Abstract
Genetic and immunological analysis of host-pathogen interactions can reveal fundamental mechanisms of susceptibility and resistance to infection. Modeling human infectious diseases among inbred mouse strains is a proven approach but is limited by naturally occurring genetic diversity. Using ENU mutagenesis, we created a recessive loss-of-function point mutation in Unc93b1 (unc-93 homolog B1 (C. elegans)), a chaperone for endosomal TLR3, TLR7, and TLR9, that we termed Letr for ‘loss of endosomal TLR response’. We used Unc93b1Letr/Letr mice to study the role of Unc93b1 in the immune response to influenza A/PR/8/34 (H1N1), an important global respiratory pathogen. During the early phase of infection, Unc93b1Letr/Letr mice had fewer activated exudate macrophages and decreased expression of CXCL10, IFN-γ, and type I IFN. Mutation of Unc93b1 also led to reduced expression of the CD69 activation marker and a concomitant increase in the CD62L naïve marker on CD4+ and CD8+ T cells in infected lungs. Finally, loss of endosomal TLR signaling resulted in delayed viral clearance that coincided with increased tissue pathology during infection. Taken together, these findings establish a role for Unc93b1 and endosomal TLRs in the activation of both myeloid and lymphoid cells during the innate immune response to influenza.
Keywords: Unc93b1, ENU mutagenesis, influenza H1N1, exudate macrophages, CD4+ and CD8+ T cells
INTRODUCTION
Influenza virus infection causes an acute respiratory tract disease with a global annual mortality of 250 000 – 500 000.1, 2 In addition to seasonal strains, humans face the persistent threat of novel strains that can emerge and spread quickly in a naïve population, potentially resulting in a pandemic.3, 4 Seasonal influenza often affects individuals with weakened immune systems including very old, very young, and immunocompromised populations.5–7 Conversely, certain highly pathogenic influenza viruses such as the 1918 H1N1 strain confer greater mortality in otherwise healthy individuals through a combination of enhanced inflammation leading to excessive tissue damage and secondary bacterial infection.8, 9 To develop effective management strategies to combat both seasonal and pandemic influenza infection, a detailed understanding of the cellular and molecular mechanisms that regulate the quality and magnitude of the host immune response is essential.
Following influenza infection, a rapid yet nonspecific innate immune response is activated in an attempt to control initial viral replication and is followed by the mobilization of professional antigen-presenting cells (APCs) including dendritic cells (DCs) and macrophages. Activated APCs express class I or class II major-histocompatibility complex (MHC) molecules as well as co-stimulatory markers such as the cluster of differentiation (CD) molecules CD40, CD80, and CD86.10 Expression of these markers, particularly by DCs, is required for the priming and activation of naïve T and B cells in the lung-associated lymph nodes (LALNs), leading to the formation of an adaptive immune response.11–15 Activated T and B cells are recruited to the site of primary infection to mediate antigen-specific immunity through cell-mediated control of viral replication and the production of specific antibodies that provide lasting protection against the infecting strain, respectively.10
Initial recognition of the influenza virus and subsequent activation of innate immunity occurs through host pattern recognition receptors (PRRs). Two of the best-studied PRRs for influenza recognition are the endosomal toll-like receptors (TLR)3 and TLR7.16–20 To initiate downstream signaling, TLR3 and TLR7 must be trafficked from the endoplasmic reticulum (ER) to the endosome by UNC93B1, an ER-resident transmembrane chaperone protein that also transports TLR9 following activation.21–23 At the endosome, TLR7 recognizes viral single-stranded RNA (ssRNA) and recruits the myeloid differentiation primary response 88 (MyD88) adaptor protein16, 20 while TLR3 recognizes double-stranded RNA (dsRNA) and recruits the toll interleukin-1 receptor-domain-containing adapter inducing interferon-β (TRIF) adaptor protein.24 One group has shown that MyD88-deficient mice are susceptible to influenza infection and have diminished recruitment of CD11b+ neutrophils and monocytes as well as reduced expression of the proinflammatory mediators IL-6, TNF-α, and interferon (IFN)-γ in the airways.25 A role for MyD88 in B cell activation and subsequent production of virus-specific IgG2a/c antibodies has also been demonstrated.18, 26 Conversely, several other investigations have demonstrated that signaling via MyD88 is not essential for survival following influenza challenge.27, 28
Despite conflicting reports as to whether a true dsRNA replication intermediate forms during influenza virus replication, TLR3 expression is upregulated in human alveolar and bronchial epithelial cells as well as mouse lungs following influenza infection, suggesting that it contributes to the host immune response.17, 19, 28, 29 TLR3 has been specifically linked to the early production of proinflammatory mediators such as IL-6, IL-8, and CCL5 in human bronchial cells during influenza infection.17, 19 Compared to wild type mice, TLR3-deficient mice have diminished production of IL-6 and IL-12 p40/p70, elevated expression of IL-9, IL-10, and IFN-γ, and fewer macrophages and CD8+ T cells in the airways.28 Somewhat unexpectedly, these mice had an improved rate of survival despite a higher lung viral load, suggesting that the TLR3-mediated host response during influenza infection is deleterious.28 On the other hand, subsequent studies of TRIF-deficient mice that also lack TLR3 signaling did not reveal a significant role for this adaptor molecule following influenza infection.25, 26 Finally, analysis of a single patient with influenza-associated encephalopathy revealed a loss-of-function missense mutation in TLR3, suggesting that this PRR may play a crucial role in protection rather than disease pathogenesis.30 While the discordant experimental findings on the role of these TLRs and adaptor molecules in influenza infection may be attributable to differences in the strain, dose, and volume of virus administered to mice with distinct genetic backgrounds, complementary human studies have also failed to clearly identify the genes that are required for protection of the host.31 Thus, on the basis of current knowledge, it is not possible to draw firm conclusions about the contributions of the TLR7-MyD88 and TLR3-TRIF signaling pathways to innate immunity against influenza infection.
Studies of inbred mouse strains as well as gene-deficient mice have been essential for elucidating key features of the host response to viral infection that are relevant to human immunity.32, 33 Owing to the limited number of available inbred and knockout strains, alternative approaches, such as the creation of random heritable mutations using the chemical N-ethyl-N-nitrosourea (ENU), have evolved in recent years.32 Rather than abrogating gene expression, ENU mutagenesis can create unique point mutations that alter protein function.34 Such mutations more closely mirror human genetic variation and can reveal specific roles for individual protein domains.34 This forward genetic approach has been successfully used to study molecules and pathways that are crucial to host defense against various pathogens.35 Here we report the creation of an ENU-induced point mutation in the mouse Unc93b1 gene that results in the complete deletion of exon 4, which encodes the 3rd and 4th transmembrane domains of the full-length protein. We have named this mutation Letr for ‘loss of endosomal TLR response’ as it confers a lack of responsiveness to stimulation with nucleic acid structures that represent pathogen-associated molecular patterns (PAMPs) known to activate the TLR3, TLR7, and TLR9 signaling pathways.
Based on the previously demonstrated function of UNC93B1 as a chaperone for endosomal TLRs21–23 as well as its contribution to the immune response against various parasites (Toxoplasma gondii, Trypanosoma cruzi, Leishmania major),36–40 DNA viruses (Mouse cytomegalovirus (MCMV), Herpes simplex virus-1 (HSV-1))41–43 and neuroadapted Sindbis virus (NSV, a positive-sense ssRNA virus),44 we hypothesized that defective UNC93B1 function would impair the innate immune response to influenza A/PR/8/34 (H1N1), a negative-sense ssRNA virus that targets the respiratory mucosa. Using our ENU-induced mutant mice that lack endosomal TLR signaling, we found that Unc93b1 contributes to the expression of type I and type II IFN (IFN-γ), recruitment of activated exudate macrophages (ExMs), and activation of T cells in the lung during the early phase of influenza infection. Importantly, despite a significant delay in viral clearance that coincided with increased tissue inflammation and epithelial reaction, the failure of Unc93b1-mediated innate immune activation did not significantly alter mortality, suggesting that alternative pathways can compensate for the lack of endosomal TLR function in this model. Collectively, these results establish a distinct role for Unc93b1 in the early activation of the innate immune response to influenza and demonstrate the importance of redundant mechanisms that protect the host against lethal infection.
RESULTS
ENU mutagenesis induced a mutation that confers defective in vitro and in vivo responses to TLR3, TLR7, and TLR9 ligands
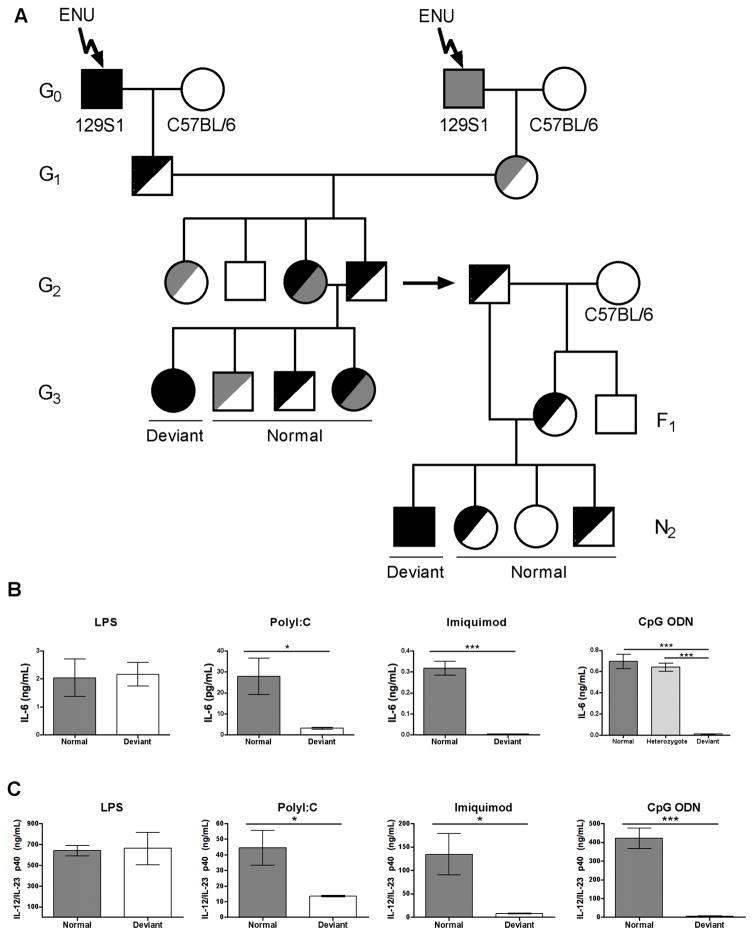
To identify novel genes or novel roles for known genes in TLR-specific immune signaling pathways, we injected male 129S1 G0 mice with ENU and screened cells from G3 mutagenized mice for defective responses to polyinosinic:polycytidylic acid (polyI:C), lipopolysaccharide (LPS), Imiquimod, and unmethylated CpG dinucleotides (CpG DNA). These structures were chosen as they are known to activate TLR3, TLR4, TLR7, and TLR9 signaling pathways respectively.45–48 The cytokines, cell types, and time points selected for analysis following in vitro and in vivo PAMP stimulation were based on data from the initial publications that identified these TLR ligands.45–48 Mutagenized G0 mice were bred to C57BL/6 female mice and the G1 progeny from independent G0 crosses were intercrossed to increase the number of recessive mutations available for screening at the G3 generation (Figure 1A). Thioglycollate-elicited peritoneal macrophages or splenocytes from G3 progeny of G2 brother-sister matings were tested for in vitro expression of IL-6 in response to PAMP stimulation. One pedigree (B28W6) showed a deviant phenotype in response to stimulation with polyI:C, Imiquimod, and CpG DNA but had normal expression following LPS stimulation (Figure 1B). Heterozygous G2 fathers from this pedigree were outcrossed to C57BL/6 female mice and the resulting F1 female progeny were backcrossed to create N2 progeny (Figure 1A). N2 mice were intercrossed and deviant lines carrying a homozygous mutation were identified by survival splenectomy and repeat in vitro phenotyping. Cells from heterozygous mice responded normally to CpG stimulation, confirming that the mutation has a recessive mode of inheritance (Figure 1B). To analyze the phenotype in vivo, progeny of deviant and normal N2 mice were injected intraperitoneally with LPS, polyI:C, Imiquimod, or CpG DNA and serum was collected three hours post-injection. Compared to the progeny of normal N2 mice, those from deviant N2 mice had significantly decreased expression of serum IL-12/IL-23 p40 in response to stimulation with polyI:C, Imiquimod, or CpG DNA but retained similar responses to LPS (Figure 1C).
