Germline or somatic inactivating mutations in BRCA1 or BRCA2 or other genes involved in the homologous recombination (HR) pathway of DNA repair collectively occur in as much as 20%–25% of advanced prostate cancers. This article discusses the current understanding of the genomic landscape of prostate cancer, focusing on the occurrence of DNA repair mutations and the therapeutic opportunities this presents.
Keywords: Prostate cancer, DNA repair, PARP inhibitors, BRCA
Abstract
Advances in DNA sequencing technology have created a wealth of information regarding the genomic landscape of prostate cancer. It had been thought that BRCA1 and BRCA2 mutations were associated with only a small fraction of prostate cancer cases. However, recent genomic analysis has revealed that germline or somatic inactivating mutations in BRCA1 or BRCA2, or other genes involved in the homologous recombination (HR) pathway of DNA repair collectively occur in as much as 20%–25% of advanced prostate cancers. A synthetic lethal therapeutic approach using poly(ADP-ribose) polymerase inhibitor therapy has been developed for BRCA mutant- and HR deficient-related cancers (those with “BRCAness”) and is being studied in multiple clinical trials. This article discusses the current understanding of the genomic landscape of prostate cancer, focusing on the occurrence of DNA repair mutations and the therapeutic opportunities that this presents.
Implications for Practice:
This review aims to update oncologists about the increased understanding of the genomes of prostate cancers and, in particular, the prevalence of mutations in DNA repair genes. These observations provide potential new therapeutic opportunities for the use of poly(ADP-ribose) polymerase inhibitors and other therapies, especially in advanced forms of the disease. Of note is the recent U.S. Food and Drug Administration breakthrough therapy designation of olaparib for the treatment of BRCA1/2- or ATM-mutated metastatic castration-resistant prostate cancer. The implications of this new knowledge for clinical practice now and in the future are discussed.
The Genomic Landscape of Prostate Cancer
More than 900,000 new cases of prostate cancer are diagnosed worldwide each year, and advanced disease remains a major treatment challenge [1, 2]. This is a genomically and clinically heterogeneous cancer, with some patients presenting with metastatic disease and others being cured after local surgery or radiation. With advances in next-generation sequencing, the genomic landscape of prostate cancer is being defined more clearly. The burden of overall mutations and copy-number alterations appears higher in metastatic prostate cancer than in localized prostate cancer. Similarly, it has been shown that mutations leading to defective DNA repair appear to be enriched in later stages of the disease, perhaps speaking to an overall poorer prognosis for patients with these mutations [3, 4].
Early studies evaluating the genomic landscape of primary prostate cancers demonstrated recurrent mutations in the FOXA1, SPOP, TP53, and PTEN genes [5]. The Cancer Genome Atlas (TCGA) Research Network has described a comprehensive analysis of 333 primary prostate cancers following radical prostatectomy and found a complex genomic landscape [4]. Almost 53% of primary tumors demonstrated ETS (erythroblast transformation specific)-family gene fusions, with TMPRSS2 being the most common fusion partner. ETS gene fusions (specifically TMPRSS2-ERG) may be associated with poly(ADP-ribose) polymerase (PARP) inhibitor sensitivity [4, 6], but the clinical significance of this potential sensitivity remains to be explored. The TCGA study also found that 19% of primary prostate cancers had mutations in DNA repair genes including 3% of patients with inactivating germline and somatic mutations in BRCA2 [4]. FANCD2, BRCA1, CKD12, and ATM made up the other mutations that were classified under DNA repair. Of note, all patients with heterozygous BRCA2 loss also had a corresponding loss of the distal RB1 tumor suppressor gene, perhaps suggesting a possible bystander effect. Although some homozygous deletions and loss-of-function mutations were seen, many of the identified mutations included heterozygous losses with unknown functional impact that will require validation biologically and in other cohorts.
