [90]. An alternative method, similar to SRM, is data-independent acquisition (DIA) in which many peptides at a time are subjected to simultaneous fragmentation [18]. Peptide identification in DIA is performed by comparing the DIA chromatographic data with a peptide spectral library from previously identified proteins [91, 92].
To identify and quantify the majority of accessible urinary proteins without fractionating the urine specimens, Muntel et. al. have developed a data-independent acquisition (DIA) workflow in which a urinary spectral library is first created from a data-dependent acquisition (DDA) analysis of numerous urine samples using multiple MS/MS instruments [90]. In Muntel's study, urine from children diagnosed with ovarian cyst (n=12), urinary tract infection (n=11), and an abdominal pain control group (n=64) were analyzed by LC-MS/MS to create a spectral library [90]. A subset of these specimens (n=23) were analyzed in the same manner to create a second spectral library [90]. When combined with a publicly available human spectral library, a comprehensive spectral library of over 10,000 proteins was generated. Using a unique urine sample (unassociated with library development), a comparison of DIA and DDA analyses was performed. The results demonstrated a marked advantage for DIA, exhibiting a nearly two-fold increase in identified peptides and proteins, and a significant reduction in the coefficient of variation (CV) for quantification [90].
4. Urinary glycoproteomics
Urine contains glycosylated proteins, one of which is the highly abundant protein, uromodulin. Glycosylation is an abundant post-translational modification occurring on asparagine (N-linked) and serine/threonine (O-linked) residues [93]. Glycosylation determines protein sub-cellular location and trafficking to microdomains [94], mediates cell adhesion, functions as a receptor in cell-to-cell communication and immune response [95]. Lectins, which are carbohydrate binding proteins, can be utilized in affinity chromatography to procure glycoprotein enriched specimen fractions. Marimuthu et. al. utilized concanavalin A, wheat germ agglutinin, and jacalin to enrich for mannose, N-acteylglucosamine, and galactose/N-acetylgalactosamine containing glycoproteins, respectively [70]. Lectin-bound proteins were eluted using equimolar mixtures of N-acetylglucosamine, melibiose and galactose. The eluate was dialyzed against wash buffer to remove free sugars and concentrated using 3kDa cutoff filters prior to mass spectrometry [70]. Following separation of the proteins by SDS-PAGE, the gel bands were excised and digested with trypsin prior to protein sequencing by LC-MS/MS. Proteins in the lectin-enriched fraction were mapped to the Human Protein Reference Database (Table 2) to assess post-translational modifications, domains, and motifs [70]. This solid phase affinity capture method categorized proteins derived from sub-cellular locations, including cytoplasmic and endoplasmic reticulum associated proteins, as well as a group of voltage gated potassium ion channels [70]. Voltage gated potassium ion channels have been linked to cell migration, angiogenesis, adhesion and apoptosis, underscoring the important diagnostic information hidden in the urinary proteome [96].
The first targeted urinary glycoproteomics study characterized desialylated glycopeptides by tandem MS experiments utilizing collision induced dissociation and electron capture dissociation fragmentation techniques [93]. 58 N- and 63 O-linked glycopeptides were categorized from 53 glycoproteins [93].
Peptides and n-glycans in body fluids can be collected separately using a single GlycoFilter without additional purification [97]. This spin filter device allows separation and capture of glycans based on pH adjustment and filtration in a step-wise pre-processing, de-N-glycosylation, and proteolysis process [97]. By incorporating stable isotope labeled oxygen (18O) into the peptide during enzymatic deglycosylation, the glycosylated peptide can be identified by the N-glycosylation consensus motif (Asn-XXX-Ser/Thr, in which XXX is any amino acid except proline) and the presence of 18O asparagine [97].
Another important source of glycosylated proteins can be found in urinary exosomes. Saraswat et. al. examined urinary exosome glycosylation patterns because the exosome glycosylation pattern influences exosome targeting and uptake [98]. 37 glycoproteins were identified in urinary exosomes by comparing the urinary exosome glycan structures to a database rather than through iterative de novo glycan structure analysis [98]. Glycoproteins were digested with trypsin prior to enrichment by Sambucus nigra agglutinin (SNA) affinity chromatography or size exclusion chromatography. SNA binds preferentially to sialic acid attached to terminal galactose in α-2,6 and to a lesser degree, α-2,3 linkages. The glycoprotein enriched fractions were analyzed using collision induced dissociation-Tandem MS [98]. From these glycoproteins, 126 N-glycopeptides from 51 N-glycosylation sites were characterized [98]. These studies highlight several different methods for enhancing identification of urinary glycoproteins.
5. Sources of variability in urine proteomics discovery
Despite technological improvements, pre-analytical variables continue to plague proteomic research. Pre-analytical variability creates barriers to inter-laboratory comparisons beyond the existing complicating factors of analytical platform differences and data reduction methods. Hydration status, diet, medications, age, and time of urine collection (first morning, second morning, random) are a few of the factors associated with inter and intra-patient urine biochemical and proteomic variability (Table 1) [11-14]. Urine specimen collection can be highly variable between studies and patient populations. Depending on the specimen requirements, collection site, and patient population (pediatric, neonatal, elderly, volunteers, healthy, etc), urine specimens may be collected via supra-pubic aspiration directly from the bladder, via an indwelling catheter, a specimen collection bag (for pediatric patients), or during voiding (mid-stream collection). The specimen collection container can also introduce variations depending on the container's construction, for example polypropylene, polystyrene, or glass, with/without a sterile interior. A “clean catch” specimen prevents excessive bacterial and cellular contaminates, however patient self-collected clean catch specimens are subject to variability in the collection technique [1]. To limit collection variability, the second morning urine specimen is typically collected, thus avoiding endogenous proteolytic activity during over-night urine retention in the bladder [17].
Urinary proteomic analysis is further complicated by differences in processing steps, such as ‘spin first’ methods to remove cellular elements by sedimentation, or ‘freeze first’ methods to stabilize proteins. Urine may contain a variety of cellular elements, organic and inorganic crystals and casts, collectively referred to as “sediment” or “pellet”. Red blood cells, leukocytes, renal and uroepithelial cells may be shed into the urine in various pathologic conditions. Urinary casts, a cylindrical structure formed in the kidney by precipitation of Tamm-Horsfall mucoprotein (uromodulin), with or without cells, lipids, or crystals, also contribute to urine sediment [99]. Freezing a urine specimen prior to centrifugation may cause cell lysis upon thawing, allowing cellular cytoplasmic proteins to contaminate the thawed urine specimen. On the other hand, spinning a specimen first removes the majority of intact cellular elements, thus depleting the proteome of these cell-derived proteins [28, 35]. Depending on the goal of the study and physiologic location of the potential biomarker (cellular versus soluble urine protein), the cellular content may be desirable or undesirable in the urinary biomarker analysis
Additional sources of pre-analytical variability introduced during urine specimen processing include centrifugation protocols (speed, time, temperature, brake settings), pH, salinity, protein concentration, and addition of protease inhibitors or chemical stabilizers [27-29, 35, 100, 101]. Urine fractionation strategies to deplete high abundance proteins and/or enrich low abundance proteins include solid phase adsorption, combinatorial peptide ligand libraries (CPLL), and hydrogel nanoparticle (NP) harvesting (Table 3) [89, 102, 103]. The effectiveness of these fractionation strategies can be manipulated by varying the physicochemical properties of the system. Changes in salinity, protein concentration, ionic strength, and/or pH can alter protein-protein interactions by a) modifying adsorption to solid phase columns, b) altering the strength of hexapeptide-protein interactions, or c) by enhancing or inhibiting hydrophobic interactions between affinity baits and proteins, or between hexapeptides and complementary proteins [6, 25, 104-107]. For example, increased salt concentration can denature antibodies thus inhibiting solid phase affinity capture [105]. Decreasing the ionic strength, increasing hydrophobic interactions, and maintaining solutions within pH 4-9 of CPLL solutions can improve protein enrichment/depletion strategies using combinatorial peptide ligand libraries (discussed further below) [6, 107].
Table 3.
Comparison of solid phase immunoadsorption, Combinatorial Peptide Ligand Libraries and hydrogel nanoparticles for separating urine proteins [22,23,25,105,107,129,166].
| Solid Phase immunoadsorption | Combinatorial Peptide Ligand Libraries | Hydrogel nanoparticles | |
|---|---|---|---|
| Separation method | Subtraction | Equilibration enrichment | Enrichment/size exclusion |
| Protein interaction principles | Antigen-antibody interaction | Physicochemical interaction (including hydrophobic, van der Waals, hydrogen bonding) between complementary hexapeptides and proteins | Hydrophobic and electrostatic interactions between the dye bait and protein |
| Affinity molecule | Monoclonal or polyclonal antibody | Hexapeptides or hexapeptides+Alcian blue dye | Organic dyes Acrylic acid Vinylsulfonic acid |
| Size exclusion | No | No | Yes |
| Thermo-reversible | No | No | Yes |
| Low abundance protein enrichment | Possible | Yes | Yes |
| Affinity | Moderate (antibody dependent) | Weak-Strong | Strong |
| Specificity | High | Moderate | Moderate (dependent on the class of dye used as the affinity bait) |
In summary, specimen collection, processing and fractionation schemas should be determined for each study prior to urine collection and processing. Clinical data and specimen processing details should be associated with each specimen, particularly for specimens obtained from a biorepository, because these clinical factors can help identify potential sources of variability and provide potential parameters for normalizing urinary biomarker data [108].
6. Determining the normal, healthy urinary proteome
Many factors not related to disease pathogenesis contribute variability to the urine proteome. Gender, age, diet, exercise, hormone status, diurnal variation, genetic variation in renal physiology and environmental factors create variability in the normal urine proteome [12, 13, 51, 109-113]. Which proteins emanate from plasma filtration versus renal secretion? How much do urinary proteins fluctuate over time in an individual? Are these proteins of pathologic significance? Can we establish expected, or ‘normal’, ranges for specific urinary proteins? The answer to these questions are a fundamental step in determining the “expected healthy urinary proteome” since normal ranges for urinary proteins have not been established except for a few high abundance, disease indicative proteins such as albumin, immunoglobulins, transferrin, and β-2-microglobulin [81]. The presence of “age dependent” urinary proteins has been associated with normal healthy states [51, 111] and diet and exercise affect the urinary proteome [10, 11, 113]. These findings indicate that large scale, longitudinal studies of individuals will be needed to establish a “healthy” living diagnostic urine profile.
Several studies have begun to address the questions posed above by characterizing urine proteins from healthy and diseased individuals [12, 13, 114-116]. To ascertain the source of urinary proteins, as plasma filtrates versus urogenital secretion, Jia et. al. compared publically available urine proteomic data sets, emanating from various institutions, methodologies, and individuals [8, 67, 117] with plasma proteome data sets (Human Proteome Organization Plasma Proteome Project and data from States et. al. [118]). The goal of this bioinformatics study was to characterize the proteins as plasma and/or urine sub-proteomes, which could potentially be used to enhance renal disease diagnostics and prognostics [3]. Gene Ontology (GO) terms statistically overrepresented in the plasma-and-urine sub-proteome versus the whole plasma proteome were searched using the Biological Networks Gene Ontology (BiNGO) program. Despite the complexities and potentially confounding differences in data sets, Jia et. al. were able to classify the proteins based on GO terms into the plasma derived sub-proteome and the urine sub-proteome [3]. The plasma sub-proteome was comprised predominately of plasma proteins filtered by the glomerulus and soluble proteins secreted by epithelial cells. The urine-only sub-proteome stemmed mainly from soluble proteins secreted by epithelial cells [3].
