Abstract
Aberrant Epidermal growth factor receptor (EGFR) signaling in non-small cell lung cancer (NSCLC) is linked to tumor progression, metastasis, and poor survival rates. Here, we report the role of Cdc42-interacting protein 4 (CIP4) in the regulation of NSCLC cell invasiveness and tumor metastasis. CIP4 was highly expressed in a panel of NSCLC cell lines and normal lung epithelial cell lines. Stable knock-down (KD) of CIP4 in lung adenocarcinoma H1299 cells, expressing wild-type EGFR, led to increased EGFR levels on the cell surface, and defects in sustained activation of Erk kinase in H1299 cells treated with EGF. CIP4 localized to leading edge projections in NSCLC cells, and CIP4 KD cells displayed defects in EGF-induced cell motility and invasion through extracellular matrix. This correlated with reduced expression and activity of matrix metalloproteinase-2 (MMP-2) in CIP4 KD cells compared to control. In xenograft assays, CIP4 silencing had no effect on tumor growth, but resulted in significant defects in spontaneous metastases to the lungs from these subcutaneous tumors. This correlated with reduced expression of the Erk target gene Zeb1, and the Zeb1 target gene MMP-2 in CIP4 KD tumors compared to control. CIP4 also enhanced rates of metastasis to the liver and lungs in an intrasplenic experimental metastasis model. In human NSCLC tumor sections, CIP4 expression was elevated ≥ 2-fold in 43% of adenocarcinomas and 32% of squamous carcinomas compared to adjacent normal lung tissues. Analysis of microarray data for NSCLC patients also revealed that high CIP4 transcript levels correlated with reduced overall survival. Together, these results identify CIP4 as a positive regulator of NSCLC metastasis, and a potential poor prognostic biomarker in lung adenocarcinoma.
Keywords: lung cancer, metastasis, EGFR, CIP4
INTRODUCTION
Non-small cell lung cancers (NSCLC) are frequently associated with EGFR over-expression,1 gene amplification,2 and acquired gain-of-function mutations.3 EGFR signaling in NSCLC promotes tumor growth, survival, and metastasis.4 For patients with EGFR mutations, EGFR inhibitors can induce rapid tumor regression.5 However, development of drug resistance and progression to metastatic disease occurs frequently.6 Although targeting downstream effectors of EGFR has shown some promise,7–9 new treatments are needed to prevent EGFR signaling to effectors that promote metastasis.
CIP4 (Cdc42-interacting protein 4) is an adaptor protein implicated in regulating EGFR trafficking and signaling in a variety of cancer cell models.10–12 CIP4 is an adaptor protein composed of F-BAR (Fer/CIP4 homology-Bin/Amphiphysin/Rvs), PKN homology region-1 (HR1), and SH3 domains.13, 14 F-BAR domains function in sensing and stabilizing membrane invaginations,15, 16 and bind phosphoinositides.11, 17–19 While Cdc42GTP is the exclusive ligand of the HR1 domain of CIP4,13 the SH3 domain interacts with a diverse array of proteins, including actin nucleation promoting factors (WASP, N-WASP, WAVE), formins (DAAM-1), Dynamin, and Cdc42/Rac GTPase-activating protein (RICH-1).17, 20–23
CIP4 has been implicated in regulating glucose metabolism,24 and allergic responses in mice.25 In neurons, CIP4 promotes formation of lamellipodia,26 and localizes to leading edge projections.18 In human breast cancer cells, CIP4 silencing led to impaired extracellular matrix degradation, cell motility and invasion.12 CIP4 also promotes chemotaxis of chronic lymphocytic leukemia cells.27 Other studies have implicated CIP4 in regulating osteosarcoma tumor progression,28 and renal cell carcinoma cell junctions.29
Here, CIP4 was studied in the context of EGFR-driven NSCLC. CIP4 expression was observed in normal human lung epithelial cells and at higher levels in NSCLC cell lines. Stable silencing of CIP4 resulted in defects in EGF-induced Erk activation, and reduced motility and invasiveness of H1299 cells. CIP4 silencing led to reduced expression and release of MMP-2, which likely contributes to the observed defects in cell invasion through ECM. In tumor xenograft assays, expression of the EMT-promoting transcription factor Zeb1 and its target gene MMP-2 were reduced in CIP4 KD primary tumors compared to control. This correlated with reduced numbers of distant metastases in mice with CIP4 KD tumors compared to control. We also profiled CIP4 expression in human NSCLC tumor tissue sections, and observed increased CIP4 expression compared to normal lung tissue. A survey of microarray studies also revealed that high CIP4 transcript levels are associated with worse overall survival in NSCLC patients.
RESULTS
CIP4 regulates EGFR signaling in lung adenocarcinoma cells
Since CIP4 was previously shown to regulate EGFR trafficking,11 and EGFR is a frequent driver of NSCLC,30 we surveyed CIP4 expression in a panel of cell lines derived from normal human lung epithelium or NSCLC tumors. Immunoblot (IB) assays revealed expression of CIP4 in an immortalized normal lung epithelial cell line (NL20), and at increased levels in the majority of lung adenocarcinoma cell lines (Figure 1a). In contrast, the levels of EGFR varied greatly likely due to gene copy number and/or mutation status (Figure 1a). We selected A549 and H1299 cells expressing wild-type EGFR for subsequent functional studies since they are effective models of EGFR-driven cell motility and tumor metastasis,31, 32 and express CIP4 at high levels. Stable knock-down (KD) of CIP4 expression was achieved using two separate shRNAs in H1299 and A549 cell lines using a lentiviral system.10 Compared to vector control cells (V), CIP4 levels were reduced by ~80% and ~70% in CIP4 KD1 and KD2 H1299 cell pools (Figure 1b). No defects in cell growth or survival were observed upon silencing of CIP4 (data not shown).
Figure 1.
CIP4 regulates EGFR trafficking in NSCLC cells. (a) CIP4 expression was analyzed in immortalized human lung epithelial NL20 cells and a panel of adenocarcinoma cell lines (A549, BH-E, H1299, H1975, HCC827, LC-T, H358, SW900, QuDB). Lysates (10 μg) were analyzed by IB with EGFR, CIP4 and β-actin antisera. (b) Stable silencing of CIP4 in H1299 cells was assessed in lysates from cells transduced with control vector (V) or CIP4 shRNA1 (KD1) or shRNA2 (KD2) lentiviruses by IB for CIP4 expression. CIP4 levels relative to β-actin were determined by densitometry and displayed below. (c) Surface biotinylation assays to assess surface levels of EGFR and efficiency of quenching with MESNA in H1299 V, KD1, and KD2 cells at 4 °C, as described in Materials and Methods. EGFR IB was performed following pulldown (PD) of biotinylated proteins using Streptavidin beads. Positions of molecular mass markers are shown on the left. Results are representative of several independent experiments.