FIGURE 1. ENU mutagenesis establishes a mouse model with defective endosomal TLR signaling.
(A) Strategy used for the creation and characterization of mice with a recessive germline mutation caused by ENU. A detailed description of the breeding scheme can be found in the materials and methods section of this manuscript. Mice were screened at the G3 generation while N2 mice and their progeny were used for sequencing, phenotype confirmation, and creation of the F2 line for subsequent experiments. Black and grey shading denotes one (half-shaded) or two (fully-shaded) copies of an ENU mutation. (B) IL-6 expression in cell culture supernatants from normal (dark grey bars), heterozygous (light grey bars) and deviant (white bars) mice. Thioglycollate-elicited peritoneal macrophages were stimulated with LPS and splenocytes were stimulated with polyI:C, Imiquimod, or CpG ODN for 24 (LPS, Imiquimod, CpG ODN) or 48 (polyI:C) hours. (C) Serum IL-12/IL-23 p40 expression three hours post-intraperitoneal injection of LPS, polyI:C, Imiquimod, or CpG ODN. Data is representative of two independent experiments (n≥3/group or n≥4 technical replicates/group). *p<0.05; ***p<0.001 determined by two-tailed unpaired t-test or one-way ANOVA with Tukey’s post-test. Main p value determined by one-way ANOVA for in vitro CpG stimulation: ***p<0.0001.
An ENU-induced single nucleotide transversion in Unc93b1 results in the loss of exon 4 due to alternative splicing
To map the gene responsible for the Letr phenotype, we performed a low-density genome scan using 375 single nucleotide polymorphisms (SNPs) among 54 mice derived from a (Letr/Letr x C3H/HeN) F1 x Letr/Letr backcross. The Letr mutation was localized to a 5.4 Mb region on chromosome 19 between markers rs31277487 and rs30935927. To test the heritability of the Letr mutation, an additional 281 mice from the same backcross were screened for IL-6 expression following in vitro stimulation of splenocytes with CpG DNA. Of these mice, 139 (49.5%) showed a normal IL-6 response while the other 142 (50.5%) animals lacked IL-6 expression with perfect concordance between the mutant phenotype and genotype for a recessive mutation.
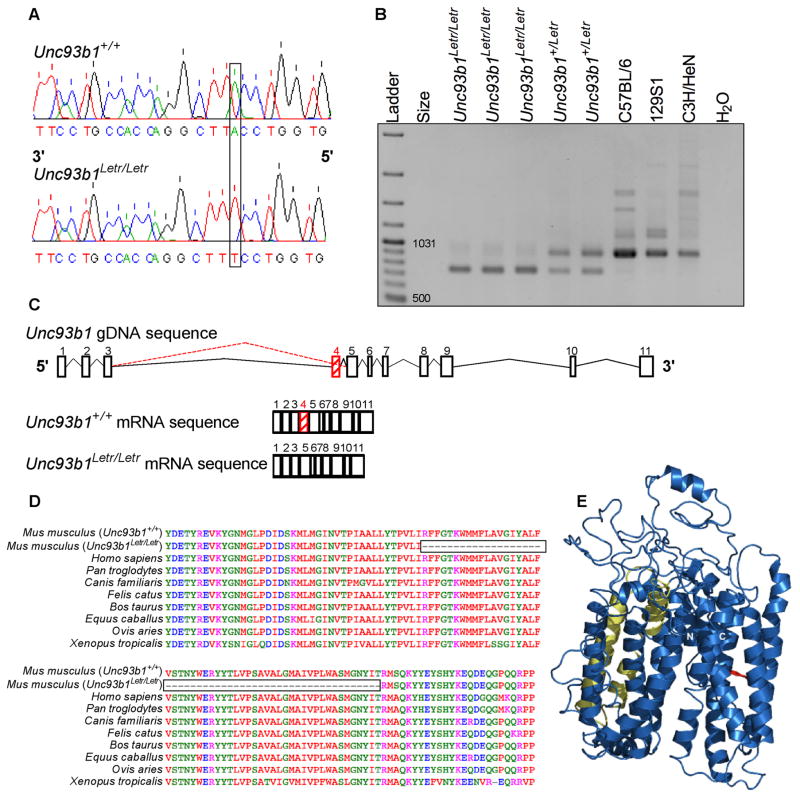
Based on a previous ENU mutagenesis study that created a mouse, termed 3d, with a similar hyporesponsive phenotype caused by a point mutation in Unc93b1 on chromosome 19,23 we took a targeted approach to identify the Letr mutation. Nucleotide sequencing of Unc93b1 genomic DNA (gDNA) from normal and deviant (Letr) mice identified a single T to A transversion at the 5′ splice donor site in the intron between exon 4 and 5 of Unc93b1 that rendered the site ineffective (Figure 2A, non-coding strand shown). Sequencing of cDNA from both Unc93b1+/+ and Unc93b1Letr/Letr mice determined that the mutation resulted in the loss of exon 4 in the 11-exon mRNA transcript (Figure 2C).
FIGURE 2. The Letr allele causes a single nucleotide transversion and alternative splicing of Unc93b1.
(A) Chromatogram of the Unc93b1+/+ (top) and Unc93b1Letr/Letr (bottom) gDNA sequence with the single T to A nucleotide change caused by the Letr mutation that is identified on the non-coding strand as A to T. (B) Gel electrophoresis of cDNA from Unc93b1Letr/Letr, Unc93b1+/Letr, and wild type inbred strains (C57BL/6, 129S1, C3H/HeN). The wild type transcript size is 918bp while the mutant transcript size is 756bp. (C) Schematic of Unc93b1 introns (lines) and exons (blocks), including the mutant (dashed red line) splicing pattern and resulting mRNA transcripts for Unc93b1+/+ and Unc93b1Letr/Letr mice. (D) Multiple species alignment of the UNC93B1 amino acid sequence surrounding and including exon 4. The boxed area delineates the missing sequence in Unc93b1Letr/Letr mice. (E) Predicted three-dimensional structure of wild type UNC93B1 with the 3d (red) and Letr (yellow) mutations mapped to their respective protein domains. The amino- and carboxy-termini are indicated as N and C, respectively.
To confirm that the ENU-induced Letr mutation caused alternative splicing, spleen RNA from Unc93b1Letr/Letr and Unc93b1+/Letr mice as well as C57BL/6, C3H/HeN, and 129S1 inbred strains was reverse transcribed and a 918 base pair (bp) region of Unc93b1 cDNA that included exon 4 was amplified and visualized by gel electrophoresis. All three inbred strains presented a single band at the predicted size while Unc93b1Letr/Letr cDNA presented a smaller band that is compatible with the 162bp deletion of exon 4 (Figure 2B). Analysis of heterozygous Unc93b1+/Letr cDNA showed both wild type and mutant Unc93b1 transcripts.
To evaluate the relative importance of exon 4 to the integrity of the UNC93B1 protein, the degree of amino acid sequence conservation was compared among diverse species. Alignment of UNC93B1 amino acid sequences encompassing exon 4 demonstrated a 54 amino acid deletion in Unc93b1Letr/Letr mice (Figure 2D). These missing amino acids were identical among all species examined, with the exception of Xenopus tropicalis that was distinguished by the substitution of nine amino acid residues. Protein modeling software was used to confirm that the wild type UNC93B1 amino acid sequence forms a 12-helix transport protein (Figure 2E).23, 49 By mapping the amino acids encoded by exon 4 to the wild type model, the splicing defect caused by the Letr mutation is predicted to eliminate the 3rd and 4th transmembrane domains of the functional UNC93B1 protein and is distinct from the 3d mutation which causes a single amino acid missense mutation (H412R) in the 9th transmembrane domain (Figure 2E).23
Allelic complementation does not rescue the hyporesponsive phenotype in Unc93b1Letr/Letr mice
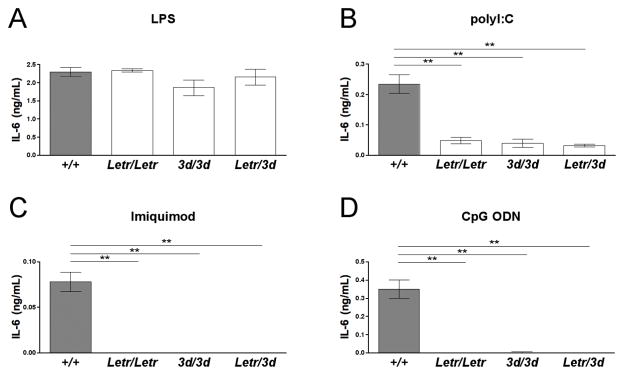
To confirm that the Letr mutation was in the Unc93b1 gene, we performed an allelic complementation study by generating Unc93b1Letr/3d F1 hybrids from Unc93b1Letr/Letr and Unc93b13d/3d mice. If the Letr mutation were elsewhere in the genome, the lack of response to PAMP stimulation would be rescued in the compound heterozygotes. All genotypes were tested for IL-6 expression following in vitro stimulation with PAMPs, as done during the initial screen. Both thioglycollate-elicited peritoneal macrophages and splenocytes stimulated with media alone produced undetectable levels of IL-6 (data not shown). Thioglycollate-elicited peritoneal macrophages from Unc93b1Letr/Letr, Unc93b13d/3d and Unc93b1Letr/3d strains had a comparable IL-6 response to Unc93b1+/+ mice following LPS stimulation (Figure 3A). Conversely, the Unc93b1Letr/Letr, Unc93b13d/3d and Unc93b1Letr/3d strains had significantly decreased or undetectable IL-6 expression compared to Unc93b1+/+ mice following stimulation with polyI:C, Imiquimod, or CpG DNA (Figure 3B–D).