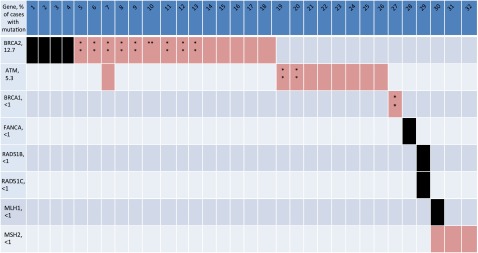
In a landmark study in 2015, the East Coast Dream Team conducted whole exome and transcriptome sequencing of 150 biopsy specimens from metastatic castration-resistant prostate cancer tumors [3]. As expected, aberrations of the androgen receptor (AR) were common and much more prevalent in advanced prostate cancer than in early-stage disease, with 71.3% of cases showing alterations in genes involved in AR signaling, including alterations in AR itself, FOXA1 (an AR transcription factor), NCOR 1/2, SPOP, and ZBTB16. PI3K, Wnt, and Rb1 pathway genes were also commonly altered. One exciting finding of this study was that 22.7% of these biopsy specimens had defects in DNA repair genes, including 8% with germline mutations in DNA repair genes, including in BRCA1/2, ATM, CDK12, FANCA, RAD51B, and RAD51C. Somatic or germline BRCA2 alterations were identified in 12.7% of cases and 5.3% of cases had germline BRCA2 loss. This finding dramatically expands the subset of patients who may benefit from PARP inhibitors to a much larger proportion of prostate cancers than previously envisioned (Fig. 1). It should also be noted that recurrent ETS fusions were observed in 56% of cases where the most common fusion was to ERG but fusions to FLI1, ETV4, and ETV5 were also observed. As mentioned, it has been shown that ETS-positive prostate cancer xenografts are sensitive to PARP inhibitors and, more recently, it was shown that ETS gene-fusion products physically interact with the enzymes PARP1 and DNA-PKcs (catalytic subunit of DNA protein kinase), both of which are required for ETS-mediated transcription and invasion.
Figure 1.
Schematic of cases with aberrations in the DNA repair pathway in metastatic castration-resistant prostate cancer [3]. Mutations in DNA repair genes were seen in 22.7% of cases. The majority of mutations were found in BRCA2 and ATM, but mutations were also observed in BRCA1, FANCA, RAD51B, RAD51C, and the mismatch repair genes MLH1 and MSH2.
Although the prevalence of somatic and germline mutations in the BRCA genes is becoming better understood, the implications for prognosis or response to therapy remain unclear. The only available data come from studies in a restricted subset of germline BRCA mutation carriers [7]. The largest of these studies showed that germline BRCA mutations are associated with an overall poorer prognosis. Castro et al. examined 2,019 patients with prostate cancer, of whom 18 were germline BRCA1 carriers, 61 were germline BRCA2 carriers, and 1,940 were noncarriers, and found that the BRCA1/2 carriers were more frequently associated with a Gleason score of at least 8 (p = .00003), T3/T4 stage (p = .003), nodal involvement (p = .0005), and metastases at diagnosis (p = .005) than prostate cancer in noncarriers. For localized prostate cancer, the 5-year metastasis-free survival was significantly higher in noncarriers (93% vs. 77% of carriers; hazard ratio: 2.7; p = .009) [7].
Understanding of the prognostic significance of prostate cancers that harbor mutations in other DNA repair genes (ATM, CDK12, FANCA, RAD51B, and RAD51C) will require long-term follow-up in large cohorts. Moreover, it is also not known whether these molecular defects are associated with any particular histologic subtype. BRCA1-associated breast cancer is much more likely to be of the triple-negative/basal phenotype. Moreover, unselected triple-negative breast cancers enrich for BRCA1 mutations; a study of 77 unselected women with triple-negative breast cancer found 15 patients (19.5%) had mutations in BRCA1 or BRCA2. This study highlights the importance of phenotypically characterizing BRCA-mutated prostate cancers tumors to aid in determining who may benefit from therapies targeting DNA repair [8].