Since CIP4 silencing in A431 epidermoid carcinoma cells resulted in defects in EGFR levels and trafficking,11 we investigated whether CIP4 KD H1299 cells had altered EGFR levels on the cell surface. Biotinylation of cell surface proteins was performed at 4 °C to prevent internalization, and biotinylated proteins were recovered by pull-down (PD) using streptavidin beads. Subsequent IB assays revealed that CIP4 KD cells had a slight increase in surface EGFR levels compared to control (Figure 1c). It is worth noting that similar defects were observed with both CIP4 shRNAs, suggesting these effects are due to loss of CIP4 and not off-target effects. As expected, addition of a quencher (MESNA) resulted in a near complete loss of biotinylated EGFR on the cell surface (Figure 1c). Surface EGFR levels were also elevated in CIP4 KD cells compared to control cells when analyzed by flow cytometry (data not shown). These results suggest that CIP4 regulates EGFR internalization or downregulation in NSCLC cells, as shown previously in other cell types.11
To test whether EGFR signaling was affected by CIP4 silencing in H1299 cells, a 20 min time course of EGF treatment was performed. As expected, EGF treatment of H1299 V and KD cells led to rapid phosphorylation of EGFR (pEGFR; Y1068) that was sustained throughout the time course (Figure 2a). CIP4 KD cells showed no overt defects in EGF-induced pEGFR levels compared to control cells, as measured by densitometry (Figure 2a). A longer time course of EGF treatment was performed to assess effects of CIP4 KD on EGFR signaling. EGF-induced phosphorylation of Akt (T308 and S473) was transient in both H1299 V and KD cells (Figure 2b). CIP4 KD cells consistently showed increased pAkt levels compared to control cells, and this correlated with increased phosphorylation of the Akt substrate mammalian target of rapamycin (mTOR) at S2448 (pmTOR; Figure 2b). In contrast, CIP4 silencing resulted in a less sustained phosphorylation of Erk (pErk; T202/Y204) at later times of EGF treatment (Figure 2b, graph depicts densitometry analyses from multiple experiments). Taken together, these results implicate CIP4 in modulating the kinetics of EGFR signaling to Erk and Akt/mTOR kinases in NSCLC cells.
Figure 2.
CIP4 modulates EGFR signaling in NSCLC cells. (a) Serum starved H1299 V and KD1 cells were treated with EGF (100 ng/ml) for 0–20 min. Lysates were subjected to IB with the antibodies indicated on the right. Densitometry was performed and relative phospho-EGFR (pY1068) levels are shown below. Positions of molecular mass markers are shown on the left. (b) Serum starved H1299 V and KD1 cells were treated with EGF (100 ng/ml) for 0–120 min. Lysates were subjected to IB with the antibodies indicated on the right. Densitometry was performed and relative phospho-Erk levels are shown in graph below (mean ± SEM; pooled data from 2 independent experiments; * indicates a significant difference between cell lines (P<0.05)). Positions of molecular mass markers are shown on the left.
CIP4 localizes to the leading edge and promotes NSCLC cell motility and invasion
EGF treatment of H1299 cells promotes lamellipodia formation and increased cell motility and invasion through ECM.31 Since CIP4 was shown to localize to lamellipodia and regulate their assembly in neurons,18 we examined CIP4 localization in H1299 cells by immunofluorescence (IF) staining and confocal microscopy. In addition to a punctate distribution in the cytoplasm, which likely reflects its localization to endosomes,11 we observed a pool of CIP4 at leading edge membrane protrusions marked by filamentous actin (F-actin; Supplementary Figure 1a). This pattern of CIP4 localization at the cell periphery was also observed in actively migrating H1299 cells, with considerable co-localization of CIP4 and N-WASP at leading edge membrane protrusions (Supplementary Figure 1b). However, it is worth noting that CIP4 puncta closest to the cell periphery showed little co-localization with N-WASP, and may correspond to interactions between the F-BAR domain and membrane phospholipids, as described in neurons.18 These results are consistent with CIP4 localization to the leading edge of motile NSCLC cells, and a potential role for CIP4 in promoting recruitment and activation of actin regulatory proteins.
To test whether CIP4 regulates NSCLC cell motility, we initially performed wound healing assays on CIP4 KD NSCLC cells treated with EGF. In comparison to the H1299 V cells which closed most of the wound area within 21 hours, KD cells displayed reduced wound closure (Figure 3a). CIP4 silencing in A549 cells also caused a significant defect in wound healing migration in response to EGF (Supplementary Figure 2). To directly visualize and compare H1299 V and KD cells during cell migration, H1299 V and KD cells were placed into opposite chambers of an Ibidi μ-dish, resulting in a uniform 500 μm gap between the cell lines (Figure 3c). Following removal of the chamber and any non-adherent cells, live cell imaging was performed for an 18 hour period by spinning disk confocal microscopy. While the majority of H1299 V cells migrated into the wound area, the response of KD cells was greatly impaired (Supplementary video 1; Figure 3d). Analysis of the migration distances of individual cells from multiple fields revealed a significant defect in KD cells compared to control (Figure 3e). Next, we compared the effects of CIP4 silencing on the invasive potential of H1299 and A549 cells using Matrigel™ coated transwell chambers. Interestingly, both H1299 KD and A549 KD cells showed severe defects in cell invasion compared to their respective controls (Figure 3f, Supplementary Figure 2d). To further investigate the role of CIP4 in cell invasive, H1299 V and KD cells were subjected to a spheroid cell invasion assay where the cells were grown as spheroids surrounded by ECM prior to inducing cell invasion with the addition of serum. Similar to the transwell assays, the H1299 KD cells displayed a dramatic reduction in the ability to invade into the ECM (Figure 3g, see Supplementary Figure 3 for representative images). Overall, these results identify CIP4 as a positive regulator of NSCLC cell motility and invasion through ECM.
