Summary
Argininosuccinate synthase 1 (ASS1) is the rate-limiting enzyme for arginine biosynthesis. ASS1 expression is lost in a range of tumor types, including 50% of malignant pleural mesotheliomas. Starving ASS1-deficient cells of arginine with arginine blockers such as ADI-PEG20 can induce selective lethality and has shown great promise in the clinical setting. We have generated a model of ADI-PEG20 resistance in mesothelioma cells. This resistance is mediated through re-expression of ASS1 via demethylation of the ASS1 promoter. Through coordinated transcriptomic and metabolomic profiling, we have shown that ASS1-deficient cells have decreased levels of acetylated polyamine metabolites, together with a compensatory increase in the expression of polyamine biosynthetic enzymes. Upon arginine deprivation, polyamine metabolites are decreased in the ASS1-deficient cells and in plasma isolated from ASS1-deficient mesothelioma patients. We identify a synthetic lethal dependence between ASS1 deficiency and polyamine metabolism, which could potentially be exploited for the treatment of ASS1-negative cancers.
Graphical Abstract
Highlights
-
•
ASS1-deficient tumors become resistant to arginine deprivation via ASS1 re-expression
-
•
ASS1-deficient cells have decreased levels of acetylated polyamine metabolites
-
•
Polyamine metabolites are decreased in ASS1-deficient cells upon arginine deprivation
-
•
ASS1 deficiency is synthetically lethal with inhibition of polyamine metabolism
Locke et al. have generated a model of ADI-PEG20 resistance in mesothelioma cells. They reveal that ASS1-deficient cells have decreased levels of acetylated polyamine metabolites, together with a compensatory increase in expression of polyamine biosynthetic enzymes. This elucidates a synthetic lethal interaction between ASS1 loss and inhibition of polyamine metabolism.
Introduction
Cancer cells often rely on subversion of normal metabolic pathways to provide the energy and building blocks for uncontrolled cell division. Such metabolic reprogramming is now appreciated as an enabling hallmark of tumorigenesis and results in the uptake of nutrients for conversion to biomass. It is becoming increasingly appreciated that cancer cells also have altered amino acid metabolism (Tsun and Possemato, 2015). Glutamine, serine, glycine, and arginine have all been implicated in driving cancer cell proliferation (Amelio et al., 2014, Lind, 2004, Wise and Thompson, 2010).
As a versatile amino acid, arginine has connections to a number of metabolic pathways pertinent to tumorigenesis, including nitric oxide, creatine, and polyamine synthesis (Jobgen et al., 2006, Leuzzi et al., 2008). The levels of the rate-limiting enzyme for arginine biosynthesis, argininosuccinate synthase 1 (ASS1), are severely reduced or absent in a number of aggressive and chemoresistant cancers (Delage et al., 2010). The mechanisms behind ASS1 loss are cancer type dependent. For example, in lymphoma (Delage et al., 2012), myxofibrosarcoma (Huang et al., 2013), nasopharyngeal carcinoma (Lan et al., 2014), bladder cancer (Allen et al., 2014), hepatocellular carcinoma (McAlpine et al., 2014), and malignant pleural mesothelioma (MPM; Szlosarek et al., 2006), methylation of the ASS1 promoter appears to mediate ASS1 repression, whereas in melanoma, the interplay between c-Myc and HIF1α controls ASS1 levels (Tsai et al., 2009). The reason for ASS1 downregulation in tumors is not fully elucidated and renders the cancer cell reliant on, or addicted to, extracellular arginine. Such arginine auxotrophy has been targeted clinically using the pegylated arginine deiminase ADI-PEG20, a mycoplasma-derived protein that degrades arginine to citrulline and ammonia (Ott et al., 2013, Synakiewicz et al., 2014, Szlosarek et al., 2013). Starvation of arginine results in specific cell death of ASS1-deficient cancer cells and provides a means to attack poor outcome and highly proliferative cancers.