To identify constituents of the typical “healthy” urinary proteome, multiplex bead immunoassays (xMAP, Luminex) have been utilized for profiling urine from apparently healthy individuals representing different ethnicities, genders, and tobacco use statuses (n=103) [115]. The multiplex analysis (211 proteins) included panels of cytokines, growth factors, and hormone receptors. Proteins were categorized based on relative abundance, after normalizing to urine creatinine concentrations: high 100ng/ml - >10ng/mL, average 10ng/ml - 100ng/mL, or low 1ng/ml - 10ng/mL abundance. Eight low molecular weight proteins were identified in the high abundance class. The average and low abundance classes exhibited diverse proteins. The average abundance proteins included proteases, soluble receptors, hormones, immunologic and growth factors [115]. The low abundance proteins consisted of cytokines, extra cellular matrix components, and matrix metalloproteinases [115]. Depending on the normalization method, the CV ranged from <10% for absolute values to 700% for urine creatinine normalized values [115]. Depending on the data normalization method, using urine creatinine, urine total protein, urine albumin, the ratio of urine albumin to urine creatinine, or β-2-microglobulin, the Pearson correlation coefficient ranged from 0.1765 for β-2-microglobulin normalized data, to 0.9457 when the data was normalized to urine total protein.
7. Data normalization issues for comparing urinary proteomes
A critical data normalization issue plaguing the urinary proteomics field was underscored in the healthy urinary proteome study [115]: What is the optimal parameter(s) to use for comparing urinary proteomic data within individuals over time, between individuals, or between laboratories and studies? Housekeeping genes, such as lactate dehydrogenase A (LDHA), have been used to normalize genomic data because LDHA is expressed at relatively constant levels and is tolerant to many, but not all, cellular perturbations [119]. To date, a single invariant urinary protein has not been identified that can be the “North Star” for guiding urinary protein normalization.
Although the correlation differences noted above in the healthy urinary proteome study accentuate some of the hurdles involved in defining/normalizing the urinary protein, Kentsis et. al. have identified a core set of “generally invariant proteins” in children (age range 1-18 years) that may serve as benchmark proteins for longitudinal evaluation of individuals [114]. Furthermore, non-protein analytes, such as creatinine [26, 45, 83, 115], neopterin [26, 120], or post-translationally modified proteins such as glycosylated hemoglobin (A1c) [121] can also be used to normalize urine proteomic data. Serum and urine creatinine levels are often used alone, or in combination with other parameters, to assess glomerular filtration because creatinine is completely filtered by the kidney and excreted in the urine [122]. For diverse patient populations with end-stage renal disease, Cystatin C alone or in combination with creatinine improved the association between the estimated Glomerular Filtration Rate (eGFR) and the risks of death and end-stage renal disease [122]. In light of the pre-analytical variables and high inter-individual and intra-individual variation in urinary proteomes, it is essential for translational research studies to select case and control specimens that represent: a) the diseased population harboring the condition being studied, b) a diseased population not harboring the condition being studied, and c) healthy individuals. In addition, it may be necessary to combine multiple parameters from cellular, biochemical, and proteomic urine analytes to develop the optimal data normalization factor.
8. Enriching urine specimens for low abundance proteins
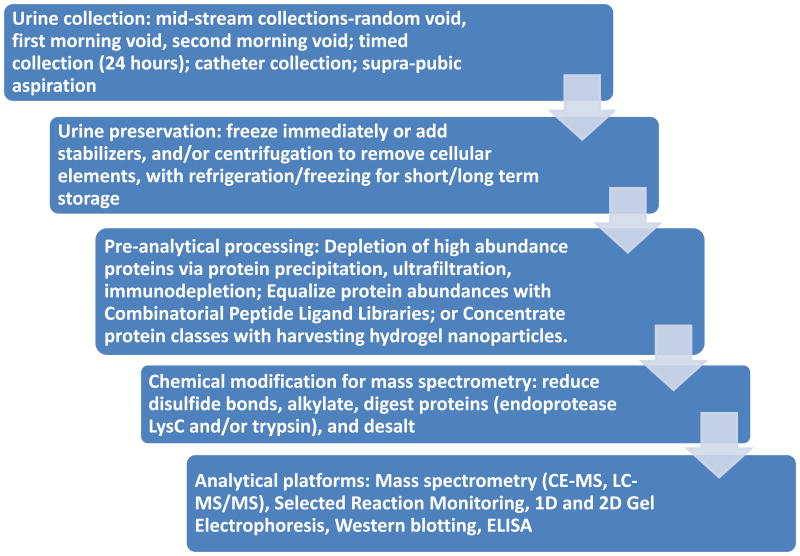
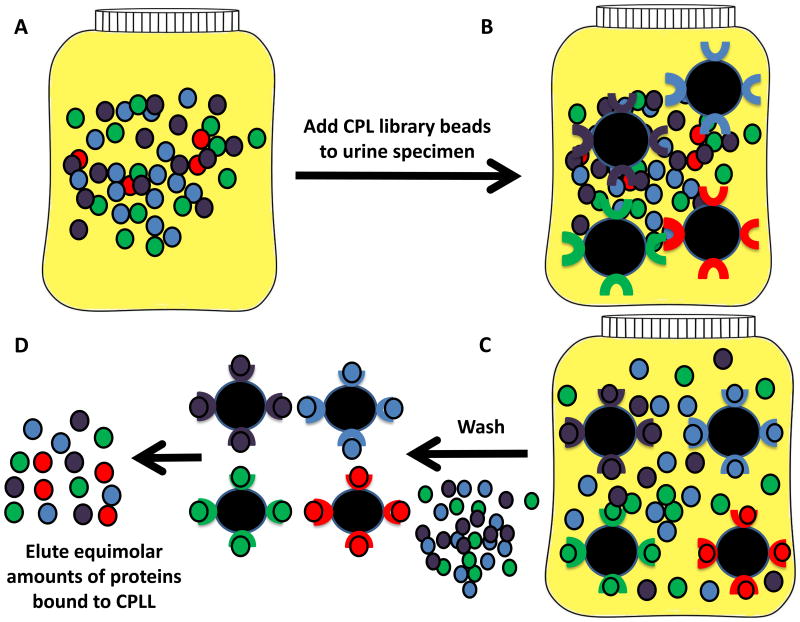
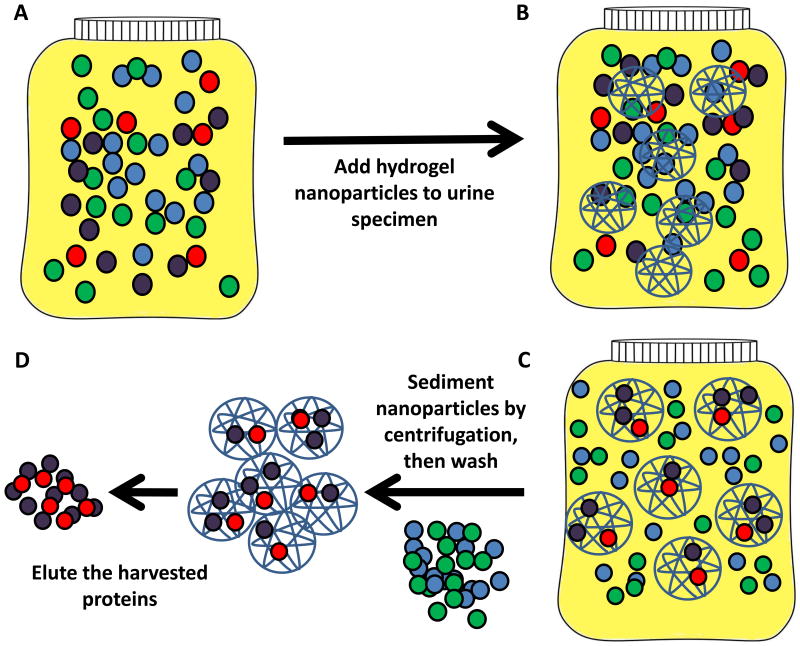
The work flow for urinary proteomics follows the general steps of sample collection, sample clarification, protein concentration/depletion/enrichment, and analysis (Figure 1). An analytical hurdle faced in proteomic analyses is the presence of high abundance proteins that can mask low abundance proteins [4, 15, 22, 23, 58, 123]. One approach to solve the problem of hidden, low abundance proteins is development of sensitive instruments, such as capillary electrophoresis time of flight mass spectrometry (CE-TOF-MS) [124], and the recently introduced capillary electrophoresis electrospray ionization mass spectrometry (CE-ESI-MS) for analysis of amino acids in urine [125]. CE-MS data has been reported to be highly sensitive (<1 fmol) with 85% recovery from the sample preparation, high resolution of monoisotopic peaks with mass accuracy <25ppm, and <100ppm for unresolved peaks (z<6) [126]. Rather than inventing new, more sensitive instruments, alternative approaches for detecting low abundance proteins rely on devising technologies for subtracting interfering high abundance proteins, enriching the low abundance protein fraction, or equalizing the abundance of both high and low abundance proteins. Two techniques for enriching low abundance proteins highlighted in this review are combinatorial peptide ligand libraries to enrich the low abundance proteins (CPLL, ProteoMiner™) (Figure 2) [22, 24, 104, 123, 127-130] and hydrogel nanoparticles (NP) for excluding high molecular weight proteins and simultaneously harvesting specific classes of proteins (Figure 3) (Table 3) [23, 57, 107, 131, 132].
Figure 1. Example workflow for deciphering the urinary proteome/peptidome.
Each step of the workflow should be optimized based on pathophysiology, and pre-analytical, analytical, post-analytical variables. Freezing urine specimens prior to centrifugation ensures that the cellular elements will be lysed and the cytoplasmic contents will contribute to the overall proteomic signature. Due to the large dynamic range of urinary protein concentrations, a variety of technologies are utilized to enhance the ability to detect low abundance urine proteins/peptides. Not all analytical platforms perform each step; however, the processes that are utilized in the test set/training set/pilot study should be identical to the processes used in the validation and verification studies.
Figure 2. Urine biomarker discovery using combinatorial peptide ligand libraries (CPLL).
CPLL consist of random hexamers of amino acids bound to beads, with one type of hexamer per bead [6, 22, 25, 72]. This approach utilizes a saturation-depletion strategy wherein high abundance proteins will saturate their ligand first, leaving the excess to be washed off, while the low abundance proteins will be able to bind continuously. This strategy minimizes the concentration difference between the low and high abundance proteins. A. Initial urine solution, B. CPLL beads incubated with urine, C. Protein binding to the hexamer bound beads occurs via van der Waals interactions, ionic interaction, and hydrogen bonding. Non-adsorbed, high abundance proteins are removed via washing, D. The hexamer-bound proteins may be recovered en masse, or by protein class, based on standard chromatography elution conditions (pH, chaotropic agent, ionic strength).