DNA Repair Mutations and PARP Inhibitors: The Story So Far
Patients with germline mutations in BRCA1 or BRCA2 have a considerably elevated risk of developing cancers of the breast and ovary; those with BRCA2 germline mutations additionally have increased risk of developing prostate and pancreatic cancers. Heterozygous germline mutations in BRCA1/2 are at risk of losing the wild-type allele, resulting in defective homologous repair that can lead to tumorigenesis. This results in a high lifetime risk for a number of cancers; large meta-analyses revealed that BRCA1 carriers have a 65% cumulative lifetime risk for breast cancer and 39% lifetime risk for ovarian cancer. BRCA2 carriers are at a smaller but still substantial 45% risk for breast cancer and an 11% lifetime risk for ovarian cancer [9]. The risk for prostate cancer is less precisely known, but in 1 study, BRCA2 conferred an 8.6-fold increased risk for prostate cancer by age 65, corresponding to an absolute risk of 15% [1].
The PARPs are a family of enzymes involved in protein modification DNA repair. Some of these enzymes, particularly PARP1, are involved in the repair of DNA single-strand breaks through base-excision repair [11, 12]. These single-strand breaks can lead to double-strand breaks at replications forks, which are normally repaired via the homologous recombination pathway involving BRCA1 and BRCA2. PARP inhibitors exploit defective DNA repair in BRCA1/2-mutated tumors and induce selective tumor cytotoxicity, relying on the concept of synthetic lethality [13, 14]. Synthetic lethality is the concept whereby a genetic mutation or compound is compatible with cell viability alone, but incompatible with viability when combined with another genetic defect or induced deficiency (i.e., PARP inhibitors and BRCA-mutated tumors).
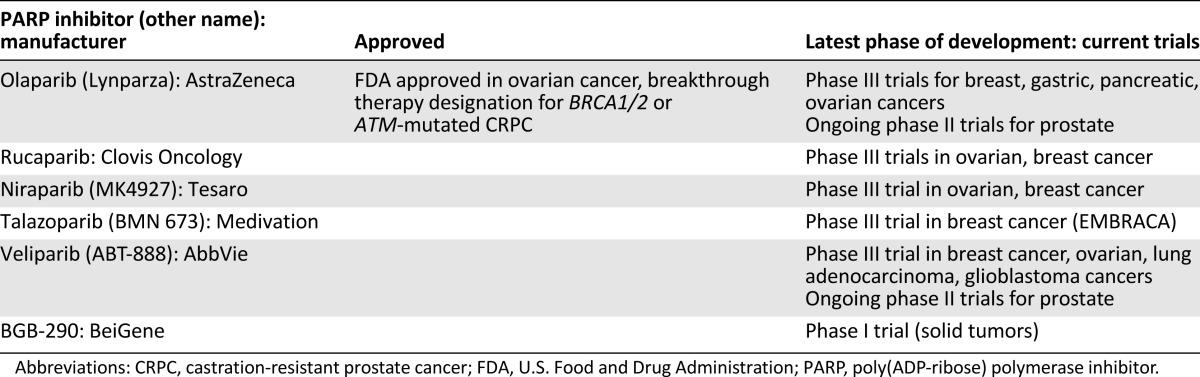
Since the discovery of the first PARP inhibitor, 3-aminobenzamide, over 30 years ago, considerable advances in this area have been made. Currently, there are at least five different PARP inhibitors in phase III clinical trials, including olaparib, rucaparib, niraparib, velaparib, and talazoparib (Table 1). In December 2014, olaparib was approved in the U.S. and Europe for the treatment of BRCA mutation-related ovarian cancer; the U.S. approval was as a third-line agent for patients who have been treated with three or more prior lines of therapy. This approval was based on a phase II clinical trial by Kaufman et al., who enrolled 298 patients with germline BRCA1/2 mutations and recurrent cancers (ovarian, breast, pancreatic, and prostate), including 193 patients with ovarian cancer [15]. Patients were treated with olaparib at a dose of 400 mg orally twice daily until disease progression or intolerance to therapy. Of the 193 patients in the ovarian cancer cohort, 137 had measurable disease at baseline and had received 3 or more prior lines of chemotherapy. In the ovarian cancer cohort, the objective response rate was 34% (the partial and complete response rates were 32% and 2%, respectively) with median response duration of 7.9 months. Median overall survival in the ovarian cancer group was 16.6 months. The most common frequent adverse events (AEs) were fatigue and anemia. Unfortunately, there were nine deaths as a result of AEs in this trial, including two patients who developed leukemia and another who developed myelodysplastic syndrome (MDS). All three of these patients had been heavily pretreated with chemotherapy before receiving olaparib, but there has been concern about the potential for olaparib to increase the risk of MDS or acute myeloid leukemia.