Figure 3.
CIP4 promotes NSCLC cell motility and invasiveness. (a) EGF-induced wound closure assay of H1299 V, KD1 and KD2 cells. Starved cells were treated with EGF (50 ng/ml). Images were taken at time 0 hr and 21 hr post treatment. (b) Quantification of the area of the wound closures for H1299 V, KD1 and KD2 cells. (c) H1299 V and KD1 cells were seeded on opposing sides of a 35 mm μ-Dish (Ibidi) and grown to confluence prior to insert removal and live cell confocal imaging. (d) Representative images of live cell migration video taken at 0, 12, and 18 hours (hr) post insert removal and addition of EGF (50 ng/ml). H1299 V cells are shown on right side of wound, and KD1 cells on the left side. (e) Quantification of total distances migrated by individual cells. Cells that actively migrated were tracked and the distances migrated were calculated (N=40 cells for each V and KD1). (f) Quantification of invasion through Matrigel™ by H1299 V and KD1 cells. A modified Boyden chamber assay was used with Matrigel™ as the matrix barrier. Cells were induced to invade over 48 hr using complete medium with EGF (50 ng/ml) as the attractant. DAPI stained cells on the lower membrane surface were counted from 4 different fields per membrane. (g) Quantification of H1299 V and KD1 cell invasion in a spheroid invasion assay. Cells were aggregated into spheroids and then induced to invade the surrounding matrix for 7 days. The area of the cell mass was calculated and considered a measurement of cell invasion. All experiments were performed 3 times in triplicate (graphs depict mean ± SEM, * indicates significant differences between cell lines (P< 0.05)).
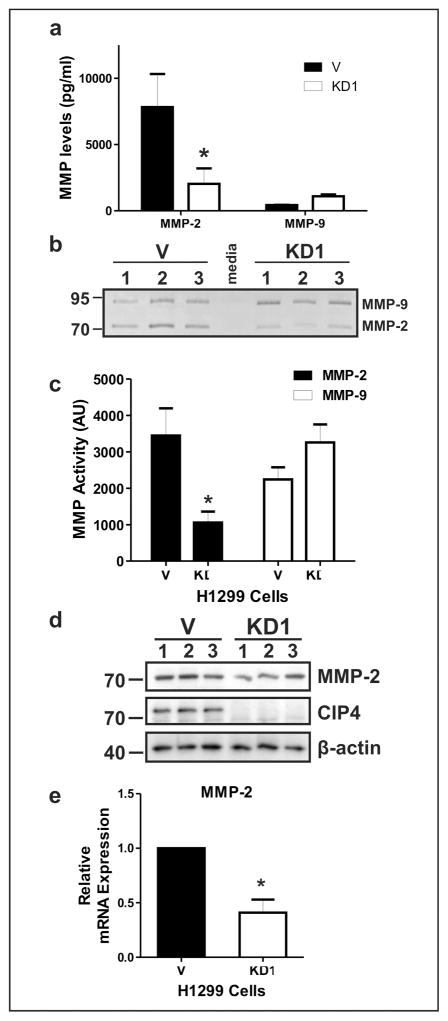
CIP4 promotes MMP-2 expression and activity in NSCLC
The effects of CIP4 silencing on cell invasion led us to investigate whether there were abnormal levels of MMP’s in the absence of CIP4. As an initial screen, conditioned media (CM) from H1299 V and KD cells were analyzed using a human MMP multiplex platform. This analysis revealed that the levels of secreted MMP-2 were greatly reduced in the CM from the KD cells whereas levels of MMP-9 were slightly elevated (Figure 4a). To test if reduced levels of secreted MMP-2 correlated with reduced enzymatic activity, the CM was analyzed using gelatin zymography. The most prominent gelatin digestion bands corresponded to the sizes of MMP-2 and MMP-9 for H1299 V CM, whereas MMP-2 activity was reduced in KD CM (Figure 4b). Quantification of these assays revealed a significant reduction in MMP-2 activity but not MMP-9 activity in CM from CIP4 KD cells (Figure 4c). To test whether CIP4 KD effects expression or secretion of MMP-2 in H1299 cells, immunoblots were performed on lysates from V and KD cells. In comparison to V control lysates, the levels of MMP-2 protein were reduced in KD cell lysates (Figure 4d). To further define how MMP-2 levels are regulated by CIP4 we measured MMP-2 transcript levels by qRT-PCR. Compared to control cells, CIP4 KD cells had a significant reduction in levels of MMP-2 transcripts (Figure 4e; GAPDH served as an input control). Also, the >60% reduction in MMP-2 transcripts likely explains the differences observed in MMP-2 protein levels and activity. Together, these data indicate that CIP4 promotes expression of MMP-2 in H1299 cells, and this pathway promotes NSCLC cell invasion.
Figure 4.
CIP4 promotes MMP-2 expression in NSCLC cells. (a) Concentrations of MMP-2 and MMP-9 proteins in CM from H1299 V and KD1 cells. Values were calculated by Eve Technologies using a human MMP multiplex assay. (b) Gelatin zymography of CM from H1299 V and KD1 cells. 10 μg protein per lane were analyzed. Positions of MMP-2 and MMP-9 activity are indicated on the right with size markers on the left. (c) Quantification of MMP-2 and MMP-9 activity by gelatin zymography. Bands of gelatin digestion for MMP-2 and MMP-9 from H1299 V and KD1 CMs were measured by densitometry. (d) IB analysis of MMP-2 in H1299 V and KD1 cell lysates (10 μg protein per lane). Positions of MMP-2, CIP4 and β-actin are shown on the right with size markers on the left. (e) Relative expression of MMP-2 transcripts in H1299 V and KD1 cells. Total RNA was isolated and analyzed by qRT-PCR using MMP-2 specific primers and normalized to GAPDH expression. All experiments were performed 3 times in triplicate (Graphs are shown as the mean ± SEM, * indicates significant differences between cell lines (P< 0.05)).
