Abstract
The progression of cancers from primary tumors to invasive and metastatic stages accounts for the overwhelming majority of cancer deaths. Understanding the molecular events which promote metastasis is thus critical in the clinic. Translational control is emerging as an important factor in tumorigenesis. The mRNA cap-binding protein eIF4E is an oncoprotein that plays an important role in cancer initiation and progression. eIF4E must be phosphorylated to promote tumor development. However, the role of eIF4E phosphorylation in metastasis is not known. Here, we show that mice in which eIF4E cannot be phosphorylated are resistant to lung metastases in a mammary tumor model, and that cells isolated from these mice exhibit impaired invasion. We also demonstrate that TGFβ induces eIF4E phosphorylation to promote translation of Snail and Mmp-3 mRNAs, and the induction of epithelial-to-mesenchymal transition (EMT). Furthermore, we describe a new model wherein EMT induced by TGFβ requires translational activation via the non-canonical TGFβ signaling branch acting through eIF4E phosphorylation.
Keywords: Cancer, eIF4E, epithelial-to-mesenchymal transition, metastasis, TGFβ
INTRODUCTION
The eukaryotic translation initiation factor 4E (eIF4E) binds to the mRNA 5′ cap structure (m7GpppN, where N is any nucleotide, and m is a methyl group), and facilitates mRNA recruitment to the ribosome. eIF4E is a subunit of the eIF4F complex, which also contains eIF4A, an RNA helicase and eIF4G, a scaffolding protein, which bridges the mRNA and the ribosome (1). In most circumstances, the formation of the eIF4F complex is rate-limiting and is dependent on eIF4E availability, as it is the least abundant of all the initiation factors (2, 3). eIF4E availability is controlled by its inhibitors, the eIF4E binding proteins (4E-BPs) (4, 5), and its activity is also regulated by phosphorylation on serine 209 by the Map Kinase Integrating Kinases (MNK1/2), downstream of both the MEK/ERK and p38 Map Kinase pathways (6–8). Phosphorylation of eIF4E requires prior docking of MNKs to the scaffolding protein eIF4G (9), indicating that eIF4E phosphorylation occurs after the formation of the eIF4F complex.
eIF4E promotes neoplastic transformation and tumorigenesis in numerous cell-based and animal models (10–12). Consistent with this activity, eIF4E is overexpressed in a wide variety of human cancers (reviewed in (13)). Consequently, there is currently much interest in its potential as a therapeutic target (14–16). The cancer promoting activity of eIF4E is dependent on its phosphorylation, as overexpression of a phosphorylation-deficient mutant (eIF4ES209A) fails to promote tumorigenicity (12, 17). Moreover, inhibiting MNKs impairs xenograft growth, and mice lacking MNKs are resistant to cancer in the Lck-Pten lymphoma model (18, 19). eIF4E promotes metastasis in various mouse models (20, 21), and is associated with poor prognosis in several types of cancer (22, 23). We have previously shown that mice bearing the eIF4ES209A mutation are resistant to prostate cancer development and that mouse embryonic fibroblasts (MEFs) isolated from these mice are resistant to neoplastic transformation (24). Interestingly, eIF4ES209A MEFs displayed impaired anchorage independent growth, consistent with a reduced metastatic potential (25). Furthermore, certain pro-metastatic factors were identified in this study as being controlled by phospho-eIF4E, notably the matrix metalloproteinases MMP-3 and MMP-9 (24). MMPs cleave several component proteins of the extracellular matrix (ECM) to promote migration and invasion (26) and induce EMT (27). These observations point to a role for eIF4E phosphorylation in metastasis, but experimental evidence for this link is lacking.
Here, we investigate the role of eIF4E phosphorylation in metastasis. We demonstrate that phosphorylation of eIF4E promotes invasion in transformed MEFs, as well as TGFβ-induced EMT in normal epithelial cells. We identify mRNAs regulated by eIF4E phosphorylation, which can mediate its pro-metastatic effects, including Snail and Mmp-3. Importantly, we validate our findings in vivo using a metastatic mouse mammary tumor model. Taken together, these results demonstrate that eIF4E phosphorylation is a key event in the metastatic process.
RESULTS
eIF4E phosphorylation promotes migration and invasion
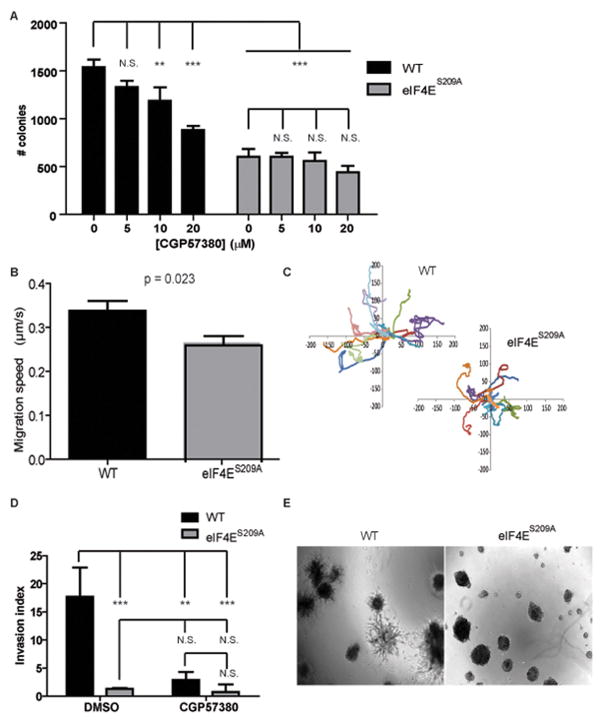
To explore whether eIF4E phosphorylation plays a role in metastasis, we utilized a cell model that we had previously developed (24). MEFs were derived from mice bearing the eIF4ES209A mutant and their WT counterparts, and transformed with c-MYC and H-RASV12. These WT and eIF4ES209A transformed MEFs display similar proliferation, cell cycle progression and levels of apoptosis, yet eIF4ES209A MEFs possess reduced tumorigenic potential (24). We chose to use this model as it is not dependent on overexpression, and avoids targeting the MNKs, which phosphorylate additional proteins such as Sprouty2 (28), cPLA2 (29) and hnRNPA1 (30, 31). To investigate the role of phospho-eIF4E in metastasis, we studied WT and eIF4ES209A transformed MEFs in assays that measure metastatic potential: anchorage independent growth (25), random migration (32), transwell invasion (33) and colony outgrowth in matrigel (34). As previously reported (24), eIF4ES209A MEFs formed 2 fold fewer colonies when plated in agarose (Fig. 1A). This effect was specific to eIF4E phosphorylation as treatment with the MNK inhibitor CGP57380 reduced colony formation in the WT MEFs in a dose-dependent manner, whereas eIF4ES209A MEFs remained insensitive to this treatment (Fig. 1A). Phosphorylation of eIF4E also promotes migration, as eIF4ES209A MEFs displayed ~20% reduction in random migration speeds, as seen by time-lapse microscopy (Fig. 1B). Accordingly, these cells traveled shorter distances than their WT counterparts (Fig. 1C). Strikingly, invasion was severely impaired in eIF4ES209A MEFs, as their invasion index was reduced by 5 fold in a transwell invasion assay (Fig. 1D). Corroborating these findings, in a matrigel colony outgrowth assay, colonies of WT MEFs exhibited a branched morphology, indicative of their invasion into the basement membrane matrix, while eIF4ES209A colonies remained spherical (Fig. 1E). Thus, eIF4E phosphorylation promotes in vitro characteristics that correlate with metastatic potential, prompting further investigation into its role in metastasis.
Figure 1. eIF4E phosphorylation enhances in vitro metastatic properties.