Approximately 50% of MPMs do not express ASS1 (Szlosarek et al., 2006), making ADI-PEG20 an attractive personalized therapeutic strategy (Delage et al., 2010) that has shown significant activity in a randomized phase II trial. Encouragingly, this trial has achieved its primary endpoint of a significant improvement in progression free survival (PFS) above the current standard of care (Szlosarek et al., 2013). This is the first biomarker-driven study and first randomized trial in a decade, since the publication of antifolates with cisplatin (Vogelzang et al., 2003), to show a 50% reduction in the risk of disease progression in MPM patients. Despite these promising initial results, resistance to ADI-PEG20 is a clinical obstacle, because neutralizing antibodies to ADI-PEG20 and re-expression of ASS1 in melanoma are dominant resistance mechanisms (Feun et al., 2008, Long et al., 2013). However, it has yet to be established how resistance to arginine deprivation occurs in MPM cells.
In this study, we examined the effects of long-term arginine deprivation on MPM cells to uncover the molecular mechanisms underlying resistance to arginine deprivation. We have generated a model of ADI-PEG20 resistance in MPM cells whereby we observe demethylation of the ASS1 promoter, allowing re-expression of the ASS1 transcript and protein. Resistance was accompanied by global changes in the levels of metabolic enzymes, resulting in greatly altered metabolic profiles. Significantly, we found that to maintain polyamine pools, the levels of acetylated polyamine metabolites were decreased in ASS1-deficient cells, indicative of reduced catabolism, together with a compensatory increase in expression of polyamine biosynthetic enzymes. Furthermore, this metabolic reprogramming elucidates a synthetic lethal interaction between ASS1 loss and polyamine metabolism, which could potentially be exploited for the treatment of ASS1-negative cancers.
Results
The Arginine Biosynthetic Pathway Is Upregulated to Confer Resistance to ADI-PEG20
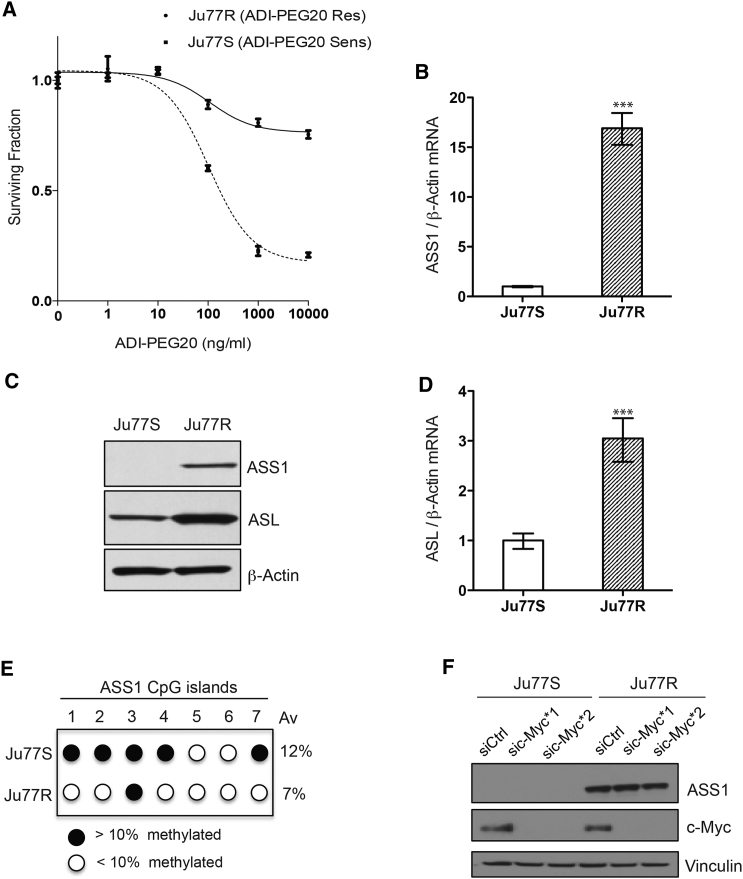
To investigate the metabolic adaptation of ASS1-deficient cells upon arginine deprivation, we generated ASS1-deficient cells resistant to the arginine-depleting drug, ADI-PEG20. The ASS1-deficient Ju77 MPM cells were cultured in ADI-PEG20, and over time, resistant colonies emerged (Figure 1A). Short-tandem repeat analysis across 18 genetic loci confirmed that the ADI-PEG20-resistant (Ju77R) cell line had not experienced genetic drift and was genetically identical to the parental ADI-PEG20-sensitive (Ju77S) cell line. To understand the mechanism behind the resistance, we analyzed the levels of the arginine biosynthetic enzymes, ASS1 and argininosuccinate lyase (ASL), in Ju77R and Ju77S cells. Ju77R cells had a 20-fold increase in ASS1 mRNA (Figure 1B) and the corresponding protein (Figure 1C) and increased levels of ASL (Figures 1C and 1D). Thus, it appears that Ju77R cells have switched on the ASS1 gene and upregulated the arginine biosynthetic pathway as an adaptive response to arginine depletion.