CIP4 promotes NSCLC metastasis in mice
To test whether CIP4 regulates NSCLC tumor progression or metastasis, H1299 V and KD subcutaneous tumors were established in the flank of Rag2−/−:IL-2Rγc−/− mice. At five weeks post injection, mice were sacrificed and tumors and several other organs were harvested. Since no overt changes in tumor size or average tumor mass was observed for CIP4 KD (Figure 5a/b), we tested whether CIP4 silencing was maintained in vivo. To do this, we analyzed primary tumor homogenates by IB, and observed a clear reduction in CIP4 levels in H1299 KD tumors compared to control (Figure 5c; β-actin served as a loading control). Next, we tested whether CIP4 KD affects MMP-2 expression during tumor progression in vivo, and observed significant reductions in MMP-2 protein and mRNA levels compared to control tumors (Figure 5c/d). Since MMP-2 expression is regulated by an EGFR/Erk/Zeb1 pathway that promotes epithelial-mesenchymal transition (EMT) in NSCLC, 33 we extended our study to Zeb1 expression. Like MMP-2, we observed a significant reduction in Zeb1 transcript levels in CIP4 KD tumors compared to control (Figure 5d). These results are consistent with CIP4 enhancing the EGFR/Erk/Zeb1/MMP-2 signaling axis in NSCLC tumors in mice. Further examination of various organs in these mice harboring subcutaneous tumors revealed GFP-positive micrometastases within the lungs (Figure 5e). Although CIP4 silencing did not eliminate lung metastases (Figure 5e), scoring of the numbers of metastases from H&E-stained lung tissue sections revealed a significant reduction in incidence compared to control mice (Figure 5e/f). To extend on these findings, an experimental metastasis model involving intrasplenic injections of H1299 V and KD cells was performed. Following splenectomy, the mice developed numerous metastases in liver and lung within 4 weeks (Figure 5g). To quantify the number of tumors, H&E staining liver and lung sections from each group were analyzed, and significantly fewer liver metastases were observed with CIP4 silencing (Figure 5h). A similar trend was observed in the lungs, and together these results provide novel evidence that CIP4 promotes NSCLC tumor metastasis.
Figure 5.
CIP4 promotes NSCLC tumor metastasis in mice. (a) Representative images of subcutaneous H1299 V and KD1 tumors 5 weeks after injection of Rag2−/−:IL-2Rγc−/− mice (scale bar is 2.5 mm). (b) Graph depicts primary tumor mass (mean ± SEM) for mice injected with H1299 V and KD1 cells (N=13 mice/group from 3 experiments). (c) CIP4 and MMP-2 expression in H1299 V and KD1 tumor homogenates was determined by IB analysis. Protein positions are indicated on the right. (d) Relative expression of MMP-2 and Zeb1 transcripts in H1299 V and KD1 tumors. Total RNA was isolated and analyzed by qRT-PCR using MMP-2 or Zeb1 specific primers and normalized to GAPDH expression. (e) Spontaneous metastases to lungs in mice harboring H1299 V and KD1 subcutaneous tumors were detected by GFP fluorescence in whole lung tissue (top), and scored from H&E stained lung sections (bottom; metastases are indicated by arrows; scale bar indicates 50 μm). (f) Graph depicts numbers of lung metastases (mean ± SEM) that were counted in a blinded fashion (N=13 mice/group from 3 experiments). (g) Experimental metastasis assays with H1299 V and KD1 cells injected in the spleen were performed and representative images of H&E stained sections are shown for liver (top) and lung (lower; scale bar indicates 50 μm). (h) Graph depicts number of liver and lung metastases after splenic injection of H1299 V and KD1 cells (mean ± SEM; N=9 mice/group from 3 experiments; * indicates significant differences between cell lines (P< 0.05)).
CIP4 is highly expressed in a subset of human NSCLC tumors and is associated with poor prognosis
Since CIP4 promoted NSCLC metastasis in mice, we investigated the expression of CIP4 in human NSCLC tumors and normal lung tissues. Initially, we surveyed CIP4 protein in homogenates prepared from 13 paired samples of NSCLC primary tumors (T) and adjacent normal (N) by IB, and observed increased CIP4 levels in most NSCLC tumors compared to normal tissues (Figure 6a). CIP4 levels were normalized to β-actin, and densitometry revealed a 2.5 ± 0.3 (mean ± SEM) fold increase in CIP4 in NSCLC tumors compared to normal lung. To gain more insights into CIP4 expression within human NSCLC tumors, we performed immunohistochemistry (IHC) staining of CIP4 after validating the specificity of CIP4 antisera by IB and IHC (Supplementary Figure 4; CIP4 Ab#2). Next, CIP4 IHC staining of human NSCLC tumor tissue microarrays was performed, and revealed low levels of CIP4 in normal lung epithelium, and increased expression in a subset of NSCLC tumors, including adenocarcinomas and squamous carcinomas (Figure 6b). Using the H-score algorithm to quantify CIP4 IHC staining, we observed >2-fold increase in CIP4 levels in tumors compared to normal tissues in 20/46 (43%) of adenocarcinomas and 11/34 (32%) of the squamous carcinomas (Figure 6c). These results indicate that CIP4 expression is weak in normal lung tissue, but in certain NSCLC cases becomes up-regulated during tumor progression. Interestingly, the web-based Kaplan-Meier Plotter analysis tool,34, 35 revealed that lung adenocarcinoma patients with high CIP4 transcript levels had worse overall survival (Figure 6d; HR=3.28, P <0.001). These data are consistent with our mouse studies that implicate CIP4 in promoting NSCLC metastasis, which is the leading cause of cancer deaths.
Figure 6.
CIP4 expression is elevated in NSCLC tumors and associated with poor overall survival. (a) CIP4 expression in human NSCLC tumors (T) and normal adjacent tissues (N). CIP4 was detected by IB analysis. The relative expression level of CIP4 for each T/N pair was determined by densitometry using β-actin as a loading control and presented as fold change. (b) Representative images of elevated CIP4 expression in NSCLC adenocarcinoma and squamous carcinoma compared to adjacent-to-cancer and normal lung tissues. CIP4 protein was detected by IHC analysis on 2 different lung cancer tissue microarrays. Scale bar indicates 20 μm. (c) H-score distribution for individual cases of NSCLC adenocarcinoma, squamous carcinoma, adjacent-to-cancer and normal tissues. (d) Kaplan-Meier plot comparing overall survival of lung adenocarcinoma patients with high or low level CIP4 expression was prepared as described in Materials and Methods.