(A), Anchorage independent growth: WT and eIF4ES209A MYC/RAS-transformed MEFs were plated in agarose and incubated for 2 weeks with the indicated concentration of CGP57380, and colonies of 8 or more cells were counted. (B–C), Random migration as monitored by time-lapse microscopy: migration speed (B) and representative migration paths (C) were determined using the Metamorph software. (D) Transwell invasion: invasion index is given as the percentage of cells having crossed the porous membrane of a Boyden chamber in the presence of a layer of matrigel versus in its absence. (E) Colony outgrowth: representative images of colony morphology for cells seeded in matrigel and incubated for 8 days. Error bars represent standard deviations (A, D) or standard errors (B). All results are representative of at least 3 independent experiments. Statistical significance determined by one-way ANOVA followed by Bonferroni’s Multiple Comparison Test (A, D) or Student T-test (B); * indicates p<0.05; ** indicates p<0.01; *** indicates p<0.001; N.S. = non-significant.
Phosphorylated eIF4E promotes the translation of metastasis-related mRNAs
To study the mechanism by which eIF4E phosphorylation favors pro-metastatic characteristics, the expression levels of the Y-box-binding protein 1 (YB1) were assessed in WT and eIF4ES209A MEFs. This protein had previously been identified as playing an important role in eIF4E-mediated invasion (35). However, YB1 levels did not vary between the two cell lines (Fig. S1), suggesting that this protein is not responsible for the invasion differences in this model. To identify candidate factors that promote the pro-metastatic phenotype of WT MEFs, mRNAs translationally regulated by eIF4E phosphorylation were analyzed. To achieve this, pathway analysis was performed on a previously generated dataset of mRNAs translationally regulated by eIF4E phosphorylation (24). Pathways significantly increased in WT cells were identified and clustered using the DAVID functional annotation database (36, 37). Enriched clusters included potentially metastasis-related functions such as chemotaxis, proteases and immune signaling, as well as a general cancer cluster and one corresponding to glutathione metabolism (Fig. 2A). Clusters corresponding to plasma membrane proteins, carbohydrate-binding proteins, leucine-rich repeat-containing proteins and zinc-binding proteins were also identified. The full list of clusters generated by DAVID and the mRNAs grouped in each cluster are presented in supplemental material (Fig. S2, S3). To validate these findings and further investigate pro-metastatic mRNAs regulated by eIF4E phosphorylation, polysome profile analysis was performed on transformed WT and eIF4ES209A MEFs serum-stimulated for 2h. qRT-PCR was used to monitor the distribution across a sucrose density gradient of mRNAs chosen for their relevance to invasion and metastasis, or because they were identified in the clustering analysis. There was no significant difference in global translation between WT and eIF4ES209A MEFs, as the polysome profiles were nearly identical (Fig. 2B). Accordingly, the distributions of housekeeping mRNAs β-actin and GAPDH across the sucrose density gradient were similar (Fig. 2C, D). In contrast, the mRNA encoding MMP-3, which had been previously identified as being sensitive to eIF4E phosphorylation (24), was significantly shifted toward light polysomes in eIF4ES209A MEFs (Fig. 2E). This is indicative of regulation of translation initiation by eIF4E phosphorylation. Similarly, the polysomal distribution of Snail mRNA, an important transcription factor in the induction of EMT (38), was also significantly shifted toward light polysomes in eIF4ES209A MEFs (Fig. 2F). However, no differences were detected for other factors tested including the EMT marker vimentin, the EMT-related transcription factor TWIST, the metalloproteinase MMP-14 and the vascular endothelial growth factor member VEGFC (Fig. S4). Our results indicate that eIF4E phosphorylation promotes the translation of mRNAs encoding regulators of EMT and invasion such as SNAIL and MMP-3.
Figure 2. eIF4E phosphorylation promotes the translation of mRNAs involved in EMT/invasion and metastasis.
(A), DAVID clustering of significantly enriched pathways from a previously generated list of phospho-eIF4E translationally-regulated mRNAs (24). Metastasis-related terms are in red boxes. (B), Polysome profile and qRT-PCR analysis of (C), β-actin, (D), GAPDH, (E), Mmp-3 and (F), Snail mRNA in polysomal fractions of Myc/Ras-transformed WT and eIF4ES209A MEFs. Levels of mRNA in each fraction are given as a percentage of the total. Error bars represent standard deviations. Results are representative of at least 3 experiments.
In keeping with the translational repression of Snail and Mmp-3 in eIF4ES209A MEFs, the expression of SNAIL and MMP-3 protein was also repressed in these cells (Fig S1 and Fig. 3A). Next, it was pertinent to demonstrate that SNAIL and MMP-3 are responsible for phospho-eIF4E’s pro-metastatic properties. To this end, we restored SNAIL and MMP-3 levels in eIF4ES209A MEFs by overexpressing eIF4EWT or by overexpressing either Snail or Mmp-3 mRNAs lacking their regulatory 5′ and 3′UTRs (Fig. 3A). With either strategy, restoration of SNAIL and MMP-3 levels rescued invasion, whereas overexpression of eIF4ES209A failed to do so (Fig. 3B). These experiments demonstrate that SNAIL and MMP-3 mediate the pro-invasive properties of phosphorylated eIF4E.
Figure 3. Overexpression of eIF4E or phospho-eIF4E targets rescues invasion.
(A) Western blot analysis and (B) transwell invasion of Myc/Ras-transformed WT and eIF4ES209A MEFs overexpressing the indicated proteins relative to WT. Error bars represent standard deviations. Results are representative of at least 3 experiments. Statistical significance determined by one-way ANOVA followed by Bonferroni’s Multiple Comparison Test; p values are given relative to eIF4ES209A cells + vector; * indicates p<0.05; N.S. = non-significant.
eIF4E is phosphorylated during TGFβ-induced EMT
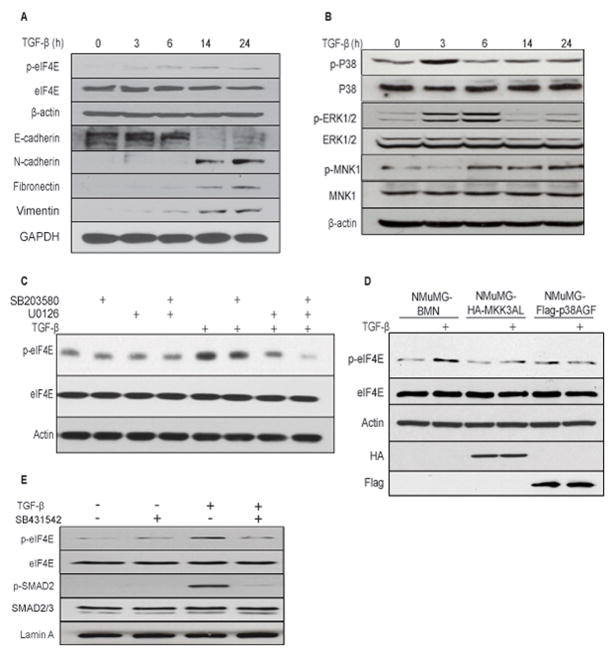
SNAIL and MMP-3 are important factors promoting cell invasiveness, metastasis and EMT (27, 38), therefore we investigated whether eIF4E phosphorylation could play a role in EMT. Since transformed WT and eIF4ES209A MEFs are inadequate for studying EMT, we employed the well-studied NMuMG cell model of TGFβ-induced EMT (39). Treatment with TGFβ engendered a strong increase in eIF4E phosphorylation in a time dependent manner (Fig. 4A). Increased phospho-eIF4E levels were accompanied by an increase in the mesenchymal markers N-cadherin, fibronectin and vimentin, as well as a decrease in the epithelial marker, E-cadherin (for a review on EMT markers, see (40)). Ostensibly, TGFβ controls the phosphorylation of eIF4E via non-canonical signaling leading to p38 and MAPK activation (41). Indeed, the phosphorylation and activation of MNK1 was preceded by that of its upstream kinases, p38 and ERK (Fig. 4B). In addition, chemical inhibition of ERK or p38 activation, using U0126 or SB203580, respectively, reduced TGFβ-stimulated phosphorylation of eIF4E (Fig. 4C), which was abrogated by a combination of both inhibitors (Fig. 4C). NMuMG cells engineered to express a dominant-negative mutant of p38 (p38AGF) or MKK3 (MKK3AL) are defective in undergoing a TGFβ-induced EMT (42). TGFβ failed to stimulate phosphorylation of eIF4E when p38 activation was blocked in p38AGF- and MKK3AL-NMuMG cell lines, consistent with non-canonical TGF-beta signaling impinging on eIF4E (Fig. 4D). TGFβ-induced eIF4E phosphorylation was dependent on the TGFβ receptor kinase activity because SB431542, an inhibitor of the TGFβ receptor kinase (43), blocked eIF4E phosphorylation (Fig. 4E). Taken together, these results demonstrate that activation of TGFβ signaling, which is frequently dysregulated in breast cancer (44), induces eIF4E phosphorylation, and that elevated phospho-eIF4E correlates with TGFβ-induced EMT.