Table 1.
PARP inhibitors in clinical development
In Europe, based in part on results of the Ledermann et al. trial [16], olaparib was approved for use in the European Medicine Agency as monotherapy maintenance treatment for women with BRCA1 and BRCA2 mutant, high-grade, serous ovarian cancer, fallopian tube, or primary peritoneal cancer who had a prior complete or partial response to platinum-based chemotherapy. This was a randomized, double-blind, placebo-controlled, phase II study involving 265 patients with platinum-sensitive, relapsed ovarian, fallopian tube, or primary peritoneal cancers. The study demonstrated a progression-free survival that was significantly longer with olaparib than with placebo (median: 8.4 months vs. 4.8 months from randomization on completion of chemotherapy; hazard ratio for progression or death: 0.35; 95% confidence interval [CI]: 0.25–0.49; p < .001).
PARP inhibitors are undergoing trials in multiple tumor types involving ovarian, breast, pancreatic, gastric, non-small cell lung cancer, melanoma, glioblastoma, and other cancers. These trials range from monotherapy trials to combination therapy trials to maintenance therapy trials [17]. Some (including TOPARP, discussed below) are expanding the inclusion criteria for PARP inhibitors beyond patients with germline BRCA1/2 carriers to those who have somatic “BRCAness” genomic signatures.
It is now apparent that platinum therapy may also be selective against particular DNA repair deficiencies. Platinum salts cause platinum DNA adducts, which may cause DNA crosslinking, requiring base-excision repair and homologous recombination for their repair. Platinum salts (carboplatin and cisplatin) have both been used for decades to treat patients with ovarian cancer, where BRCA1 and BRCA2 mutation is common. The Triple-Negative Breast Cancer Trial (TNT trial) was a proof-of-concept phase III trial that showed that patients with advanced, triple-negative breast cancer with BRCA1/2 mutations had a greater response and progression-free survival to carboplatin than with docetaxel [18].
Despite the initial promise of PARP inhibitors as potentiators of chemotherapy, clinical experience has been that dose escalation is problematic because of hematologic toxicity. For example, in a completed phase I clinical trial of cisplatin, gemcitabine, and olaparib in patients with advanced solid tumors, investigators saw significant hematologic toxicity that hampered their ability to dose escalate to efficacious doses of olaparib. The protocol needed to be amended to only enroll patients treated with 2 or fewer prior myelosuppressive regimens and treated patients with low doses of olaparib (100 mg once daily on days 1–4 of the cycle) [19]. Similarly, our group has had significant difficulties finding a maximum tolerated dose in a carboplatin/talazoparib trial, due to significant hematologic toxicity at multiple dose levels (NCT02358200, https://clinicaltrials.gov/ct2/show/NCT02358200). A number of trials are now evaluating PARP inhibitors in combination with agents that have considerably less hematologic toxicity but, hopefully, synergism, nonetheless, including combinations with immunotherapy, histone deacetylase (HDAC) inhibitors, AKT inhibitors, as well as androgen receptor-directed therapy in prostate cancer (http://www.clinicaltrials.gov).