FIGURE 3. Lack of complementation confirms that the Letr mutation is in Unc93b1.
IL-6 expression of (A) thioglycollate-elicited peritoneal macrophages or (B-D) splenocytes from Unc93b1+/+, Unc93b1Letr/Letr, Unc93b13d/3d, and Unc93b1Letr/3d F1 mice following in vitro stimulation with (A) LPS, (B) polyI:C, (C) Imiquimod, or (D) CpG ODN for 24 (LPS, Imiquimod, CpG ODN) or 48 (polyI:C) hours. Data is representative of duplicate stimulations (n≥3/group). **p<0.01 determined by one-way ANOVA with Dunnett’s post-test comparing all mutant strains to the Unc93b1+/+ strain. Main p values determined by one-way ANOVA: (A) ns (B–D) ***p<0.001.
Unc93b1Letr/Letr mice have fewer activated ExMs in the lungs following influenza infection
UNC93B1 functions as a chaperone for TLR3, TLR7, and TLR9 and plays a role in host defense against various pathogens including the DNA viruses MCMV and HSV-1 and the RNA virus NSV.21–23, 41–44 To determine if Unc93b1 also mediates innate immunity against RNA viral infection of the respiratory mucosa, we characterized the response of Unc93b1Letr/Letr mice to experimental challenge with influenza A/PR/8/34 (H1N1). TLR3, TLR7, and the MyD88 adaptor protein, have been implicated in the immune response to influenza infection, albeit with variable outcomes.16–18, 20, 25, 26, 28 Unc93b1Letr/Letr mice provide a unique opportunity to determine the combined consequence of defective endosomal TLR3 and TLR7 activation in the presence of a functional MyD88 molecule that also participates in IL-1R- and IL-18R-mediated signaling.50
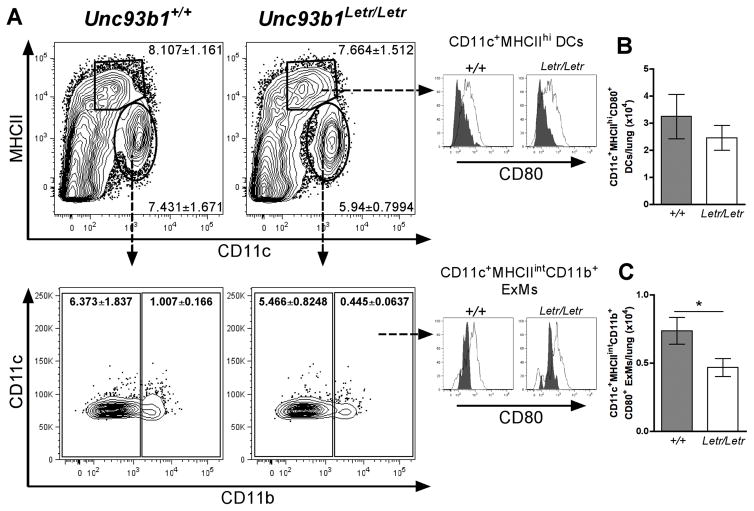
As a role for Unc93b1 has been linked to antigen presentation during T. gondii infection,38 we sought to determine whether the loss of Unc93b1 function would affect the activation of lung macrophages and DCs during influenza infection.51 Based on the literature, day 3 was selected as a representative time point for characterization of early immune differences following experimental influenza challenge.27, 28, 52 Initially, CD11c and MHCII were used to distinguish macrophages (CD11c+MHCIIint) from DCs (CD11c+MHCIIhi) and CD11b expression was then used to separate the former population into resident alveolar macrophages (AMs) (CD11b−) and monocyte-derived ExMs (CD11b+) (Figure 4A). CD80 expression was used as a marker of cell activation (Figure 4A). Following influenza infection, the absolute number of DCs, macrophages, and ExMs was not significantly different between Unc93b1+/+ and Unc93b1Letr/Letr mice in either the LALNs or lungs (Supplementary Figure 1A–C). Subsequent analysis of cellular activation status revealed that the loss of Unc93b1 did not affect the number of CD80+ DCs at day 3 post-infection (Figure 4B); however, there were significantly fewer CD80+ ExMs in the lungs of Unc93b1Letr/Letr compared to Unc93b1+/+ mice at this time point (Figure 4C). Simultaneous analysis of the LALNs showed that there was no significant difference in the number of CD80+ DCs while the number of CD80+ ExMs was too low for reliable detection (Supplementary Figure 1D), suggesting that the differential activation seen in the Unc93b1Letr/Letr mice was restricted to the site of infection.
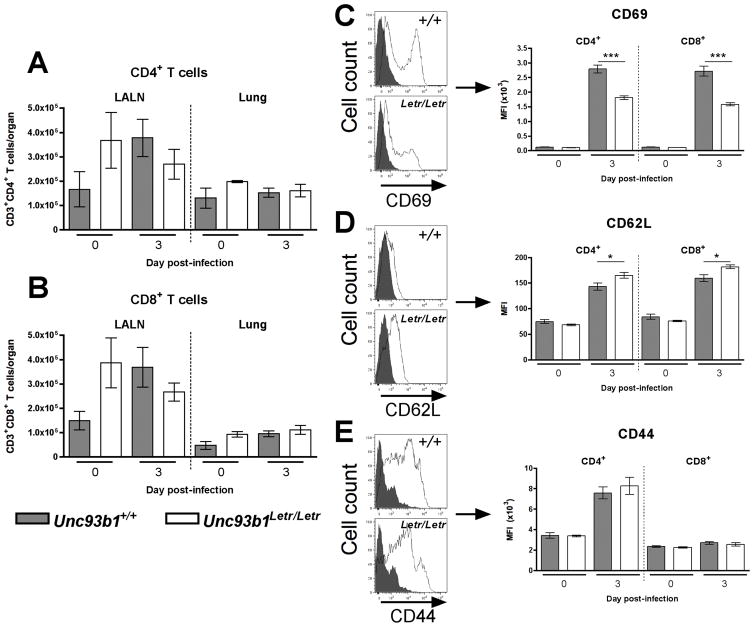
FIGURE 4. Unc93b1Letr/Letr lungs have fewer activated ExMs but not DCs following infection.
(A) Representative dot plots of CD11c+MHCIIhi DCs, CD11c+MHCIIint macrophages, CD11b− AMs, and CD11b+ ExMs. For precise analysis, all samples were gated on live, CD45+, B220−, Ly6G− cells to eliminate potentially confounding B cell and neutrophil populations prior to identification of specific myeloid cell subsets. Numbers on each dot plot represent the percentage (mean ± SEM) of total live, CD45+ cells in each subset at day 3 post-infection. Histograms represent CD80 expression on DCs (top) and ExMs (bottom) in Unc93b1+/+ and Unc93b1Letr/Letr lungs at day 0 (grey peaks) and day 3 (white peaks). Absolute number of (B) CD80+ DCs and (C) CD80+ ExMs in the lungs of Unc93b1+/+ (grey bars) and Unc93b1Letr/Letr (white bars) mice at day 3 post-infection. Data is representative of two independent experiments (n=6/group). *p<0.05 determined by two-tailed unpaired t-test.
Loss of Unc93b1 function reduces CD4+ and CD8+ T cell activation in the lungs following influenza infection
As Unc93b1 also has a role in the activation of CD4+ and CD8+ T cells during T. gondii infection,38 we analyzed the number and activation status of these lymphoid cell subsets in our model. Activated CD4+ and CD8+ T cells produce an antigen-specific response that contributes to control of lung viral load. Specifically, CD8+ T cells function primarily through direct lysis of virally infected cells, while CD4+ T cells are classically known as helper cells that enhance CD8+ T cell and B cell activation and, more recently, have been shown to have a direct cytolytic function as well.53–55
Flow cytometry analysis at day 0, 3 and 7 post-infection did not reveal a difference in the total number of CD4+ or CD8+ T cells observed in the LALNs or lungs between Unc93b1+/+ and Unc93b1Letr/Letr mice (Figure 5A, B and Supplementary Figure 2G). To determine whether the loss of Unc93b1 altered T cell activation during influenza infection, the surface expression of an early activation marker (CD69) as well as markers of a naïve (CD62L) and effector (CD44) T cell phenotype were evaluated at day 3 and 7 post-infection. Representative expression plots for T cell activation markers are displayed as histograms (Figure 5C–E, left panel). The basal level of all CD4+ and CD8+ T cell activation markers examined was similar between Unc93b1+/+ and Unc93b1Letr/Letr mice (Figure 5C–E, right panel). At day 3 post-influenza challenge both CD4+ and CD8+ T cells in the lungs of Unc93b1Letr/Letr mice had significantly less expression of CD69 and significantly increased expression of CD62L compared to Unc93b1+/+ mice (Figure 5C, D). No significant differences in the expression of CD44 on CD4+ and CD8+ T cells derived from Unc93b1+/+ or Unc93b1Letr/Letr mice were observed at day 3 post-infection (Figure 5E). Finally, the difference in lung CD4+ and CD8+ T cell activation between Unc93b1+/+ and Unc93b1Letr/Letr strains was not observed at day 7 in the lungs (Supplementary Figure 2A–C) nor was there a difference in the activation of CD4+ and CD8+ T cells in the LALNs at day 3 or 7 post-infection (Supplementary Figure 2D–F).
FIGURE 5. Early activation of CD4+ and CD8+ T cells is diminished in Unc93b1Letr/Letr lungs following infection.
Absolute number of live, CD45+, CD3+, CD49b− (A) CD4+ and (B) CD8+ T cells in the LALNs and lungs of Unc93b1+/+ (grey bars) and Unc93b1Letr/Letr (white bars) mice. Mean fluorescence intensity (MFI) of (C) CD69, (D) CD62L, and (E) CD44 on Unc93b1+/+ and Unc93b1Letr/Letr lung CD4+ and CD8+ T cells. Representative histograms for each activation marker show expression at day 0 (grey peak) and day 3 (white peak). Data is representative of two independent experiments for infected mice and one experiment for uninfected mice (n=4–6/group). *p<0.05; ***p<0.001 determined by two-tailed unpaired t-test.