DISCUSSION
This study identifies CIP4 as a positive regulator of EGFR-driven lung adenocarcinoma cell invasion and tumor metastasis in vivo. We observed less sustained activation of Erk kinase downstream of EGFR in NSCLC cells with CIP4 KD. Since Erk promotes Zeb1 expression, and upregulation of EMT genes including MMP-2,33, 36 we tested effects of CIP4 KD on this axis. Indeed, CIP4 silencing in NSCLC tumor xenograft assays led to reduced expression of Zeb1 and MMP-2 in primary tumors, and this correlated with reduced metastases in these mice. These results are consistent with defects in EGF-induced cell migration and invasion in CIP4 KD NSCLC cells in vitro. In human NSCLC tissues, CIP4 protein levels were high in a subset of lung adenocarcinomas and squamous carcinomas compared to normal lung tissues. Interestingly, high CIP4 transcript levels were significantly associated with reduced overall survival in patients with lung adenocarcinoma. Together, these results provide novel evidence that CIP4 promotes EGFR-driven NSCLC cell invasion, tumor metastasis, and that high CIP4 expression may be a biomarker linked to poor prognosis.
Several recent studies have functionally linked CIP4 to EGFR trafficking,11 and downstream signaling to pathways controlling cell motility and invasiveness.10, 12, 27, 28 These functions of CIP4 are likely due to its role as a hub for signaling to actin regulatory proteins at sites of membrane curvature.14 This includes coordinating actin polymerization surrounding membrane invaginations during clathrin-mediated endocytosis by CIP4 and related F-BAR proteins.17, 19, 37 In addition, recent studies identified CIP4 localization to membrane protrusions, including lamellipodia and invadopodia.10, 12, 26, 27 In cortical neurons, CIP4 localization to lamellipodia was dependent on its F-BAR domain and PI3K activity, while negatively regulated by Arp2/3-induced actin branching.18 It is likely that these pathways regulate CIP4 localization dynamics to lamellipodia in NSCLC cells. We also observed co-localization of CIP4 and N-WASP in puncta below the leading edge membranes in H1299 cells, which may correspond to its role as an activator of N-WASP.12, 27 Studies are ongoing to identify whether CIP4 regulates the dynamics of lamellipodia formation, as recently described in neurons and CLL,18, 27 and identify the relevant binding partners of CIP4 at the leading edge of NSCLC cells.
Here, we show that CIP4 silencing in NSCLC cell lines (H1299 and A549) causes defects in EGF-induced cell motility and invasion through ECM. This is a similar phenotype to a previous study of CIP4 function in MDA-MB-231 breast cancer cells.12 However, unlike MDA-MB-231 cells, our NSCLC cell models lacked detectable invadopodia and ability to focally degrade ECM (data not shown). In MDA-MB-231 cells expressing activated Src, the internalization of MT1-MMP occurs via a CIP4-dependent pathway regulated by phosphorylation of CIP4 by Src.10 Although H1299 cells lack MT1-MMP, they express high levels of MMP-2 and -9 and are highly invasive.38, 39 By profiling levels of secreted MMP’s in CIP4 KD NSCLC cells we identified a significant reduction in MMP-2 levels. This difference was also observed at the level of MMP-2 gene expression, which may be explained by less sustained Erk activation in CIP4 KD cells. It is worth noting that recent studies identify an EGFR/Erk pathway as critical for MMP-2 expression and the invasive phenotype of NSCLC cells.33, 36 Since MMP-2 is also part of the EMT signature, this could be consistent with recent studies linking CIP4 to promoting EMT in kidney epithelial cells and mesothelial cells.40, 41 Since E-cadherin can antagonize EGFR-driven EMT in NSCLC cells,33 and the Drosophila CIP4 ortholog is implicated in E-cadherin internalization,42 this may contribute to the defects we have observed in EGFR signaling and MMP-2 expression. Although H1299 cells do not express E-cadherin, we have observed defects in Zeb1 expression in CIP4 KD tumors. Since MMP-2 is a Zeb1 target gene, this likely explains the reduced levels of MMP-2 in CIP4 KD NSCLC cells and tumors. Thus, CIP4 is an emerging regulator of EMT in NSCLC that contributes to metastasis.
In our study, CIP4 was shown to enhance MMP-2 expression and the incidence of metastases in NSCLC tumor-bearing mice. These findings are highly consistent with CIP4 promoting NSCLC cell motility and invasion through ECM in vitro. This is the first report of CIP4 function in NSCLC tumor progression and metastasis, and potentially relevant to other EGFR-driven cancer models. Recently, CIP4 has been identified as a positive regulator of osteosarcoma tumor growth and lung metastasis.28 Another study suggests CIP4 may have opposing roles in cancers depending on differential methylation of the trip10/cip4 gene promoter.43 They found that ectopic expression of CIP4 in neuroblastoma cells enhanced tumor growth; while the opposite effects were observed in an ovarian cancer cell model.43 The contrasting functions of CIP4 in tumor progression may also reflect differences in driver mutations and signaling pathways associated with these cancer types. In our study, CIP4 promotes metastasis of EGFR-driven NSCLC tumors to the lungs in mice harboring subcutaneous tumors. Since CIP4 silencing also reduced the numbers of liver and lung metastases in an intrasplenic model, we believe that CIP4 functions in promoting extravasation and seeding efficiency of circulating tumor cells. It is interesting to note, that extravasation of T cells was also defective in CIP4 knock-out mice during contact hypersensitivity reactions.25 In addition, CIP4 also promotes Zeb1 and MMP-2 expression in primary tumors, likely contributing to early steps in the cancer metastasis process.
Our study also identified high CIP4 protein levels in a subset of human lung adenocarcinomas and squamous carcinoma tissue specimens. Since there was limited clinical data associated with these TMAs, we queried the microarray databases linked to lung adenocarcinoma patient outcomes. Interestingly, low levels of CIP4 was associated with >70% overall survival compared to high CIP4 levels and <20% overall survival probability. Similar analyses of CIP4 family members (Toca-1 and FBP17), or binding partners (Cdc42 and N-WASP), failed to show an association with overall survival (data not shown). It will be important to further test the potential association between CIP4 and MMP-2 levels with NSCLC clinical parameters and patient outcomes. Also, mechanisms controlling CIP4 expression in NSCLC tumors should be explored. Future studies aimed at disrupting CIP4-mediated signaling networks in these cancers may protect against metastasis and improve patient outcomes.