Figure 3. Protein enrichment strategy using hydrogel nanoparticles.
Porous hydrogel nanoparticles contain a dye, which contains functional groups and/or metal ions, that enhance binding of specific classes of proteins. The porous hydrogel simultaneously excludes high molecular weight proteins such as albumin, while protecting the harvested proteins from endogenous enzymatic digestion [23, 57, 107]. The chemical bait can be selected to preferentially sequester classes of proteins, such as negatively charged or positively charged proteins. A. Urine containing two potential biomarkers (red and purple circles) and high abundance proteins (green and blue circles). B. Nanoparticle incubation with the urine specimen. C. Nanoparticles concentrate the low abundance proteins. D. Following centrifugation to sediment the nanoparticles, the excluded, high abundance proteins are washed off and the concentrated, low abundance proteins are eluted from the particles.
One means of enriching a sample for your proteins of interest is to deplete the unwanted proteins. Bulk methods including ultrafiltration and immunodepletion have been widely used to deplete high abundance proteins, for example albumin, transferrin, immunoglobulins, and haptoglobin. Ultrafiltration, also known as filter aided separation, uses column chromatography to eliminate proteins above a specific molecular weight, usually in the range of >10-30 kDa. The proteins retained in the column are washed with buffers to remove salts and chemicals. The column-retained proteins are digested and the resulting peptides are eluted from the column [103]. Ultrafiltration effectively concentrates large volumes of urine prior to mass spectrometry [88, 109, 116, 126, 133]. Precipitation of proteins via ice cold ethanol or trichloroacetic acid allows concentration of soluble urine proteins [12, 111, 134]. Specific classes of protein can be depleted using immunodepletion [102]. Filip et.al. evaluated commercially available immunodepletion columns (Seppro IgY 14, ProteoPrep and SpinTrap) and an ion-exchange column (ProteoSep) for their ability to enhance detection of low abundance urine proteins using LC-MS/MS [102]. Fractionated and unfractionated specimens from patients with chronic kidney disease and healthy patients were compared by 1-D gel electrophoresis and nano-LC QTOF mass spectrometry. The immunodepletion columns were superior for depleting albumin fractions compared to the ion-exchange column. Fewer proteins were detected in the urine from chronic kidney disease patients, suggesting that the low abundance proteins were most likely masked by abundant urinary proteins that weren't depleted by the plasma protein depletion columns, or the mass spectrometer sensitivity was inadequate [102].
9. Combinatorial peptide ligand libraries
A second means of enriching a specimen for low abundance proteins is to selectively absorb proteins under capacity-limiting conditions [25]. Combinatorial peptide ligand libraries consist of hexamer peptides bound to a resin bead, with each bead containing millions of copies of a unique hexapeptide and a library of beads comprising many different hexapeptides on their respective beads [25]. This solid-phase hexapeptide library enriches low abundance proteins and also decreases the concentration of high abundance proteins, while representing all protein moieties in the original urine specimen [22]. CPLL were designed on the principles of affinity chromatography in which a chemical carrier is attached to a solid support to allow binding of a complimentary protein ligand. The physicochemical properties of a protein are determined by the amino acid sequence and their spatial configuration [25]. CPLL-ligand interactions vary from weak to strong depending on the type of physicochemical interaction (van der Waals interaction, hydrogen bonding, hydrophobic, ionic) [22]. Thus a mixture of many hexapeptide ligands could theoretically bind every protein in a complex milieu such as serum or urine. Each hexapeptide binds its respective protein ligand until saturation is reached, thus equalizing the concentration between low and high abundance proteins [130]. Increasing the volume of sample in relation to a fixed number of hexapeptide beads enhances the number of low abundance proteins [135]. 1M ammonium sulfate may be used to enhance hydrophobic hexapeptide-complementary protein interactions on CPLL [136]. Unbound/weakly bound proteins are commonly removed by washing, and the hexapeptide-bound proteins are eluted using appropriate conditions for the protein of interest/detection method (Figure 2). A potential limitation of using CPLL to enrich low abundance proteins is the low specificity of CPLL binding [135], however this issue has been addressed by standardizing the elution protocols [135], optimizing the elution schemes [7, 104, 128, 135], and performing pilot studies for different biological hypotheses.
Proteins bound to the CPLL may be directly trypsinized from the beads for direct LC-MS/MS analysis [137, 138]. One must use more trypsin than that used for in-gel digestion to allow the trypsin to reach all areas within the hexapeptide coated bead. Direct CPLL trypsinization ensures that even minute amounts of proteins are digested for analysis [137, 138]. A more common strategy is to eluate/desorb the bound proteins. Proteins can be eluted either globally from all beads at the same time, or specific classes of proteins may be eluted from the beads by selecting specific elution conditions (pH, chaotropic agent, ionic strength) [72]. Elution schemes for CPLL consist of a) disruption of hydrogen and ionic bond interactions with 100mM Tris, pH 7.4, 100mM lysine, pH 7.4, or a mixture of Arginine, Lysine, Histidine and Glutamine [104], b) disruption of all types of protein-ligand interactions with detergent and a reducing agent (2-4% sodium dodecyl sulfate and 2.5-3% dithiothreitol) [135], c) dissociation of ionic and electrostatic interactions with guanidine hydrochloride [129], or d) disruption of hydrophobic interactions with urea-thiourea-cysteic acid [129]. Native proteins can be enriched in urine using the four amino acid mixture [104]. It should be noted that bile salts and urinary pigments (Urobilin, urochrome, uroerythrin) [139] present in urine were observed to bind strongly to CPLL, thus quenching albumin binding; however this same lack of albumin binding may not occur in kidney disease in which urinary pigment is decreased [104]. Urinary pigments were observed to be increased in patients in the third trimester of pregnancy, during high fever, Grave's Disease, and short periods of fasting [139]. Fortunately, butanol extraction can remove the interfering urine pigments [7, 129].
An optimized protocol for separation of urinary exosomes, utilizing CPLL to enrich low abundance proteins, and linear trap quadarople (LTQ) Orbitrap mass spectrometry has been developed by Santucci et. al. [7, 106]. Second morning urine specimens were stabilized with protease inhibitors on ice, clarified by centrifugation, and frozen. Following thawing, the specimens were spun at high speed, dialyzed against 25mM sodium phosphate, and refrozen. To isolate exosomes, supernatant from the high speed centrifugation was reduced with dithiothreitol to prevent exosome aggregation with uromodulin, followed by ultracentrifugation. The supernatant from the ultracentrifugation was dialyzed against water, acidified and precipitated with butanol. The butanol extraction phase was lyophilized for CPLL treatment [7]. Using this protocol, they detected 1724 new urinary proteins, many of which were found in exosomes [7].
The newest iteration of CPLL contain Alcian blue dye coupled to succinylated hexamers of hexapeptides to enhance protein binding [6]. By adding the copper containing phthalocyanine dye, Alcian blue, to the succinylated hexamers of CPLL, 115 urinary proteins with nucleotide, carbohydrate or cation binding, or catalytic activity were detected [6]. Combining CPLL equilibration techniques and physicochemical dye interactions into one capture moiety has revealed 38 previously undetected urine proteins, increasing the total urinary proteome to 4430 gene products [6]. By substituting the dye or altering the functional group of the hexapeptide libraries via different chemical transformations, a plethora of new protein harvesting agents can be synthesized to explore the urinary proteome.
10. Hydrogel nanoparticles
Hydrogel nanoparticles (NP) are three-dimensional hydrophilic copolymers of poly (N-isopropylacrylamide) (pNIPAm) and other compounds that change conformation from an extended coil to a globular structure in aqueous solutions [23, 140]. Hydrogel nanoparticles are synthesized via monomer polymerization with a cross-linking agent, which itself is a monomer with functional moieties that can also be polymerized [23]. These responsive hydrogels change solvation properties in response to temperature, pH, and ionic strength, providing added functionality to the polymer [23]. Hydrogel nanoparticles swell at lower temperatures, thus increasing their porosity at room temperature (500-700nm diameter) [23, 140]. This thermo-reversible property of hydrogel pNIPAm copolymers with acrylic acid has been extensively tested as a drug delivery vehicle [140].
Capitalizing on the NP modifiable functionality, Luchini et. al. synthesized hydrogel nanoparticles for molecular sieving, in solution, to sequester rather than release proteins [23, 107, 141]. The hydrogel nanoparticles can be easily modified by changing the polymer:cross linking agent ratio, thus controlling the nanoparticle size and effective porosity independently of temperature, while still maintaining the thermos-response properties of the hydrogel [23]. These characteristics allow the NP to rapidly harvest molecules out of solution due to the nanoparticle's large, tunable surface area, porosity, hydrophilicity, buoyancy, and dual hydrophobic and hydrophilic chemical moieties that can be substituted in the polymer [23, 107, 141]. In addition, NIPAm and acrylic acid copolymer nanoparticles were synthesized to incorporate an organic dye within the nanoparticle structure as a bait molecule for capturing low abundance proteins out of solution [23, 107, 141]. These common histology and affinity chromatography dyes have a high affinity for select classes of proteins based on the corresponding dye/protein physiochemical properties and hydrophobic interactions [23, 107, 141]. The nanoparticle pore size acts as a molecular sieve, effectively preventing large proteins such as albumin and immunoglobulins from entering the pores [107]. Another desirable property of the NP for harvesting low abundance proteins is their ability to protect their protein cargo from endogenous enzymatic digestion [23].
The ratio of nanoparticles to specimen volume can be adjusted to effectively capture 100% of the low abundance proteins in the specimen, hence amplifying the sensitivity of downstream analysis, including standard mass spectrometry, immunoassays, or western blotting [142]. In one step, in solution, hydrogel nanoparticles perform molecular size sieving, exclude high abundance proteins, capture classes of proteins based on the dye's functional group properties, and protect the captured proteins from enzymatic degradation [142]. Hydrogel nanoparticles are now commercially available in large quantities with uniform size distribution (NanoTrap ®) for harvesting and concentrating proteins in body fluids [143].
Hydrogel nanoparticle protein harvesting has been used to concentrate human growth hormone in serum and urine specimens, allowing the first estimate of growth hormone levels in human urine [132, 144, 145]. Growth hormone levels are important during clinical evaluation of growth hormone insufficiency. Growth hormone is secreted in pulses from the somatrotroph cells of the pituitary gland resulting in wide fluctuations in blood levels, with peak levels of 50-100ng/mL and less than 0.03ng/mL at baseline [144]. Pathophysiologic factors including gender, age, sleep, physical exertion, nutritional and metabolic factors, influence growth hormone secretion. The normal serum concentration of growth hormone is 1-10ng/mL, which is below the limit of detection of clinical grade immunoassays (50pg/mL) [144]. Growth hormone is efficiently filtered through the glomerulus, reabsorbed, and degraded in the proximal tubular cells; consequently the growth hormone concentration in urine is considerably less than in plasma [144]. Nanoparticles containing Cibacron Blue F3GA, a sulfonated triazine dye as the affinity bait, were used to harvest and concentrate urinary growth hormone from a healthy donor. The urine contained an estimated 0.175pg/mL of growth hormone, with 1.34 ng/mL in the donor matched serum specimen, thus confirming the ability of the hydrogel nanoparticles to concentrate very low abundance proteins [144].