Unc93b1Letr/Letr mice have a selective decrease in type I/II IFN and CXCL10 expression during influenza infection
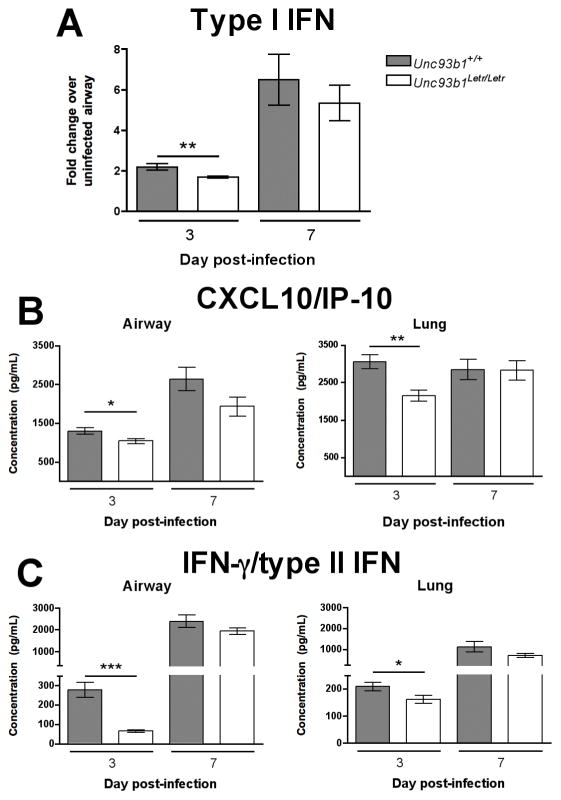
Following influenza infection, the lungs produce a variety of soluble mediators that are involved in the recruitment and activation of innate and adaptive immune cells as well as the destruction of virally infected host cells.10 To determine whether the diminished cellular activation in Unc93b1Letr/Letr mice was associated with an altered pattern of inflammatory mediator secretion in the airway or lungs, the expression of representative cytokines and chemokines that have been implicated in the host response to influenza was determined at serial time points. At day 3 post-infection the expression of type I IFN was significantly lower in the airways of Unc93b1Letr/Letr mice compared to the Unc93b1+/+ strain; however, at day 7 post-infection a comparable increase was observed in both strains (Figure 6A). Significantly lower expression of CXCL10 (Figure 6B) and IFN-γ (Figure 6C) was also observed at day 3 post-infection in the airway and lungs of Unc93b1Letr/Letr mutants compared to the Unc93b1+/+ strain. The expression of other proinflammatory mediators in the airway, including the cytokines IL-6 and TNF-α, and the chemokines CCL2, CCL3, CXCL1, and CXCL2 was not significantly different at day 3 or day 7 post-infection (Supplementary Figure 3). These findings demonstrate that soluble inflammatory mediator production at the site of infection was selectively altered by the loss of Unc93b1 function.
FIGURE 6. Defective Unc93b1-dependent signaling leads to reduced expression of type I/II IFN and CXCL10.
(A) Relative type I IFN expression in the airways of Unc93b1+/+ (grey bars) and Unc93b1Letr/Letr (white bars) mice at day 3 and 7 following influenza infection. Data is expressed as fold change compared to uninfected samples. Airway and lung expression of (B) CXCL10 and (C) IFN-γ from Unc93b1+/+ and Unc93b1Letr/Letr mice at day 3 and 7 post-infection. Data is pooled from two independent experiments (n≥9/group). *p<0.05; **p<0.01; ***p<0.001 determined by two-tailed unpaired t-test.
Unc93b1Letr/Letr mice have delayed viral clearance and increased tissue inflammation following influenza infection
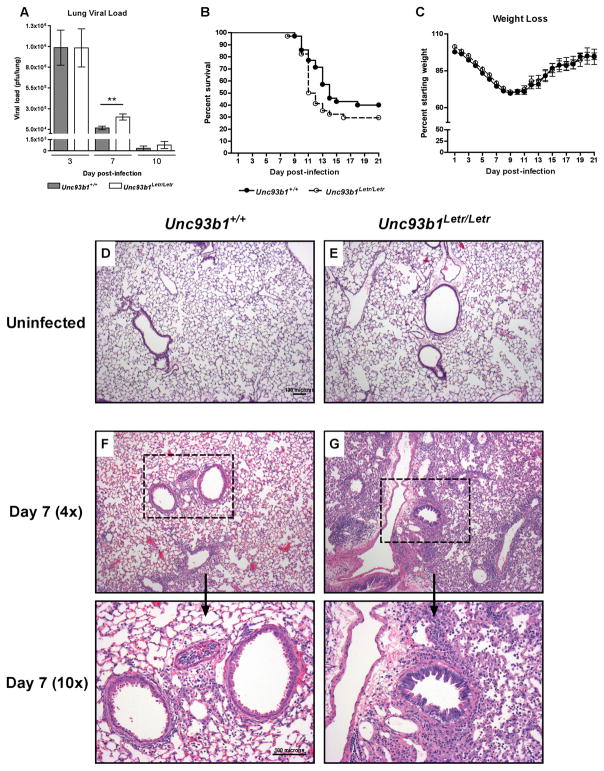
To determine if the significant differences in immune cell activation and inflammatory mediator expression caused by the loss of Unc93b1 function had an impact on host outcome following influenza infection, a comparative analysis of lung viral load, inflammation, and mortality was conducted between Unc93b1+/+ and Unc93b1Letr/Letr strains after intranasal challenge with 400 pfu of the virulent influenza A/PR/8/34 (H1N1) strain. In both genotypes, the peak lung viral load was observed at day 3 post-infection and decreased thereafter (Figure 7A).28, 56 Notably, at day 7 post-infection, the lung viral load in Unc93b1Letr/Letr mice was significantly higher compared to Unc93b1+/+ mice (Figure 7A).
FIGURE 7. Unc93b1 plays a role in mediating viral clearance and tissue inflammation but does not influence survival following influenza A/PR/8/34 (H1N1) infection.
(A) Lung viral load in Unc93b1+/+ (grey bars) and Unc93b1Letr/Letr (white bars) mice at day 3, 7, and 10 following influenza infection. (B) Survival and (C) weight loss of Unc93b1+/+ (solid line) and Unc93b1Letr/Letr (dashed line) mice over a 21-day period following influenza infection. Representative lung sections from (D, E) uninfected or (F, G) infected lungs at day 7 post-infection of (D, F) Unc93b1+/+ and (E, G) Unc93b1Letr/Letr mice. Day 7 post-infection is visualized at 4x and 10x magnification. Data is pooled from independent experiments. (A) n≥10/group, (B) n≥34/group, (C) n≥27/group. **p<0.01 determined by (A) two-tailed unpaired t-test or (B) Logrank test.
In response to influenza challenge, both mouse strains showed a similar pattern of weight loss with a nadir at day 9 followed by recovery of initial weight in surviving animals by day 21 post-infection (Figure 7C).52 Under these experimental conditions, Unc93b1+/+ mice had a median survival time of 14 days and an overall mortality of 60% while Unc93b1Letr/Letr mice had a median survival time of 11.5 days and an overall mortality of 70% (Figure 7B). These data suggest that the loss of Unc93b1 may be associated with an earlier onset of death but does not significantly affect the overall survival rate following severe influenza infection. Finally, analysis of lung tissue sections stained with hematoxylin and eosin (H&E) at day 7 post-infection demonstrated that Unc93b1Letr/Letr mice had increased inflammation and signs of epithelial reactivity compared to Unc93b1+/+ mice (Figure 7F, G). The differential tissue pathology was coincident with a significantly higher viral load in the Unc93b1Letr/Letr mice (Figure 7A), suggesting that inflammation in the Unc93b1Letr/Letr lungs may reflect a heightened response to increased viral replication. Analysis of lung tissue sections from uninfected mice did not show any difference between genotypes (Figure 7D, E). Taken together, these data indicate that impaired Unc93b1 function predisposes the host to a more severe initial disease course following influenza challenge; however, this difference does not significantly alter the ability of the Unc93b1Letr/Letr mice to survive the infection.
DISCUSSION
The intricate balance between host immunity and pathogen virulence determines the outcome of an infection. ENU mutagenesis is an advantageous strategy for interrogation of the immune response to infection because it efficiently creates random heritable point mutations throughout the genome that may not occur through natural variation in inbred strains.32 ENU can also cause distinct single nucleotide alterations within a gene and create mutations that may closely resemble human conditions.32 Using this technique we screened for a recessive defect in TLR signaling in vitro to target immune processes that play an important role in microbial infection.35 We discovered an alternative splicing mutation in Unc93b1, a gene that has been implicated in host immunity to diverse pathogens, and have identified a specific role for this gene in the innate immune response to influenza. Our findings demonstrate that Unc93b1 contributes to the early activation of ExMs and CD4+ and CD8+ T cells in the lungs as well as expression of type I IFN, type II IFN, and CXCL10 in the airways. Mutation of Unc93b1 also delayed lung viral clearance and promoted tissue inflammation during infection, although it did not significantly alter survival. These findings establish the specific contribution of Unc93b1-dependent endosomal TLR activation during the initial host inflammatory response and highlight the role of complementary immune mechanisms that mediate survival and disease resolution following severe influenza infection.
Using a low-density genome scan, linkage analysis, and nucleotide sequencing, we determined that the Letr mutation causes a T to A transversion at the 5′ splice donor site in the intron between exon 4 and 5 of the Unc93b1 gene that results in the deletion of exon 4 in the spliced mRNA transcript. The Letr mRNA encodes a predicted protein that lacks the 3rd and 4th transmembrane domains of the full-length UNC93B1. The 54 amino acid residues eliminated by the Letr mutation are highly conserved between Mus musculus, Homo sapiens, and other diverse organisms, suggesting that the two missing transmembrane helices encode important protein domains. Functional analysis using in vitro and in vivo PAMP stimulation clearly demonstrate that the ENU-induced Letr mutation is recessive and confers a complete loss of endosomal TLR function with no detectable pattern of codominance. Residual IL-6 production following polyI:C stimulation in mice or cells carrying the Letr mutation is most likely attributable to recognition by the cytosolic melanoma differentiation-associated protein 5 (MDA5) pathway.57
Due to the inherently random nature of ENU mutagenesis, one cannot control the genes, or sites within a gene, that are altered. A major advantage of this hypothesis-free approach is that screening for a particular phenotype can lead to the discovery of previously unknown genes as well as novel functions for known genes.32 An earlier large-scale ENU mutagenesis study also identified a loss-of-function mutation, termed 3d, in the Unc93b1 gene and clearly established its role in immune responsiveness.23 In contrast to the ENU-induced splicing mutation of Unc93b1 that we describe here, the 3d mutation causes a missense mutation in exon 9 that leads to a single amino acid substitution (H412R) in the 9th transmembrane domain of UNC93B1. Despite the different location and unique consequences of these two ENU-induced mutations, both produce a loss-of-function phenotype that confirms the essential contribution of the affected transmembrane-spanning domains to protein integrity. In vitro studies have shown that the 3d mutation inhibits UNC93B1 binding and trafficking of endosomal TLRs from the ER to the endosome following PAMP stimulation.21, 22 A similar defect in stimulus-dependent trafficking to the plasma membrane was also observed with naturally occurring deletions of multiple transmembrane domains of the SLC7A7 transport protein.58 Although we have not formally demonstrated that TLR binding and translocation is inhibited by the Letr mutation, in light of the predicted deletion of two transmembrane domains of the UNC93B1 protein, it is tempting to speculate that a related mechanism occurs as a result of this ENU-induced allele. Alternatively, deletion of two transmembrane domains of UNC93B1 could also cause a severe structural defect that abrogates protein production, as observed following the loss of a single transmembrane span from the 10-transmembrane domain glucose-6-phosphate protein.59
Studies using Unc93b13d/3d mice have demonstrated an enhanced pathogen load and increased susceptibility to T. gondii, T. cruzi, L. major, and MCMV infection in association with reduced expression of inflammatory mediators including IFN-γ (T. cruzi, MCMV, L. major), IFN-α (MCMV) and IL-12p40 (T. gondii, T. cruzi).23, 37, 38, 40, 42 The current report using Unc93b1Letr/Letr mice extends the role of endosomal TLR-mediated inflammation to early host defense against influenza. Thus, the Unc93b1Letr/Letr and Unc93b13d/3d mice represent alternative models for the study of diverse host-pathogen interactions. In contrast, Fukui et al. have shown that a point mutation in the cytoplasmic amino terminus of UNC93B1 (D34A) causes lethal inflammation. Specifically, Unc93b1D34A/D34A mice show preferential and constitutive transport of TLR7 over TLR9, resulting in a severe autoimmune phenotype in the absence of external stimulation.60, 61 A comparison of the 3d, Letr, and D34A mutations provides crucial insights into the structure-function relationship of UNC93B1 and demonstrates that unique point mutations of the Unc93b1 gene can confer a deficient or overexuberant immune phenotype.