Figure 1.
The Arginine Biosynthetic Pathway Is Upregulated in Ju77R Cells
(A) Ju77S and Ju77R MPM cells were treated with increasing concentrations of ADI-PEG20 (0, 1, 10, 100, 1,000, and 10,000 ng/ml). After 4 days of treatment, cell viability was measured using an ATP-based luminescence assay.
(B) Real-time qPCR analysis of RNA extracted from Ju77S and Ju77R cells. mRNA expression was measured using ASS1 and β-actin TaqMan probes. β-actin was used as a control. ∗∗∗p < 0.0005.
(C) Western blot analysis of protein extracted from Ju77S and Ju77R cells. Protein expression was analyzed using anti-ASS1, anti-ASL, and β-actin antibodies. β-actin is used as a loading control.
(D) Real-time qPCR analysis of RNA extracted from Ju77S and Ju77R cells. mRNA expression was measured using ASL and β-actin TaqMan probes. β-actin was used as a control. ∗∗∗p < 0.0005.
(E) Methylation analysis across seven CpG islands in the ASS1 promoter. Genomic DNA was extracted from Ju77S and Ju77R cells, and bisulfite was converted. DNA was pyrosequenced across the seven CpG islands in the ASS1 promoter.
(F) Western blot analysis of Ju77S and Ju77R cells transfected with a non-targeting control siRNA (siCtrl) or two siRNA oligos targeting c-Myc. Protein was extracted after 72 hr, and expression was analyzed using anti-ASS1, anti-c-Myc, and vinculin antibodies. Vinculin is used as a loading control.
All experiments were carried out in triplicate. For (A), (B), and (D), error bars represent SEM. See also Figure S1.