MATERIALS AND METHODS
Cell lines and antibodies
Normal lung epithelial cell line NL20 and NSCLC cell lines were previously described.44 NCI-H1299 cells (American Type Culture Collection, ATCC) were grown in DMEM (Sigma) supplemented with 10% FBS (PAA); A549 cells (kindly provided by Dr. Susan Cole, Queen’s University) were grown in RPMI (Sigma) supplemented with 5% FBS and 1% L-glutamine. CIP4 antibodies include rabbit anti-CIP4 Ab#1 used for immunoblotting,11 and rabbit anti-CIP4 Ab#2 used for immunohistochemistry (raised and affinity purified using the peptide QDTPIYTEFDEDFEE, Open Biosystems). Commercial antibodies included: CIP4, N-WASP, pS2448-mTOR, pS473-Akt, pT308-Akt and Akt1/2 were from Cell Signaling Technology; EGFR, pY1068-EGFR and PE-conjugated mouse anti-human EGF receptor were from BD Bioscience; ERK1, pERK, β-actin, MMP-2, α-actinin, and EGFR were from Santa Cruz Biotechnology.
Cell lysis and immunoblotting
NSCLC cell lines in growth media were lysed using NP-40 Lysis Buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM Na3VO4, 100 μM phenylmethylsulfonyl fluoride). For some experiments, cells were starved of serum and treated with EGF (100 ng/ml; Peprotech) prior to cell lysis. Immunoblot (IB) analysis was performed with antibodies: CIP4 (1:1000), EGFR (1:2500), pY1068-EGFR (1:1000), ERK1 (1:1000), pERK (1:1000), β-actin (1:1000), pS2448-mTOR (1:1000), α-actinin (1:1000), MMP-2 (1:1000), pS473-Akt (1:1000), pT308-Akt (1:1000) and Akt1/2 (1:1000). Horseradish peroxidase-conjugated sheep anti-mouse immunoglobulin (1:10,000; GE Healthcare) or goat anti-rabbit immunoglobulin (1:10,000; GE Healthcare) and enhanced chemiluminescence (ECL Western Blotting Substrate; Thermo Scientific) were used for detection.
Lentivirus production and stable CIP4 silencing
Stable CIP4 knock-down (KD) was achieved in H1299 and A549 cell lines using a pLKO.1-based lentiviral system (Open Biosystems), as described previously.10 Viral supernatants were titred on H1299 cells, and added to H1299 and A549 cells at a multiplicity of infection (MOI) of ~3:1 and selected using puromycin (2 μg/ml). For tumor studies, control and CIP4 KD cells were transduced with pWPXLd to provide a GFP marker.
Surface biotinylation assays
H1299 V, KD1 and KD2 cells were grown in 35 mm dishes to 80–90% confluency. Cells were starved O/N and then placed on ice for 20–30 min to inhibit receptor internalization and surface biotinylation assays performed as described previously.10
Cell migration and invasion assays
Scratch wound migration assays were performed on H1299 V and KD cells as previously described.10 For A549 cells, a 35 mm μ-Dish (Ibidi) was used with complete media containing EGF (50 ng/ml) for 9 hours. For live cell migration assays, H1299 V and KD1 cells were grown to confluence in either side of a 35 mm μ-Dish (Ibidi), serum starved overnight, and culture insert removed prior to imaging on a Quorum WaveFX-X1 spinning disc confocal microscope (Quorum Technologies Inc.). Images were captured every 15 minutes for 18 hours. Videos were created using MetaMorph Microscopy Automation & Image Analysis Software (Molecular Devices). The migration distance of individual cells was measured using Image J software with Manual Tracker and Click Forward add-ons. Transwell invasion assays were performed as previously described.10 A549 cells were treated with TGF-β1 (2 ng/ml) prior to these assays to enhance their invasiveness. After 48 hrs, DAPI-stained cells were imaged by epifluorescence microscopy (4 fields/insert) and scored using Image Pro Plus.
Spheroid cell invasion assay
The invasive ability of the H1299 V and KD cells was investigated using the Cultrex 96 Well 3-D Spheroid BME Cell Invasion Assay according to the manufacturer’s instructions (cat # 3500-096-K; Trevigen). Briefly, 3000 cells were resuspended in spheroid formation ECM solution and gently pelleted in a 96 well round bottom spheroid formation plate. After 3 days, spheroids were imaged. To induce invasion, the invasion matrix and medium were added to each well. Cells invaded the surrounding matrix for 7 days and images were captured. The area of each cell mass (pre- and post-invasion) was measured. The difference in the areas was considered to be a measurement of cell invasion.
Gelatin zymography and MMP array
Gelatin zymography was performed to determine MMP-2 and MMP-9 activity in conditioned media (0.5% FBS/DMEM). Samples were analyzed in SDS-PAGE gels containing 0.2% (w/v) gelatin as previously described.12 Conditioned media from H1299 V and KD cells were also analyzed using a Human MMP & TIMP Array Discovery Assay (Eve Technologies Corporation, Calgary AB, Canada).
RNA purification, cDNA synthesis and qRT-PCR
Total RNA was purified from cultured cells and tumor tissues using Trizol (Life technologies). First strand cDNA was synthesized using the iScript Select cDNA Synthesis Kit (Bio-Rad). Quantitative PCR was performed using iQ SYBR Green Supermix Kit (Bio-Rad). Primer sequences were: MMP-2 forward 5′-AGCTCCCGGAAAAGATTGATG-3′, MMP-2 reverse 5′-CAGGGTGCTGGCTGAGTAGAT-3′, Zeb1 forward 5′-GTAGAGGATCAGAATGACTCTG-3′, Zeb1 reverse 5′-CCAGAATGTAATCGCATGTGTTC-3′, GAPDH forward 5′-GCCTTCCGTGTCCCCACTGC-3′, GAPDH reverse 5′-CAATGCCAGCCCCAGCGTCA-3′.
Immunofluorescence
H1299 cells growing on glass coverslips coated with 10 μg/ml fibronectin (Roche) were fixed (4% PFA), permeabilized (0.2% Triton-X100), and incubated in blocking buffer (5% goat serum, 5% BSA) for 30 min. Primary antibodies (mouse α-CIP4, 1:100; rabbit α-N-WASP, 1:100) were detected with appropriate secondary antibodies (Invitrogen). Images were acquired by confocal microscopy.
