Abstract
Antibodies capable of neutralizing divergent influenza A viruses could form the basis of a universal vaccine. Here, from subjects enrolled in an H5N1 DNA/MIV-prime-boost influenza vaccine trial, we sorted hemagglutinin-cross reactive memory-B cells and identified three antibody classes, each capable of neutralizing diverse subtypes of group 1 and 2-influenza A viruses. Co-crystal structures with hemagglutinin revealed that each class utilized characteristic germline genes and convergent sequence motifs to recognize overlapping epitopes in the hemagglutinin stem. All six analyzed subjects had sequences from at least one multidonor class, and -in half the subjects- multidonor-class sequences were recovered from >40% of cross reactive-B cells. By contrast, these multidonor-class sequences were rare in published antibody datasets. Vaccination with a divergent hemagglutinin can thus increase the frequency of B cells encoding broad influenza A-neutralizing antibodies; we propose the sequence signature-quantified prevalence of these B cells as a metric to guide universal influenza A-immunization strategies.
INTRODUCTION
Influenza A viruses can be categorized into two phylogenetic groups (group 1 and group 2), each containing diverse subtypes (Figure 1A). Currently, group 1 influenza viruses from the H1 subtype (1918 and 2009 H1N1 pandemics), and the group 2 H3 subtype (1968 H3N2 pandemic), co-circulate and cause seasonal infections in over 10% of the human population each year. Other subtypes have emerged or threaten to re-emerge including the group 1 H2 subtype, endemic in humans from 1957–1968, the group 1 H5 subtype, which includes lethal avian strains (Subbarao et al., 1998), and the group 1 H6 and H9 and the group 2 H7 and H10 subtypes, which have been associated with human infections and fatalities in recent years (Butt et al., 2005; Morens et al., 2013). Frequent zoonotic cross-overs that may cause pandemics of unpredictable frequency and severity highlight the need for a universal influenza vaccine that is capable of protecting against divergent influenza A viruses.
Figure 1. H5N1-vaccine recipients have cross-reactive B cells that utilize the same genetic elements and neutralize group 1 and 2-influenza A viruses.
(A) Phylogenetic tree depicting influenza A subtypes, generated using HA sequences (one per subtype) with program MEGA6. Scale bar indicates distance per fractional nucleotide change. (B) Neutralization by serum from 63 vaccinees, sampled two weeks after final H5N1 immunization and assessed against vaccine strain (A/Indonesia/5/2005) and heterologous group 2 HA strains (H3N2: A/Hong Kong/1-4-MA21-1/1968; H7N7: A/Netherlands/219/2003). Ten subjects were selected for flow cytometric (FACS) characterization, as highlighted in key. Dotted line indicates the limit of detection. (C) FACS analysis of PBMC samples isolated from H5N1-vaccine recipients two weeks after final vaccination and co-stained with HA probes H5 (A/Indonesia/5/2005) and H3 (A/Perth/16/2009). Sizable populations of H5-H3 cross-reactive memory B cells observed in six of ten subjects (Fig. S2). (D) Clonal diversity of H5-H3 cross-reactive B cells. The HV repertoire from each subject is shown as a pie chart; with each slice representing a unique HV clone or clonally related family. Total number of HV sequences recovered per subject is indicated by the number at the center of each pie chart. (E) Genetic and functional characteristics of selected antibodies recovered from H5-H3 cross-reactive B cells. Structurally characterized antibodies indicated by ☼. See also Figures S1–4 and Tables S1–S4.
Potential approaches to a universal influenza vaccine involve the elicitation of neutralizing antibodies that recognize the influenza hemagglutinin (HA) from multiple subtypes, thereby providing protection from divergent influenza viruses. One means to accomplish this involves ontogeny-based strategies, which seek to identify antibodies of reproducible classes and to induce similar antibodies by vaccination (Jardine et al., 2015; Lingwood et al., 2012; Pappas et al., 2014). We consider antibodies to be of the same class when they recognize the same region, employ the same structural mode of recognition, and develop through similar recombination and maturation pathways (Kwong and Mascola, 2012). Reproducible classes, which are observed in multiple individuals, represent immunological solutions to the challenge of broad influenza A neutralization that might be available to the general human population.
The influenza A-neutralizing stem-directed antibodies that utilize the HV1-69 germline gene are one such multidonor class (Ekiert et al., 2009; Sui et al., 2009). In terms of reproducibility, the HV1-69-derived antibodies have the additional advantage of utilizing heavy chain-only recognition, and prior studies have shown their vaccine-induced elicitation (Khurana et al., 2013; Ledgerwood et al., 2011; Ledgerwood et al., 2013; Sui et al., 2009; Wheatley et al., 2015; Whittle et al., 2014). However, HV1-69-derived antibodies generally do not neutralize both group 1 and 2 strains of influenza A, and, only a single HV1-69-derived antibody has been identified (CR9114) capable of neutralizing both group 1 and 2 strains of influenza A (Dreyfus et al., 2012). Other broadly neutralizing antibodies have been identified, such as FI6v3 and 39.29, both of which derive from the HV3-30 germline gene; however, co-crystal structures with HA reveal different modes of recognition (Corti et al., 2011; Nakamura et al., 2013), and FI6v3 and 39.29 are thus not members of the same class. Indeed, a reproducible antibody class capable of neutralizing both group 1 and 2 influenza A viruses has not been observed in multiple donors.
We previously showed that subjects enrolled in the phase I clinical trial, VRC 310 – who received an A/Indonesia/05/2005 monovalent inactivated virus (MIV) vaccine primed by an H5 DNA plasmid vaccine (Ledgerwood et al., 2011; Ledgerwood et al., 2013) (Table S1A) – showed transient expansion of H1- and H5-cross reactive memory B cells specific to the HA stem (Wheatley et al., 2015; Whittle et al., 2014). To determine whether these memory B cells might encode multidonor class antibodies capable of neutralizing group 1 and 2 influenza A virus, we sorted memory B cells that reacted with both H5 (group 1) and H3 (group 2) HAs. Immunoglobulin transcripts from post-vaccination cross-reactive memory B cells were sequenced, and the encoded antibodies synthesized and characterized. Specifically, we assessed breadth and potency of influenza A virus neutralization, determined representative crystal structures in complex with HA, analyzed sequence convergence based upon V(D)J-gene recombination and somatic hypermutation (SHM), and tested sequence signatures for their ability to identify group 1 and 2-neutralizing antibodies. Our findings reveal reproducible immunological pathways to achieve broadly reactive antibodies and support a B cell ontogeny-based approach to obtaining a universal influenza A vaccine.
RESULTS
Identification of memory B cells cross-reactive with group 1 and 2 influenza A HAs
We studied ten subjects from the VRC 310 H5N1 vaccine trial who displayed a range of vaccine-elicited serum H5N1 neutralization activity, as well as varied but detectable responses against group 2 strains A/Hong Kong/1-4-MA21-1/1968 (H3N2) or A/Netherlands/219/2003 (H7N7) (Figures 1B and S1, Tables S1B and S2). We used recombinant group 1-specific (H5) and group 2-specific (H3) HA probes – modified to prevent sialic acid binding (HAΔSA) (Wheatley et al., 2015; Whittle et al., 2014) – to co-stain and sort PBMCs isolated two weeks post H5N1 MIV boost (Figure 1C and S2). We recovered sequences of memory B cell immunoglobulin gene transcripts from six of the ten studied subjects (Figure 1D).
The sequence repertoire of each subject was generally dominated by clonally related transcripts comprising a small number of clonal expansions. Transcripts derived from diverse HV genes including HV 1-18, 3-23, 3-64D, 4-30, 4-34 and 6-1 (Figure 1D, E). Notably, transcripts from HV1-69, which often dominate group 1-specific stem-reactive antibodies (Lingwood et al., 2012; Pappas et al., 2014; Wheatley et al., 2015), comprised only 2.5% of this set of group 1 (H5+)-group 2 (H3+) double-positive memory B cells.