Following influenza challenge, macrophages and DCs are among the first cells recruited to the respiratory tract.62, 63 Pulmonary macrophages may be classified into two types; AMs that reside in naïve lungs and elicit an early response along with respiratory epithelial cells, and monocyte-derived ExMs that are recruited from the bloodstream in a CCR2-dependent manner and differentiate into effector phagocytes that can lyse infected cells.63, 64 Inflammatory DC subsets are also recruited from the bloodstream, take up antigen at the site of infection, and traffic in a CCR7-dependent manner to the LALNs to activate naïve T cells.10 Both ExMs and inflammatory DCs express CD80; however, ExMs are relatively poor activators of naïve T cells.63, 65 At an early time point after influenza infection we detected fewer activated CD80+ ExMs, but not CD80+ DCs, in the lungs of Unc93b1Letr/Letr mice. This data reaffirms a role for Unc93b1 in APC activation following infection38 and is consistent with a previous study in which a lack of endosomal TLR signaling did not affect co-stimulatory molecule expression on DCs.66
Residual APC activation in our model can be attributed to alternative pathogen recognition mechanisms such as the nucleotide-binding domain and leucine-rich repeat containing receptor family, pyrin domain containing 3 (NLRP3) inflammasome in macrophages and DCs67 or the cytosolic retinoic acid-inducible gene 1 (RIG-I) pathway in DCs.68, 69 Recognition of viral infection in conventional DCs (cDCs) preferentially utilizes RIG-I signaling66, 68 while plasmacytoid DCs (pDCs) rely on TLR7.16, 20 Since pDCs are the main producers of type I IFN, it is plausible that the delayed expression of type I IFN in the airways of Unc93b1Letr/Letr mice is due to defective TLR7-mediated activation of this specialized cell type. Conversely, the fact that cDCs produce type I IFN through a RIG-I-dependent mechanism68, 70 may explain why the type I IFN response was diminished but not completely abolished in Unc93b1Letr/Letr mice. Only the combined loss of MyD88 and mitochondrial antiviral signaling protein (MAVS), the adaptor molecule for RIG-I and MDA5, has been shown to completely abolish type I IFN signaling following influenza infection in vivo.18
At day 2 or day 3 post-infection with influenza, T cells in the LALNs downregulate CD62L and CCR7, proliferate, and subsequently traffic to the site of infection where they express various activation markers and acquire the capacity to secrete IFN-γ.56, 71 Consistent with an impaired activation state, CD4+ and CD8+ T cells in the lungs of Unc93b1Letr/Letr mice showed a decrease in CD69 and a concomitant increase in CD62L expression at day 3 post-infection. Importantly, TLRs have previously been shown to directly activate T cells.72 Tlr1, Tlr2, Tlr3, Tlr6 and Tlr7 are expressed on both human peripheral blood and mouse C57BL/6 T cells73–76 and direct stimulation with TLR3 or TLR7 ligands promotes T cell survival, CD38 and CD69 expression, and IFN-γ production.72, 73, 77, 78 Studies in murine lymphocytic choriomeningitis virus and T. gondii infection have demonstrated that the selective loss of MyD88 in T cells leads to decreased survival and IFN-γ production79, 80 and, in HIV-infected patients, direct TLR7 activation of CD8+ T cells in vitro increased T cell proliferation and IFN-γ production.81 As DC activation in Unc93b1Letr/Letr mice appeared to be normal, the delayed lymphocyte activation phenotype in our model may represent an intrinsic defect in CD4+ and CD8+ T cell activation caused by impaired Unc93b1 function. Nevertheless, at day 7 post-infection, the CD4+ and CD8+ T cell activation profile in Unc93b1Letr/Letr lungs was comparable to the wild type strain and most likely reflects increased infiltration of primed CD4+ and CD8+ T cells from the LALNs.56, 71 Indeed, the formation of antigen-specific CD8+ T cells and IFN-γ +CD4+ T cells at later time points post-infection can occur in the absence of TLR7 activation.26, 82 Studies in MyD88-deficient mice have also demonstrated that signaling through this adaptor protein is not required for antigen-specific CD8+ T cell activation later in the infection, while evidence for the importance of MyD88 in CD4+ T cell activation is variable.18, 26
Immune activation and cell migration following influenza infection is mediated by the expression of a large number of proinflammatory cytokines and chemokines.10 In our model the loss of Unc93b1 function led to a reduction in the expression of IFN-γ, CXCL10, and Type I IFN at day 3 post-infection. NK cells can produce IFN-γ in response to IFN-α production by macrophages.83 Accordingly, the diminished IFN-γ response in the Unc93b1Letr/Letr mice at this early time point may be attributable to fewer activated ExMs in the lung. At day 7 post-infection the comparable level of IFN-γ between Unc93b1+/+ and Unc93b1Letr/Letr mice is most likely attributable to production by antigen-specific T cells.56, 71 CXCL10 primarily attracts lymphocytes to the lungs and can be produced in response to IFN-γ and by cells of both the innate and adaptive immune response, including ExMs.84, 85 The reduced expression of CXCL10 in Unc93b1Letr/Letr mice at day 3 post-infection may also be linked to fewer activated ExMs with comparable induction at day 7 post-infection mediated by infiltrating CD4+ and CD8+ T cells.56, 71 The selective defect in proinflammatory mediator expression by Unc93b1Letr/Letr mice following influenza infection is similar to observations in Unc93b13d/3d mice following T. gondii infection as well as patients with HSV-1 encephalitis and natural UNC93B1 mutations38, 41.
In contrast to studies of Unc93b13d/3d mice that have shown increased mortality following infection with MCMV, NSV, T. cruzi, and T. gondii,23, 37, 38, 44 the Unc93b1Letr/Letr mice and their wild type counterparts had a comparable rate of survival following influenza infection. Nonetheless, during the innate immune response to influenza, Unc93b1Letr/Letr mice exhibited several early immune defects including reduced myeloid cell recruitment, lymphoid cell activation, and soluble inflammatory mediator production that collectively led to impaired viral clearance as well as increased tissue inflammation and epithelial reactivity during the course of infection. The different disease outcome in previous studies of Unc93b1 and the current report indicates that there is a limited requirement for Unc93b1 in the generation of protective immunity against influenza. Indeed, previous studies have shown that influenza activates a series of inflammatory pathways that could provide compensatory immunity in Unc93b1Letr/Letr mice. For example, the cytosolic RIG-I signaling pathway, while not essential for survival, has been implicated in type I IFN expression from myeloid DCs, fibroblasts, and human lung epithelial cells following influenza infection.18, 57, 68 NLRP3-dependent activation of caspase-1 is also essential for survival of an influenza infection and the lack of either component leads to decreased recruitment of monocytes and neutrophils to the airways and lower expression of IL-1β, IL-18, CXCL1, and CXCL2 at day 3 post-infection.27, 52 Remarkably, in the absence of pattern recognition through TLR7 and MAVS, IL-1R signaling in dendritic cells was shown to be necessary and sufficient for the generation of virus-specific CD8+ T cell immunity against influenza.82 Finally, our data highlight the differences in host response following systemic challenge with a purified PAMP that potently activates a single TLR and mucosal infection with a respiratory virus that activates multiple PRRs with complementary and overlapping immune functions.
In summary, we have demonstrated that Unc93b1Letr/Letr mice have defective endosomal TLR activation that diminishes innate immune responses at the site of infection during the early phase of influenza infection. Our study suggests two potential mechanisms by which Unc93b1 may contribute to innate immunity against influenza. First, Unc93b1-dependent type I IFN expression may stimulate ExMs that, in turn, enhance initial IFN-γ production by NK cells and induces CXCL10. Second, Unc93b1 may mediate endosomal TLR signaling on CD4+ and CD8+ T cells that triggers their activation at the site of infection. Further detailed investigations of Unc93b1Letr/Letr mice should reveal the exact mechanisms through which Unc93b1 regulates the innate immune response to influenza and, potentially, other viral respiratory pathogens.
MATERIALS AND METHODS
Ethics statement
All experiments using mice were performed according to the guidelines of the Canadian Council on Animal Care and approved by the Animal Use Care Committee of the McGill University Health Centre (Montréal, Canada).
Mice, virus, cell lines
C57BL/6 and 129S1 mice used for ENU mutagenesis and C3H/HeN mice used for breeding were purchased from Harlan Laboratories (Indianapolis, IN). Unc93b13d/3d mice were obtained from the Mutant Mouse Regional Resource Center (UC Davis, Davis, CA). Unc93b1+/+ and Unc93b1Letr/Letr F2 mice on the C57BL/6 background were bred in a specific pathogen-free facility (McGill University Health Centre). The influenza A/PR/8/34 (H1N1) virus was a generous gift from V. von Messling (INRS-Institut Armand-Frappier, Montréal, Canada). Madin-Darby canine kidney (MDCK) cells used for plaque assays were a generous gift from M. Divangahi (McGill University, Montréal, Canada). The B16-Blue IFNα/β cell line was purchased (InvivoGen, San Diego, CA).
ENU mutagenesis and breeding
Mutagenesis of 8–12-week-old G0 male 129S1 mice was conducted as previously described using 150mg/kg of ENU.86 Following fertility recovery (8–12 weeks post-injection) mutagenized G0 males were crossed to C57BL/6 females in a three-generation breeding scheme for recessive ENU mutations.87 G1 progeny from distinct G0 pairs were mated and G2 brother-sister matings were performed to produce 1200 G3 progeny for initial phenotypic screening. G2 fathers of G3 mice that displayed a deviant phenotype were outcrossed to C57BL/6 females and the resulting progeny were backcrossed to their G2 fathers. All backcross progeny were re-phenotyped using survival splenectomy and deviant male and female animals were selected for creation of homozygous lines. Deviant animals were outcrossed to C3H/HeN mice and the F1 progeny were backcrossed to the deviant parent to generate 54 segregating mice used to map the genetic location of the mutation.