Tumor xenograft assays
H1299 V and KD1 cells (106 cells/100 μl DMEM) were subcutaneously injected into the left flank of 6–8 week male Rag2−/−:IL-2Rγc−/− mice as previously described.31 For experimental metastasis assays, intrasplenic injections of Rag2−/−:IL-2Rγc−/− mice with H1299 V and KD1 cells (50,000 cells/50 μl DMEM) were performed on exteriorized spleens. After 3–5 minutes, the splenic vein and artery were ligated and the spleen was removed. After 4 weeks the mice were sacrificed and tissues analyzed as above. All experiments were approved by the Queen’s University Animal Care Committee in accordance with Canadian Council for Animal Care regulations.
Human lung tissue samples and immunohistochemistry
Tumor and adjacent normal lung tissues were acquired at initial diagnosis of 13 NSCLC patients (ages 48–75; stage II–III; Ontario Tumor Bank), and subjected to IB analysis. Human lung cancer tissue microarrays (LC1005, LC1501, US Biomax, Inc.) were stained using the Discovery XT Staining System (Ventana Medical Systems, Inc.). Antigens were retrieved with an EDTA pH 8.0 solution and incubated with rabbit anti-CIP4 Ab #2 (1:10). CIP4 immunohistochemistry (IHC) staining was visualized using DAB and a hematoxylin counterstain. Microarrays were scanned using the Aperio CS digital slide scanner (Queen’s Laboratory for Molecular Pathology) and analyzed with ImageScope software (Aperio). Tumor-specific H-scores were calculated based on positive pixel intensity according to the formula: (% weak positive X 1) + (% positive X 2) + (% strong positive X 3). For analysis of CIP4 transcripts in lung adenocarcinoma microarray studies, a Kaplan-Meier curve for overall survival was created using Kaplan-Meier Plotter (www.kmplot.com) with patients grouped according to high and low expression of Trip10. Hazard Ratio (with 95% confidence interval) and logrank P values were calculated and are displayed on the graph.
Statistical Analysis
All experiments were performed at least 3 times, and graphs depict mean±SEM. Paired Student’s t-test were performed to compare V and KD cell lines, with significant differences defined by P<0.05.
Supplementary Material
Acknowledgments
The authors thank Stephanie Everingham for technical assistance, Jeff Mewburn for help with time lapse confocal microscopy, and Lee Boudreau (Queen’s Laboratory for Molecular Pathology) for help optimizing IHC staining. Thanks also to Lois Mulligan, Peter Greer and Chris Nicol for sharing reagents and equipment. This research was supported by operating grants from Canadian Cancer Society Research Institute and Canadian Institutes for Health Research to AWBC (MOP119562).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
References
- 1.Rusch V, Klimstra D, Venkatraman E, Pisters PW, Langenfeld J, Dmitrovsky E. Overexpression of the epidermal growth factor receptor and its ligand transforming growth factor alpha is frequent in resectable non-small cell lung cancer but does not predict tumor progression. Clin Cancer Res. 1997;3:515–522. [PubMed] [Google Scholar]
- 2.Hirsch FR, Herbst RS, Olsen C, et al. Increased EGFR gene copy number detected by fluorescent in situ hybridization predicts outcome in non-small-cell lung cancer patients treated with cetuximab and chemotherapy. J Clin Oncol. 2008;26:3351–3357. doi: 10.1200/JCO.2007.14.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Zhao M, Pu J, Forrester JV, McCaig CD. Membrane lipids, EGF receptors, and intracellular signals colocalize and are polarized in epithelial cells moving directionally in a physiological electric field. FASEB J. 2002;16:857–859. doi: 10.1096/fj.01-0811fje. [DOI] [PubMed] [Google Scholar]
- 5.Rosell R, Moran T, Carcereny E, et al. Non-small-cell lung cancer harbouring mutations in the EGFR kinase domain. Clin Transl Oncol. 2010;12:75–80. doi: 10.1007/S12094-010-0473-0. [DOI] [PubMed] [Google Scholar]
- 6.Yang M, Shan B, Li Q, et al. Overcoming erlotinib resistance with tailored treatment regimen in patient-derived xenografts from naive Asian NSCLC patients. Intl J Cancer. 2013;132:E74–84. doi: 10.1002/ijc.27813. [DOI] [PubMed] [Google Scholar]
- 7.Downward J. Targeting RAS and PI3K in lung cancer. Nat Med. 2008;14:1315–1316. doi: 10.1038/nm1208-1315. [DOI] [PubMed] [Google Scholar]
- 8.Ishikawa D, Takeuchi S, Nakagawa T, et al. mTOR Inhibitors Control the Growth of EGFR Mutant Lung Cancer Even after Acquiring Resistance by HGF. PloS One. 2013;8:e62104. doi: 10.1371/journal.pone.0062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol. 2009;6:587–595. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- 10.Hu J, Mukhopadhyay A, Truesdell P, et al. Cdc42-interacting protein 4 is a Src substrate that regulates invadopodia and invasiveness of breast tumors by promoting MT1-MMP endocytosis. J Cell Sci. 2011;124:1739–1751. doi: 10.1242/jcs.078014. [DOI] [PubMed] [Google Scholar]
- 11.Hu J, Troglio F, Mukhopadhyay A, et al. F-BAR-containing adaptor CIP4 localizes to early endosomes and regulates Epidermal Growth Factor Receptor trafficking and downregulation. Cell Signal. 2009;21:1686–1697. doi: 10.1016/j.cellsig.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Pichot CS, Arvanitis C, Hartig SM, et al. Cdc42-interacting protein 4 promotes breast cancer cell invasion and formation of invadopodia through activation of N-WASp. Cancer Res. 2010;70:8347–8356. doi: 10.1158/0008-5472.CAN-09-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aspenstrom P. A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr Biol. 1997;7:479–487. doi: 10.1016/s0960-9822(06)00219-3. [DOI] [PubMed] [Google Scholar]