Bacterial antigens can be harvested by hydrogel nanoparticles to enhance the sensitivity of western blot detection. Lyme Disease is transmitted via ticks infected with Borrelia spirochetes, commonly B. burgdorferi in the eastern United States. Diagnosis is typically made based on the presence of erythema migrans (bulls eye) rash and/or positive serologic test for antibodies to Borrelia antigens [57]. Lyme Disease can be difficult to diagnose in the early stages due to vague or overlapping symptoms, lack of the classic erythema migrans rash, or indeterminate serology results, thus potentially delaying treatment. As a proof-of-principle assay for developing a urine-based Lyme Disease assay, hydrogel nanoparticles were used to concentrate B. burgdorferi proteins spiked into urine [56]. NIPAm-acrylic acid nanoparticles containing the nondisperse dye Acid Black 48 were shown to sequester and concentrate B. burgdorferi outer surface proteins in urine, as well as from an individual Ixodes scapularis tick [56]. A non-invasive urine test to detect the presence of the B. burgdorferi antigens prior to development of antibodies could transform early diagnosis and treatment for Lyme Disease [57].
Following the proof-of-principle study, the dye bait was optimized for concentrating Borrelia antigens [57]. NP containing the anthraquinone dye Remazol Brilliant Blue were used to enhance the sensitivity of detection of B. burgdorferi via western blotting with a monoclonal antibody against a specific C-terminal OspA domain [57]. Urine specimens from 151 patients suspected of early stage or recurrent Lyme disease and 117 healthy controls were evaluated for the presence of urinary OspA [57]. Hydrogel nanoparticle harvesting of urinary proteins was able to confirm the presence of Borrelia OspA in untreated patients who had a characteristic erythema migrans rash. Furthermore, 41% of patients with suspected chronic Lyme Borrellia infection also were shown to harbor Borrelia OspA in their urine [57]. Clinical evaluation of the urine nanoparticle procedure is on-going.
Hydrogel nanoparticle protein harvesting has also been applied for diagnosing congenital transmission of Chagas Disease, caused by the parasite Trypanosoma cruzi [55]. Early detection of congenital Chagas Disease is difficult because of the low sensitivity of microscopic test and poor patient follow-up [55]. Using hydrogel nanoparticles containing the azo dye Trypan blue as the affinity bait, T. cruzi antigens were detectable in urine specimens by western blot with a monoclonal antibody against T. cruzi lipophosphoglycan, thus enabling an early diagnosis for congenital transmission of Chagas Disease [55]. The sensitivity of the NP enhanced western blot was 91.3%, with a specificity of 96.5% [55].
These aforementioned studies using combinatorial peptide libraries or hydrogel nanoparticle harvesting are heralding in a new era of urinary proteomics in which hormones, infectious disease antigens, and other very low abundance proteins and protein classes can be selectively enriched, identified, and quantified with existing immunoassays and mass spectrometers.
11. Urine proteome assays under development
Non-invasive assays for monitoring immediate blood glucose levels and long-term glucose control have the potential to dramatically impact the quality of life for diabetic patients. The non-invasive diagnostic trend encourages better diet/medication compliance and could decrease health care costs. Recently, non-invasive methods for monitoring glucose levels using mid-infrared spectroscopy of interstitial fluid between the layers of skin have been developed [146]. Urine is also being explored as a surrogate to blood for monitoring diabetes-related biomarkers. Zhang et. al. applied matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) and LC-MS/MS to urine specimens from patients with type 2 diabetes (n=49) and non-diabetic controls (n=49) to identify differentially expressed urinary proteins [121]. Urine proteins were concentrated using weak cation exchange chromatography with magnetic beads. A fragment of fibrinogen alpha chain precursor and a prothrombin precursor were upregulated in the diabetic patient specimens [121]. The authors speculate that the coagulation-associated peptides may reflect hypercoagulable states and impending diabetic vascular complications. Of note, the fibrinogen alpha chain precursor and prothrombin precursor were not statistically significant when the peptides were compared to blood glucose levels, but were significant when the data was compared to the glycosylated hemoglobin values [121]. Glycosylated hemoglobin (A1c) levels reflect long-term (over the course of 90 days) glucose control [147]. This difference in statistically significant data emphasizes the necessity of normalizing/comparing urinary proteomic data to a relevant biochemical assay in order to discern potentially relevant information.
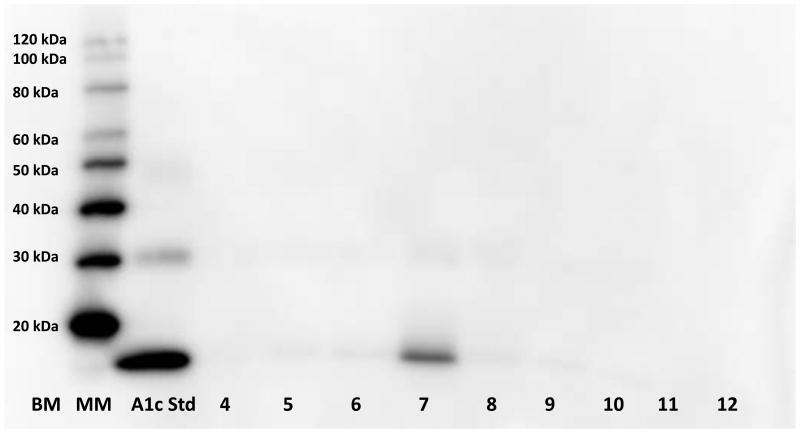
Although glycosylated hemoglobin assays reflect long-term glucose levels, the standard assay uses whole blood collected by venipuncture [147]. We are developing a non-invasive urine assay based on hydrogel nanoparticle sequestration of A1c derived from urinary erythrocytes. The first step in developing this assay was to screen various dyes for the ability to capture glycosylated proteins, thus facilitating enrichment of A1c. We found a copper containing azo dye, Reactive Black 31 (RB31, CAS # 12731-63-4), to be the optimal chemical bait for capturing glycosylated hemoglobin in urine (Figure 4). The utility of RB31 dye was confirmed for protein harvesting by using synthetic urine (Surine) containing spiked-in serum. Compared to the starting solution without nanoparticles, the RB31 dye containing nanoparticles were able to concentrate proteins in this serum-Surine solution (Figure 5). The ultimate goal of a urine-based glycosylated hemoglobin assay is to develop a urine-based point-of-care test for measuring the ratio of A1c to total hemoglobin. Of course, this assay is in the preliminary stages of development and must undergo analytical validation for sensitivity, specificity, accuracy and precision as well as clinical validation for patients with diabetes [148].
Figure 4. Proof of principle for developing a urine-based glycosylated hemoglobin (A1c) assay.
Different dyes were incorporated into hydrogel nanoparticles to determine which nanoparticle bait (dye) was optimal for harvesting A1c. 100ng A1c (LEE Biosolutions cat#325-10, 15kD) was added to 5mL of Surine™ (synthetic urine negative control, Sigma-Aldrich) prior to adding the nanoparticles. Following elution from the nanoparticles, western blotting with anti-A1c (LifespanBio, LS-C194149-250) was used to demonstrate A1c capture by the nanoparticles. Reactive Black 31 (Lane 7) was found to be the optimal dye for harvesting A1c. Lane 1: BenchMark pre-stained protein ladder, Lane 2: Magic Mark chemiluminescent protein standard, Lane 3: 50ng A1c, Lane 4: Trypan Blue, Lane 5: Congo Red, Lane 6: Direct Blue 2, Lane 7: Reactive Black 31, Lane 8: Reactive Blue 221, Lane 9: Remazol Brilliant Blue, Lane 10: Sequential harvesting with Trypan Blue then Reactive Blue 221 nanoparticles, Lane 11: Surine™ sans nanoparticles, Lane 12: empty. (Lanes 4-10=proteins eluted from the respective nanoparticle).
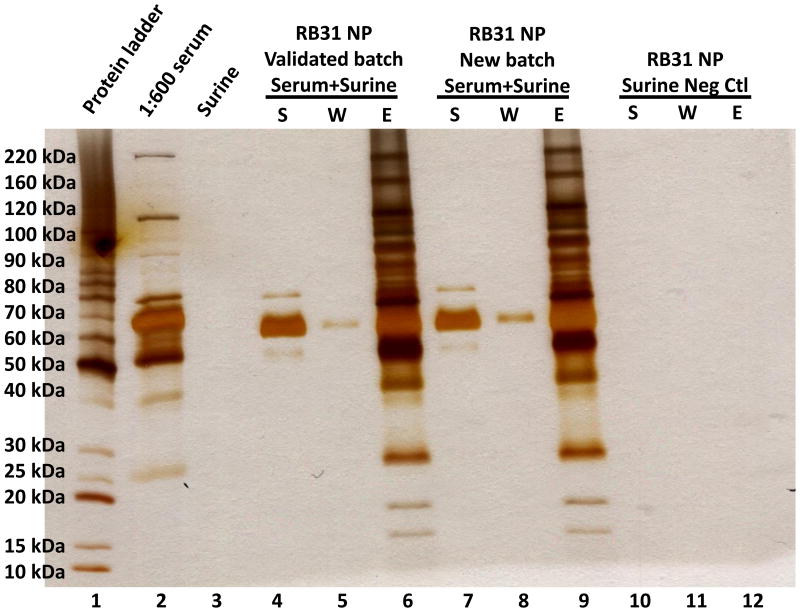
Figure 5. Validation of Reactive Black 31 nanoparticles (RB31 NP) for harvesting proteins.
Serum (1:600 dilution) was spiked into synthetic urine (Surine™) to validate the protein harvesting ability of the Reactive Black 31 dye containing nanoparticles. The high abundance proteins remain in the supernatant, showing that they are not captured by the nanoparticles. A previously validated batch of nanoparticles was compared to a new batch and Surine™ (as a negative control). Lane 1: Benchmark unstained protein ladder, Lane 2: initial solution (1:600 serum), Lane 3: Surine™, Lane 4: supernatant of a previously validated RB31 NP batch, Lane 5: wash, Lane 6: elution, Lane 7: new batch of RB31 NP supernatant, Lane 8: new batch of RB31 NP wash, Lane 9: new batch of RB31 NP elution, Lane 10: supernatant negative control (Surine™), Lane 11: Surine™ wash, Lane 12: Surine™ elution.
12. Expert Commentary
Technological advancements in instrumentation and specimen processing for low abundance proteins are the catalysts driving the elucidation of the urinary proteome. Masking of low abundance proteins by high abundance proteins is not unique to urine specimens and fractionation techniques that have been developed for serum protein analysis are now being applied to urine [149].. These technological ehancements are providing insights into inter-individual and intra-individual variation of the urinary proteome. We are now glimpsing the reality of establishing normal values, or at least typical values, for urine proteins in certain populations [11, 113, 115].