Figure 4. eIF4E is phosphorylated downstream of MAPK signaling during TGFβ-induced EMT.
Western blot analysis of (A) EMT markers and (B) MAPK signaling in a time-course experiment of NMuMG cells treated with 5ng/mL TGFβ. Western blot analysis of the effect of ERK and p38 MAPK inhibitors (C), expression of dominant-negative MAPK signal transducers (D), and TGFβ receptor inhibition (E) on TGFβ-induced eIF4E phosphorylation in NMuMG cells. Results are representative of at least independent experiments.
eIF4E phosphorylation promotes TGFβ-induced EMT
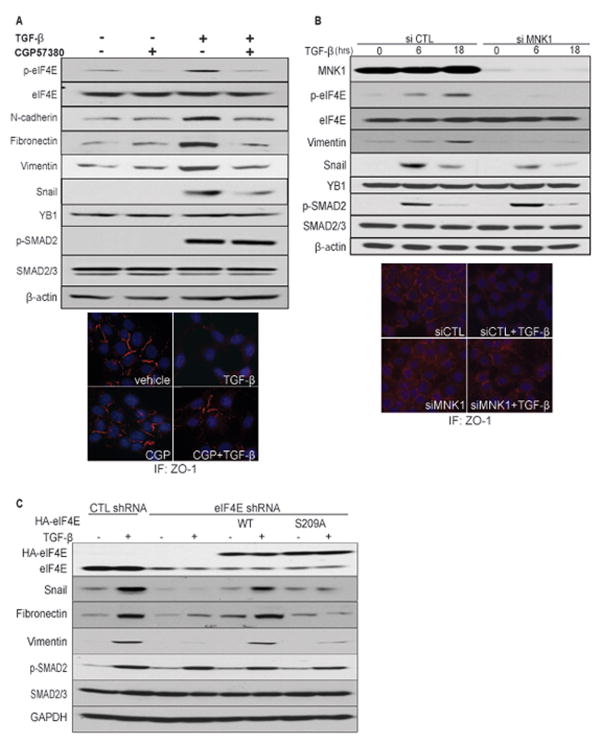
To determine whether eIF4E phosphorylation is a driver or a side-effect of EMT induction, we used a chemical inhibitor of the MNKs, CGP57380 (45). Treatment with this inhibitor blocked TGFβ-stimulated eIF4E phosphorylation and SNAIL expression (Fig. 5A), without affecting Snail mRNA levels (Fig. S5). Importantly, CGP57380 abrogated the molecular and morphological changes associated with EMT. Specifically, this inhibitor blocked the upregulation of the mesenchymal markers vimentin, fibronectin and N-cadherin by TGFβ (Fig. 5A, upper panel), and favoured the maintenance of tight junctions, as seen by ZO-1 expression, characteristic of an epithelial phenotype (Fig. 5A, lower panel and Fig. S6). The effect of pharmacologically targeting MNK activity on TGFβ-stimulated EMT was not due to inhibition of the Smad2 transcriptional pathway, since TGFβ treatment resulted in comparable phosphorylation of Smad2 in the presence or absence CGP57380 (Fig. 5A). To provide further evidence for the role of eIF4E phosphorylation in EMT, we knocked down MNK1 by siRNA. MNK1 was targeted because it is the isoform that responds to MAPK signaling, while MNK2 is relatively insensitive to upstream signaling and maintains a basal level of activation (46). Knock down of MNK1 prevented TGFβ-stimulated eIF4E phosphorylation as well as the TGFβ-induced expression of SNAIL and vimentin (Fig. 5B, upper panel) and maintained the expression of ZO-1 (Fig. 5B, lower panel and Fig. S6), further demonstrating that MNK1-mediated eIF4E phosphorylation promotes EMT. As seen with the MNK inhibitor treatment, the effects of MNK1 knock down were not due to changes in SMAD2 activation (Fig. 5B), and Snail mRNA levels were unaffected (Fig. S5). Consistent with experiments performed using MEFs, YB1 levels were unaffected by eIF4E phosphorylation in NMuMG cells (Fig. 5A, B).
Figure 5. eIF4E phosphorylation is required for TGFβ-induced EMT.
(A) Western blot analysis (upper panel) and immunofluorescence (lower panel) of EMT markers in NMuMG cells treated with 5ng/mL TGFβ, with or without 20μM CGP57380. (B) Western blot analysis (upper panel) and immunofluorescence (lower panel) of EMT markers in NMuMG cells treated with 5ng/mL TGFβ in the presence of scrambled siRNA or siMNK1. (C) Western blot analysis of EMT markers in MCF10A cells expressing the indicated shRNAs and eIF4E variants. Statistical significance was determined by one-way ANOVA followed by the Newman–Keuls post-hoc test. *** p<0.001; n.s. = non-significant.
Considering that the MNKs have targets other than eIF4E, notably Sprouty2 (28), cPLA2 (29) and hnRNPA1 (30), we performed a knockdown/add-back experiment to investigate the impact of phospho-eIF4E on EMT in a MNK-independent model. We stably expressed either eIF4EWT or eIF4ES209A in MCF10A cells, followed by shRNA silencing of the endogenous eIF4E. The shRNA was specific to the human eIF4E and therefore did not inhibit expression of the exogenous murine eIF4E (Fig. 5C). Knockdown of eIF4E impaired SNAIL expression and blocked TGFβ-mediated EMT, as indicated by reduced induction of vimentin and fibronectin (Fig. 5C). Strikingly, when endogenous eIF4E was knocked down, only eIF4EWT but not eIF4ES209A rescued SNAIL, vimentin and fibronectin expression (Fig. 5C). Importantly, only cells expressing eIF4EWT, either the endogenous or exogenous form, expressed appreciable levels of SNAIL, vimentin and fibronectin (Fig. 5C). In contrast, cells expressing the S209A mutant form of eIF4E had low amounts of the latter EMT markers, similarly to cells expressing low levels of eIF4E (Fig. 5C). The effects we observe in the MCF10A model system indicate that defects in the MNK/eIF4E axis do not impinge upon canonical TGFβ signaling, since TGFβ-induced phosphorylation of Smad2 remained unchanged under all experimental conditions (Fig. 5C). Thus, these experiments demonstrate a critical role for translational control by phosphorylated eIF4E in TGFβ-induced EMT.
eIF4E phosphorylation correlates with EMT and invasion in vivo
We validated our findings in vivo in a widely used model to study EMT and metastasis. This model consists of a series of isogenic mammary cancer cell lines (67NR, 168FARN, and 4T07) that are progressively more aggressive and present an increasingly mesenchymal phenotype (47). Specifically, when inoculated into the mouse mammary fat pad, the 67NR cell line forms only non-invasive primary tumors, 168FARN cells are locally invasive and detectable in the lymph node, while the 4T07 cells metastasize to the lung. In mammary tumors formed by these cell lines, increased MNK1 activation and eIF4E phosphorylation coincided with the acquisition of invasive properties and with the expression of SNAIL, as this protein was low in 67NR tumors, but increased in locally invasive 168FARN and metastatic 4T07 tumors (Fig. S7). The correlation between SNAIL expression, eIF4E phosphorylation and the acquisition of invasive properties of these cells, in vivo, provides further evidence for the importance of eIF4E phosphorylation in the metastatic process.