Group 1 and 2-neutralizing antibodies from different vaccinees are genetically similar
Immunoglobulin sequences recovered from H5+ and H3+ cross-reactive memory B cells (Table S3) showed surprising similarity. Notably, many immunoglobulin sequences from different donors derived from the same genetic elements (Figure 1E). To analyze commonalities of immunoglobulin transcripts between subjects, we considered the following seven genetic elements: inferred heavy variable (HV), heavy diversity (HD), and heavy joining (HJ) genes, and 3rd-heavy chain complementarity-determining region (CDR H3) length for heavy chain-gene transcript, and inferred light variable (LV) and light joining (LJ) genes, and CDR L3 length for corresponding light chain transcripts (Figure 1E). Frequentist analysis indicated the presence of four or more of the same genetic elements in separate lineages to be statistically significant (P ≤ 0.001) (Figure 1E), and representative antibodies were cloned and expressed from such lineages. Of note, our antibody nomenclature specifies donor, lineage, and clone; e.g., antibody 56.a.09 (Figure 1E, third row) is named for subject (56), lineage within this subject (a), and clone within this lineage (09). Most of the expressed antibodies bound HA, and all antibodies that bound HA competed with the antigen-binding fragment (Fab) of the stem-directed antibodies CR9114 (Dreyfus et al., 2012) or F10 (Sui et al., 2009) (Figure 1E), and negative stain-electron microscopy (EM) indicated binding to the HA stem (Figure S3). Most of these antibodies neutralized viruses from both group 1 and 2 HA, including H1, H3, H5 and H7 subtypes, with select antibodies also demonstrating neutralization of viruses from subtypes H2, H9, and H10 (Figures 1E and S4, Table S4). Notably, in both pseudovirus and microneutralization assays of influenza A viruses, the breadth and potency for several of the newly identified antibodies was comparable to that of antibody CR9114 (Dreyfus et al., 2012).
To understand the immunological basis of these multidonor humoral responses against influenza A, we analyzed the recombination, SHM, and structural constraints, which drove the generation and development of these antibodies.
A multidonor class of broadly neutralizing antibodies with HV6-1+HD3-3 germline genes
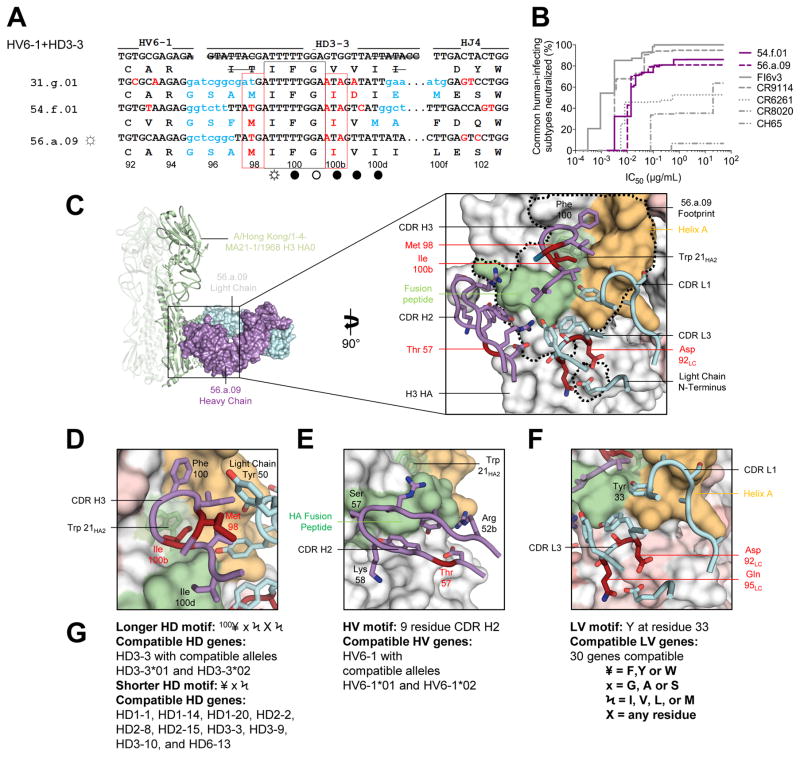
Three memory B cell lineages, from subjects 31, 54 and 56, shared heavy chain sequences derived from recombination of HV6-1, HD3-3, and HJ4 or HJ5 to yield highly similar amino acid sequences in CDR H3 (Figure 1E, top row; Table S3). In each case, the heavy chain was paired with a light chain sequence from KV3-20, KJ2 or KJ3 resulting in a CDR L3 of 9 amino acids. Similar affinity maturation patterns were observed: a Val100bIleHC alteration of an HD-gene encoded section of the CDR H3 was completely conserved in all three lineages (Figure 2A) (for clarity, each residue number is followed by a subscript denoting parent molecule: HC for heavy chain; LC for light chain; HA1, HA2 or HA for either HA subunit or HA in general). Notably, these antibodies displayed neutralization breadth and potency that rivaled that of CR9114 and exceeded that of the group-specific CR6261 or CR8020 or of the head-directed antibody CH65 (Ekiert et al., 2009; Ekiert et al., 2011; Whittle et al., 2011) (Figures 2B and S4, Table S4).
Figure 2. A multidonor HV6-1+HD3-3 class of broadly neutralizing antibodies.
(A) Immunoglobulin heavy chains utilizing germline genes HV6-1, HD3-3 and HJ4 or HJ5. Germline HV, HD, and HJ gene-encoded nucleotide and amino-acid residues are shown in black, with junction-encoded residues in light blue and residues that have undergone SHM in red. Nucleotides removed by exonuclease trimming indicated with a line through the letters. Conserved HD3-3 encoded residues (IFG) highlighted by a black box; recurrent SHM-derived Ile100bHC highlighted by a red box. HA contacts indicated with open circles (○) denoting antibody main-chain-only contacts, open circles with rays (☼) denoting antibody side-chain-only contacts, and filled circles (●) denoting both main-chain and side-chain contacts. (B) Neutralization breadth-potency curve for HV6-1+HD 3-3 antibodies, with breadth shown as percentage of pseudoviruses neutralized at each IC50 cutoff, and virus panel comprising 15 strains that includes influenza A subtypes known to infect humans (H1, H2, H3, H5, H7, H9). (C) Co-crystal structure of Fab 56.a.09 in complex with an H3 HA monomer (A/Hong Kong/1-4-MA21-1/1968). Fab heavy and light chains colored purple and cyan respectively and depicted in surface representation, while the H3 HA is depicted in ribbon and shown as a trimer (Figure S5 shows HA crystal packing). Inset: Interacting CDR loops of the 56.a.09 Fab are shown in ribbon and sticks and colored as in panel (A) with the antibody footprint outlined. (D) A five-amino acid motif within the CDR H3 inserts into the conserved Trp21 pocket of H3. (E) HV6-1encoded CDR H2 depicted with HA fusion peptide; HV6-1 is the only HV gene which encodes a nine residue CDR H2. (F) Light chain interactions that contribute to the antibody-binding surface; these are not specific to KV3-20. (G) Analysis of antibody gene compatibility, highlighting additional CDR H3 residues that may be compatible with HA binding, e.g. 100HC could be a F, Y, or W residue (¥), 101HC could be a G, A, or S residue (x). See also Figures S3–S7 and Tables S3–S7.
To provide insight into the structural basis for the similarity between these HV6-1+HD3-3 antibodies, we determined the crystal structure of the antigen-binding fragment (Fab) for antibody, 56.a.09, alone, and in complex with A/Hong Kong/1-4-MA21-1/1968 (H3N2) HA at 3.3 Å resolution (Figure 2C and S5, Tables S5 and S6). Unexpectedly, the crystallized HA was not trimeric, with the asymmetric unit for the crystallized Fab-H3 complex comprising an HA head of one protomer interacting with the HA stem of an adjacent protomer in a “head-to-stem” dimeric arrangement (Figure S5B, C). Despite this non-trimeric arrangement, the Cα-RMSD between the 56.a.09-bound HA and the ligand-free HA was less than 1 Å, and, for clarity, we thus depict the 56.a.09 bound complex as a typical HA trimer (Figure 2C).