Now that we can delve deeper into the body fluid proteome, we need to step back and reconsider the potential effects of specimen processing, i.e. the ‘spin first versus freeze first’ urine stabilization methods and fractionation methods, on the urinary proteome. This is particularly important when comparing studies for biomarker discovery in healthy versus disease states. Studies investigating pre-analytical variables are not typically considered newsworthy, but their importance impacts every clinical study seeking an answer to a biological question [28, 31, 35, 100, 101]. The main pre-analytical question that should be addressed for each urinary proteomic study is: Do the biochemical, uroepithelial cells, or bacterial cells present in the urine specimen contribute to the pathophysiology of the condition to be studied? Are these urinary proteins that we can now quantify derived from the plasma ultra-filtrate, or are they degradative products from the urogenital environment?
We shouldn't dismiss routine clinical biochemical and cytological urinalysis of urine specimens now that we can harvest, concentrate, identify, and quantify specific proteins/peptides. Urinalysis, including microscopic examination of the urine sediment, prior to proteomic biomarker discovery could reveal sources of false positive or false negative biomarkers [150]. Urine test strip (dipstick) assays are commercially available, rapid, and sensitive [27]. Furthermore, biochemical anlaytes measurable via a test strip, such as total protein, glucose, specific gravity, urobilinogen, glucose, ketones, blood, nitrite and leukocyte esterase, can potentially be used in data normalization and as specimen rejection criteria. Integration of clinical diagnostic assays with biomarker discovery will be the next leap in correlating urinary proteins/peptides with pathophysiology.
Urinary proteomic translational research has benefited greatly from dedicated clinicians and scientists who have recognized the need for: a) more studies that advance from initial testing in pilot studies, to controlled and validated studies, and finally progressing to large scale clinical trials, b) non-redundant urinary proteomic database access and maintenance, and c) adoption of clinical laboratory consensus statements and/or laboratory standards for comparability of results across analysis platforms and between laboratories [37, 151, 152]. These standards and guidelines facilitate discoveries by providing a framework for specimen collection, processing, analysis, and data reduction. Further innovations in urine specimen processing, exemplified by GlycoFilter sample preparation [97], CPLL-Alcian blue dye constructs [6], and hydrogel-dye nanoparticles [57], will reveal the true extent of urinary biomolecules and the plethora of molecular interactions waiting to be discovered, correlated, and integrated with treatment options for many different types of diseases/conditions.
13. Five-year View
Embracing new technology platforms and specimen processing methods to measure and detect the unseen or previously undetectable proteins, post-translational modifications, nucleic acids, exosomes, and infectious agents in urine will be a key driver in urinary proteomics. One size does not fit all, especially in clinical applications. Specimen processing procedures will need to be adjusted based on the clinical hypothesis, goal of the study, source of the molecule to be measured, the analytical platform, and/or the parameter for normalizing data.. Multi-omic biomarker panels may become more commonplace as biochemical, proteomic, genomic, and metabolomic tools and data are developed and integrated into translational research studies to generate a molecular portrait for diagnosis, prognosis, and/or treatment efficacy.
We need to rethink how we are analyzing the urinary proteome. Rather than pooling specimens from multiple individuals, or from the same individual over time, we need to envision a future in which we can monitor/measure an individual patient's urinary proteome longitudinally. The individual patient would serve as their own baseline and we could monitor longitudinal fluctuations of inflammatory markers which are harbingers of cancer, infection, renal failure, etc. Improvements in point-of-care testing using mobile imaging such as with a cell phone camera [153, 154] are providing the technology to develop individual urine health monitoring devises. With miniaturized specimen processing modules and mobile imaging, we can envision ‘urine health trackers’ similar to the wearable wrist-style electronic health monitors.
14. Key Issues.
The depth and breadth of the urinary proteome is increasing due to advances in analytical sensitivity and innovations in specimen processing to diminish high abundance urinary proteins that mask potential biomarkers within the low abundance proteome.
Specimen processing impacts the content and quantity of urinary proteins/peptides.
Specimen processing should be based on the clinical question being addressed in the study. Do the biochemicals, uroepithelial cells, or bacterial cells present in the urine specimen contribute to the pathophysiology of the condition to be studied? Are these urinary proteins that we can now quantify derived from the plasma ultra-filtrate, or are they degradative products from the urogenital environment?
The urinary proteome exhibits inter and intra-individual variations. Population data from apparently healthy individuals is needed to develop normal ranges of urinary proteins/peptides.
Pre-analytical variables need to be addressed in urinary proteome studies prior to starting the study.
Urinary proteomics offers opportunities for development and validation of non-invasive diagnostics for monitoring immune function, as well as cardiovascular, renal, and infectious diseases.
Further innovations in specimen processing will reveal the true extent of urinary biomolecules and the plethora of molecular interactions waiting to be discovered, correlated, and integrated with treatment options for many different types of diseases/conditions.
Data normalization for urine proteomics presents challenges not encountered in serum/plasma proteomic studies because urine volume is highly variable between and within individuals, unlike the relatively stable blood volume. Urine proteomic studies will likely require a multi-analyte normalization method that incorporates excretion, secretion, and filtration parameters to be able to compare urinary proteomic data.
Acknowledgments
This work was supported in part by George Mason University, the National Institutes of Health (NIH) Innovative Molecular Analysis Technologies (IMAT) program through a grant to Lance Liotta (1R33CA173359-01), and a grant from the Gates Foundation to L. Liotta (Nanotrap sensitivity enhancement of LAM in urine). Lance Liotta is Principle Investigator for the IMAT and Gates Foundation grants and kindly provided editorial advice for this manuscript. The funding sources did not have any role in the study design; the collection, analysis and interpretation of data; manuscript preparation; or the decision to submit the paper for publication. Conflict of interest statement: VE is an inventor of hydrogel nanoparticle technologies discussed in this article and, as a university employee, may receive patent royalties per university policies. VE and MH receive salary from NIH IMAT grant 1R33CA173359-01. Author contributions: MH contributed experimental data. MH, JD, and VE wrote the manuscript.
References
- 1.Faulkner WR, King JW. Renal function. In: Tietz NW, editor. Fundamentals in Clinical Chemistry. W.B. Suanders Co; Philadelphia: 1976. pp. 975–1014. [Google Scholar]
- 2.Decramer S, Gonzalez de Peredo A, Breuil B, et al. Urine in clinical proteomics. Mol Cell Proteomics. 2008;7(10):1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Jia L, Zhang L, Shao C, et al. An attempt to understand kidney's protein handling function by comparing plasma and urine proteomes. PLoS One. 2009;4(4):e5146. doi: 10.1371/journal.pone.0005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 5.Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 6.Candiano G, Santucci L, Petretto A, et al. Widening and diversifying the proteome capture by combinatorial peptide ligand libraries via Alcian Blue dye binding. Anal Chem. 2015;87(9):4814–4820. doi: 10.1021/acs.analchem.5b00218. [DOI] [PubMed] [Google Scholar]
- 7.Santucci L, Candiano G, Petretto A, et al. From hundreds to thousands: Widening the normal human Urinome (1) J Proteomics. 2015;112:53–62. doi: 10.1016/j.jprot.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 8.Pieper R, Gatlin CL, McGrath AM, et al. Characterization of the human urinary proteome: a method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics. 2004;4(4):1159–1174. doi: 10.1002/pmic.200300661. [DOI] [PubMed] [Google Scholar]
- 9.Chatziharalambous D, Lygirou V, Latosinska A, et al. Analytical Performance of ELISA Assays in Urine: One More Bottleneck towards Biomarker Validation and Clinical Implementation. PLoS One. 2016;11(2):e0149471. doi: 10.1371/journal.pone.0149471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binder H, Wirth H, Arakelyan A, et al. Time-course human urine proteomics in space-flight simulation experiments. BMC Genomics. 2014;15(Suppl 12):S2. doi: 10.1186/1471-2164-15-S12-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y. Variations of human urinary proteome. Adv Exp Med Biol. 2015;845:91–94. doi: 10.1007/978-94-017-9523-4_9. [DOI] [PubMed] [Google Scholar]
- 12.Khristenko NA, Larina IM, Domon B. Longitudinal Urinary Protein Variability in Participants of the Space Flight Simulation Program. J Proteome Res. 2016;15(1):114–124. doi: 10.1021/acs.jproteome.5b00594. [DOI] [PubMed] [Google Scholar]
- 13.Larina IM, Pastushkova L, Tiys ES, et al. Permanent proteins in the urine of healthy humans during the Mars-500 experiment. J Bioinform Comput Biol. 2015;13(1):1540001. doi: 10.1142/S0219720015400016. [DOI] [PubMed] [Google Scholar]
- 14.Nagaraj N, Mann M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J Proteome Res. 2011;10(2):637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- 15.Davis MT, Spahr CS, McGinley MD, et al. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. II. Limitations of complex mixture analyses. Proteomics. 2001;1(1):108–117. doi: 10.1002/1615-9861(200101)1:1<108::AID-PROT108>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 16.Spahr CS, Davis MT, McGinley MD, et al. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. I. Profiling an unfractionated tryptic digest. Proteomics. 2001;1(1):93–107. doi: 10.1002/1615-9861(200101)1:1<93::AID-PROT93>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Klein J, Eales J, Zurbig P, Vlahou A, Mischak H, Stevens R. Proteasix: a tool for automated and large-scale prediction of proteases involved in naturally occurring peptide generation. Proteomics. 2013;13(7):1077–1082. doi: 10.1002/pmic.201200493. [DOI] [PubMed] [Google Scholar]
- 18.Bantscheff M, Lemeer S, Savitski MM, Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem. 2012;404(4):939–965. doi: 10.1007/s00216-012-6203-4. [DOI] [PubMed] [Google Scholar]