eIF4E phosphorylation promotes tumor onset and metastasis in PyMT mammary tumor model
To assess the role of eIF4E phosphorylation in metastatic progression using a genetic model, eIF4ES209A mice were crossed with MMTV-PyMT mice. In this mammary tumor model, tumors metastasize to the lungs with 100% penetrance (48). eIF4ES209A-PyMT mice exhibited a significant delay in tumor onset, developing palpable tumors on average 2 weeks later than their WT counterparts (Fig. 6A), confirming the importance of eIF4E phosphorylation in tumorigenesis (24). The penetrance of mammary tumor development was also reduced in eIF4ES209A-PyMT mice, as approximately 10% percent remained tumor-free. Nonetheless, once established, both WT and eIF4ES209A tumors grew at similar rates (Fig. 6B). End-point tumors displayed similar proliferation, as evaluated by ki67 staining of tumor sections (Fig. 6C), and similar apoptosis, as measured by caspase 3 activation (Fig. 6D). Importantly, the metastatic potential of eIF4ES209A tumors was reduced, as eIF4ES209A-PyMT mice displayed a two-fold reduction in lung metastases as compared to WT-PyMT mice (Fig. 6E, F). Metastatic burden was assessed at equivalent tumor burden rather than at a defined age; it therefore does not reflect the late onset of eIF4ES209A tumors. Thus, eIF4E phosphorylation promotes the metastatic properties of eIF4E, in addition to its tumorigenic activity.
Figure 6. eIF4ES209A mice are resistant to mammary tumor development and metastasis.
(A) Onset and (B) growth of mammary tumors in WT and eIF4ES209A mice expressing the PyMT transgene. Quantification of (C) Ki67 and (D) cleaved caspase 3 immunohistochemical staining in tumor sections from WT and eIF4ES209A mice. (E) Metastasis count and (F) representative lung images of WT and eIF4ES209A mice at experimental end point. Arrows point to lung metastases. Statistical significance determined by Student T-test; p-values are indicated, N.S. = non-significant. The number of mice used is indicated in the figure.
Based on our findings that eIF4E phosphorylation promotes the translation of mRNAs encoding SNAIL and MMP-3, we analyzed the expression of these factors by IHC in tumor samples from PyMT-WT and PyMT-eIF4ES209A mice. Phosphorylation of eIF4E was elevated in WT tumors, as expected due to activation of the MAPK pathway by the PyMT oncogene (49), but remained undetectable in eIF4ES209A tumors (Fig. 7A). SNAIL and MMP-3 exhibited localized expression patterns in both WT and eIF4ES209A tumors (Fig. 7A). Indeed, as previously reported, these proteins were mainly expressed at tumor/stroma boundaries (50, 51). Importantly, WT tumors displayed more extensive areas of SNAIL and MMP-3 expression (Fig. 7A), resulting in a higher percentage of SNAIL and MMP-3 positive cells (Fig. 7B,C). Taken together, these data support an important role for the phosphorylation of eIF4E in promoting invasion and metastasis through translational upregulation of its targets such as SNAIL and MMP-3.
Figure 7. eIF4E phosphorylation correlates with the amounts of SNAIL and MMP-3 in MMTV-PyMT tumors.
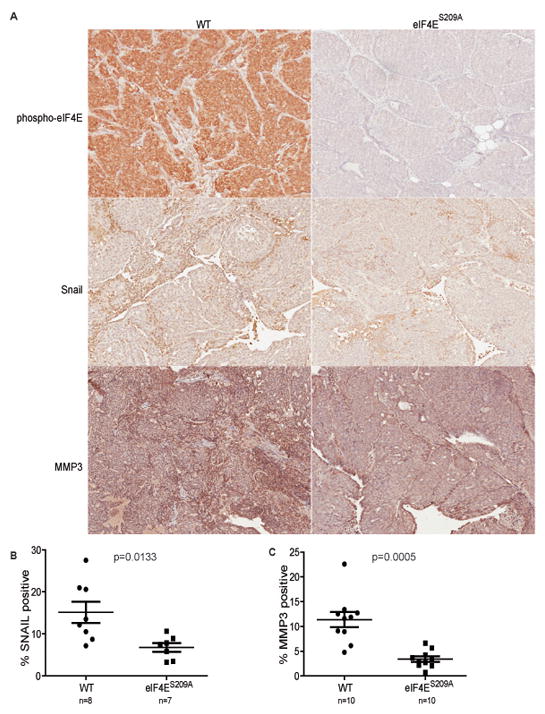
(A) Representative images of IHC analysis of eIF4E phosphorylation, SNAIL and MMP-3 expression in primary MMTV-PyMT tumors. (B) SNAIL and (C) MMP-3 quantification of IHC analysis of primary MMTV-PyMT tumors. Statistical significance determined by Student T-test; p-values are indicated. The number of mice used is indicated in the figure.
DISCUSSION
eIF4E is an oncoprotein that is overexpressed in many human malignancies and its expression levels have been associated with poor survival in cancers of the breast, prostate, head and neck, pharynx, lung, bladder, liver, oesophagus and stomach (23, 52–63). In one study on prostate cancer, the contribution of various factors dysregulating eIF4E has been investigated; eIF4E overexpression and 4E-BP1 phosphorylation correlated with poor survival, whereas 4E-BP1 overexpression exhibited inverse correlation (23). In prostate cancer cell lines, targeting eIF4E using active site mTOR inhibitors has been shown to inhibit the translation of pro-metastatic mRNAs such as YB1, vimentin, MTA1 and CD44 (35). For breast cancer, the most recent mechanistic insight into the role of eIF4E in vivo was obtained using the MMTV-PyMT model to show that eIF4E promotes the translation of VEGFA and MMP9 (21), corroborating the observations obtained with human samples (64–66). However, the role of eIF4E phosphorylation in metastatic cancer has so far been overlooked. While eIF4E phosphorylation is known to be elevated in many human cancers (67) and has been reported in some cases to correlate with poor outcome (68–71), the role of phospho-eIF4E in metastasis had yet to be established.
Here, we show that eIF4E phosphorylation promotes metastatic progression, in addition to its role in tumor development. We provide mechanistic insight into the role of eIF4E phosphorylation in metastasis, by showing that phospho-eIF4E is required for the translation of Snail and Mmp-3 mRNAs, two factors that regulate EMT and invasion. Importantly, our investigation into the role of the MNK/eIF4E axis as an essential step during metastasis and TGFβ-induced EMT uncovered an unanticipated intersection of canonical and non-canonical TGFβ signaling to cooperatively promote EMT. We propose that a subset of mRNAs induced by canonical TGFβ signaling is better translated when eIF4E becomes phosphorylated via MNK1, which lies immediately downstream of the non-canonical TGFβ signaling pathway (see model, Fig. 8).
Figure 8. Model.
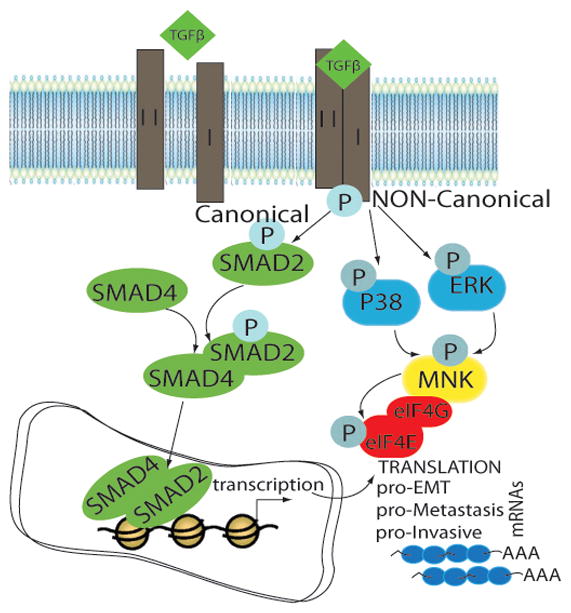
Non-canonical TGFβ signaling promotes the phospho-eIF4E dependent translation of EMT/metastasis-promoting transcripts, which are induced transcriptionally by the canonical SMAD pathway.