Antibody 56.a.09 recognized a conserved region on the HA stem in a manner that avoided glycans at residues Asn21HA1 (conserved on group 1 viruses) and Asn38HA1 (conserved on group 2 viruses), providing a structural explanation for its extraordinary group 1 and 2 neutralization breadth. Antibody 56.a.09 bound primarily with its heavy chain (934 Å2 buried surface area (BSA) versus 386 Å2 BSA for the light chain). Heavy chain binding involved the HD3-3-encoded CDR H3 (Figure 2C) with Phe100HC and Gly100aHC contributing ~240 Å2 of BSA and the SHM-altered Val100bIleHC inserting directly into the Trp21HA2 pocket (contributing over 100 Å2 of interactive surface). In addition, Met98HC interacted with a conserved aromatic residue present on all light chains, helping to orient the CDR H3 (Figure 2D). Heavy-chain binding also involved the HV6-1 germline-encoded CDR H2, which uniquely encodes a 9 amino acid CDR H2, contributed 182 Å2 of BSA, was unmutated in contact residues in all three subjects (Figure S6), and interacted with the conserved fusion peptide (Figure 2E). With respect to light chain, the largely unmutated KV3-20-derived V genes (4–6% SHM) observed in lineages from these three subjects interacted with the HA stem through both CDR L1 and CDR L3 (Figure 2F, Table S6), with Tyr33LC contributing the largest BSA among all light chain residues (73.1 Å2).
HV6-1+HD3-3-derived antibodies were thus found in three independent donors, shared genetic elements in both the heavy and light chain, displayed convergent affinity maturation, and appeared to share the same mode of recognition (structure-function analysis of recognition interface and SHM are provided in Figure S7). Furthermore, we tested for functional complementation: swapping of heavy and light chains of the three HV6-1+HD3-3-derived antibodies resulted in six functional antibodies from nine possible pairings (all three pairing with the heavy chain of antibody 31.g.01 failed to express) (Table S7). Overall, these results indicate the HV6-1+HD3-3-derived antibodies form a multidonor class. Structural analysis indicated numerous light chains to be compatible with binding, and approximately 99% of the human population (Genomes Project et al., 2012) possess alleles of the HV6-1 and HD3-3 genes compatible with the class elicitation and recognition described here (Figures 2D–G).
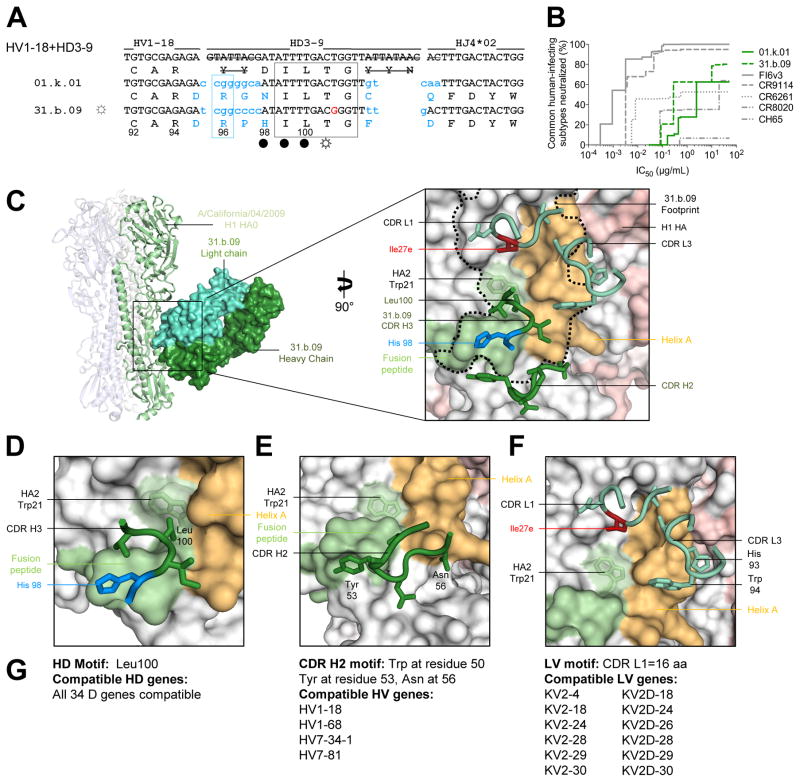
A second multidonor class of broadly neutralizing antibodies with HV1-18+HD3-9 germline genes
Two memory B cell lineages from subjects 1 and 31 shared immunoglobulin heavy chain sequence derived from recombination of HV1-18 with HD3-9 and HJ4 to yield highly similar amino acid sequences in a CDR H3 of 15-amino acids (Figures 1E and S6). Notably an Arg96HC residue was encoded by N-nucleotide addition in both cases (Figure 3A). In each donor, the heavy chain was paired with a light chain derived from KV2-30. Encoded immunoglobulins were expressed and shown to neutralize primarily group 1 strains of influenza A, although a few group 2 strains were neutralized (Figure S4, Table S4). Overall neutralization from these HV1-18+HD3-9 antibodies appeared more similar to the group 1-specific antibody CR6261, than to the very broad CR9114 or HV6-1-derived antibodies; nevertheless, neutralization breadth encompassed ~50% of influenza A subtypes that commonly infect humans (Figure 3B).
Figure 3. A multidonor HV1-18+HD3-9 class of broadly neutralizing antibodies.
(A) Immunoglobulin heavy chains utilizing germline genes HV1-18, HD3-9 and HJ4, with sequences annotated as described in Figure 2A. (B) Neutralization breadth-potency curve for HV1-18+HD 3-9 antibodies on a panel of influenza A viruses that includes subtypes known to infect humans. (C) Co-crystal structure of Fab 31.b.09 in complex with an H1 trimer (A/California/04/2009). Fab heavy and light chains colored dark green and light green respectively and depicted in surface representation, while the H1 HA is depicted in ribbon and colored blue, green, and white. Inset: Interacting CDR loops of the 31.b.09 Fab are shown in ribbon and sticks and colored as in panel (A) with the antibody footprint outlined. (D) A conserved motif within the CDR H3 inserts into the highly conserved Trp21 pocket of HA while also interacting with the fusion peptide. (E) The HV1-18 encoded CDR H2 also interacts with the opposing side of the fusion peptide. (F) Light chain interactions from CDR L3 and CDR L1 also contribute to the antibody binding surface area. (G) Analysis of antibody gene compatibility. See also Figures S3–S7 and Tables S3–S7.
We determined the crystal structure of Fab 31.b.09 in complex with A/California/04/2009 H1 (Figure 3C, Tables S5 and S6). Similar to the 56.a.09-H3 complex structure, the crystallized hemagglutinin in the Fab 31.b.09 complex was not a trimer, but a molecular dimer (Figure S5). Despite this unexpected non-trimeric arrangement, the Cα-RMSD between the 31.b.09-bound HA and the ligand-free HA in the stem region was 0.6 Å, and for clarity we depict the 31.b.09 bound complex in a more typical trimeric arrangement (Figure 3C). The HV1-18+HD3-9-derived antibody 31.b.09 bound an epitope that overlapped the HV6-1+HD3-3 class epitope, but with antibody rotated ~105° degrees (mostly involving a rotation perpendicular to the trimer axis) (Figure 3C). Antibody 31.b.09 bound with both heavy and light chains (343 Å2 BSA for heavy chain and 540 Å2 BSA for the light chain). Heavy chain interactions were generated through CDR H2 and H3 loops. In CDR H2 (127 Å2 BSA), the HV1-18 germline encoded Tyr53HC recognized the fusion peptide of HA2 and Asn56HC recognized helix A of HA1, while the CDR H3 (216 Å2 BSA) was positioned over the fusion peptide-helix A interface with Ile99HC and Leu100HC inserting into the hydrophobic groove between these two conserved elements (Figure 3C–E). Light chain interactions involved CDR L1 and L3, which recognized helix A (Figure 3F). We tested functional complementation: swapping of heavy and light chains between two antibodies of the putative class resulted in functional antibodies for all four of the possible pairings (Table S7). Overall the results indicated the HV1-18+HD3-9-derived antibodies form a multidonor class. The light chain had a 16 amino acid CDR L1, which could be encoded by 12 other LV genes, and residue Ile27eLC, which forms a critical contact, was a product of SHM. The multiple alternative D gene alleles that could be used to generate CDR H3s compatible with recognition by this multidonor class, in combination with a large number of compatible light chains, led to a calculated distribution of potential HV1-18 combinations in the human population of close to 100% (Genomes Project et al., 2012) (Figure 3G).