TOPARP: Proof of Concept of PARP Inhibitor Efficacy in Prostate Cancer
In 2009, Fong et al. published results of a phase I trial in which 60 patients were enrolled and given escalating doses of olaparib monotherapy. This cohort notably included one patient with BRCA2-mutated, castration-resistant prostate cancer who had a greater than 50% decrease in prostate-specific antigen (PSA) level and resolution of bone metastases. The patient remained in the trial for over 2 years [20]. This laid the groundwork for the TOPARP (Phase II Trial of Olaparib in Patients With Advanced Castration-Resistant Prostate Cancer) published in October 2015 by Mateo et al. [21]. In this phase II clinical trial, 50 patients with metastatic castration-resistant prostate cancer were treated with olaparib, 400 mg twice a day. Of the 50 patients enrolled, 49 (98%) had received prior treatment with docetaxel, 49 (98%) had received abiraterone or enzalutamide, and 29 (58%) had received cabazitaxel. Paired biopsy procedures were mandated before and during treatment and next-generation sequencing identified deleterious mutations, homozygous deletions, or both in DNA repair genes (including BRCA1/2, Fanconi anemia genes, ATM, and CHEK2) in 16 of the 49 evaluable patients (33%). Of these patients, 88% had a response to olaparib, indicating the specificity for response in patients with DNA repair mutations was 94%.
In this study, response criteria were broad, and included a) an objective response by RESIST or b) at least a 50% decrease in PSA level or c) a confirmed reduction in circulating tumor cell count. Presumably these response criteria were chosen as a route to the development of a more-sensitive biomarker suite. Nonetheless, radiologic progression-free survival and overall survival for patients who had DNA repair defects or “biomarker positivity” were 9.8 months and 13.8 months, respectively, versus a 2.7 months and 7.5 months, respectively, in those patients who were biomarker negative (p < .001 and p = .05 for respective analyses). Anemia (in 10 of 50 patients; 20%) and fatigue (in 6 of 50; 12%) were the most common grade 3 and 4 adverse effects, in keeping with other studies of olaparib.
Of interest, investigators saw a response in a patient with bilallelic somatic mutations in HDAC2, which has not traditionally been reported as a mutation associated with PARP inhibitor (PARPi) response, although it is known that HDAC1 and HDAC2 participate in DNA repair [20, 22]. Based on this trial, olaparib has been granted breakthrough therapy designation for BRCA1/2- or ATM-mutated castration-resistant prostate cancer. This trial also laid the groundwork for further studies in which biomarker positivity will likely serve as an eligibility criterion, as well as studies evaluating PARP inhibitors in earlier stages of prostate cancer and in combination with other prostate cancer therapies, including AR-targeted agents. An international, randomized phase II clinical trial is evaluating the combination of olaparib with abiraterone versus placebo with abiraterone in patients with metastatic castration-resistant prostate cancer. The estimated enrollment is 150 patients, with completion estimated around October 2016 (NCT01972217). Also underway is another multicenter, phase II clinical trial evaluating the combination of veliparib with abiraterone versus abiraterone with placebo in patients with metastatic castration-resistant prostate cancer; estimated enrollment is 150 patients (NCT 01576172).
Biomarkers for BRCAness and PARPi Responsiveness in Prostate Cancer
As demonstrated in the TOPARP trial, the proportion of patients who may preferentially respond to PARP inhibitors or platinum chemotherapies is not limited to patients with germline BRCA1/2 mutations. As a consequence, there are numerous efforts to identify tumors with “BRCAness” (i.e., those that may display functional defects similar to those in BRCA-mutated tumors). In prostate cancer, it has become increasingly obvious that there is a large subset of tumors with somatic mutations in genes involved in HR that may confer sensitivity to PARP inhibitors, including ATM, FANC, CDK12, RAD51B, RAD51C, PALB2, ATR, CHEK1, CHEK2, DSS1, and ETS gene fusions (TMPRSS2:ERG) [3, 6, 21, 23]. Thus, one approach for future clinical trials would be to aggregate these genetic mutations into one large BRCAness panel that would serve as the trial inclusion criteria.
As demonstrated in the TOPARP trial, the proportion of patients who may preferentially respond to PARP inhibitors or platinum chemotherapies is not limited to patients with germline BRCA1/2 mutations. As a consequence, there are numerous efforts to identify tumors with “BRCAness” (i.e., those that may display functional defects similar to those in BRCA-mutated tumors).