- 19.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389(4):1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 20.Wittke S, Fliser D, Haubitz M, et al. Determination of peptides and proteins in human urine with capillary electrophoresis-mass spectrometry, a suitable tool for the establishment of new diagnostic markers. J Chromatogr A. 2003;1013(1-2):173–181. doi: 10.1016/s0021-9673(03)00713-1. [DOI] [PubMed] [Google Scholar]
- 21.Wittke S, Mischak H, Walden M, Kolch W, Radler T, Wiedemann K. Discovery of biomarkers in human urine and cerebrospinal fluid by capillary electrophoresis coupled to mass spectrometry: towards new diagnostic and therapeutic approaches. Electrophoresis. 2005;26(7-8):1476–1487. doi: 10.1002/elps.200410140. [DOI] [PubMed] [Google Scholar]
- 22.Castagna A, Cecconi D, Sennels L, et al. Exploring the hidden human urinary proteome via ligand library beads. J Proteome Res. 2005;4(6):1917–1930. doi: 10.1021/pr050153r. [DOI] [PubMed] [Google Scholar]
- 23.Luchini A, Geho DH, Bishop B, et al. Smart hydrogel particles: biomarker harvesting: one-step affinity purification, size exclusion, and protection against degradation. Nano Lett. 2008;8(1):350–361. doi: 10.1021/nl072174l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Righetti PG, Boschetti E, Lomas L, Citterio A. Protein Equalizer Technology : the quest for a “democratic proteome”. Proteomics. 2006;6(14):3980–3992. doi: 10.1002/pmic.200500904. [DOI] [PubMed] [Google Scholar]
- 25.Thulasiraman V, Lin S, Gheorghiu L, et al. Reduction of the concentration difference of proteins in biological liquids using a library of combinatorial ligands. Electrophoresis. 2005;26(18):3561–3571. doi: 10.1002/elps.200500147. [DOI] [PubMed] [Google Scholar]
- 26.Danish LM, Heistermann M, Agil M, Engelhardt A. Validation of a Novel Collection Device for Non-Invasive Urine Sampling from Free-Ranging Animals. PLoS One. 2015;10(11):e0142051. doi: 10.1371/journal.pone.0142051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Froom P, Bieganiec B, Ehrenrich Z, Barak M. Stability of common analytes in urine refrigerated for 24 h before automated analysis by test strips. Clin Chem. 2000;46(9):1384–1386. [PubMed] [Google Scholar]
- 28.Hepburn S, Cairns DA, Jackson D, et al. An analysis of the impact of pre-analytical factors on the urine proteome: Sample processing time, temperature, and proteolysis. Proteomics Clin Appl. 2015;9(5-6):507–521. doi: 10.1002/prca.201400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ling XB, Mellins ED, Sylvester KG, Cohen HJ. Urine peptidomics for clinical biomarker discovery. Adv Clin Chem. 2010;51:181–213. doi: 10.1016/s0065-2423(10)51007-2. [DOI] [PubMed] [Google Scholar]
- 30.Norden AG, Rodriguez-Cutillas P, Unwin RJ. Clinical urinary peptidomics: learning to walk before we can run. Clin Chem. 2007;53(3):375–376. doi: 10.1373/clinchem.2006.084038. [DOI] [PubMed] [Google Scholar]
- 31.Olszowy P, Buszewski B. Urine sample preparation for proteomic analysis. J Sep Sci. 2014;37(20):2920–2928. doi: 10.1002/jssc.201400331. [DOI] [PubMed] [Google Scholar]
- 32.Lassek C, Burghartz M, Chaves-Moreno D, et al. A metaproteomics approach to elucidate host and pathogen protein expression during catheter-associated urinary tract infections (CAUTIs) Mol Cell Proteomics. 2015;14(4):989–1008. doi: 10.1074/mcp.M114.043463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall T, Williams KM. Total protein determination in urine: elimination of a differential response between the coomassie blue and pyrogallol red protein dye-binding assays. Clin Chem. 2000;46(3):392–398. [PubMed] [Google Scholar]
- 34.Thomsen HS, Morcos SK. Contrast media and the kidney: European Society of Urogenital Radiology (ESUR) guidelines. Br J Radiol. 2003;76(908):513–518. doi: 10.1259/bjr/26964464. [DOI] [PubMed] [Google Scholar]
- 35.Court M, Selevsek N, Matondo M, et al. Toward a standardized urine proteome analysis methodology. Proteomics. 2011;11(6):1160–1171. doi: 10.1002/pmic.201000566. [DOI] [PubMed] [Google Scholar]
- 36.Mischak H, Allmaier G, Apweiler R, et al. Recommendations for biomarker identification and qualification in clinical proteomics. Sci Transl Med. 2010;2(46) doi: 10.1126/scitranslmed.3001249. 46ps42. [DOI] [PubMed] [Google Scholar]
- 37.Mischak H, Ioannidis JP, Argiles A, et al. Implementation of proteomic biomarkers: making it work. Eur J Clin Invest. 2012;42(9):1027–1036. doi: 10.1111/j.1365-2362.2012.02674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez-Suarez E, Siwy J, Zurbig P, Mischak H. Urine as a source for clinical proteome analysis: from discovery to clinical application. Biochim Biophys Acta. 2014;1844(5):884–898. doi: 10.1016/j.bbapap.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto T, Langham RG, Ronco P, Knepper MA, Thongboonkerd V. Towards standard protocols and guidelines for urine proteomics: a report on the Human Kidney and Urine Proteome Project (HKUPP) symposium and workshop, 6 October 2007, Seoul, Korea and 1 November 2007, San Francisco, CA, USA. Proteomics. 2008;8(11):2156–2159. doi: 10.1002/pmic.200800138. [DOI] [PubMed] [Google Scholar]
- 40.Mischak H, Vlahou A, Ioannidis JP. Technical aspects and inter-laboratory variability in native peptide profiling: the CE-MS experience. Clin Biochem. 2013;46(6):432–443. doi: 10.1016/j.clinbiochem.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 41.Albalat A, Mischak H, Mullen W. Clinical application of urinary proteomics/peptidomics. Expert Rev Proteomics. 2011;8(5):615–629. doi: 10.1586/epr.11.46. [DOI] [PubMed] [Google Scholar]
- 42.Farrah T, Deutsch EW, Omenn GS, et al. State of the human proteome in 2013 as viewed through PeptideAtlas: comparing the kidney, urine, and plasma proteomes for the biology- and disease-driven Human Proteome Project. J Proteome Res. 2014;13(1):60–75. doi: 10.1021/pr4010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katz R. Biomarkers and surrogate markers: an FDA perspective. NeuroRx. 2004;1(2):189–195. doi: 10.1602/neurorx.1.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mischak H. How to get proteomics to the clinic? Issues in clinical proteomics, exemplified by CE-MS. Proteomics Clin Appl. 2012;6(9-10):437–442. doi: 10.1002/prca.201200027. [DOI] [PubMed] [Google Scholar]
- 45.Mischak H, Delles C, Vlahou A, Vanholder R. Proteomic biomarkers in kidney disease: issues in development and implementation. Nat Rev Nephrol. 2015;11(4):221–232. doi: 10.1038/nrneph.2014.247. [DOI] [PubMed] [Google Scholar]
- 46.de Bono JS, Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467(7315):543–549. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 47.Jameson JL, Longo DL. Precision medicine--personalized, problematic, and promising. N Engl J Med. 2015;372(23):2229–2234. doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
- 48.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frantzi M, Latosinska A, Merseburger AS, Mischak H. Recent progress in urinary proteome analysis for prostate cancer diagnosis and management. Expert Rev Mol Diagn. 2015;15(12):1539–1554. doi: 10.1586/14737159.2015.1104248. [DOI] [PubMed] [Google Scholar]
- 50.Raimondo F, Corbetta S, Chinello C, Pitto M, Magni F. The urinary proteome and peptidome of renal cell carcinoma patients: a comparison of different techniques. Expert Rev Proteomics. 2014;11(4):503–514. doi: 10.1586/14789450.2014.926222. [DOI] [PubMed] [Google Scholar]
- 51.Wu J, Gao Y. Physiological conditions can be reflected in human urine proteome and metabolome. Expert Rev Proteomics. 2015;12(6):623–636. doi: 10.1586/14789450.2015.1094380. [DOI] [PubMed] [Google Scholar]
- 52.Kentsis A, Lin YY, Kurek K, et al. Discovery and validation of urine markers of acute pediatric appendicitis using high-accuracy mass spectrometry. Ann Emerg Med. 2010;55(1):62–70 e64. doi: 10.1016/j.annemergmed.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kharbanda AB, Rai AJ, Cosme Y, Liu K, Dayan PS. Novel serum and urine markers for pediatric appendicitis. Acad Emerg Med. 2012;19(1):56–62. doi: 10.1111/j.1553-2712.2011.01251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young BL, Mlamla Z, Gqamana PP, et al. The identification of tuberculosis biomarkers in human urine samples. Eur Respir J. 2014;43(6):1719–1729. doi: 10.1183/09031936.00175113. [DOI] [PubMed] [Google Scholar]
- 55.Castro-Sesquen YE, Gilman RH, Galdos-Cardenas G, et al. Use of a novel chagas urine nanoparticle test (chunap) for diagnosis of congenital chagas disease. PLoS Negl Trop Dis. 2014;8(10):e3211. doi: 10.1371/journal.pntd.0003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Douglas TA, Tamburro D, Fredolini C, et al. The use of hydrogel microparticles to sequester and concentrate bacterial antigens in a urine test for Lyme disease. Biomaterials. 2011;32(4):1157–1166. doi: 10.1016/j.biomaterials.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Magni R, Espina BH, Shah K, et al. Application of Nanotrap technology for high sensitivity measurement of urinary outer surface protein A carboxyl-terminus domain in early stage Lyme borreliosis. J Transl Med. 2015;13:346. doi: 10.1186/s12967-015-0701-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bauca JM, Martinez-Morillo E, Diamandis EP. Peptidomics of urine and other biofluids for cancer diagnostics. Clin Chem. 2014;60(8):1052–1061. doi: 10.1373/clinchem.2013.211714. [DOI] [PubMed] [Google Scholar]
- 59.Pan S, Brentnall TA, Chen R. Proteomics analysis of bodily fluids in pancreatic cancer. Proteomics. 2015;15(15):2705–2715. doi: 10.1002/pmic.201400476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Theodorescu D, Schiffer E, Bauer HW, et al. Discovery and validation of urinary biomarkers for prostate cancer. Proteomics Clin Appl. 2008;2(4):556–570. doi: 10.1002/prca.200780082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H, Aina OH, Lam KS, et al. Identification of a bladder cancer-specific ligand using a combinatorial chemistry approach. Urol Oncol. 2012;30(5):635–645. doi: 10.1016/j.urolonc.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown CE, McCarthy NS, Hughes AD, et al. Urinary proteomic biomarkers to predict cardiovascular events. Proteomics Clin Appl. 2015;9(5-6):610–617. doi: 10.1002/prca.201400195. [DOI] [PubMed] [Google Scholar]
- 63.Kuznetsova T, Mischak H, Mullen W, Staessen JA. Urinary proteome analysis in hypertensive patients with left ventricular diastolic dysfunction. Eur Heart J. 2012;33(18):2342–2350. doi: 10.1093/eurheartj/ehs185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang ZY, Thijs L, Petit T, et al. Urinary Proteome and Systolic Blood Pressure as Predictors of 5-Year Cardiovascular and Cardiac Outcomes in a General Population. Hypertension. 2015;66(1):52–60. doi: 10.1161/HYPERTENSIONAHA.115.05296. [DOI] [PubMed] [Google Scholar]