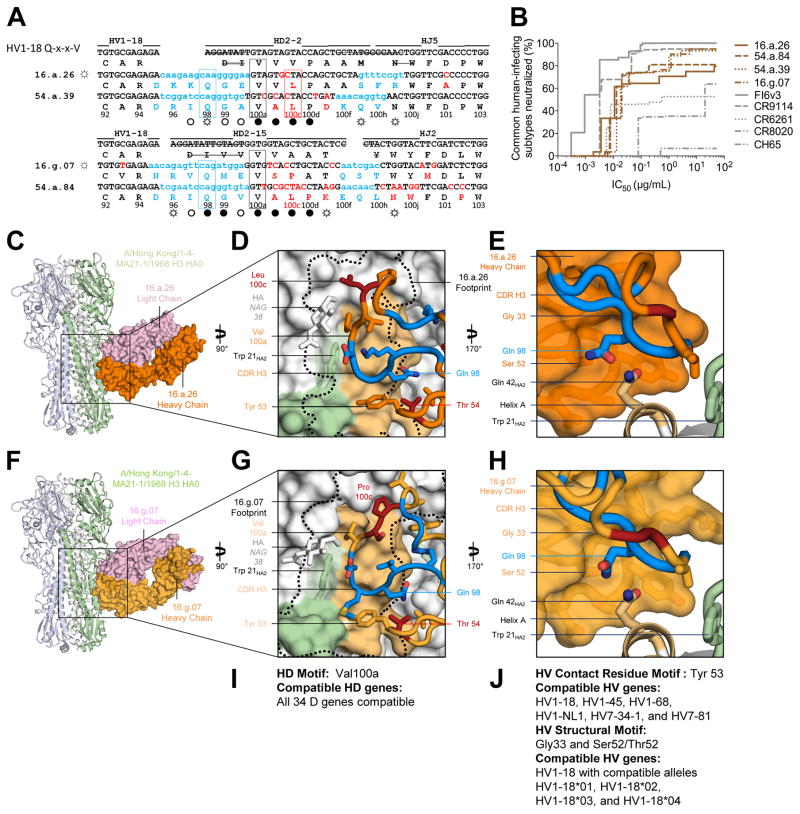
A third multidonor class of broadly neutralizing antibodies with HV1-18 germline gene and Q-x-x-V motif
Multiple B cell lineages in two subjects (16 and 54) produced distinctive HV1-18-derived immunoglobulins sharing five genetic elements and having a CDR H3 of 21 amino acids derived from recombination with either HD2-2 or HD2-15 genes (Figure 4A, Table S3). This set of immunoglobulins all shared an SHM-derived Thr54HC, a Gln98HC encoded by P-nucleotide addition, and a germline HD-encoded aliphatic residue at position100aHC (Q-x-x-V motif). Neutralization breadth and potency for this set of immunoglobulins were similar to that of antibody CR9114 (Figure S4, Table S4), neutralizing all common human-infecting subtypes of influenza A except H2, and substantially exceeded the breadth of group 1-specific or group 2-specific stem antibodies or the head-directed antibody CH65 (Figure 4B). To understand the basis of their recognition, we crystallized representative antibodies with HA. Co-crystal structures of representative HV1-18+HD2-2 (16.a.26) and HV1-18+HD2-15 (16.g.07) Fabs in complex with the A/Hong Kong/1-4-MA21-1/1968 (H3N2) HA revealed highly similar epitopes (Figure 4C–H, Tables S5, and S6). Antibody recognition occurred primarily through heavy chain recognition of the conserved HA stem. With 16.a.26, the heavy chain contributed 514 Å2 of BSA while the light chain contributed 283 Å2 of BSA, and with 16.g.07, the heavy chain contributed 646 Å2 of BSA while the light chain contributed 329 Å2 of BSA.
Figure 4. A multidonor HV1-18 class of broadly neutralizing antibodies with Q-x-x-V motif.
(A) Immunoglobulin heavy chains utilizing germline genes HV1-18, HD2-2 or HD2-15 and HJ5 or HJ2, with sequences annotated as described in Figure 2A. (B) Neutralization breadth-potency curve for HV1-18 (Q-x-x-V) class antibodies on a panel of influenza A viruses that includes subtypes known to infect humans. (C) Co-crystal structure of Fab 16.a.26 (HV1-18, HD2-2, HJ5) in complex with H3-HK68. Fab heavy and light chains colored orange and lavender respectively and depicted in surface representation while HA is depicted in ribbon and shown as a trimer. (D) The 16.a.26 CDR H3 is depicted with junction-encoded and mutated residues colored as in panel (A) and germline-encoded residues in orange with the antibody footprint on the HA outlined in black. (E) Antibody heavy chain depicted in surface representation with CDR H3 loop in ribbon. 16.a.26 Gln98HC occupies a turn in the CDR H3, which interacts with the conserved residue Gln42HA2. (F) Co-crystal structure of Fab 16.g.07 (HV1-18, HD2-15, and HJ2) in complex with H3-HK68 depicted as in panel (C). (G) The 16.g.07 CDR H3 depicted as in panel (D) with the antibody footprint on the HA outlined in black. (H) The 16.g.07 CDR H3 is depicted as in panel (E) with Gln98HC contacting Gln42HA2. (I) Analysis of D-gene compatibility. (J) Analysis of V-gene compatibility. See also Figures S3–S7 and Tables S3–S7.
Heavy chain interactions were generated primarily through CDR H2 (~150 Å2 BSA for 16.a.26 and ~200 Å2 BSA for 16.g.07) and CDR H3 (~300 Å2 BSA for 16.a.26 and ~450 Å2 BSA for 16.g.07) with the HV1-18 germline-encoded Tyr53HC and the SHM-derived Thr54HC recognizing the N-terminal region of HA1 and the hydrophobic groove between helix A and the fusion peptide of HA2 (Figures 4D, G, Table S6). The CDR H3s of these antibodies also bound the conserved hydrophobic groove next to helix A and adjacent to Trp21HA2. The conserved Val100aHC inserted into a pocket present on both group 1 and 2 HAs, just above Trp21HA2 and proximal to Ile48HA2. Despite differences between residues encoded by HD2-2 (16.a.26) and HD2-15 (16.g.07), the CDR H3s from both antibodies were oriented perpendicular to the Fab axis and interacted similarly with HA. The conserved Gln98HC interacted with Gln42HA2 by utilizing a germline-encoded pocket unique to HV1-18-encoded antibodies, which was formed by antibody framework residues Gly33HC and Ser 52HC (Figure 4E, H). The location of Gln98HC within the framework pocket likely stabilized the perpendicular orientation of the CDR H3 relative to the antibody-framework regions, and may allow for CDR H3-motifs derived from diverse D genes to bind to this conserved hydrophobic groove. In this regard, we note that, sequences from subject 01 with an HV1-18 germline gene and a Gln98-x-x-Val100a motif in a 17 amino acid CDR H3 were able to neutralize H3 and H5 strains of influenza. Although light chains of this class contribute about one-third of the total buried surface area, analysis of antibodies of this class revealed light chain sequences to derive from diverse LV genes, with only KV3-11 appearing twice. Also, the recently reported HV1-18-derived group 1 and group 2-neutralizing antibody CT149 uses a Gln98HC-x-x-Val100aHC motif with a 19 amino acid CDR H3 to bind the HA stem (Wu et al., 2015) and recognized HA in a manner highly similar to both 16.a.26 and 16.g.07. We tested the functional complementation for antibodies 16.a.26, 16.g.07, 54.a.39, 54.a.84, and CT149 from donors 16, 54, and SH-K1 (the source of antibody CT149). Swapping of heavy and light chains between these five antibodies resulted in 10 functional antibodies from the 25 possible pairings (Table S7). Functionality correlated strongly with HV gene identity and CDR H3 length (Table S7), suggesting a requirement for specific heavy-light chain pairings. Despite this requirement, the HV1-18 antibodies with 54Thr and CDR H3 98Q-x-x-V motif appeared to form a multidonor class. Thus the recombination of HV1-18 with many alternative D gene segments yields a Q-x-x-V motif in the CDR H3, which may comprise a common solution for neutralization of group1 and group 2 influenza A viruses. The many alternative HD gene alleles that can be used to generate CDR H3s for this multidonor class in combination with no obvious light chain bias led to a calculated distribution of potential HV1-18 combinations in the human population of close to 100% (Genomes Project et al., 2012) (Figure 4I, J).