MNK1 has previously been linked to TGFβ signaling and was reported to promote the translation of SMAD2 (72). However, no differences in the level of SMAD2 expression or activation were observed in our experiments. Cell differences aside, the decrease in SMAD2 mRNA upon MNK1 silencing observed by Grzmil et al. occurred at 48h. Furthermore, as stated in their paper, a corresponding decrease in SMAD2 protein upon MNK1 siRNA knockdown was only detected after 72h. All of the experiments here were performed after MNK1 siRNA silencing for 24h or less. These differences may explain the discrepancies between the two studies.
Our data demonstrate that eIF4E phosphorylation is important for EMT and invasion, which are early steps in metastatic progression. This raises the interesting possibility of using eIF4E phosphorylation as a biomarker for tumors at high risk of metastasizing. In support of this, several studies reported a correlation between high phospho-eIF4E levels and cancer progression and/or poor prognosis (68–71). However, it will be important to consider the levels of eIF4E phosphorylation in the context of total eIF4E expression, as well as expression and phosphorylation status of the 4E-BPs, as each of these markers can affect patient outcome, as seen in prostate cancer (23).
It would be important to explore the effects of reducing eIF4E phosphorylation at various stages of tumor progression. In particular, the dependency of tumors on eIF4E phosphorylation for invasion raises the possibility of its therapeutic targeting in metastatic cancer. Cercosporamide inhibits eIF4E phosphorylation and tumor growth in xenograft models, as well as the growth of metastatic lung colonies in a B16 experimental metastasis assay (19). In this study, treatment with cercosporamide was initiated 24h after injection of the B16 cells, after the initial colonization of the lungs; these experiments could not have assessed the potential of preventing invasion by blocking eIF4E phosphorylation. Therefore, cercosporamide may possess important unexplored properties as a metastasis preventive agent, in addition to its inhibition of established metastatic colonies.
There are currently several inhibitors in clinical trials targeting eIF4E expression via antisense oligonucleotides, its availability through mTOR inactivation or its recruitment to the eIF4F complex by blocking the eIF4E-eIF4G interaction (16). However, the application of some of these inhibitors as well as currently used therapeutic agents such as rapamycin (73), gemcitabine (71), cytarabine (74) and herceptin (75) lead to increased eIF4E phosphorylation and concomitant resistance to treatment. Therefore, inhibitors of eIF4E phosphorylation may be useful in preventing resistance to some anti-cancer drugs, in addition to blocking metastatic progression.
IHC studies of tumor heterogeneity have identified phospho-eIF4E as one of the few biomarkers with high expression throughout the whole tumor sample (Ramón y Cajal, S. Rojo, S. Pons, B. Castellana, B. Hernandez-Losa, J. Sonenberg, N. Robichaud, N. De Mattos-Arruda, L. Cortes, J. Peg, V. Submitted manuscript). In contrast most commonly studied signal transduction markers such as pAKT, pmTOR and pMAPK displayed focal or “patchy” expression in human breast cancers, indicating that large sections of most tumors would be unresponsive to inhibitors of these kinases. These findings further argue for the necessity of developing inhibitors of eIF4E phosphorylation for clinical use.
MATERIALS AND METHODS
Reagents
TGFβ was purchased from PeproTech (USA). Insulin was from Sigma. Scrambled control siRNA and MNK1-specific siRNAs were from IDT (USA).
Mice and tissue preparation
eIF4ES209A mice (24) were bred with MMTV-PyMT, kindly provided by W.J. Muller (Goodman Cancer Research Centre) (48). Tumor onset in the mammary glands of control and experimental virgin mice was determined by palpation 3 times per week. Tumor growth was monitored by caliper measurements once per week or every 2 days near the experimental end point (single tumor above 2 cm3 or total burden above 6 cm3), at which point lung and tumor samples were collected. Step sections covering the entire lung were obtained and metastases were counted on all slides. The slide with the highest number of metastases was considered representative and used in further analyses. Lung and tumor samples were fixed in 10% formalin for paraffin imbedding and sectioning at 6 μm thickness. Pulmonary metastases were counted on H&E-stained 50 μm step-sections. Serial sections of tumor samples were obtained for immunohistochemistry. Immunohistochemistry was performed using the Vectastain ABC peroxidase system (Vector Labs, USA). All experiments involving animals were conducted in accordance with McGill University (Montreal, QC, Canada) animal care guidelines.
Cell lines
67NR, 168FARN and 4T07 cells were obtained from Fred Miller (Karmanos Institute, Detroit, USA). Cells (5×105) were injected into each 2nd mammary fat pad of BALB/c mice. Thirty days later, mice were sacrificed and the primary tumor harvested. Tumors obtained were immediately snap-frozen and pulverized under liquid nitrogen and stored at −80°C until further use. Extracts were prepared by suspending the specimens in RIPA lysis buffer (76) and used for immunoblotting.
Wild-type and eIF4ES209A transformed MEFs were obtained as described (24). For rescue experiments, MEFs were transfected with pcDNA3.1 constructs using lipofectamine 2000 and selected with G418 sulfate. Normal Murine Mammary Gland (NMuMG) epithelial cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, insulin, 2mM glutamine, 100 U/ml penicillin, and streptomycin. MCF10A cells were cultured in DMEM supplemented with 5% horse serum, hydrocortisone (0.5 μg/ml), insulin (10 μg/ml), epidermal growth factor (20 ng/ml), and penicillin-streptomycin (100 μg/ml each). NMuMG-BMN, NMuMG-HA-MKK3AL, and NMuMG-Flag-p38AGF cells were kindly provided by A. Bakin (Roswell Park Cancer Institute) and cultured in the same media as parental NMuMG cells. MCF10A stable cells expressing eIF4E or eIF4ES209A were infected with retrovirus produced in phoenix 293T cells, and subsequently selected with G418 sulfate and propagated. Cells were subsequently infected with lentiviral particles containing non-target control shRNA or human eIF4E shRNA (Sigma, USA). siRNAs were introduced into NMuMG using RNAiMAX (Life Technologies, USA) in accordance with the manufacturer’s protocol.
Western blot analysis
Cell monolayers were washed with PBS and harvested with trypsin. Washed cells were pelleted and lysed in RIPA lysis buffer containing protease and phosphatase inhibitors (Roche). The lysate was cleared by centrifugation and protein concentration was determined with the Bio-Rad protein concentration assay solution (Bio-Rad, Mississauga, ON). 25 to 150 micrograms of protein were separated by 10–12% SDS-PAGE and electroblotted onto a nitrocellulose membrane. Membranes were incubated overnight at 4°C with the indicated primary antibodies, and the following day incubated with the appropriate secondary anti-rabbit or anti-mouse antibodies for 1–3h at room temperature. Membranes were developed with the enhanced chemiluminescence Western Blot Detection Kit. Antibodies are described in the supplemental data (Table S1).
Fluorescence microscopy
Cells were cultured on glass coverslips in 12-well plates for 24h, then fixed with ethanol:acetic acid at −20°C, incubated with antibodies against the epithelial marker ZO-1 and then incubated with Alexa fluor 488 conjugated goat anti-rabbit IgG (Life Technologies, USA). Nuclei were stained with Hoechst. The mounted samples were scanned with a Leica DM LB2 microscope. Differences in ZO1 immunofluorescence were quantified by selecting a defined area corresponding to edges between two cells and by calculating the imageJ parameter “RawIntDen”, which is the sum of the pixel values in the selected area.
Migration and invasion
Random migration was monitored by time-lapse microscopy and analysed using MetaMorph Automation & Image Analysis Software (Molecular Devices, USA); 30,000 cells were plated in matrigel-coated 6 well plates and stimulated with 10% serum. Transwell migration assays were performed using 24-well cell culture inserts (BD Biosciences, USA) as described (77). Invasion index was obtained as the percentage of transwell migration with/without coating of the inserts with 50μL 5% growth factor-reduced (GFR) matrigel (BD Biosciences, USA). For colony outgrowth assays, 2000 cells were plated into 50μL GFR matrigel in 96-well plates and incubated for 7 days.