Previously, we demonstrated that expression of ASS1 in mesothelioma is regulated by methylation at CpG islands in the ASS1 promoter, which represses transcription (Szlosarek et al., 2006). To investigate whether changes in ASS1 promoter methylation underlie re-expression of ASS1 in Ju77R cells, we performed bisulfite pyrosequencing across seven CpG sites. Upon comparison with the parental Ju77S cell line, Ju77R cells had decreased methylation at six of seven CpG sites, and there was an overall 5% reduction in promoter methylation (Figure 1E). The low level of methylation in the Ju77S parental line (12%) was quite surprising, given the absence of ASS1 mRNA (Figure 1B) and sensitivity to ADI-PEG20 (Figure 1A). We therefore compared methylation in other ASS1-negative and ASS1-positive MPM cells to see whether this was correlated to ADI-PEG20 sensitivity and ASS1 mRNA levels. We found an inverse correlation between promoter methylation and mRNA level, which in turn correlated with ADI-PEG20 sensitivity (Figures S1A and S1B). The cell line most sensitive to ADI-PEG20, MSTO-211H (MSTO), had 10% overall promoter methylation, compared to 2% for the most resistant cell line, H226. Thus, it is possible that 10% methylation represents a threshold for methylation-induced transcriptional silencing of the ASS1 gene in MPM cells. Previously, it has been shown in melanoma cells that ASS1 levels can be upregulated by c-Myc to mediate resistance to ADI-PEG20 (Long et al., 2013, Tsai et al., 2009). To investigate whether c-Myc was involved in the re-expression of ASS1 in Ju77R cells, we analyzed c-Myc levels in the Ju77S and Ju77R cells (Figure 1F). No difference in the expression levels of c-Myc was observed between these cell lines. To further confirm that c-Myc is not driving the re-expression of ASS1 in Ju77R cells, we used two small interfering RNA (siRNA) oligos targeting c-Myc and examined ASS1 levels (Figure 1F). We did not observe change in ASS1 levels in the absence of c-Myc expression. Altogether, our data suggest that re-expression of ASS1 in the Ju77R cells is via demethylation of the ASS1 promoter and not through regulation by c-Myc. The Ju77R cell line represents a cellular model of resistance to ADI-PEG20 and demonstrates ASS1 promoter demethylation in response to arginine deprivation.
ASS1 Loss Is Associated with Polyamine Metabolic Reprogramming
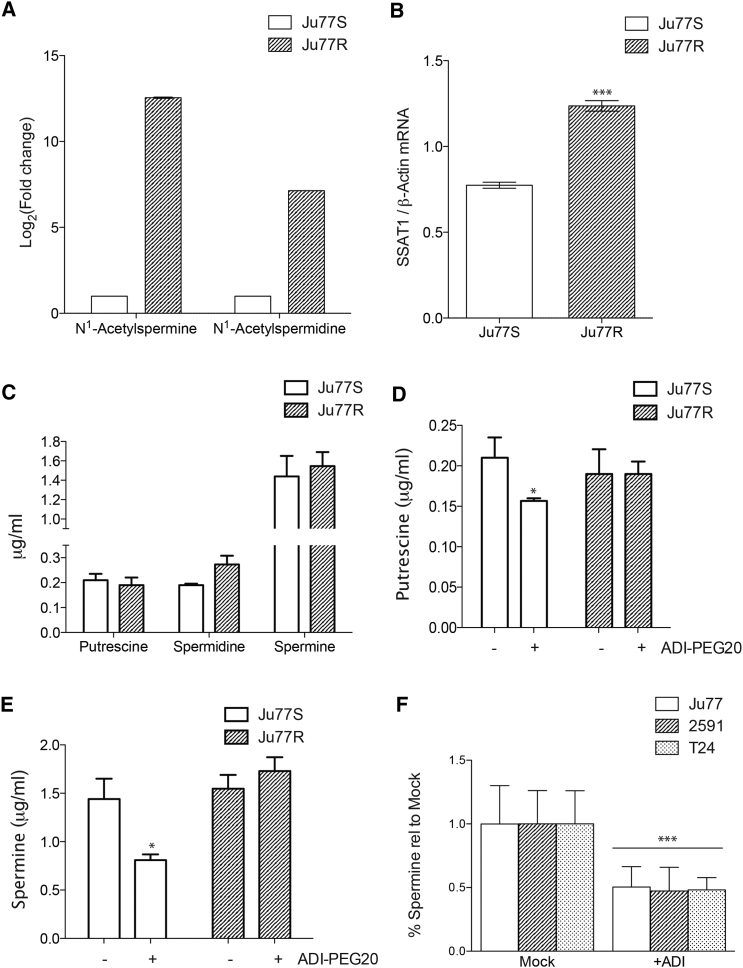
To further investigate the consequences of ADI-PEG20 resistance in the Ju77R cells, we assessed the metabolic response to ASS1 re-expression and resistance to ADI-PEG20 by performing untargeted metabolomic analysis using liquid chromatography-mass spectrometry (LC-MS) in Ju77S and Ju77R cells. A total of 5,220 different ion masses were detected by LC-MS, which produced two groups by principle-component analysis (PCA), indicating significant metabolic changes in the Ju77R cells (Figure S2A). The metabolomic analysis showed that the levels of the acetylated polyamines N1-acetylspermidine and N1-acetylspermine were significantly lower in the ASS1-deficient Ju77S cells in comparison to the ASS1-proficient Ju77R cells (Figure 2A). Polyamines are polycations derived from arginine that support numerous biological processes essential for cell survival. As such, their levels are under tight homeostatic control through regulated synthetic and catabolic pathways. Acetylation is the rate-limiting step in polyamine catabolism and maintains intracellular levels through export from the cell or recycling to putrescine (Kramer et al., 2008). The low levels of acetylated polyamines in Ju77S cells indicated reduced catabolism, so we measured the levels of the key enzyme responsible for polyamine acetylation, spermidine/spermine N1-acetyltransferase 1 (SSAT1). We observed a significant decrease in expression of SSAT1 in the ASS1-deficient cells (Figure 2B), consistent with our metabolomic analysis.