Multidonor and lineage-unique antibodies that neutralize group 1 and 2 viruses recognize similar epitopes
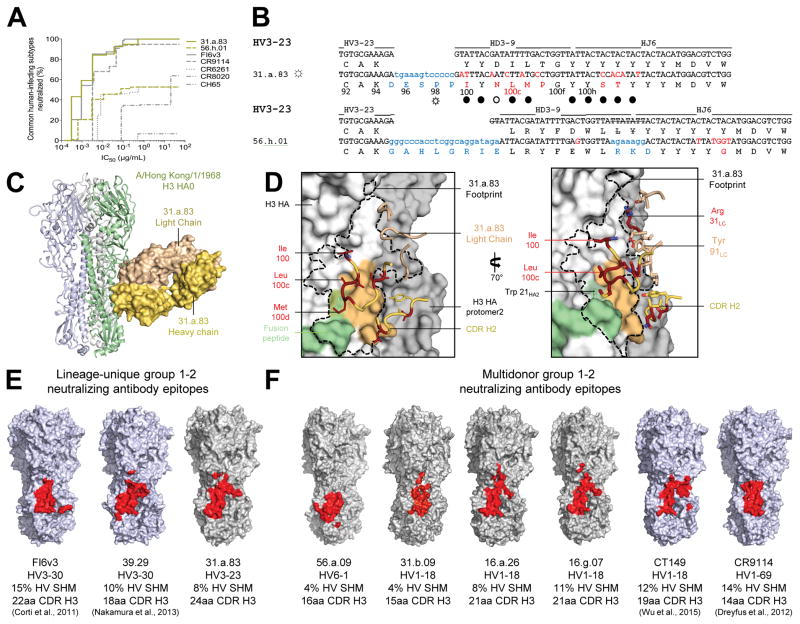
For the six subjects analyzed, the most highly expanded lineage was lineage “a” from subject 31, from which we sequenced 104 clones (Figure 1E, Table S2). Clone 83 of this lineage (antibody 31.a.83) derived from HV3-23, HD3-9 and HJ6 and displayed the highest neutralization breadth of the antibodies we sequenced and expressed, neutralizing all of the common influenza A subtypes that infect humans (Figure 5A). Structural characterization of antibody 31.a.83 in complex with HA-HK68 (Table S5) revealed binding to the conserved HA stem, primarily through CDR H3 residues, many of which were somatically hypermutated (Figure 5B–D, Table S6). Antibody 31.a.83 utilized hydrophobic residues to contact an epitope adjacent to helix A and involving the Trp21 pocket with its CDR H3 parallel to helix A in a manner that avoided the conserved N-glycan at Asn38HA2 present in group 2 HAs.
Figure 5. A conserved site of group 1 and 2-influenza A virus vulnerability targeted by multidonor and lineage-unique antibodies.
(A) Neutralization breadth-potency curve for HV3-23-derived antibodies, on a panel of influenza A viruses that includes subtypes known to infect humans. (B) Immunoglobulin heavy chains utilizing germline genes HV3-23, HD3-9 and HJ6, with sequences annotated as described in Figure 2A. (C) Co-crystal structure of Fab 31.a.83 in complex with HA (H3-HK68). (D) Epitope for antibody 31.a.83 (outlined in black) with heavy chain depicted in yellow and junction-encoded residues (highlighted in blue), mutated residues (in red), and germline-encoded residues (in gold) and with light chain depicted in tan. (E–F) Antibodies capable of neutralizing group 1 and 2 influenza A viruses recognize overlapping epitopes within the HA stem (colored red). The HA surface is colored blue for structures determined previously (FI6v3 PDB ID:3ZTN; 39.29 PDB ID:4KVN; CT149 PDB ID:4R8W; CR9114 PDB ID:4FQI), and gray for structures determined in the current study. See also Figures S3–S6 and Tables S3–S7.
A second lineage, “h” from subject 56 (lineage 56.h), also derived from the same germline genes as antibody 31.a.83 (Figure 5B and S6). However, critical CDR H3 contact residues of antibody 31.a.83 were not conserved and a 4-amino acid shift relative to the V-gene was observed (Figure 5B). Moreover, the representative antibody we expressed and analyzed from this second lineage, antibody 56.h.01, neutralized H1, H2 and H9 subtypes, but not H3, H5, or H7 subtypes (Figures 1E and S4, Table S4). Together, these observations indicated lineages 31.a and 56.h to have different modes of recognition, providing an example of influenza A-targeting antibodies that derived from the same heavy chain-VDJ genes, but did not use the same mode of recognition nor share convergent development.
Thus antibody lineages may be multidonor (common or public), meaning that they are observed in different individuals and share the same genetic elements and mode of recognition (Henry Dunand and Wilson, 2015; Zhou et al., 2013), or unique (uncommon or private), meaning that they have only been observed a single time. We observed no substantial difference between epitopes of multidonor and unique antibodies capable of neutralizing group 1 and 2-influenza A viruses (Figures 5E, F), nor did we observe segregation in antibody approach to HA used by multidonor or unique antibodies (Figures S3E, F). Antibodies from multidonor lineages did, however, have lower SHM (averaging 8% for multidonor versus 11% for the unique). The lower SHM suggested that multidonor antibodies may undergo more parsimonious development. We note that unique lineages accounted for the majority of cross-reactive B cells in 4 of the 6 analyzed VRC 310 donors (1, 31, 36 and 56); in light of the positive functional characteristics of several of unique lineages (e.g. 31.a.83), it seems likely that these antibodies would contribute to a protective response. As the number of donors with sequenced cross-reactive memory B cells increases, we would expect some of the antibodies described here as “unique” to be observed in other donors.