Plasmids and constructs
eIF4E and eIF4ES209A constructs were described (10, 24). pcDNA3.1-FLAG-eIF4EWT was described (78) and used to obtain pcDNA3.1-FLAG-eIF4ES209A by quick-change PCR mutagenesis with the following primers:
5′-GACACAGCTACTAAGGCAGGCTCCACCACTAAAAATAGGTTTGTTGTTTAAGAAG-3′,
5′-GTGGTGGAGCCTGCCTTAGTAGCTGTGTCTGCGTGGGACTGATAACC-3′.
MMP-3 cDNA was obtained from Origene (USA) in pCMV6-XL4 and cloned into pcDNA3.1 using the NotI restriction sites. pCDNA3.1-HA-SNAIL was obtained from Addgene (USA).
Polysome profile analysis, RNA isolation and RT-qPCR
Polysome profile analysis was carried out as described (78). RNA from each fraction was isolated using easy-BLUE kit (FroggaBio, Canada) and treated with DNaseTurbo (Ambion, USA) according to the manufacturer’s instructions. Reverse transcription PCR (RT-PCR) of 100ng RNA of each fraction was carried out using SuperScript III First-Strand Synthesis System (Invitrogen). qPCRs were carried out in a Mastercycler Realplex2 (Eppendorf) system using iQ Sybr green Supermix (Bio-Rad) according to the manufacturer’s instructions.
Bioinformatics analysis
To identify genes whose translation is sensitive to eIF4E-phosphorylation we reanalyzed a previously generated data set (24). To enable analysis of differential translation using the anota algorithm (79), we simulated a third replicate as described (80). This analysis results in an increase in false positives, but provides a ranking of genes that can be used for robust analysis, such as analysis of overrepresentation of genes belonging to specific pathways. To identify differential translation we used anota with the following settings: maxSlope=1.5, minSlope=(−0.5), maxRvmPAdj=0.15, selDeltaPT=log2(1.5) and minEff=log2(1.5). Identified differentially translated genes were further analyzed using DAVID (81).
Supplementary Material
Acknowledgments
This work was by supported by The Susan G. Komen Breast Cancer Foundation (IIR12224057), and the Canadian Cancer Society (2010-700377) to N.S. W.H.M. was supported by the Cancer Research Society (2012-17280). LF was supported by PCFA#YI-0310. N.S. is a Howard Hughes Medical Institute Senior International Scholar. W.H.M. is a Chercheur National of Fonds de la Recherche en Santé du Quebec (FRSQ). N.R. was supported by scholarships from the Fonds de la Recherche en Santé du Québec (20874), the Canadian Institutes of Health Research (220151) and the Vanier Canada Graduate Scholarship (267839). We thank W.J. Muller and I. Topisirovic for advice; C. Zakaria, A. Sylvestre, S. Perreault and C. Lister for technical assistance, N. Siddiqui for critical reading of the manuscript and S. Ramón y Cajal (Vall d’Hebron University Hospital, Barcelona, Spain) for his support and insights into tumor heterogeneity. We thank the Animal Facility and the Histology Facility at the Goodman Cancer Research Centre for mouse work and tissue processing.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
References
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–45. doi: 10.1016/j.cell.2009.01.042. Epub 2009/02/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiremath LS, Webb NR, Rhoads RE. Immunological detection of the messenger RNA cap-binding protein. The Journal of biological chemistry. 1985;260(13):7843–9. Epub 1985/07/05. [PubMed] [Google Scholar]
- 3.Duncan R, Milburn SC, Hershey JW. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. The Journal of biological chemistry. 1987;262(1):380–8. Epub 1987/01/05. [PubMed] [Google Scholar]
- 4.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371(6500):762–7. doi: 10.1038/371762a0. Epub 1994/10/27. [DOI] [PubMed] [Google Scholar]
- 5.Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. The Journal of biological chemistry. 1998;273(22):14002–7. doi: 10.1074/jbc.273.22.14002. Epub 1998/06/05. [DOI] [PubMed] [Google Scholar]
- 6.Flynn A, Proud CG. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. The Journal of biological chemistry. 1995;270(37):21684–8. doi: 10.1074/jbc.270.37.21684. Epub 1995/09/15. [DOI] [PubMed] [Google Scholar]
- 7.Joshi B, Cai AL, Keiper BD, Minich WB, Mendez R, Beach CM, et al. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. The Journal of biological chemistry. 1995;270(24):14597–603. doi: 10.1074/jbc.270.24.14597. Epub 1995/06/16. [DOI] [PubMed] [Google Scholar]
- 8.Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Molecular and cellular biology. 1999;19(3):1871–80. doi: 10.1128/mcb.19.3.1871. Epub 1999/02/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. The EMBO journal. 1999;18(1):270–9. doi: 10.1093/emboj/18.1.270. Epub 1999/01/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345(6275):544–7. doi: 10.1038/345544a0. Epub 1990/06/07. [DOI] [PubMed] [Google Scholar]
- 11.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nature medicine. 2004;10(5):484–6. doi: 10.1038/nm1042. Epub 2004/04/21. [DOI] [PubMed] [Google Scholar]
- 12.Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, et al. Dissecting eIF4E action in tumorigenesis. Genes & development. 2007;21(24):3232–7. doi: 10.1101/gad.1604407. Epub 2007/12/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E--from translation to transformation. Oncogene. 2004;23(18):3172–9. doi: 10.1038/sj.onc.1207549. Epub 2004/04/20. [DOI] [PubMed] [Google Scholar]
- 14.Lee T, Pelletier J. Eukaryotic initiation factor 4F: a vulnerability of tumor cells. Future medicinal chemistry. 2012;4(1):19–31. doi: 10.4155/fmc.11.150. Epub 2011/12/16. [DOI] [PubMed] [Google Scholar]
- 15.Jia Y, Polunovsky V, Bitterman PB, Wagner CR. Cap-dependent translation initiation factor eIF4E: an emerging anticancer drug target. Medicinal research reviews. 2012;32(4):786–814. doi: 10.1002/med.21260. Epub 2012/04/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malina A, Mills JR, Pelletier J. Emerging therapeutics targeting mRNA translation. Cold Spring Harbor perspectives in biology. 2012;4(4):a012377. doi: 10.1101/cshperspect.a012377. Epub 2012/04/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Topisirovic I, Ruiz-Gutierrez M, Borden KL. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer research. 2004;64(23):8639–42. doi: 10.1158/0008-5472.CAN-04-2677. Epub 2004/12/03. [DOI] [PubMed] [Google Scholar]
- 18.Ueda T, Sasaki M, Elia AJ, Chio II, Hamada K, Fukunaga R, et al. Combined deficiency for MAP kinase-interacting kinase 1 and 2 (Mnk1 and Mnk2) delays tumor development. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(32):13984–90. doi: 10.1073/pnas.1008136107. Epub 2010/08/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konicek BW, Stephens JR, McNulty AM, Robichaud N, Peery RB, Dumstorf CA, et al. Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer research. 2011;71(5):1849–57. doi: 10.1158/0008-5472.CAN-10-3298. Epub 2011/01/15. [DOI] [PubMed] [Google Scholar]
- 20.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23(18):3189–99. doi: 10.1038/sj.onc.1207545. Epub 2004/04/20. [DOI] [PubMed] [Google Scholar]
- 21.Nasr Z, Robert F, Porco JA, Muller WJ, Pelletier J. eIF4F suppression in breast cancer affects maintenance and progression. Oncogene. 2012 doi: 10.1038/onc.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettersson F, Yau C, Dobocan MC, Culjkovic-Kraljacic B, Retrouvey H, Puckett R, et al. Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17(9):2874–84. doi: 10.1158/1078-0432.CCR-10-2334. Epub 2011/03/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graff JR, Konicek BW, Lynch RL, Dumstorf CA, Dowless MS, McNulty AM, et al. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer research. 2009;69(9):3866–73. doi: 10.1158/0008-5472.CAN-08-3472. Epub 2009/04/23. [DOI] [PubMed] [Google Scholar]