- 65.Bakun M, Senatorski G, Rubel T, et al. Urine proteomes of healthy aging humans reveal extracellular matrix (ECM) alterations and immune system dysfunction. Age (Dordr) 2014;36(1):299–311. doi: 10.1007/s11357-013-9562-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nkuipou-Kenfack E, Bhat A, Klein J, et al. Identification of ageing-associated naturally occurring peptides in human urine. Oncotarget. 2015;6(33):34106–34117. doi: 10.18632/oncotarget.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7(9):R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonzales PA, Pisitkun T, Hoffert JD, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20(2):363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101(36):13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marimuthu A, O'Meally RN, Chaerkady R, et al. A comprehensive map of the human urinary proteome. J Proteome Res. 2011;10(6):2734–2743. doi: 10.1021/pr2003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Desiere F, Deutsch EW, King NL, et al. The PeptideAtlas project. Nucleic Acids Res. 2006;34(Database issue):D655–658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guerrier L, Thulasiraman V, Castagna A, et al. Reducing protein concentration range of biological samples using solid-phase ligand libraries. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;833(1):33–40. doi: 10.1016/j.jchromb.2005.12.048. [DOI] [PubMed] [Google Scholar]
- 73.Shaheen SP, Levinson SS. Serum free light chain analysis may miss monoclonal light chains that urine immunofixation electrophoreses would detect. Clin Chim Acta. 2009;406(1-2):162–166. doi: 10.1016/j.cca.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 74.Nakayama A, Kubota R, Sakatsume M, et al. Cellulose Acetate Membrane Electrophoresis Based Urinary Proteomics for the Identification of Characteristic Proteins. J Clin Lab Anal. 2015 doi: 10.1002/jcla.21863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petricoin EF, 3rd, Espina V, Araujo RP, et al. Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res. 2007;67(7):3431–3440. doi: 10.1158/0008-5472.CAN-06-1344. [DOI] [PubMed] [Google Scholar]
- 76.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4(7):873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 77.Li QR, Fan KX, Li RX, et al. A comprehensive and non-prefractionation on the protein level approach for the human urinary proteome: touching phosphorylation in urine. Rapid Commun Mass Spectrom. 2010;24(6):823–832. doi: 10.1002/rcm.4441. [DOI] [PubMed] [Google Scholar]
- 78.Fu Q, Grote E, Zhu J, et al. An Empirical Approach to Signature Peptide Choice for Selected Reaction Monitoring: Quantification of Uromodulin in Urine. Clin Chem. 2016;62(1):198–207. doi: 10.1373/clinchem.2015.242495. [DOI] [PubMed] [Google Scholar]
- 79.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Prakash A, Rezai T, Krastins B, et al. Interlaboratory reproducibility of selective reaction monitoring assays using multiple upfront analyte enrichment strategies. J Proteome Res. 2012;11(8):3986–3995. doi: 10.1021/pr300014s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grant GH, Kachmar JF. The proteins of body fluids. In: Tietz NW, editor. Fundamentals of Clinical Chemistry. W.B. Saunders Copmany; Philadelphia: 1976. pp. 298–376. [Google Scholar]
- 82.Gudbjartsson DF, Holm H, Indridason OS, et al. Association of variants at UMOD with chronic kidney disease and kidney stones-role of age and comorbid diseases. PLoS Genet. 2010;6(7):e1001039. doi: 10.1371/journal.pgen.1001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Graham LA, Padmanabhan S, Fraser NJ, et al. Validation of uromodulin as a candidate gene for human essential hypertension. Hypertension. 2014;63(3):551–558. doi: 10.1161/HYPERTENSIONAHA.113.01423. [DOI] [PubMed] [Google Scholar]
- 84.Padmanabhan S, Melander O, Johnson T, et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010;6(10):e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kottgen A, Glazer NL, Dehghan A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009;41(6):712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rampoldi L, Kottgen A, Devuyst O. The effect of common uromodulin variants on urinary protein level and gene transcription. Kidney Int. 2013;84(2):410–411. doi: 10.1038/ki.2013.162. [DOI] [PubMed] [Google Scholar]
- 87.Carty DM, Siwy J, Brennand JE, et al. Urinary proteomics for prediction of preeclampsia. Hypertension. 2011;57(3):561–569. doi: 10.1161/HYPERTENSIONAHA.110.164285. [DOI] [PubMed] [Google Scholar]
- 88.Stalmach A, Johnsson H, McInnes IB, et al. Identification of urinary peptide biomarkers associated with rheumatoid arthritis. PLoS One. 2014;9(8):e104625. doi: 10.1371/journal.pone.0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cantley LG, Colangelo CM, Stone KL, et al. Development of a Targeted Urine Proteome Assay for kidney diseases. Proteomics Clin Appl. 2016;10(1):58–74. doi: 10.1002/prca.201500020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Muntel J, Xuan Y, Berger ST, et al. Advancing Urinary Protein Biomarker Discovery by Data-Independent Acquisition on a Quadrupole-Orbitrap Mass Spectrometer. J Proteome Res. 2015;14(11):4752–4762. doi: 10.1021/acs.jproteome.5b00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bruderer R, Bernhardt OM, Gandhi T, et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol Cell Proteomics. 2015;14(5):1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gillet LC, Navarro P, Tate S, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11(6) doi: 10.1074/mcp.O111.016717. O111 016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Halim A, Nilsson J, Ruetschi U, Hesse C, Larson G. Human urinary glycoproteomics; attachment site specific analysis of N- and O-linked glycosylations by CID and ECD. Mol Cell Proteomics. 2012;11(4) doi: 10.1074/mcp.M111.013649. M111 013649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huet G, Gouyer V, Delacour D, et al. Involvement of glycosylation in the intracellular trafficking of glycoproteins in polarized epithelial cells. Biochimie. 2003;85(3-4):323–330. doi: 10.1016/s0300-9084(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 95.Bhatia PK, Mukhopadhyay A. Protein glycosylation: implications for in vivo functions and therapeutic applications. Adv Biochem Eng Biotechnol. 1999;64:155–201. doi: 10.1007/3-540-49811-7_5. [DOI] [PubMed] [Google Scholar]
- 96.Pardo LA, Stuhmer W. The roles of K(+) channels in cancer. Nat Rev Cancer. 2014;14(1):39–48. doi: 10.1038/nrc3635. [DOI] [PubMed] [Google Scholar]
- 97.Zhou H, Froehlich JW, Briscoe AC, Lee RS. The GlycoFilter: a simple and comprehensive sample preparation platform for proteomics, N-glycomics and glycosylation site assignment. Mol Cell Proteomics. 2013;12(10):2981–2991. doi: 10.1074/mcp.M113.027953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saraswat M, Joenvaara S, Musante L, Peltoniemi H, Holthofer H, Renkonen R. N-linked (N-) glycoproteomics of urinary exosomes. Mol Cell Proteomics. 2015;14(2):263–276. doi: 10.1074/mcp.M114.040345. [Corrected] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fairley JK, Owen JE, Birch DF. Protein composition of urinary casts from healthy subjects and patients with glomerulonephritis. Br Med J (Clin Res Ed) 1983;287(6408):1838–1840. doi: 10.1136/bmj.287.6408.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ammerlaan W, Trezzi JP, Mathay C, Hiller K, Betsou F. Method validation for preparing urine samples for downstream proteomic and metabolomic applications. Biopreserv Biobank. 2014;12(5):351–357. doi: 10.1089/bio.2014.0013. [DOI] [PubMed] [Google Scholar]
- 101.Mischak H, Kolch W, Aivaliotis M, et al. Comprehensive human urine standards for comparability and standardization in clinical proteome analysis. Proteomics Clin Appl. 2010;4(4):464–478. doi: 10.1002/prca.200900189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Filip S, Vougas K, Zoidakis J, et al. Comparison of Depletion Strategies for the Enrichment of Low-Abundance Proteins in Urine. PLoS One. 2015;10(7):e0133773. doi: 10.1371/journal.pone.0133773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yu Y, Pieper R. Urine sample preparation in 96-well filter plates to characterize inflammatory and infectious diseases of the urinary tract. Adv Exp Med Biol. 2015;845:77–87. doi: 10.1007/978-94-017-9523-4_8. [DOI] [PubMed] [Google Scholar]
- 104.Candiano G, Santucci L, Bruschi M, et al. “Cheek-to-cheek” urinary proteome profiling via combinatorial peptide ligand libraries: A novel, unexpected elution system. J Proteomics. 2012;75(3):796–805. doi: 10.1016/j.jprot.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 105.Koves K, Arimura A. Solid-phase adsorption method for removing undesired antibodies from polyclonal antiserum. J Histochem Cytochem. 1989;37(6):903–908. doi: 10.1177/37.6.2723405. [DOI] [PubMed] [Google Scholar]
- 106.Santucci L, Candiano G, Petretto A, et al. From hundreds to thousands: Widening the normal human Urinome. Data Brief. 2014;1:25–28. doi: 10.1016/j.dib.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tamburro D, Fredolini C, Espina V, et al. Multifunctional core-shell nanoparticles: discovery of previously invisible biomarkers. J Am Chem Soc. 2011;133(47):19178–19188. doi: 10.1021/ja207515j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vaught JB, Caboux E, Hainaut P. International efforts to develop biospecimen best practices. Cancer Epidemiol Biomarkers Prev. 2010;19(4):912–915. doi: 10.1158/1055-9965.EPI-10-0058. [DOI] [PubMed] [Google Scholar]
- 109.Castagna A, Channavajjhala SK, Pizzolo F, Olivieri O. Hormone-dependent changes in female urinary proteome. Adv Exp Med Biol. 2015;845:103–120. doi: 10.1007/978-94-017-9523-4_11. [DOI] [PubMed] [Google Scholar]
- 110.Castagna A, Olivieri O, Milli A, et al. Female urinary proteomics: New insight into exogenous and physiological hormone-dependent changes. Proteomics Clin Appl. 2011;5(5-6):343–353. doi: 10.1002/prca.201000105. [DOI] [PubMed] [Google Scholar]
- 111.Froehlich JW, Vaezzadeh AR, Kirchner M, et al. An in-depth comparison of the male pediatric and adult urinary proteomes. Biochim Biophys Acta. 2014;1844(5):1044–1050. doi: 10.1016/j.bbapap.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kohler M, Schanzer W, Thevis M. Effects of exercise on the urinary proteome. Adv Exp Med Biol. 2015;845:121–131. doi: 10.1007/978-94-017-9523-4_12. [DOI] [PubMed] [Google Scholar]
- 113.Li M, Zhao M, Gao Y. Effect of transient blood glucose increases after oral glucose intake on the human urinary proteome. Proteomics Clin Appl. 2015;9(5-6):618–622. doi: 10.1002/prca.201400174. [DOI] [PubMed] [Google Scholar]
- 114.Kentsis A, Monigatti F, Dorff K, Campagne F, Bachur R, Steen H. Urine proteomics for profiling of human disease using high accuracy mass spectrometry. Proteomics Clin Appl. 2009;3(9):1052–1061. doi: 10.1002/prca.200900008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nolen BM, Orlichenko LS, Marrangoni A, et al. An extensive targeted proteomic analysis of disease-related protein biomarkers in urine from healthy donors. PLoS One. 2013;8(5):e63368. doi: 10.1371/journal.pone.0063368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Siwy J, Schanstra JP, Argiles A, et al. Multicentre prospective validation of a urinary peptidome-based classifier for the diagnosis of type 2 diabetic nephropathy. Nephrol Dial Transplant. 2014;29(8):1563–1570. doi: 10.1093/ndt/gfu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oh J, Pyo JH, Jo EH, et al. Establishment of a near-standard two-dimensional human urine proteomic map. Proteomics. 2004;4(11):3485–3497. doi: 10.1002/pmic.200401018. [DOI] [PubMed] [Google Scholar]