In vitro and in vivo phenotyping using PAMPs
Whole spleens were removed, placed in 4mL of RPMI (Gibco, Burlington, Canada) supplemented with 1% L-glutamine (Gibco), 1% penicillin/streptomycin (Gibco), 10% fetal bovine serum (FBS) (Gibco), and 0.1% β-mercaptoethanol (Gibco) and mechanically disrupted using the frosted end of a microscope slide (Fisher Scientific, Toronto, Canada). A single cell suspension was created by multiple passages through an 18G 1” and 21G 1” needle using a 5mL syringe (BD Biosciences, Mississauga, Canada). To obtain macrophages, mice were injected intraperitoneally with 1mL of thioglycollate, prepared according to the manufacturer’s instructions (BD Biosciences). Seventy-two hours later, cells were recovered by intraperitoneal lavage with 10mL of incomplete RPMI. Following treatment with ACK lysis buffer (0.82% NH4Cl, 0.1% KHCO3, 0.0038% Na2EDTA, pH 7.4), and collection by centrifugation, all cells were enumerated with a Coulter Z1 Particle Counter (Becton Coulter, Mississauga, Canada) and 2x105 cells were plated on 96-well tissue culture plates (Sarstedt, Montréal, Canada). Peritoneal cells were incubated for 24 hours in RPMI supplemented with 1% L-glutamine, 1% penicillin/streptomycin and 10% FBS to allow for macrophage adherence, washed with sterile phosphate-buffered saline (PBS) (Wisent, St-Bruno, Canada) and stimulated with 100ng/mL of LPS from Escherichia coli 055:B548 (Sigma-Aldrich, Oakville, Canada). Splenocytes were stimulated immediately after plating with 1μM unmethylated CpG oligodeoxynucleotide (CpG ODN)47 (AlphaDNA, Montréal, Canada), 5μg/mL Imiquimod86 (InvivoGen) or 50–150μg/mL polyI:C (InvivoGen). Cell supernatants were harvested 24 or 48 hours after stimulation. For in vivo characterization, mice were injected intraperitoneally with 0.5mg/mL of LPS from Escherichia coli 055:B548, 150μg/mL Imiquimod, 2.5μg/g polyI:C in 20mg/mL D(+)-Galactosamine hydrochloride (DGalN)45 (Sigma-Aldrich), or 50nmol/mL CpG ODN in 20mg/mL DGalN47; delivered in 1mL of sterile PBS. Mice were euthanized three hours post-injection, blood was collected by cardiac puncture and serum was separated using the Z-Gel microtubes (Sarstedt). Cytokine and chemokine quantification was conducted using commercial ELISA kits (R&D systems, Minneapolis, MN).
Genotyping
gDNA from a panel of 54 (Letr/Letr x C3H/HeN) F1 x Letr/Letr mice was submitted for typing of 375 informative SNPs using a low-density Illumina genotyping platform and the Unc93b1 gene was sequenced using gDNA and cDNA derived from normal and deviant mice (The Centre for Applied Genomics, Toronto, Canada). The primers used for gDNA sequencing were: EseqF 5′-GTAGTTGTAAAGAAGTGTGGCA-3′, EseqR 5′-GCTAAAGTTGGCAAAGAAGT-3′, FseqF 5′-CACCAGAGGCCATGTCCAA-3′, FseqR 5′-GGCTGAGAAGACAATGGACTA-3′, GseqF 5′-CAAACTGCCACCCCAAAG-3′, GseqR 5′-GTTTGCTGGGTGACTGAGTG-3′. The primers used for cDNA sequencing were: UncP12 (reverse): 5′-CCATGAGCACGCTCTCTACA-3′, UncP22 (reverse): 5′-GCCTGCCACCAGTGATAGAT-3′, UncP23 (reverse): 5′-AAGGGCAGCTGGAAGATGT-3′, UncP24 (forward): 5′-ACATCTTCCAGCTGCCCTTC-3′, UncP31 (forward): 5′-CACACTCCTGGGCATCCTAT-3′, UncP38 (reverse): 5′-CCATGTTGCCATACTTCACCT-3′, UncP39 (forward): 5′-CCGGACATCGATAGCAAGAT-3′, BGH (reverse): 5′-TAGAAGGCACAGTCGAGG-3′, T7 (forward): 5′-TAATACGACTCACTATAGGG-3′ (AlphaDNA).
Unc93b1 PCR and gel electrophoresis
RNA was extracted from spleens using the RNeasy Plus Mini kit (Qiagen, Toronto, Canada) with DNase digestion using the RNase-Free DNase Set (Qiagen) according to the manufacturer’s instructions. Reverse transcription was carried out using the High Capacity cDNA Archive Kit (Applied Biosystems, Burlington, Canada). PCR was performed on 20ng of cDNA with 10μL 5x Herculase Buffer and 0.5μL Herculase Polymerase (Agilent, Mississauga, Canada), 5μL dNTP (Invitrogen, Burlington, Canada), and 100ng of each primer (forward: 5′-GGTGCCCAGAACTGCAAG-3′ reverse: 5′-CCATGAGCACGCTCTCTACA-3′) in a final volume of 50μL. The PCR cycling conditions were 94°C for 10 minutes, 35 cycles of 94°C for 30 seconds/52°C for 30 seconds/72°C for 1 minute, and 72°C for 7 minutes. The PCR product was visualized on a 1% agarose gel in TAE buffer containing 0.5μg/mL of ethidium bromide.
Analysis of Unc93b1 sequence and protein structure
The relative size of the Unc93b1 introns and exons was determined from the Ensembl database (ENSMUST00000162708). UNC93B1 amino acid sequences were derived from the NCBI nucleotide database for the following species Mus musculus (NM_019449.2), Homo sapiens (NM_030930.2), Pan troglodytes (XM_003313185.1), Canis familiaris (XM_540813.3), Felis catus (XM_003993751.1), Bos taurus (NM_001193147.1), Equus caballus (XM_001916968.2), Ovis aries (XM_004023404.1), and Xenopus tropicalis (NM_001100253.1). The amino acid sequences were aligned using Clustal Omega software, version 1.2 (European Molecular Biology Laboratory, Heidelberg, Germany).88 To determine the predicted three-dimensional protein structure, the wild type UNC93B1 amino acid sequence was entered into the Protein Homology/analogY Recognition Engine (Phyre), version 2.0 (Structural Bioinformatics Group, Imperial College, London, UK).89 The predicted model, including the location of the 3d and Letr mutations, was visualized using the PyMol Molecular Graphics System, version 1.7 (Schrödinger, LLC).90
In vivo influenza infection
6-week-old Unc93b1+/+ and Unc93b1Letr/Letr F2 mice were anaesthetized by intraperitoneal injection of 150mg/kg of ketamine (Bioniche, Pointe-Claire, Canada) and 10mg/kg of xylazine (Bayer, Toronto, Canada) in sterile PBS and infected intranasally with 400 pfu of influenza A/PR/8/34 (H1N1). Infected mice were monitored daily for signs of sickness and sacrificed if they appeared moribund.
Flow cytometry
Lungs were extracted, placed in 5mL of incomplete DMEM, finely chopped with a no.23 scalpel and incubated with a further 5mL of RPMI supplemented with 300U/mL of Collagenase (Sigma-Aldrich) for 1 hour at 37°C in 5% CO2. The tissue was then passed multiple times through a 16G 1” needle and a 100μM nylon strainer (BD Biosciences) and collected by centrifugation. LALN cells were prepared as described for splenocytes. Red blood cells were lysed with ACK buffer and cells were counted using a hemocytometer. Cells were stained with Fixable Viability Dye-efluor780 (eBioscience, San Diego, CA) and, following a wash in sterile PBS, Fc receptors were blocked using the anti-mouse CD16/CD32 (clone 93) antibody (eBioscience). Cells were then fluorescently labeled using cocktails of fluorescence-conjugated antibodies: rat anti-mouse CD45-V500 (clone 30-F11), rat anti-mouse Ly6G-Alexa Fluor 700 (clone 1A8), rat anti-mouse CD45R/B220-PECF594 (clone RA3-6B2) (BD Biosciences); anti-mouse/human CD11b-Brilliant Violet 711 (clone M1/70), anti-mouse CD11c-Brilliant Violet 605 (clone N418), anti-mouse CD80-PE (clone 16-10A1), anti-mouse CD8a-Alexa Fluor 700 (clone 53-6.7), anti-mouse CD62L-Brilliant Violet 605 (clone MEL-14), anti-mouse CD69-PE/Cy7 (clone H1.2F3) (BioLegend, San Diego, CA); anti-mouse MHC class II (I-A/I-E)-PE/Cy7 (clone M5/114.15.2), anti-mouse CD3e-FITC (clone 145-2C11), anti-mouse CD4-eFluor450 (clone GK1.5), anti-mouse CD49b-allophycocyanin (clone DX5), anti-mouse CD44-PE (clone IM7) (eBioscience). Data was collected on the LSRFortessa using FacsDiva software version 6.2 (BD Biosciences) and analyzed using FlowJo, version 9.1 (Tree Star Inc., Ashland, OR) with fluorescence-minus-one gating controls.
Bronchoalveolar lavage and lung inflammatory mediator quantification
Following euthanasia, bronchoalveolar lavage (BAL) fluid was collected from the airways by inserting a 22G 1” catheter into the mouse trachea and slowly injecting and removing four-0.5mL aliquots of ice-cold sterile PBS. Cells were removed from BAL fluid by centrifugation. Lungs were perfused with 10mL of sterile PBS via the right ventricle, excised, and placed in 2mL of sterile PBS supplemented with 1x Halt protease inhibitor cocktail (Fisher Scientific). Lungs were processed using a rotor-stator homogenizer (Fisher Scientific) and samples were centrifuged to pellet insoluble material. BAL fluid and lung homogenate were stored at −80°C until further analysis. ELISAs were used for protein quantification according to the manufacturer’s instructions (R&D Systems). Type I IFN was quantified using B16-Blue cells that were cultured using standard techniques in RPMI media supplemented with 1% L-glutamine, 0.5% penicillin/streptomycin, 10% FBS and 0.2% Normocin (InvivoGen). After two passages, cells were washed with sterile PBS and cultured in the same media with the addition of 0.1% Zeocin (InvivoGen) for an additional two or three passages. 20μL/well of BAL sample, recombinant IFN-β (positive control) (PBL Interferon Source, Piscataway, NJ) or sterile PBS (negative control) was added to a 96-well tissue culture plate and 7.56×104 B16-Blue cells (in a volume of 180μL) were added to these wells and incubated at 37°C in 5% CO2 for 24 hours. 20μL of this culture supernatant was added to a new 96-well plate containing 180μL of Quanti-Blue medium, prepared according to the manufacturer’s instructions (InvivoGen), and incubated at 37°C for 3.5 hours. Absorbance was measured at 655nm and type I IFN quantified as the fold change in expression relative to uninfected airway.