Other groups are working on deriving transcriptional signatures of BRCAness, including Larsen et al., who analyzed 55 familial BRCA1- or BRCA2-mutant breast tumors and 128 sporadic breast tumors to derive a transcriptional signature that predicted BRCA-mutated tumors in an independent dataset [24]. Another group has developed a gene expression profile to predict response to anthracycline and cyclophosphamide chemotherapy for breast cancer. This was done by first creating a DNA gene expression microarray from 21 patients with Fanconi anemia and 11 control subjects to identify pathways associated with DNA repair processes [25]. The activity of this pathway was then analyzed in cohorts of BRCA1/2 carriers as well as those with sporadic breast cancer. From this work, the group developed a 44-gene expression signature used to prospectively identify DNA repair-deficient subgroups. This gene expression signature predicted complete pathologic response versus residual disease after neoadjuvant DNA-damaging chemotherapy (5-fluorouracil, anthracycline, and cyclophosphamide) with an odds ratio of 3.96 (95% CI: 1.67–9.41; p =. 002). Other groups are investigating functional biomarkers of DNA repair, such as immunohistochemical detection of nuclear RAD51 localization (a key marker of homologous recombination), which, if absent, speaks to DNA repair deficiency, while others are exploring PARP enzymatic assays. The challenge to these explorations is that they require biopsy specimens to be collected after treatment to look for treatment efficacy. Of course, one can also use larger studies designed like TOPARP in which larger populations are exposed to PARP inhibitors and responders are sequenced to get a broader understanding of which tumors and patients possess “BRCAness.”
Current Implications and Future Directions
Given the enrichment of DNA repair deficiencies in advanced disease, for patients who demonstrate resistance to abiraterone or enzalutamide or a robust response to platinum chemotherapy, genomic sequencing of metastatic sites promises to be a powerful tool. The broad availability of commercial next generation genomic sequencing has simplified this process for clinicians, who can now submit a variety of tumor samples, ranging from paraffin embedded tumor samples to whole blood. More challenging, however, is determining which of the various findings can be used to inform an effective treatment plan.
Given the enrichment of DNA repair deficiencies in advanced disease, for patients who demonstrate resistance to abiraterone or enzalutamide or a robust response to platinum chemotherapy, genomic sequencing of metastatic sites promises to be a powerful tool.
From a diagnostic perspective, patients found to have somatic mutations in DNA repair (in particular, the BRCA1/2, FANC, and PALB2 genes) should also be referred for comprehensive germline testing to fully understand their genetic risk and to inform their family members. Such testing and counseling are now available at many centers of excellence throughout the world. In addition to family assessment, targeted assessment for other cancers known to arise in this syndrome should be conducted.
As for therapeutic approaches, a variety of patient and disease factors need to be taken into consideration. The first factor to consider is disease state. It is unclear when PARP inhibitors or DNA damaging agents (such as platinums) should be introduced to the treatment paradigm, given our recent understanding that DNA repair mutations can present in localized prostate cancer. For patients who have known DNA repair mutations, studies need to be done to identify the best clinical space (localized prostate cancer vs. metastatic castration-sensitive disease vs. metastatic castration-resistant prostate cancer) to introduce PARP inhibitors or DNA-damaging chemotherapy. Beyond platinums and PARP inhibitors, there are other DNA damaging agents that also need to be evaluated, including anthracyclines and topoisomerase inhibitors. It is similarly uncertain at this point if primary prostate biopsy or prostatectomy specimens should be analyzed by genomic sequencing, or if should this be reserved for the patients with refractory metastatic disease progressing past abiraterone, enzalutamide, and chemotherapy
Nonetheless, in patients with DNA repair defects, platinum chemotherapies are likely a good first choice for metastatic disease until PARP inhibitors gain U.S. Food and Drug Administration approval. Looking ahead, along with combining PARP inhibitors with chemotherapy and AR-targeted agents, there is considerable interest in combining PARP inhibitors with immunotherapy, AKT inhibitors, and HDAC inhibitors. Further studies to understand the phenotypic profile of DNA repair mutations in prostate cancer and the prognostic significance of these genomic defects will help clarify how to best treat and care for patients with prostate cancer.