- 118.States DJ, Omenn GS, Blackwell TW, et al. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol. 2006;24(3):333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 119.Eisenberg E, Levanon EY. Human housekeeping genes, revisited. Trends Genet. 2013;29(10):569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 120.Hamerlinck FF. Neopterin: a review. Exp Dermatol. 1999;8(3):167–176. doi: 10.1111/j.1600-0625.1999.tb00367.x. [DOI] [PubMed] [Google Scholar]
- 121.Zhang M, Fu G, Lei T. Two urinary peptides associated closely with type 2 diabetes mellitus. PLoS One. 2015;10(4):e0122950. doi: 10.1371/journal.pone.0122950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shlipak MG, Matsushita K, Arnlov J, et al. Cystatin C versus creatinine in determining risk based on kidney function. N Engl J Med. 2013;369(10):932–943. doi: 10.1056/NEJMoa1214234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Candiano G, Musante L, Bruschi M, et al. Repetitive fragmentation products of albumin and alpha1-antitrypsin in glomerular diseases associated with nephrotic syndrome. J Am Soc Nephrol. 2006;17(11):3139–3148. doi: 10.1681/ASN.2006050486. [DOI] [PubMed] [Google Scholar]
- 124.Theodorescu D, Fliser D, Wittke S, et al. Pilot study of capillary electrophoresis coupled to mass spectrometry as a tool to define potential prostate cancer biomarkers in urine. Electrophoresis. 2005;26(14):2797–2808. doi: 10.1002/elps.200400208. [DOI] [PubMed] [Google Scholar]
- 125.Rodrigues KT, Mekahli D, Tavares MF, Van Schepdael A. Development and validation of a CE-MS method for the targeted assessment of amino acids in urine. Electrophoresis. 2016 doi: 10.1002/elps.201500534. [DOI] [PubMed] [Google Scholar]
- 126.Good DM, Zurbig P, Argiles A, et al. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol Cell Proteomics. 2010;9(11):2424–2437. doi: 10.1074/mcp.M110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Boschetti E, Giorgio Righetti P. Hexapeptide combinatorial ligand libraries: the march for the detection of the low-abundance proteome continues. Biotechniques. 2008;44(5):663–665. doi: 10.2144/000112762. [DOI] [PubMed] [Google Scholar]
- 128.Candiano G, Dimuccio V, Bruschi M, et al. Combinatorial peptide ligand libraries for urine proteome analysis: investigation of different elution systems. Electrophoresis. 2009;30(14):2405–2411. doi: 10.1002/elps.200800762. [DOI] [PubMed] [Google Scholar]
- 129.Righetti PG, Candiano G, Citterio A, Boschetti E. Combinatorial peptide ligand libraries as a “Trojan Horse” in deep discovery proteomics. Anal Chem. 2015;87(1):293–305. doi: 10.1021/ac502171b. [DOI] [PubMed] [Google Scholar]
- 130.Righetti PG, Castagna A, Antonioli P, Boschetti E. Prefractionation techniques in proteome analysis: the mining tools of the third millennium. Electrophoresis. 2005;26(2):297–319. doi: 10.1002/elps.200406189. [DOI] [PubMed] [Google Scholar]
- 131.Fredolini C, Meani F, Luchini A, et al. Investigation of the ovarian and prostate cancer peptidome for candidate early detection markers using a novel nanoparticle biomarker capture technology. AAPS J. 2010;12(4):504–518. doi: 10.1208/s12248-010-9211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fredolini C, Tamburro D, Gambara G, et al. Nanoparticle technology: amplifying the effective sensitivity of biomarker detection to create a urine test for hGH. Drug Test Anal. 2009;1(9-10):447–454. doi: 10.1002/dta.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Olivieri O, Castagna A, Guarini P, et al. Urinary prostasin: a candidate marker of epithelial sodium channel activation in humans. Hypertension. 2005;46(4):683–688. doi: 10.1161/01.HYP.0000184108.12155.6b. [DOI] [PubMed] [Google Scholar]
- 134.Guo Z, Zhang Y, Zou L, et al. A Proteomic Analysis of Individual and Gender Variations in Normal Human Urine and Cerebrospinal Fluid Using iTRAQ Quantification. PLoS One. 2015;10(7):e0133270. doi: 10.1371/journal.pone.0133270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Santucci L, Candiano G, Bruschi M, et al. Combinatorial peptide ligand libraries for the analysis of low-expression proteins: Validation for normal urine and definition of a first protein MAP. Proteomics. 2012;12(4-5):509–515. doi: 10.1002/pmic.201100404. [DOI] [PubMed] [Google Scholar]
- 136.Santucci L, Candiano G, Petretto A, et al. Combinatorial ligand libraries as a two-dimensional method for proteome analysis. J Chromatogr A. 2013;1297:106–112. doi: 10.1016/j.chroma.2013.04.065. [DOI] [PubMed] [Google Scholar]
- 137.Fonslow BR, Carvalho PC, Academia K, et al. Improvements in proteomic metrics of low abundance proteins through proteome equalization using ProteoMiner prior to MudPIT. J Proteome Res. 2011;10(8):3690–3700. doi: 10.1021/pr200304u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Meng R, Gormley M, Bhat VB, Rosenberg A, Quong AA. Low abundance protein enrichment for discovery of candidate plasma protein biomarkers for early detection of breast cancer. J Proteomics. 2011;75(2):366–374. doi: 10.1016/j.jprot.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 139.Drabkin DL. The normal pigment of the urine: I. The relationship of urinary pigment output to diet and metabolism. Journal of Biological Chemistry. 1927;75:443–479. [Google Scholar]
- 140.Klouda L. Thermoresponsive hydrogels in biomedical applications: A seven-year update. Eur J Pharm Biopharm. 2015;97(Pt B):338–349. doi: 10.1016/j.ejpb.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 141.Patanarut A, Luchini A, Botterell PJ, et al. Synthesis and characterization of hydrogel particles containing Cibacron Blue F3G-A. Colloids Surf A Physicochem Eng Asp. 2010;362(1-3):8–19. doi: 10.1016/j.colsurfa.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Luchini A, Fredolini C, Espina BH, et al. Nanoparticle technology: addressing the fundamental roadblocks to protein biomarker discovery. Curr Mol Med. 2010;10(2):133–141. doi: 10.2174/156652410790963268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Magni R, Espina BH, Liotta LA, Luchini A, Espina V. Hydrogel nanoparticle harvesting of plasma or urine for detecting low abundance proteins. J Vis Exp. 2014;(90):e51789. doi: 10.3791/51789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fredolini C, Meani F, Reeder KA, et al. Concentration and Preservation of Very Low Abundance Biomarkers in Urine, such as Human Growth Hormone (hGH), by Cibacron Blue F3G-A Loaded Hydrogel Particles. Nano Res. 2008;1(6):502–518. doi: 10.1007/s12274-008-8054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Luchini A, Tamburro D, Magni R, et al. Application of Analyte Harvesting Nanoparticle Technology to the Measurement of Urinary HGH in Healthy Individuals. J Sports Med Doping Stud. 2012;2(6) doi: 10.4172/2161-0673.1000e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pleitez M, von Lilienfeld-Toal H, Mantele W. Infrared spectroscopic analysis of human interstitial fluid in vitro and in vivo using FT-IR spectroscopy and pulsed quantum cascade lasers (QCL): Establishing a new approach to non invasive glucose measurement. Spectrochim Acta A Mol Biomol Spectrosc. 2012;85(1):61–65. doi: 10.1016/j.saa.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 147.Nathan DM, Singer DE, Hurxthal K, Goodson JD. The clinical information value of the glycosylated hemoglobin assay. N Engl J Med. 1984;310(6):341–346. doi: 10.1056/NEJM198402093100602. [DOI] [PubMed] [Google Scholar]
- 148.Group F-NBW. BEST (Biomarkers, EndpointS, and other Tools) 2016 [Google Scholar]
- 149.Lowenthal MS, Mehta AI, Frogale K, et al. Analysis of albumin-associated peptides and proteins from ovarian cancer patients. Clin Chem. 2005;51(10):1933–1945. doi: 10.1373/clinchem.2005.052944. [DOI] [PubMed] [Google Scholar]
- 150.Wronska DB, Krajewska M, Lygina N, et al. Peptide-conjugated glass slides for selective capture and purification of diagnostic cells: Applications in urine cytology. Biotechniques. 2014;57(2):63–71. doi: 10.2144/000114195. [DOI] [PubMed] [Google Scholar]
- 151.Institute CLS. GP16-A3 Urinalysis; Approvide Guideline-Third Edition. 2009;56 [Google Scholar]
- 152.Institute CLS. C62-A Liquid chromatography-Mass spectrometry methods, Approved guidelines. 2014;88 [Google Scholar]
- 153.Coskun AF, Nagi R, Sadeghi K, Phillips S, Ozcan A. Albumin testing in urine using a smart-phone. Lab Chip. 2013;13(21):4231–4238. doi: 10.1039/c3lc50785h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang S, Zhao X, Khimji I, et al. Integration of cell phone imaging with microchip ELISA to detect ovarian cancer HE4 biomarker in urine at the point-of-care. Lab Chip. 2011;11(20):3411–3418. doi: 10.1039/c1lc20479c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bermes EW, Erviti V, Forman DT. Sttistics, normal values, and quality control. In: Tietz NW, editor. Fundamentals of Clinical Chemistry. W.B. Saunders Co; Philadelphia: 1976. pp. 60–102. [Google Scholar]
- 156.Berglund L, Bjorling E, Oksvold P, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008;7(10):2019–2027. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- 157.Jupp S, Klein J, Schanstra J, Stevens R. Developing a kidney and urinary pathway knowledge base. J Biomed Semantics. 2011;2(Suppl 2):S7. doi: 10.1186/2041-1480-2-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li SJ, Peng M, Li H, et al. Sys-BodyFluid: a systematical database for human body fluid proteome research. Nucleic Acids Res. 2009;37(Database issue):D907–912. doi: 10.1093/nar/gkn849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Omenn GS, States DJ, Adamski M, et al. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5(13):3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 160.Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas--a tool for pathology. J Pathol. 2008;216(4):387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 161.Shifman MA, Li Y, Colangelo CM, et al. YPED: a web-accessible database system for protein expression analysis. J Proteome Res. 2007;6(10):4019–4024. doi: 10.1021/pr070325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Siwy J, Mullen W, Golovko I, Franke J, Zurbig P. Human urinary peptide database for multiple disease biomarker discovery. Proteomics Clin Appl. 2011;5(5-6):367–374. doi: 10.1002/prca.201000155. [DOI] [PubMed] [Google Scholar]
- 163.Ternent T, Csordas A, Qi D, et al. How to submit MS proteomics data to ProteomeXchange via the PRIDE database. Proteomics. 2014;14(20):2233–2241. doi: 10.1002/pmic.201400120. [DOI] [PubMed] [Google Scholar]
- 164.Uhlen M, Bjorling E, Agaton C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4(12):1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 165.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(Database issue):D204–212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Righetti PG, Fasoli E, Boschetti E. Combinatorial peptide ligand libraries: the conquest of the ‘hidden proteome’ advances at great strides. Electrophoresis. 2011;32(9):960–966. doi: 10.1002/elps.201000589. [DOI] [PubMed] [Google Scholar]