Figure 2.
Polyamine Metabolic Reprogramming in ASS1-Deficient Cells
(A) Untargeted metabolic analysis was performed on the Ju77S and Ju77R cells. The acetylated polyamines N1-acetylspermine and N1-acetylspermidine were identified as significantly decreased in the Ju77S cells. The graph represents the log2-fold change in comparison to levels in the Ju77S cells.
(B) Real-time qPCR analysis of RNA extracted from Ju77S and Ju77R cells. mRNA expression was measured using SSAT1 and β-actin TaqMan probes. β-actin was used as a control. ∗∗∗p < 0.0005.
(C) Targeted metabolic analysis was performed on the Ju77S and Ju77R cells upon comparison to the commercially available polyamine metabolites putrescine, spermidine, and spermine. The graph represents the absolute level of each metabolite (in micrograms per milliliter).
(D and E) Targeted metabolic analysis was performed on the Ju77S and Ju77R cells before and after 24 hr of treatment with ADI-PEG20 (750 ng/ml) upon comparison to the commercially available polyamine metabolites putrescine (D) and spermine (E). The graph represents the absolute level of each metabolite (in micrograms per milliliter). ∗p < 0.05.
(F) Targeted metabolic analysis was performed on the ASS1-deficient Ju77, 2591, and T24 cells before and after 24 hr of treatment with ADI-PEG20 (750 ng/ml) upon comparison to the commercially available polyamine metabolite spermine. The graph represents the level in comparison to mock (PBS) treated cells. ∗∗∗p < 0.0005.
See also Figure S2A.
It has previously been shown that levels of SSAT1 can influence polyamine levels by depleting spermidine and spermine with an increase in N1-acetylspermidine, which is subsequently oxidatively degraded to putrescine (Mandal et al., 2013). We therefore carried out targeted metabolomics in Ju77S and Ju77R cells to determine whether SSAT1 levels affected polyamine pools. In this targeted approach, we quantified a range of commercially obtained polyamine metabolites (putrescine, spermine, and spermidine) and compared the spectra with data derived from our analysis. The basal levels of putrescine and spermine were similar in both cell lines; however, there was a trend toward decreased baseline spermidine levels in Ju77S cells (Figure 2C). This finding suggests that in the absence of arginine biosynthesis, ASS1-deficient cells are unable to maintain levels of spermidine. However, upon ADI-PEG20 treatment, putrescine and spermine levels were significantly reduced in Ju77S cells but not in Ju77R cells (p < 0.05; Figures 2D and 2E). These results suggest that in Ju77S cells, exogenous arginine transported into the cell is used at least partly for polyamine synthesis. It is therefore possible that Ju77R cells are able to resist arginine deprivation-induced changes in polyamine levels through their ability to endogenously synthesize arginine and subsequently have a reduced reliance on extracellular arginine. To determine whether these changes in polyamine synthesis following arginine depletion were specific to the Ju77 cell lines, we performed targeted metabolomics on two other ASS1-negative cell lines, in addition to Ju77 cells. All ASS1-deficient cells showed a significant reduction in spermine levels upon arginine depletion (p < 0.0005; Figure 2F). This reduction in polyamine levels suggests exogenous arginine is required to maintain polyamine biosynthesis in MPM cells lacking ASS1.