- 14.Aspenstrom P, Fransson A, Richnau N. Pombe Cdc15 homology proteins: regulators of membrane dynamics and the actin cytoskeleton. Trends Biochem Sci. 2006;31:670–679. doi: 10.1016/j.tibs.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Frost A, Perera R, Roux A, et al. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimada A, Niwa H, Tsujita K, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 17.Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Saengsawang W, Taylor KL, Lumbard DC, et al. CIP4 coordinates with phospholipids and actin-associated proteins to localize to the protruding edge and produce actin ribs and veils. J Cell Sci. 2013;126:2411–2423. doi: 10.1242/jcs.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol. 2006;172:269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aspenstrom P, Richnau N, Johansson AS. The diaphanous-related formin DAAM1 collaborates with the Rho GTPases RhoA and Cdc42, CIP4 and Src in regulating cell morphogenesis and actin dynamics. Exp Cell Res. 2006;312:2180–2194. doi: 10.1016/j.yexcr.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Richnau N, Aspenstrom P. Rich, a rho GTPase-activating protein domain-containing protein involved in signaling by Cdc42 and Rac1. J Biol Chem. 2001;276:35060–35070. doi: 10.1074/jbc.M103540200. [DOI] [PubMed] [Google Scholar]
- 22.Takano K, Toyooka K, Suetsugu S. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J. 2008;27:2817–2828. doi: 10.1038/emboj.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian L, Nelson DL, Stewart DM. Cdc42-interacting protein 4 mediates binding of the Wiskott-Aldrich syndrome protein to microtubules. J Biol Chem. 2000;275:7854–7861. doi: 10.1074/jbc.275.11.7854. [DOI] [PubMed] [Google Scholar]
- 24.Feng Y, Hartig SM, Bechill JE, Blanchard EG, Caudell E, Corey SJ. The Cdc42-interacting protein-4 (CIP4) gene knock-out mouse reveals delayed and decreased endocytosis. J Biol Chem. 2010;285:4348–4354. doi: 10.1074/jbc.M109.041038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koduru S, Kumar L, Massaad MJ, et al. Cdc42 interacting protein 4 (CIP4) is essential for integrin-dependent T-cell trafficking. Proc Natl Acad Sci USA. 2010;107:16252–16256. doi: 10.1073/pnas.1002747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saengsawang W, Mitok K, Viesselmann C, et al. The F-BAR protein CIP4 inhibits neurite formation by producing lamellipodial protrusions. Curr Biol. 2012;22:494–501. doi: 10.1016/j.cub.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malet-Engra G, Viaud J, Ysebaert L, et al. CIP4 Controls CCL19-Driven Cell Steering and Chemotaxis in Chronic Lymphocytic Leukemia. Cancer Res. 2013;73:3412–3424. doi: 10.1158/0008-5472.CAN-12-3564. [DOI] [PubMed] [Google Scholar]
- 28.Koshkina NV, Yang G, Kleinerman ES. Inhibition of Cdc42-interacting protein 4 (CIP4) impairs osteosarcoma tumor progression. Curr Cancer Drug Targets. 2013;13:48–56. [PubMed] [Google Scholar]
- 29.Tsuji E, Tsuji Y, Fujiwara T, Ogata S, Tsukamoto K, Saku K. Splicing variant of Cdc42 interacting protein-4 disrupts beta-catenin-mediated cell-cell adhesion: expression and function in renal cell carcinoma. Biochem Biophys Res Comm. 2006;339:1083–1088. doi: 10.1016/j.bbrc.2005.11.117. [DOI] [PubMed] [Google Scholar]
- 30.Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J. 2010;277:301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 31.Ahn J, Truesdell P, Meens J, et al. Fer Protein-Tyrosine Kinase Promotes Lung Adenocarcinoma Cell Invasion and Tumor Metastasis. Mol Cancer Res. 2013;11:952–963. doi: 10.1158/1541-7786.MCR-13-0003-T. [DOI] [PubMed] [Google Scholar]
- 32.Huang MS, Wang TJ, Liang CL, et al. Establishment of fluorescent lung carcinoma metastasis model and its real-time microscopic detection in SCID mice. Clin Exp Met. 2002;19:359–368. doi: 10.1023/a:1015562532564. [DOI] [PubMed] [Google Scholar]
- 33.Bae GY, Choi SJ, Lee JS, et al. Loss of E-cadherin activates EGFR-MEK/ERK signaling, which promotes invasion via the ZEB1/MMP2 axis in non-small cell lung cancer. Oncotarget. 2013;4:2512–2522. doi: 10.18632/oncotarget.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 36.Dong QZ, Wang Y, Tang ZP, et al. Derlin-1 is overexpressed in non-small cell lung cancer and promotes cancer cell invasion via EGFR-ERK-mediated up-regulation of MMP-2 and MMP-9. Am J Pathol. 2013;182:954–964. doi: 10.1016/j.ajpath.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 37.Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kao SJ, Su JL, Chen CK, et al. Osthole inhibits the invasive ability of human lung adenocarcinoma cells via suppression of NF-kappaB-mediated matrix metalloproteinase-9 expression. Toxicol Applied Pharmacol. 2012;261:105–115. doi: 10.1016/j.taap.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, Dong Q, Wang Y, Qu L, Qiu X, Wang E. Downregulation of Mig-6 in nonsmall-cell lung cancer is associated with EGFR signaling. Mol Carcinogen. 2012;51:522–534. doi: 10.1002/mc.20815. [DOI] [PubMed] [Google Scholar]
- 40.Bai S, Zeng R, Zhou Q, et al. Cdc42-interacting protein-4 promotes TGF-Beta1-induced epithelial-mesenchymal transition and extracellular matrix deposition in renal proximal tubular epithelial cells. Intl J Biol Sci. 2012;8:859–869. doi: 10.7150/ijbs.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Bi M, Zhong F, Jiao X, Zhang D, Dong Q. Role of CIP4 in high glucose induced epithelial--mesenchymal transition of rat peritoneal mesothelial cells. Ren Fail. 2013;35:989–995. doi: 10.3109/0886022X.2013.808957. [DOI] [PubMed] [Google Scholar]
- 42.Leibfried A, Fricke R, Morgan MJ, Bogdan S, Bellaiche Y. Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Curr Biol. 2008;18:1639–1648. doi: 10.1016/j.cub.2008.09.063. [DOI] [PubMed] [Google Scholar]
- 43.Hsu CC, Leu YW, Tseng MJ, et al. Functional characterization of Trip10 in cancer cell growth and survival. J Biomed Sci. 2011;18:12. doi: 10.1186/1423-0127-18-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Z, Hao Y, Liu N, Raptis L, Tsao MS, Yang X. TAZ is a novel oncogene in non-small cell lung cancer. Oncogene. 2011;30:2181–2186. doi: 10.1038/onc.2010.606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.