Sequence signatures for multidonor antibody classes
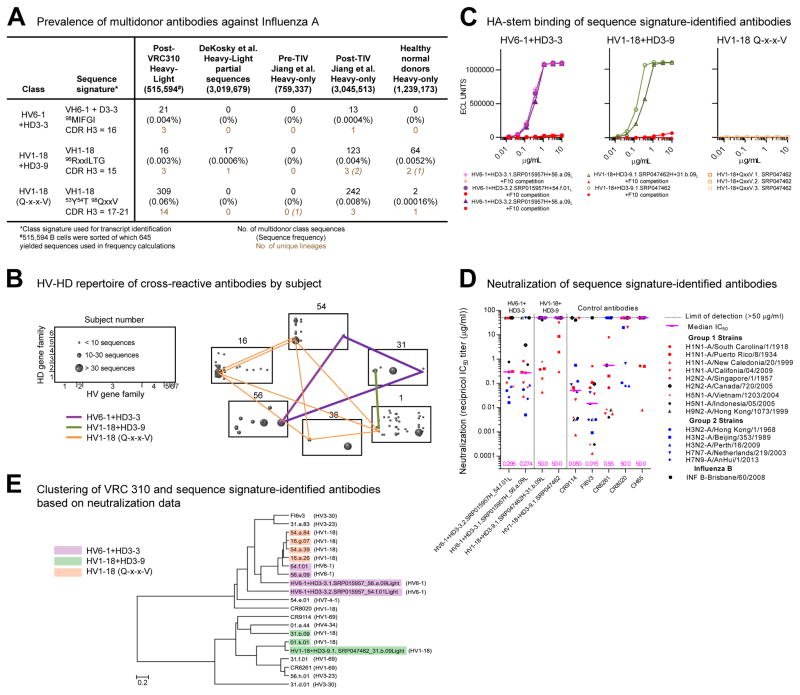
We analyzed the multidonor antibodies identified here for class specific-sequence signatures (Figure 6A) as a means to quantify class transcripts and to identify potential class members in other subjects. Notably, despite a sequence signature requiring residue 54HC to be altered by affinity maturation, the HV1-18 (Q-x-x-V) class accounted for over half of the H5+ and H3+ cross-reactive B cells we sequenced (Figure 6A) and could be found in half the analyzed vaccinees (Figure 6B).
Figure 6. Sequence signatures of multidonor antibody lineages.
(A) Multidonor class antibodies capable of neutralizing group 1 and group 2-influenza A viruses: sequence signature, number, and frequency of signature-identified class members. The number of singleton transcripts from 454-derived NGS data is reported in italicized font; these were omitted from transcript and lineage quantifications as described in Supplementary Methods. (B) HV-HD germline-gene origin plots. One box shown per subject with antibody frequencies plotted as circles at their HV (horizontal), HD (vertical) coordinates. The size of the plotted circle corresponds to the number of antibody sequences as shown in key at left. Multidonor antibodies in different subjects connected by lines colored according to each multidonor class. (C) Binding and competition MSD-ECLIA for antibodies identified in (DeKosky et al., 2015; Jiang et al., 2013) databases; competition with stem antibody F10 highlighted in red. (D) Neutralization of influenza pseudoviruses. Median IC50 for each antibody is indicated by a horizontal line with value (μg/ml) shown at the base of the graph. (E) Clustering of antibodies based on their neutralization fingerprints on a 15-virus panel. See also Figures S6 and Tables S3–S4, S7.
For other subjects, we searched published human antibody datasets, both those with paired heavy-light sequences (DeKosky et al., 2015), as well as those with heavy-chain only sequences (Figure 6A, Table S1C). Searches with the HV6-1+HD3-3 signature did not yield sequence matches in paired heavy-light sequences, but in the published heavy chain-only datasets, we found 13 matches, which appeared to derive from a single lineage (Figure S6). We chose both consensus as well as the sequence closest to consensus to synthesize and reconstitute with light chains of HV6-1+HD3-3 class antibodies from subjects 54 and 56. One of these reconstituted antibodies did not express, but the other three did and bound H1 HA in a manner that could be competed with antibody F10 (Figure 6C). We tested two of these antibodies, and both neutralized group 1 and 2-influenza A strains (Figure 6D, Table S3), and the neutralization signatures (Georgiev et al., 2013) of the synthesized antibodies clustered in a dendrogram with known HV6-1+HD3-3 class antibodies (Figure 6E).
With the HV1-18+HD3-9 signature, we found 17 matches in published paired heavy-light chain sequences, which appeared to derive from a single lineage. We synthesized consensus sequences and reconstituted published heavy and light chains as well as the published heavy chain and light chain of this class from subject 31. Both of these reconstituted antibodies bound H1 hemagglutinin in a manner that could be competed with antibody F10 (Figure 6C), neutralized group 1 influenza A strains (Figure 6D, Table S4), and clustered in a neutralization dendrogram with known HV1-18+HD3-9 class antibodies (Figure 6E). The neutralization breadth of these antibodies was lower than those isolated from VRC 310 subjects, likely due to the use of germline sequences to complete the CDR L1 and CDR L2 regions of this antibody (the somatic mutation of 27ELC to Ile is required for optimal recognition (Figure 3F)).
With the HV1-18 (Q-x-x-V) signature, searches did not yield sequence matches in paired heavy-light sequences, but in published heavy chain-only datasets (Table S1C), we found 244 matches, which appeared to derive from 4 lineages (Figure 6A and S6). We synthesized four sequences (consensus or closest NGS read) and reconstituted with the five light chains used previously in swapping experiments (Table S7). None of these reconstituted antibodies bound a set of HAs (Figure 6C) or neutralized any of the 15 viruses in our neutralization panel. Analysis of the tested heavy chains indicated that their CDR H3 length matched that of CT149 in three of four cases, but was below the 78% identity threshold that correlated with function in heavy-light complementation of this class (Table S7).
Altogether the results indicate sequence signatures with sufficient specificity to identify other functional class members by sequence alone could be obtained for two multidonor classes: HV6-1+HD3-3 and HV1-18+HD3-9. The sequence signature for the third class, HV1-18 (Q-x-x-V), was complicated by incompatibility of some heavy-light pairs from this class; nonetheless, sequence searches for this third class did place an upper limit on the prevalence of this multidonor antibody class in the searched databases.
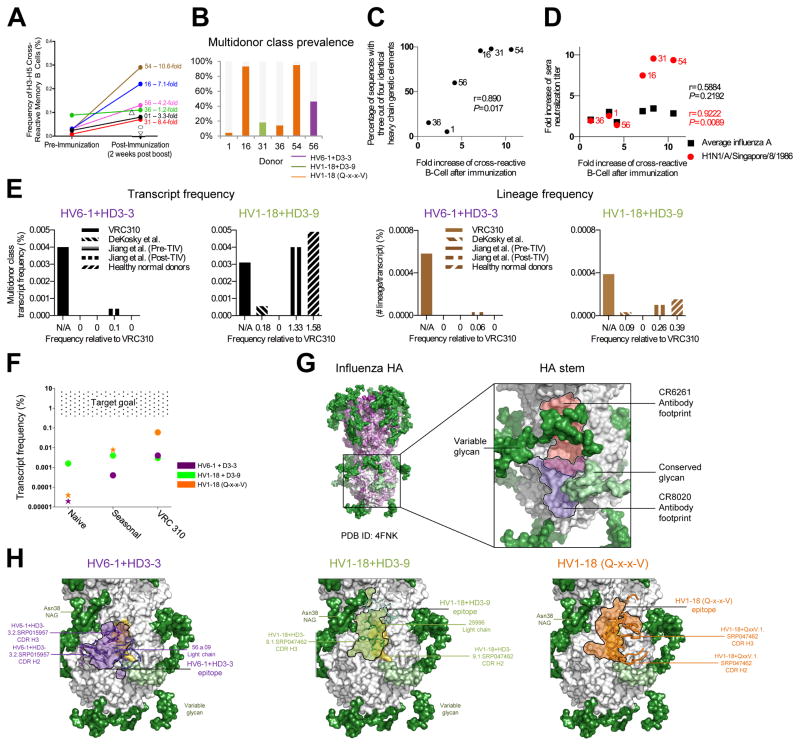
Vaccine induction of multidonor broadly neutralizing antibodies
In the VRC 310 trial, we observed a significant expansion (P=0.0284) of H5+H3+ memory B cells following H5 DNA prime-MIV boost, ranging from an increase of 1.2- to 10.6-fold (Figure 7A). Notably subjects with the largest increases and the highest frequencies of multidonor antibodies had the largest percentages of antibodies belonging to the three multidonor classes identified here (Figure 7B). Our initial observation of a high number of transcripts with multiple genetic commonalities may be explained by the preferential expansion of multidonor class transcripts; indeed, the fold-increase in cross-reactive B cells by subject correlated with the percentage of antibody sequences with inter-subject genetic commonalities (Figure 7C). Importantly, the frequency of cross-reactive memory B cells post-VRC 310 vaccination correlated with the sequence signature-identified prevalence of multidonor class antibodies (P=0.0045) (Figure S6D). Moreover, while we did not see significant correlation between fold increase in overall sera titer versus an increase in cross-reactive memory B cells, we did observe a significant correlation in titers with the H1N1 virus, A/Singapore/8/1986, which we previously found to be especially sensitive to neutralization by stem-directed antibodies (Lingwood et al., 2012) (Figure 7D).