Viral quantification by plaque assay
Lungs were extracted from mice at multiple time points post-infection and placed in incomplete DMEM (Wisent) at a 20% weight/volume ratio. Lungs were homogenized using a rotor-stator homogenizer and supernatants were collected by centrifugation. MDCK cells were cultured using standard techniques in DMEM supplemented with 1% L-glutamine, 1% penicillin/streptomycin and 10% FBS. Three-six cell passages were conducted before one million cells were plated/well on a 6-well plate and incubated overnight at 37°C in 5% CO2. Plaque assays were performed in a similar manner to a previous protocol.91 Briefly, cells were washed twice with sterile PBS and 100μL of serially diluted lung homogenate in incomplete DMEM was added. Following a 30 minute incubation at 37°C in 5% CO2, sample was removed and cells were covered with 3mL of a 1:1 mixture of 1.6% agarose and 2x MEM (Gibco) supplemented with 6% of a 7.5% NaHCO3 solution (BioShop Canada Inc., Burlington, Canada), 1% L-glutamine, 2% penicillin/streptomycin and 0.1% TPCK-trypsin (Sigma-Aldrich). After a 48 hour incubation at 37°C in 5% CO2, 3mL of a 3:1 methanol (Sigma-Aldrich) and acetic acid (Fisher Scientific) solution was added. Four hours following, the solution and agarose plugs were removed and plates were allowed to dry for 24 hours. Plaques were visualized by staining with a 0.2% Crystal Violet solution in 20% ethanol (Sigma-Aldrich).
Lung histology
Lungs were flushed with 10mL of PBS via the right ventricle. A catheter was inserted into the trachea and lungs were allowed to expand to a pressure of 25cm using 10% buffered formalin acetate (Fisher Scientific). Inflated lungs were then placed in 5mL of 10% buffered formalin acetate and subsequently embedded in paraffin, sectioned at 5μm, and stained with H&E at the Histology Facility of the Goodman Cancer Research Centre (McGill University, Montréal, Canada). Representative photographs of lung sections were taken using a BX51 microscope (Olympus, Richmond Hill, Canada), QICAM Fast 1394 digital CCD camera (QImaging, Surrey, Canada) and Image-Pro Plus software version 7.0.1.658 (Media Cybernetics, Rockville, MD).
Statistical analysis
Survival curve analysis was performed using the Logrank test. For comparison between multiple columns a 1-way ANOVA with Dunnett’s or Tukey’s post-hoc test was used. A two-tailed unpaired t-test was conducted for all other pairwise comparisons. Statistical analysis was performed using GraphPad Prism 4 (GraphPad Software, La Jolla, CA).
Supplementary Material
Acknowledgments
We thank Gregory Boivin and François Coulombe for advice with experimental procedures, Marie-Line Goulet for advice on experimental design as well as helpful comments on the manuscript, and Dr. David Logan Burk for help with three-dimensional protein modeling. We also acknowledge Dr. Veronika von Messling and Dr. Maziar Divangahi for providing materials and advice on working with the influenza virus.
This work was supported by a CIHR Team Grant in Mouse Mutagenesis and Infectious Diseases (CTP-87520), a Canada Research Chair in Host Resistance to Respiratory Infections (STQ), and the Fonds de Recherche du Québec – Santé. EIL is a recipient of awards from the CIHR Québec Respiratory Health Training Program, the McGill University Health Centre, and the McGill University Faculty of Medicine.
Footnotes
Conflict of Interest
The authors declare no potential conflicts of interest that are relevant to this manuscript.
Supplementary information for this article is available.
References
- 1.Fauci AS. Seasonal and pandemic influenza preparedness: science and countermeasures. J Infect Dis. 2006;194(Suppl 2):S73–6. doi: 10.1086/507550. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Influenza Fact Sheet No.211. 2009. [Google Scholar]
- 3.Fraser C, Donnelly CA, Cauchemez S, Hanage WP, Van Kerkhove MD, Hollingsworth TD, et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science. 2009;324(5934):1557–61. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459(7249):931–9. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker WH, Mullooly JP. Impact of epidemic type A influenza in a defined adult population. Am J Epidemiol. 1980;112(6):798–811. doi: 10.1093/oxfordjournals.aje.a113052. [DOI] [PubMed] [Google Scholar]
- 6.Toapanta FR, Ross TM. Impaired immune responses in the lungs of aged mice following influenza infection. Respir Res. 2009;10:112. doi: 10.1186/1465-9921-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webster RG. Immunity to influenza in the elderly. Vaccine. 2000;18(16):1686–9. doi: 10.1016/s0264-410x(99)00507-1. [DOI] [PubMed] [Google Scholar]
- 8.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198(7):962–70. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perrone LA, Plowden JK, Garcia-Sastre A, Katz JM, Tumpey TM. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4(8):e1000115. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohlmeier JE, Woodland DL. Immunity to respiratory viruses. Annu Rev Immunol. 2009;27:61–82. doi: 10.1146/annurev.immunol.021908.132625. [DOI] [PubMed] [Google Scholar]
- 11.Ho AW, Prabhu N, Betts RJ, Ge MQ, Dai X, Hutchinson PE, et al. Lung CD103+ dendritic cells efficiently transport influenza virus to the lymph node and load viral antigen onto MHC class I for presentation to CD8 T cells. J Immunol. 2011;187(11):6011–21. doi: 10.4049/jimmunol.1100987. [DOI] [PubMed] [Google Scholar]
- 12.Ingulli E, Funatake C, Jacovetty EL, Zanetti M. Cutting edge: antigen presentation to CD8 T cells after influenza A virus infection. J Immunol. 2009;182(1):29–33. doi: 10.4049/jimmunol.182.1.29. [DOI] [PubMed] [Google Scholar]
- 13.Kim TS, Braciale TJ. Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PLoS One. 2009;4(1):e4204. doi: 10.1371/journal.pone.0004204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vermaelen KY, Carro-Muino I, Lambrecht BN, Pauwels RA. Specific migratory dendritic cells rapidly transport antigen from the airways to the thoracic lymph nodes. J Exp Med. 2001;193(1):51–60. doi: 10.1084/jem.193.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waithman J, Mintern JD. Dendritic cells and influenza A virus infection. Virulence. 2012;3(7):603–8. doi: 10.4161/viru.21864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303(5663):1529–31. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 17.Guillot L, Le Goffic R, Bloch S, Escriou N, Akira S, Chignard M, et al. Involvement of toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded RNA and influenza A virus. J Biol Chem. 2005;280(7):5571–80. doi: 10.1074/jbc.M410592200. [DOI] [PubMed] [Google Scholar]
- 18.Koyama S, Ishii KJ, Kumar H, Tanimoto T, Coban C, Uematsu S, et al. Differential role of TLR- and RLR-signaling in the immune responses to influenza A virus infection and vaccination. J Immunol. 2007;179(7):4711–20. doi: 10.4049/jimmunol.179.7.4711. [DOI] [PubMed] [Google Scholar]
- 19.Le Goffic R, Pothlichet J, Vitour D, Fujita T, Meurs E, Chignard M, et al. Cutting Edge: Influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J Immunol. 2007;178(6):3368–72. doi: 10.4049/jimmunol.178.6.3368. [DOI] [PubMed] [Google Scholar]
- 20.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A. 2004;101(15):5598–603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177(2):265–75. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452(7184):234–8. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 23.Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7(2):156–64. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 24.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4(2):161–7. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 25.Seo SU, Kwon HJ, Song JH, Byun YH, Seong BL, Kawai T, et al. MyD88 signaling is indispensable for primary influenza A virus infection but dispensable for secondary infection. J Virol. 2010;84(24):12713–22. doi: 10.1128/JVI.01675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heer AK, Shamshiev A, Donda A, Uematsu S, Akira S, Kopf M, et al. TLR signaling fine-tunes anti-influenza B cell responses without regulating effector T cell responses. J Immunol. 2007;178(4):2182–91. doi: 10.4049/jimmunol.178.4.2182. [DOI] [PubMed] [Google Scholar]
- 27.Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30(4):556–65. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Goffic R, Balloy V, Lagranderie M, Alexopoulou L, Escriou N, Flavell R, et al. Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2006;2(6):e53. doi: 10.1371/journal.ppat.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80(10):5059–64. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hidaka F, Matsuo S, Muta T, Takeshige K, Mizukami T, Nunoi H. A missense mutation of the Toll-like receptor 3 gene in a patient with influenza-associated encephalopathy. Clin Immunol. 2006;119(2):188–94. doi: 10.1016/j.clim.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Horby P, Nguyen NY, Dunstan SJ, Baillie JK. The role of host genetics in susceptibility to influenza: a systematic review. PLoS One. 2012;7(3):e33180. doi: 10.1371/journal.pone.0033180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acevedo-Arozena A, Wells S, Potter P, Kelly M, Cox RD, Brown SD. ENU mutagenesis, a way forward to understand gene function. Annu Rev Genomics Hum Genet. 2008;9:49–69. doi: 10.1146/annurev.genom.9.081307.164224. [DOI] [PubMed] [Google Scholar]
- 33.Buer J, Balling R. Mice, microbes and models of infection. Nat Rev Genet. 2003;4(3):195–205. doi: 10.1038/nrg1019. [DOI] [PubMed] [Google Scholar]
- 34.Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A. Mouse ENU mutagenesis. Hum Mol Genet. 1999;8(10):1955–63. doi: 10.1093/hmg/8.10.1955. [DOI] [PubMed] [Google Scholar]
- 35.Richer E, Qureshi ST, Vidal SM, Malo D. Chemical mutagenesis: a new strategy against the global threat of infectious diseases. Mamm Genome. 2008;19(5):309–17. doi: 10.1007/s00335-008-9114-0. [DOI] [PubMed] [Google Scholar]