- 24.Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(32):14134–9. doi: 10.1073/pnas.1005320107. Epub 2010/08/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Price JE, Fan D, Zhang RD, Bucana CD, Fidler IJ. Correlation of growth capacity of human tumor cells in hard agarose with their in vivo proliferative capacity at specific metastatic sites. Journal of the National Cancer Institute. 1989;81(18):1406–12. doi: 10.1093/jnci/81.18.1406. Epub 1989/09/20. [DOI] [PubMed] [Google Scholar]
- 26.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147(5):992–1009. doi: 10.1016/j.cell.2011.11.016. Epub 2011/11/29. [DOI] [PubMed] [Google Scholar]
- 27.Lochter A, Galosy S, Muschler J, Freedman N, Werb Z, Bissell MJ. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. The Journal of cell biology. 1997;139(7):1861–72. doi: 10.1083/jcb.139.7.1861. Epub 1998/02/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DaSilva J, Xu L, Kim HJ, Miller WT, Bar-Sagi D. Regulation of sprouty stability by Mnk1-dependent phosphorylation. Molecular and cellular biology. 2006;26(5):1898–907. doi: 10.1128/MCB.26.5.1898-1907.2006. Epub 2006/02/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hefner Y, Borsch-Haubold AG, Murakami M, Wilde JI, Pasquet S, Schieltz D, et al. Serine 727 phosphorylation and activation of cytosolic phospholipase A2 by MNK1-related protein kinases. The Journal of biological chemistry. 2000;275(48):37542–51. doi: 10.1074/jbc.M003395200. Epub 2000/09/09. [DOI] [PubMed] [Google Scholar]
- 30.Guil S, Long JC, Caceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Molecular and cellular biology. 2006;26(15):5744–58. doi: 10.1128/MCB.00224-06. Epub 2006/07/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buxade M, Parra JL, Rousseau S, Shpiro N, Marquez R, Morrice N, et al. The Mnks are novel components in the control of TNF alpha biosynthesis and phosphorylate and regulate hnRNP A1. Immunity. 2005;23(2):177–89. doi: 10.1016/j.immuni.2005.06.009. Epub 2005/08/23. [DOI] [PubMed] [Google Scholar]
- 32.Byers HR, Etoh T, Doherty JR, Sober AJ, Mihm MC., Jr Cell migration and actin organization in cultured human primary, recurrent cutaneous and metastatic melanoma. Time-lapse and image analysis. The American journal of pathology. 1991;139(2):423–35. Epub 1991/08/11. [PMC free article] [PubMed] [Google Scholar]
- 33.Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aaronson SA, Kozlowski JM, et al. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer research. 1987;47(12):3239–45. Epub 1987/06/15. [PubMed] [Google Scholar]
- 34.Sommers CL, Byers SW, Thompson EW, Torri JA, Gelmann EP. Differentiation state and invasiveness of human breast cancer cell lines. Breast cancer research and treatment. 1994;31(2–3):325–35. doi: 10.1007/BF00666165. Epub 1994/01/01. [DOI] [PubMed] [Google Scholar]
- 35.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485(7396):55–61. doi: 10.1038/nature10912. Epub 2012/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. Epub 2009/01/10. [DOI] [PubMed] [Google Scholar]
- 37.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. Epub 2008/11/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119(6):1420–8. doi: 10.1172/JCI39104. Epub 2009/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell research. 2009;19(2):156–72. doi: 10.1038/cr.2009.5. Epub 2009/01/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. The Journal of clinical investigation. 2009;119(6):1429–37. doi: 10.1172/JCI36183. Epub 2009/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell research. 2009;19(1):128–39. doi: 10.1038/cr.2008.328. Epub 2008/12/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bakin AV, Rinehart C, Tomlinson AK, Arteaga CL. p38 mitogen-activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. Journal of cell science. 2002;115(Pt 15):3193–206. doi: 10.1242/jcs.115.15.3193. Epub 2002/07/16. [DOI] [PubMed] [Google Scholar]
- 43.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Molecular pharmacology. 2002;62(1):65–74. doi: 10.1124/mol.62.1.65. Epub 2002/06/18. [DOI] [PubMed] [Google Scholar]
- 44.Imamura T, Hikita A, Inoue Y. The roles of TGF-beta signaling in carcinogenesis and breast cancer metastasis. Breast Cancer. 2012;19(2):118–24. doi: 10.1007/s12282-011-0321-2. Epub 2011/12/06. [DOI] [PubMed] [Google Scholar]
- 45.Knauf U, Tschopp C, Gram H. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Molecular and cellular biology. 2001;21(16):5500–11. doi: 10.1128/MCB.21.16.5500-5511.2001. Epub 2001/07/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheper GC, Morrice NA, Kleijn M, Proud CG. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Molecular and cellular biology. 2001;21(3):743–54. doi: 10.1128/MCB.21.3.743-754.2001. Epub 2001/01/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer research. 1992;52(6):1399–405. Epub 1992/03/15. [PubMed] [Google Scholar]
- 48.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Molecular and cellular biology. 1992;12(3):954–61. doi: 10.1128/mcb.12.3.954. Epub 1992/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dilworth SM. Polyoma virus middle T antigen and its role in identifying cancer-related molecules. Nature reviews Cancer. 2002;2(12):951–6. doi: 10.1038/nrc946. Epub 2002/12/03. [DOI] [PubMed] [Google Scholar]
- 50.Franci C, Takkunen M, Dave N, Alameda F, Gomez S, Rodriguez R, et al. Expression of Snail protein in tumor-stroma interface. Oncogene. 2006;25(37):5134–44. doi: 10.1038/sj.onc.1209519. Epub 2006/03/29. [DOI] [PubMed] [Google Scholar]
- 51.Heppner KJ, Matrisian LM, Jensen RA, Rodgers WH. Expression of most matrix metalloproteinase family members in breast cancer represents a tumor-induced host response. The American journal of pathology. 1996;149(1):273–82. Epub 1996/07/01. [PMC free article] [PubMed] [Google Scholar]
- 52.Li BD, McDonald JC, Nassar R, De Benedetti A. Clinical outcome in stage I to III breast carcinoma and eIF4E overexpression. Annals of surgery. 1998;227(5):756–6l. doi: 10.1097/00000658-199805000-00016. discussion 61–3. Epub 1998/05/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li BD, Gruner JS, Abreo F, Johnson LW, Yu H, Nawas S, et al. Prospective study of eukaryotic initiation factor 4E protein elevation and breast cancer outcome. Annals of surgery. 2002;235(5):732–8. doi: 10.1097/00000658-200205000-00016. discussion 8–9. Epub 2002/05/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holm N, Byrnes K, Johnson L, Abreo F, Sehon K, Alley J, et al. A prospective trial on initiation factor 4E (eIF4E) overexpression and cancer recurrence in node-negative breast cancer. Annals of surgical oncology. 2008;15(11):3207–15. doi: 10.1245/s10434-008-0086-9. Epub 2008/08/23. [DOI] [PubMed] [Google Scholar]