Figure 7. Vaccine induction of antibodies capable of neutralizing group 1 and 2 influenza A viruses.
(A) Frequencies of H5-H3 cross-reactive memory B cells pre- and post-H5N1 vaccination. Subjects for whom pre-immunization samples were no longer available are indicated with open symbols; subject name and fold increase shown for others. (B) Frequency of multidonor class sequences by donor and multidonor class. (C) Fold-increase in cross-reactive B cells relative to prevalence of heavy chain sequences with three (out of a possible four) of the same heavy chain genetic elements in at least one sequence in any of the six analyzed subjects (Pearson correlation with the total number of sequences provided in Figure 1D). (D) Fold-increase in cross-reactive B cells relative to the fold-increase in sera neutralization titer for all tested influenza A strains (shown in black) (see Figure 1, Figure S1), or the single H1N1-A/Singapore/8/1986 strain (shown in red). (E) Bar graphs of transcript frequencies (left) and lineage frequencies (right). Left: frequency of multidonor-class transcripts by dataset; Right: frequency of multidonor class lineages for each dataset. (F) Transcript frequency versus dataset and goal. Stars depict upper-bound estimates, and circles depict frequencies confirmed by neutralization. (G) Multidonor antibodies displayed in ribbon with class-conserved contact residues shown in stick. Antibody epitopes shown in purple (HV6-1+HD3-3), green (HV1-18+HD3-9), and orange (HV1-18 with Q-x-x-V) with black outlines. Glycans shown in surface representation and colored by conservation: conserved (light green) or variable (dark green). See also Figures S3, S6 and Tables S3–S4.
To quantify the frequency of multidonor class antibodies in unvaccinated subjects, we examined NGS-determined memory B cell transcripts from healthy normal donors (Figure 6A). To compare frequencies from VRC 310-vaccinated versus unvaccinated subjects, we equated the total number of sorted memory B cells with the total number of transcripts and observed a substantially higher transcript frequency (and to a lesser degree, a higher lineage frequency) for multidonor class sequences upon H5N1 vaccination in the VRC 310 trial (Figure 7E).
We also examined published antibody sequences from subjects immunized with the 2009 or 2010 seasonal influenza vaccine (Jiang et al., 2013) (Figure 6A, 7E and S6). Overall, transcripts matching the HV1-18+HD3-9 signature were ~10x more prevalent prior to vaccination than the other multidonor transcripts; however, this class appeared not to expand upon either seasonal or VRC 310 vaccination. By contrast, transcripts matching HV6-1+HD3-3 and HV1-18 (Q-x-x-V) signatures appeared to be present at low frequency prior to vaccination, to increase upon seasonal vaccination, and to increase up to 1000-fold upon VRC 310 vaccination (Figure 7F).
DISCUSSION
The vaccine induction of broadly neutralizing antibodies against influenza A virus has been a long standing immunological goal. While the human immune system can generate antibodies capable of neutralizing group 1 and 2 strains of influenza (Corti et al., 2011; Ekiert et al., 2009; Sui et al., 2009; Whittle et al., 2011), clear evidence of their induction by vaccination had not been reported. In this study, we used a sequence-based structural approach to identify three multidonor classes of antibodies capable of neutralizing group 1 and group 2-influenza A viruses in VRC 310-vaccinated subjects. One multidonor class utilized HV6-1+HD3-3 germline genes (Figure 2); a second class utilized HV1-18+HD3-9 germline genes and a 15-amino acid CDR H3 (Figure 3); and a third class utilized HV1-18 germline gene and a Gln98HC-x-x-Val100aHC motif (Figure 4). For each class, we delineated sequence signatures (Figure 6). Despite the lack of serological indicators of vaccine-induced improvement in cross neutralization with VRC 310-vaccinated subjects (Figure S1), we observed a significant vaccine-induced expansion of cross-reactive memory B cells (Figure 7A) and a clear increase in the frequency of transcripts for two of the multidonor antibody classes (Figure 7F). These findings reveal the vaccine induction of broadly neutralizing influenza A antibodies.
Stereotypic antibody signatures have been reported for some bacterial polysaccharide antigens (Adderson et al., 1993), CD4-induced, V1V2-directed and VRC01-classes of HIV-1-neutralizing antibodies (Gorman et al., 2016; Huang et al., 2004; Zhou et al., 2013), and both stem and head-directed influenza neutralizing antibodies (Pappas et al., 2014; Schmidt et al., 2015). Thus, despite the potential repertoire of human immunoglobulins being large (Boyd et al., 2009) and SHM further increasing diversity to the point where highly similar modes of antigen recognition might be expected very infrequently, our findings reveal that multidonor classes can be induced by vaccination in humans.
The coexistence of multiple vaccine-induced pathways to generate influenza group 1 and group 2 neutralizing antibodies is encouraging for efforts aimed at achieving analogous responses in genetically diverse human populations. The stem-directed antibodies induced here potently neutralize in pseudotype assays (Figure S4), but less potently in live influenza A virus assays (Table S4). While determination of the in vivo concentrations of stem antibodies required for protective efficacy may require passive infusion trials in humans, it seems likely that a protective response will require increased titers of group 1 and 2 neutralizing antibodies than achieved in the VRC 310 trial. Our NGS-based measurements indicated VRC 310 vaccination to boost the frequency of transcripts from two multidonor classes substantially towards the target goal, and we even observed increases after seasonal vaccination (Figure 7F). In this regard, sequence signature-based quantification may provide a suitably sensitive technology to detect transcript frequencies of group 1 and 2 neutralizing antibodies, to measure their expression in appropriate memory B cell subsets and long-lived plasma cells, and to assess their durability. Appropriate SHM is an additional aspect, and we analyzed the recognition of germline-reverted versions of each of the three multidonor classes as well as the effect of mutational analysis of critical contacts (Figures S7). Further studies aimed at increasing the prevalence of group 1 and 2-neutralizing influenza antibody lineages –guided by the sequence signatures identified here – may provide a means to achieve the protective efficacy required of a universal influenza A vaccine. In this regard, it is helpful to know that 100–1000-fold increases in the transcript frequencies for two multidonor classes of influenza A-neutralizing antibodies could be achieved through immunization with a divergent influenza (Figures 6A and 7F), likely by enhancing immune focus to the HA stem (Figure 7G), an approach utilized by “stem-only” or “headless” immunogens (Impagliazzo et al., 2015; Yassine et al., 2015) and by chimeric HA immunogens (Krammer et al., 2015). It will be fascinating to evaluate how these immunogens, in various vectored, subunit, and prime-boost combinations, will fare at further inducing, maintaining, or expanding the multidonor broadly neutralizing antibodies identified here.
EXPERIMENTAL PROCEDURES
Ethics statement and VRC 310 study design
The VRC 310 study protocol and associated procedures were approved by the NIAID Institutional Review Board. All participants provided written informed consent in accordance with the Declaration of Helsinki. The VRC 310 study (ClinicalTrials.gov identifier NCT01086657) (Ledgerwood et al., 2011; Ledgerwood et al., 2013) was conducted by the Vaccine Research Center, National Institutes of Health (NIH) (see Table S1 and Supplemental Experimental Procedures for details).