- 36.Andrade WA, do Souza MC, Ramos-Martinez E, Nagpal K, Dutra MS, Melo MB, et al. Combined action of nucleic acid-sensing Toll-like receptors and TLR11/TLR12 heterodimers imparts resistance to Toxoplasma gondii in mice. Cell Host Microbe. 2013;13(1):42–53. doi: 10.1016/j.chom.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caetano BC, Carmo BB, Melo MB, Cerny A, dos Santos SL, Bartholomeu DC, et al. Requirement of UNC93B1 reveals a critical role for TLR7 in host resistance to primary infection with Trypanosoma cruzi. J Immunol. 2011;187(4):1903–11. doi: 10.4049/jimmunol.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melo MB, Kasperkovitz P, Cerny A, Konen-Waisman S, Kurt-Jones EA, Lien E, et al. UNC93B1 mediates host resistance to infection with Toxoplasma gondii. PLoS Pathog. 2010;6(8):e1001071. doi: 10.1371/journal.ppat.1001071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pifer R, Benson A, Sturge CR, Yarovinsky F. UNC93B1 is essential for TLR11 activation and IL-12-dependent host resistance to Toxoplasma gondii. J Biol Chem. 2011;286(5):3307–14. doi: 10.1074/jbc.M110.171025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schamber-Reis BL, Petritus PM, Caetano BC, Martinez ER, Okuda K, Golenbock D, et al. UNC93B1 and nucleic acid-sensing Toll-like receptors mediate host resistance to infection with Leishmania major. J Biol Chem. 2013;288(10):7127–36. doi: 10.1074/jbc.M112.407684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casrouge A, Zhang SY, Eidenschenk C, Jouanguy E, Puel A, Yang K, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314(5797):308–12. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 42.Crane MJ, Gaddi PJ, Salazar-Mather TP. UNC93B1 mediates innate inflammation and antiviral defense in the liver during acute murine cytomegalovirus infection. PLoS One. 2012;7(6):e39161. doi: 10.1371/journal.pone.0039161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang JP, Bowen GN, Zhou S, Cerny A, Zacharia A, Knipe DM, et al. Role of specific innate immune responses in herpes simplex virus infection of the central nervous system. J Virol. 2012;86(4):2273–81. doi: 10.1128/JVI.06010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esen N, Blakely PK, Rainey-Barger EK, Irani DN. Complexity of the microglial activation pathways that drive innate host responses during lethal alphavirus encephalitis in mice. ASN Neuro. 2012;4(4):207–21. doi: 10.1042/AN20120016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732–8. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 46.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3(2):196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 47.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–5. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11(4):443–51. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 49.Kashuba VI, Protopopov AI, Kvasha SM, Gizatullin RZ, Wahlestedt C, Kisselev LL, et al. hUNC93B1: a novel human gene representing a new gene family and encoding an unc-93-like protein. Gene. 2002;283(1–2):209–17. doi: 10.1016/s0378-1119(01)00856-3. [DOI] [PubMed] [Google Scholar]
- 50.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9(1):143–50. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 51.Short KR, Brooks AG, Reading PC, Londrigan SL. The fate of influenza A virus after infection of human macrophages and dendritic cells. J Gen Virol. 2012;93(Pt 11):2315–25. doi: 10.1099/vir.0.045021-0. [DOI] [PubMed] [Google Scholar]
- 52.Thomas PG, Dash P, Aldridge JR, Jr, Ellebedy AH, Reynolds C, Funk AJ, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30(4):566–75. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown DM, Dilzer AM, Meents DL, Swain SL. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J Immunol. 2006;177(5):2888–98. doi: 10.4049/jimmunol.177.5.2888. [DOI] [PubMed] [Google Scholar]
- 54.Brown DM, Lee S, de Garcia-Hernandez ML, Swain SL. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J Virol. 2012;86(12):6792–803. doi: 10.1128/JVI.07172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hufford MM, Kim TS, Sun J, Braciale TJ. Antiviral CD8+ T cell effector activities in situ are regulated by target cell type. J Exp Med. 2011;208(1):167–80. doi: 10.1084/jem.20101850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roman E, Miller E, Harmsen A, Wiley J, Von Andrian UH, Huston G, et al. CD4 effector T cell subsets in the response to influenza: heterogeneity, migration, and function. J Exp Med. 2002;196(7):957–68. doi: 10.1084/jem.20021052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 58.Mykkanen J, Torrents D, Pineda M, Camps M, Yoldi ME, Horelli-Kuitunen N, et al. Functional analysis of novel mutations in y(+)LAT-1 amino acid transporter gene causing lysinuric protein intolerance (LPI) Hum Mol Genet. 2000;9(3):431–8. doi: 10.1093/hmg/9.3.431. [DOI] [PubMed] [Google Scholar]
- 59.Chen LY, Lin B, Pan CJ, Hiraiwa H, Chou JY. Structural requirements for the stability and microsomal transport activity of the human glucose 6-phosphate transporter. J Biol Chem. 2000;275(44):34280–6. doi: 10.1074/jbc.M006439200. [DOI] [PubMed] [Google Scholar]
- 60.Fukui R, Saitoh S, Kanno A, Onji M, Shibata T, Ito A, et al. Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity. 2011;35(1):69–81. doi: 10.1016/j.immuni.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 61.Fukui R, Saitoh S, Matsumoto F, Kozuka-Hata H, Oyama M, Tabeta K, et al. Unc93B1 biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA- but against RNA-sensing. J Exp Med. 2009;206(6):1339–50. doi: 10.1084/jem.20082316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herold S, von Wulffen W, Steinmueller M, Pleschka S, Kuziel WA, Mack M, et al. Alveolar epithelial cells direct monocyte transepithelial migration upon influenza virus infection: impact of chemokines and adhesion molecules. J Immunol. 2006;177(3):1817–24. doi: 10.4049/jimmunol.177.3.1817. [DOI] [PubMed] [Google Scholar]
- 63.Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J Immunol. 2008;180(4):2562–72. doi: 10.4049/jimmunol.180.4.2562. [DOI] [PubMed] [Google Scholar]
- 64.Herold S, Steinmueller M, von Wulffen W, Cakarova L, Pinto R, Pleschka S, et al. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J Exp Med. 2008;205(13):3065–77. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delamarre L, Pack M, Chang H, Mellman I, Trombetta ES. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science. 2005;307(5715):1630–4. doi: 10.1126/science.1108003. [DOI] [PubMed] [Google Scholar]
- 66.Lopez CB, Moltedo B, Alexopoulou L, Bonifaz L, Flavell RA, Moran TM. TLR-independent induction of dendritic cell maturation and adaptive immunity by negative-strand RNA viruses. J Immunol. 2004;173(11):6882–9. doi: 10.4049/jimmunol.173.11.6882. [DOI] [PubMed] [Google Scholar]
- 67.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206(1):79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23(1):19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 69.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 70.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 71.Lawrence CW, Braciale TJ. Activation, differentiation, and migration of naive virus-specific CD8+ T cells during pulmonary influenza virus infection. J Immunol. 2004;173(2):1209–18. doi: 10.4049/jimmunol.173.2.1209. [DOI] [PubMed] [Google Scholar]
- 72.Caron G, Duluc D, Fremaux I, Jeannin P, David C, Gascan H, et al. Direct stimulation of human T cells via TLR5 and TLR7/8: flagellin and R-848 up-regulate proliferation and IFN-gamma production by memory CD4+ T cells. J Immunol. 2005;175(3):1551–7. doi: 10.4049/jimmunol.175.3.1551. [DOI] [PubMed] [Google Scholar]
- 73.Gelman AE, Zhang J, Choi Y, Turka LA. Toll-like receptor ligands directly promote activated CD4+ T cell survival. J Immunol. 2004;172(10):6065–73. doi: 10.4049/jimmunol.172.10.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdorfer B, Giese T, et al. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol. 2002;168(9):4531–7. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- 75.Kabelitz D. Expression and function of Toll-like receptors in T lymphocytes. Curr Opin Immunol. 2007;19(1):39–45. doi: 10.1016/j.coi.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 76.Kulkarni R, Behboudi S, Sharif S. Insights into the role of Toll-like receptors in modulation of T cell responses. Cell Tissue Res. 2011;343(1):141–52. doi: 10.1007/s00441-010-1017-1. [DOI] [PubMed] [Google Scholar]
- 77.Funderburg N, Luciano AA, Jiang W, Rodriguez B, Sieg SF, Lederman MM. Toll-like receptor ligands induce human T cell activation and death, a model for HIV pathogenesis. PLoS One. 2008;3(4):e1915. doi: 10.1371/journal.pone.0001915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tabiasco J, Devevre E, Rufer N, Salaun B, Cerottini JC, Speiser D, et al. Human effector CD8+ T lymphocytes express TLR3 as a functional coreceptor. J Immunol. 2006;177(12):8708–13. doi: 10.4049/jimmunol.177.12.8708. [DOI] [PubMed] [Google Scholar]
- 79.LaRosa DF, Stumhofer JS, Gelman AE, Rahman AH, Taylor DK, Hunter CA, et al. T cell expression of MyD88 is required for resistance to Toxoplasma gondii. Proc Natl Acad Sci U S A. 2008;105(10):3855–60. doi: 10.1073/pnas.0706663105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rahman AH, Cui W, Larosa DF, Taylor DK, Zhang J, Goldstein DR, et al. MyD88 plays a critical T cell-intrinsic role in supporting CD8 T cell expansion during acute lymphocytic choriomeningitis virus infection. J Immunol. 2008;181(6):3804–10. doi: 10.4049/jimmunol.181.6.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song Y, Zhuang Y, Zhai S, Huang D, Zhang Y, Kang W, et al. Increased expression of TLR7 in CD8(+) T cells leads to TLR7-mediated activation and accessory cell-dependent IFN-gamma production in HIV type 1 infection. AIDS Res Hum Retroviruses. 2009;25(12):1287–95. doi: 10.1089/aid.2008.0303. [DOI] [PubMed] [Google Scholar]
- 82.Pang IK, Ichinohe T, Iwasaki A. IL-1R signaling in dendritic cells replaces pattern-recognition receptors in promoting CD8(+) T cell responses to influenza A virus. Nat Immunol. 2013;14(3):246–53. doi: 10.1038/ni.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matikainen S, Paananen A, Miettinen M, Kurimoto M, Timonen T, Julkunen I, et al. IFN-alpha and IL-18 synergistically enhance IFN-gamma production in human NK cells: differential regulation of Stat4 activation and IFN-gamma gene expression by IFN-alpha and IL-12. Eur J Immunol. 2001;31(7):2236–45. [PubMed] [Google Scholar]
- 84.Zhou J, Law HK, Cheung CY, Ng IH, Peiris JS, Lau YL. Differential expression of chemokines and their receptors in adult and neonatal macrophages infected with human or avian influenza viruses. J Infect Dis. 2006;194(1):61–70. doi: 10.1086/504690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tighe RM, Liang J, Liu N, Jung Y, Jiang D, Gunn MD, et al. Recruited exudative macrophages selectively produce CXCL10 after noninfectious lung injury. Am J Respir Cell Mol Biol. 2011;45(4):781–8. doi: 10.1165/rcmb.2010-0471OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Richer E, Prendergast C, Zhang DE, Qureshi ST, Vidal SM, Malo D. N-ethyl-N-nitrosourea-induced mutation in ubiquitin-specific peptidase 18 causes hyperactivation of IFN-alphass signaling and suppresses STAT4-induced IFN-gamma production, resulting in increased susceptibility to Salmonella typhimurium. J Immunol. 2010;185(6):3593–601. doi: 10.4049/jimmunol.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cook MC, Vinuesa CG, Goodnow CC. ENU-mutagenesis: insight into immune function and pathology. Curr Opin Immunol. 2006;18(5):627–33. doi: 10.1016/j.coi.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 88.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols. 2009;4(3):363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 90.The PyMol Molecular Graphics System. version 1.7. Schrödinger, LLC; [Google Scholar]
- 91.Brown EG. Increased virulence of a mouse-adapted variant of influenza A/FM/1/47 virus is controlled by mutations in genome segments 4, 5, 7, and 8. J Virol. 1990;64(9):4523–33. doi: 10.1128/jvi.64.9.4523-4533.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.