Footnotes
For Further Reading: Elena Castro, Christos Mikropoulos, Elizabeth K. Bancroft et al. The PROFILE Feasibility Study: Targeted Screening of Men With a Family History of Prostate Cancer. The Oncologist 2016;21:716–722.
Implications for Practice: Prostate biopsy is a feasible and safe approach to prostate cancer screening in men with a family history and detects a high proportion of prostate cancer that needs radical treatment. Calculating a polygenic risk score using prostate cancer risk single nucleotide polymorphisms could be a potential future screening tool for prostate cancer.
Author Contributions
Conception/Design: Mallika Dhawan, Charles J. Ryan, Alan Ashworth
Collection and/or assembly of data: Mallika Dhawan, Charles J. Ryan, Alan Ashworth
Data analysis and interpretation: Mallika Dhawan, Charles J. Ryan, Alan Ashworth
Manuscript writing: Mallika Dhawan, Charles J. Ryan, Alan Ashworth
Final approval of manuscript: Mallika Dhawan, Charles J. Ryan, Alan Ashworth
Disclosures
Alan Ashworth: AstraZeneca, Institute of Cancer Research (IP). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer [published correction appears in Cell 2015;162(2):454] Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner J, Ateeq B, Li Y, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer [published correction appears in Cancer Cell 2013;23(4):557] Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31:1748–1757. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Angulo AM, Timms KM, Liu S, et al. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res. 2011;17:1082–1089. doi: 10.1158/1078-0432.CCR-10-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoniou A, Pharoah PD, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: A combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kote-Jarai Z, Leongamornlert D, Saunders E, et al. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: Implications for genetic testing in prostate cancer patients. Br J Cancer. 2011;105:1230–1234. doi: 10.1038/bjc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amé JC, Spenlehauer C, de Murcia G. The PARP superfamily. BioEssays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 12.Dantzer F, de La Rubia G, Ménissier-De Murcia J, et al. Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry. 2000;39:7559–7569. doi: 10.1021/bi0003442. [DOI] [PubMed] [Google Scholar]
- 13.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 14.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 15.Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 17.Clinicatrials.gov. Available at http://www.clinicaltrials.gov. Accessed January 2016.
- Tutt A, Ellis P, Kilburn L et al. The TNT trial: A randomized phase III trial of carboplatin (C) compared with docetaxel (D) for patients with metastatic or recurrent locally advanced triple negative or BRCA1/2 breast cancer (CRUK/07/012) [abstract S3-01]. Presented at the 2014 San Antonio Breast Cancer Symposium. December 11, 2014, San Antonio, Texas. [Google Scholar]
- 19.Rajan A, Carter CA, Kelly RJ, et al. A phase I combination study of olaparib with cisplatin and gemcitabine in adults with solid tumors. Clin Cancer Res. 2012;18:2344–2351. doi: 10.1158/1078-0432.CCR-11-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 21.Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao OS, Goodman OB., Jr Synergistic loss of prostate cancer cell viability by coinhibition of HDAC and PARP. Mol Cancer Res. 2014;12:1755–1766. doi: 10.1158/1541-7786.MCR-14-0173. [DOI] [PubMed] [Google Scholar]
- 23.McCabe N, Turner NC, Lord CJ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-Ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 24.Larsen MJ, Kruse TA, Tan Q, et al. Classifications within molecular subtypes enables identification of BRCA1/BRCA2 mutation carriers by RNA tumor profiling. PLoS One. 2013;8:e64268. doi: 10.1371/journal.pone.0064268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulligan JM, Hill LA, Deharo S, et al. Identification and validation of an anthracycline/cyclophosphamide-based chemotherapy response assay in breast cancer. J Natl Cancer Inst. 2014;106:djt335. doi: 10.1093/jnci/djt335. [DOI] [PMC free article] [PubMed] [Google Scholar]