- 55.McClusky DR, Chu Q, Yu H, Debenedetti A, Johnson LW, Meschonat C, et al. A prospective trial on initiation factor 4E (eIF4E) overexpression and cancer recurrence in node-positive breast cancer. Annals of surgery. 2005;242(4):584–90. doi: 10.1097/01.sla.0000184224.55949.90. discussion 90–2. Epub 2005/09/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crew JP, Fuggle S, Bicknell R, Cranston DW, de Benedetti A, Harris AL. Eukaryotic initiation factor-4E in superficial and muscle invasive bladder cancer and its correlation with vascular endothelial growth factor expression and tumour progression. British journal of cancer. 2000;82(1):161–6. doi: 10.1054/bjoc.1999.0894. Epub 2000/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seki N, Takasu T, Mandai K, Nakata M, Saeki H, Heike Y, et al. Expression of eukaryotic initiation factor 4E in atypical adenomatous hyperplasia and adenocarcinoma of the human peripheral lung. Clinical cancer research: an official journal of the American Association for Cancer Research. 2002;8(10):3046–53. Epub 2002/10/11. [PubMed] [Google Scholar]
- 58.Seki N, Takasu T, Sawada S, Nakata M, Nishimura R, Segawa Y, et al. Prognostic significance of expression of eukaryotic initiation factor 4E and 4E binding protein 1 in patients with pathological stage I invasive lung adenocarcinoma. Lung Cancer. 2010;70(3):329–34. doi: 10.1016/j.lungcan.2010.03.006. Epub 2010/07/14. [DOI] [PubMed] [Google Scholar]
- 59.Nathan CO, Franklin S, Abreo FW, Nassar R, De Benedetti A, Glass J. Analysis of surgical margins with the molecular marker eIF4E: a prognostic factor in patients with head and neck cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1999;17(9):2909–14. doi: 10.1200/JCO.1999.17.9.2909. Epub 1999/11/24. [DOI] [PubMed] [Google Scholar]
- 60.Wu M, Liu Y, Di X, Kang H, Zeng H, Zhao Y, et al. EIF4E over-expresses and enhances cell proliferation and cell cycle progression in nasopharyngeal carcinoma. Med Oncol. 2013;30(1):400. doi: 10.1007/s12032-012-0400-z. Epub 2013/01/02. [DOI] [PubMed] [Google Scholar]
- 61.Wang XL, Cai HP, Ge JH, Su XF. Detection of eukaryotic translation initiation factor 4E and its clinical significance in hepatocellular carcinoma. World journal of gastroenterology: WJG. 2012;18(20):2540–4. doi: 10.3748/wjg.v18.i20.2540. Epub 2012/06/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salehi Z, Mashayekhi F. Expression of the eukaryotic translation initiation factor 4E (eIF4E) and 4E-BP1 in esophageal cancer. Clinical biochemistry. 2006;39(4):404–9. doi: 10.1016/j.clinbiochem.2005.11.007. Epub 2005/12/27. [DOI] [PubMed] [Google Scholar]
- 63.Chen CN, Hsieh FJ, Cheng YM, Lee PH, Chang KJ. Expression of eukaryotic initiation factor 4E in gastric adenocarcinoma and its association with clinical outcome. Journal of surgical oncology. 2004;86(1):22–7. doi: 10.1002/jso.20037. Epub 2004/03/30. [DOI] [PubMed] [Google Scholar]
- 64.Nathan CO, Carter P, Liu L, Li BD, Abreo F, Tudor A, et al. Elevated expression of eIF4E and FGF-2 isoforms during vascularization of breast carcinomas. Oncogene. 1997;15(9):1087–94. doi: 10.1038/sj.onc.1201272. Epub 1997/08/28. [DOI] [PubMed] [Google Scholar]
- 65.Scott PA, Smith K, Poulsom R, De Benedetti A, Bicknell R, Harris AL. Differential expression of vascular endothelial growth factor mRNA vs protein isoform expression in human breast cancer and relationship to eIF-4E. British journal of cancer. 1998;77(12):2120–8. doi: 10.1038/bjc.1998.356. Epub 1998/07/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Byrnes K, White S, Chu Q, Meschonat C, Yu H, Johnson LW, et al. High eIF4E, VEGF, and microvessel density in stage I to III breast cancer. Annals of surgery. 2006;243(5):684–90. doi: 10.1097/01.sla.0000216770.23642.d8. discussion 91–2. Epub 2006/04/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fan S, Ramalingam SS, Kauh J, Xu Z, Khuri FR, Sun SY. Phosphorylated eukaryotic translation initiation factor 4 (eIF4E) is elevated in human cancer tissues. Cancer biology & therapy. 2009;8(15):1463–9. doi: 10.4161/cbt.8.15.8960. Epub 2009/06/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshizawa A, Fukuoka J, Shimizu S, Shilo K, Franks TJ, Hewitt SM, et al. Overexpression of phospho-eIF4E is associated with survival through AKT pathway in non-small cell lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16(1):240–8. doi: 10.1158/1078-0432.CCR-09-0986. Epub 2009/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng J, Li J, Xu L, Xie G, Wen Q, Luo J, et al. Phosphorylated Mnk1 and eIF4E Are Associated with Lymph Node Metastasis and Poor Prognosis of Nasopharyngeal Carcinoma. PloS one. 2014;9(2):e89220. doi: 10.1371/journal.pone.0089220. Epub 2014/02/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferrandiz-Pulido C, Masferrer E, Toll A, Hernandez-Losa J, Mojal S, Pujol RM, et al. mTOR Signaling Pathway in Penile Squamous Cell Carcinoma: pmTOR and peIF4E Over Expression Correlate with Aggressive Tumor Behavior. The Journal of urology. 2013 doi: 10.1016/j.juro.2013.06.015. Epub 2013/06/15. [DOI] [PubMed] [Google Scholar]
- 71.Adesso L, Calabretta S, Barbagallo F, Capurso G, Pilozzi E, Geremia R, et al. Gemcitabine triggers a pro-survival response in pancreatic cancer cells through activation of the MNK2/eIF4E pathway. Oncogene. 2013;32(23):2848–57. doi: 10.1038/onc.2012.306. Epub 2012/07/17. [DOI] [PubMed] [Google Scholar]
- 72.Grzmil M, Morin P, Jr, Lino MM, Merlo A, Frank S, Wang Y, et al. MAP kinase-interacting kinase 1 regulates SMAD2-dependent TGF-beta signaling pathway in human glioblastoma. Cancer research. 2011;71(6):2392–402. doi: 10.1158/0008-5472.CAN-10-3112. Epub 2011/03/17. [DOI] [PubMed] [Google Scholar]
- 73.Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer research. 2005;65(16):7052–8. doi: 10.1158/0008-5472.CAN-05-0917. Epub 2005/08/17. [DOI] [PubMed] [Google Scholar]
- 74.Yin B, Morgan K, Hasz DE, Mao Z, Largaespada DA. Nfl gene inactivation in acute myeloid leukemia cells confers cytarabine resistance through MAPK and mTOR pathways. Leukemia. 2006;20(1):151–4. doi: 10.1038/sj.leu.2404033. Epub 2005/11/25. [DOI] [PubMed] [Google Scholar]
- 75.Astanehe A, Finkbeiner MR, Krzywinski M, Fotovati A, Dhillon J, Berquin IM, et al. MKNK1 is a YB-1 target gene responsible for imparting trastuzumab resistance and can be blocked by RSK inhibition. Oncogene. 2012;31(41):4434–46. doi: 10.1038/onc.2011.617. Epub 2012/01/18. [DOI] [PubMed] [Google Scholar]
- 76.RIPA buffer (05-01) Cold Spring Harbor Protocols. 2006;2006(4) pdb.rec10617. [Google Scholar]
- 77.Ling C, Su VM, Zuo D, Muller WJ. Loss of the 14-3-3sigma tumor suppressor is a critical event in ErbB2-mediated tumor progression. Cancer discovery. 2012;2(1):68–81. doi: 10.1158/2159-8290.CD-11-0189. Epub 2012/05/16. [DOI] [PubMed] [Google Scholar]
- 78.Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, et al. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer research. 2012;72(24):6468–76. doi: 10.1158/0008-5472.CAN-12-2395. Epub 2012/10/27. [DOI] [PubMed] [Google Scholar]
- 79.Larsson O, Sonenberg N, Nadon R. anota: Analysis of differential translation in genome-wide studies. Bioinformatics. 2011;27(10):1440–1. doi: 10.1093/bioinformatics/btr146. Epub 2011/03/23. [DOI] [PubMed] [Google Scholar]
- 80.Larsson O, Sonenberg N, Nadon R. Identification of differential translation in genome wide studies. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(50):21487–92. doi: 10.1073/pnas.1006821107. Epub 2010/12/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, et al. Extracting biological meaning from large gene lists with DAVID. In: Baxevanis Andreas D, et al., editors. Current protocols in bioinformatics. Unit 13. Chapter 13. 2009. p. 1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.