Expression of HA probes, flow cytometry, cell sorting, and sequencing
HA constructs consisting of the extracellular domain of HA modified to ablate sialic acid binding and C-terminally fused to i) a T4-fibritin trimerization motif, ii) a biotinylatable AviTag sequence, and iii) a hexahistidine affinity tag were synthesized (Genscript), cloned into a CMV-expression plasmid, and expressed as previously described (Whittle et al., 2014). Cryopreserved PBMC samples were stained and sorted on a FACS Aria II using fluorescently labelled recombinant H1 (A/New Caledonia/20/1999), H5 (A/Indonesia/05/2005) or H3 (A/Perth/16/2009) probes; single memory B cells binding to both H5 and H3 probes were sorted, and sequencing of immunoglobulin genes by multiplex PCR was performed as previously described (Whittle et al., 2014). (See Supplemental Experimental Procedures for details).
Production of pseudotyped lentiviral vectors and influenza A viruses and measurement of antibody neutralizing activity
Influenza HA pseudotyped lentiviral vectors expressing a luciferase reporter gene were produced and used to infect 293A cells. All influenza viruses used in the microneutralization assays were expanded in MDCK cells in the presence of TPCK-treated trypsin (Sigma) and titrated in MDCK cells. (See Supplemental Experimental Procedures for details.)
Structural analysis and sequence bioinformatics
Both negative stain-EM and X-ray crystallography were used to characterize antibodies from the VRC 310 trial and their complexes with HA. An Antibodyomics1.0 pipeline, modified to analyze both 454 and Illumina output, was used to analyze B cell transcripts for the presence of sequence signatures specific to multidonor antibodies. Frequentist probabilities were used to determine likelihoods of sequence convergence. (See Supplemental Experimental Procedures for details.)
Supplementary Material
Acknowledgments
We thank D. Ambrosak and R. Nguyen for assistance with flow cytometry, J. Chrzas, J. Gonczy, U. Chinte, and staff at Southeast Regional Collaborative Access Team (SER-CAT) for help with X-ray diffraction data collection, G. Georgiou and S.R. Quake for human antibody sequences, J. Stuckey for assistance with figures, and members of the Structural Biology Section, Structural Bioinformatics Core Section, and Virology Laboratory of the Vaccine Research Center for helpful comments. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, and the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH. This work was supported in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E. Use of insertion device 22 (SER-CAT) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-109-Eng-38.
Footnotes
ACCESSION NUMBERS
Coordinates and structure factors have been deposited with the Protein Data Bank (PDB) under access codes 5K9J, 5K9K, 5K9O, 5K9Q, 5KAN, and 5KAQ. Sequences for all of the antibodies described in this manuscript have been deposited in Genbank under accession numbers KX386124- KX387227. NGS data have been deposited to Short Reads Archive (SRP026397 and SRP073039).
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and seven tables and can be found with this article online at http://dx.doi.org/xxx.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests. Reprints permission information are available at www.cell.com.
AUTHOR CONTRIBUTIONS
M.G.J., A.K.W., P.V.T., G.-Y.C., C.S., P.D.K., J.R.M and A.B.M. conceived, designed, and coordinated the study; M.G.J., A.K.W., P.V.T., G.-Y.C., C.S., L.S., P.D.K., J.R.M., and A.B.M wrote and revised the manuscript and figures; B.S.G., and J.E.L. carried out VRC 310 trial and provided subject samples; R.A.K., and M.R., provided support for B cell analysis and sorting; J.C.B., M.K., and J.R.W. designed HA constructs; M.G.J., A.K.W., P.V.T., R.T.B., A.D., R.A.G., M.K., W.-P.K., K.L., S.N.N., M.S.P., E.S.Y., B.Z., Y.Z., M.A., S.D., C.R.L., A.R., L.W., X.W., H.M.Y., C.S., Y.M., Y.T., U.B., NISC CSP performed experiments; M.G.J., G.Y.-C., C.S., C.-H.S., and P.D.K. carried out bioinformatics analysis. M.G.J., A.K.W., P.V.T., G.-Y.C., C.S., R.T.B., I.S.G., M.K., W.-P.K., K.L., T.B., S.D., Y.T., U.B., J.C.M., K.S., D.C.D., L.S., P.D.K., J.R.M and A.B.M. analyzed data. All authors read and approved the manuscript. Detailed author contributions are provided in Supplemental Information.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adderson EE, Shackelford PG, Quinn A, Wilson PM, Cunningham MW, Insel RA, Carroll WL. Restricted immunoglobulin VH usage and VDJ combinations in the human response to Haemophilus influenzae type b capsular polysaccharide. Nucleotide sequences of monospecific anti-Haemophilus antibodies and polyspecific antibodies cross-reacting with self antigens. The Journal of clinical investigation. 1993;91:2734–2743. doi: 10.1172/JCI116514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Science translational medicine. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, et al. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. Journal of clinical microbiology. 2005;43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nature medicine. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, et al. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science. 2013;340:751–756. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- Gorman J, Soto C, Yang MM, Davenport TM, Guttman M, Bailer RT, Chambers M, Chuang GY, DeKosky BJ, Doria-Rose NA, et al. Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nature structural & molecular biology. 2016;23:81–90. doi: 10.1038/nsmb.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Dunand CJ, Wilson PC. Restricted, canonical, stereotyped and convergent immunoglobulin responses. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2015;370 doi: 10.1098/rstb.2014.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He XS, Dekker CL, Zheng NY, Huang M, Sullivan M, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Science translational medicine. 2013;5:171ra119. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Wu J, Dimitrova M, King LR, Manischewitz J, Graham BS, Ledgerwood JE, Golding H. DNA priming prior to inactivated influenza A(H5N1) vaccination expands the antibody epitope repertoire and increases affinity maturation in a boost-interval-dependent manner in adults. The Journal of infectious diseases. 2013;208:413–417. doi: 10.1093/infdis/jit178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Palese P, Steel J. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Current topics in microbiology and immunology. 2015;386:301–321. doi: 10.1007/82_2014_408. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledgerwood JE, Wei CJ, Hu Z, Gordon IJ, Enama ME, Hendel CS, McTamney PM, Pearce MB, Yassine HM, Boyington JC, et al. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect Dis. 2011;11:916–924. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledgerwood JE, Zephir K, Hu Z, Wei CJ, Chang L, Enama ME, Hendel CS, Sitar S, Bailer RT, Koup RA, et al. Prime-boost interval matters: a randomized phase 1 study to identify the minimum interval necessary to observe the h5 DNA influenza vaccine priming effect. The Journal of infectious diseases. 2013;208:418–422. doi: 10.1093/infdis/jit180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, McTamney PM, Yassine HM, Whittle JR, Guo X, Boyington JC, Wei CJ, Nabel GJ. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489:566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens DM, Taubenberger JK, Fauci AS. H7N9 avian influenza A virus and the perpetual challenge of potential human pandemicity. mBio. 2013;4 doi: 10.1128/mBio.00445-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura G, Chai N, Park S, Chiang N, Lin Z, Chiu H, Fong R, Yan D, Kim J, Zhang J, et al. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe. 2013;14:93–103. doi: 10.1016/j.chom.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, Fernandez-Rodriguez B, Agatic G, Giacchetto-Sasselli I, Pellicciotta G, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014 doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, Haynes BF, Harrison SC. Viral receptor-binding site antibodies with diverse germline origins. Cell. 2015;161:1026–1034. doi: 10.1016/j.cell.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, et al. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature structural & molecular biology. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley AK, Whittle JR, Lingwood D, Kanekiyo M, Yassine HM, Ma SS, Narpala SR, Prabhakaran MS, Matus-Nicodemos RA, Bailer RT, et al. H5N1 Vaccine-Elicited Memory B Cells Are Genetically Constrained by the IGHV Locus in the Recognition of a Neutralizing Epitope in the Hemagglutinin Stem. J Immunol. 2015;195:602–610. doi: 10.4049/jimmunol.1402835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. Journal of virology. 2014;88:4047–4057. doi: 10.1128/JVI.03422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Cho M, Shore D, Song M, Choi J, Jiang T, Deng YQ, Bourgeois M, Almli L, Yang H, et al. A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus. Nat Commun. 2015;6:7708. doi: 10.1038/ncomms8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nature medicine. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.