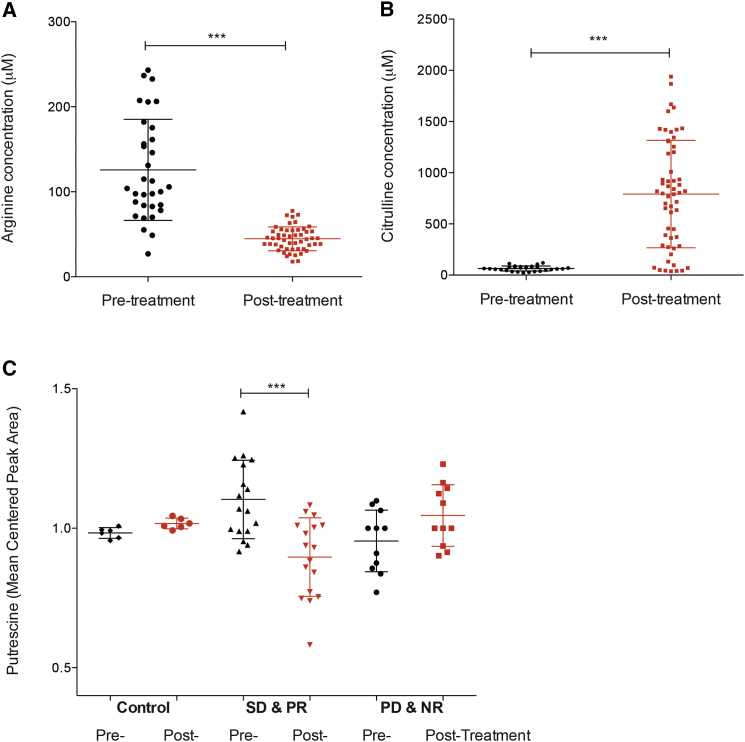
Next, we investigated whether these changes in polyamine metabolites were also occurring in ASS1-deficient mesothelioma patients treated with ADI-PEG20. To this end, we obtained plasma samples from the ADAM (arginine deiminase and mesothelioma) trial, which is a randomized phase II study of ADI-PEG20 and best supportive care versus best supportive care alone in patients with ASS1-deficient mesothelioma patients (ClinicalTrials.gov no. NCT01279967). Our analysis included ASS1-deficient, control mesothelioma patients receiving best supportive care alone (n = 6) and ASS1-deficient mesothelioma patients either with stable disease or partial response (SD/PR; n = 17) or with mixed response or progressive disease (MR/PD; n = 11) after ADI-PEG20 treatment and best supportive care. As expected, we observed a significant decrease in arginine levels (Figure 3A) and a subsequent significant increase in citrulline levels (Figure 3B) after ADI-PEG20 treatment in plasma from ASS1-deficient mesothelioma patients. Upon analysis, we could detect the polyamine putrescine, but not spermine and spermidine, from all plasma samples. These observations correlate with other studies indicating low levels of spermine and spermidine compared to putrescine in plasma (Liu et al., 2013, Magnes et al., 2014). We observed a significant reduction of putrescine in plasma samples from ASS1-deficient mesothelioma patients with SD/PR after ADI-PEG20 treatment and best supportive care (p < 0.0002; Figure 3C). No decrease in putrescine levels was observed in ASS1-deficient control patients or ASS1-deficient mesothelioma patients with MR/PD after treatment. These data strongly support our in vitro data that polyamine levels are decreased upon ADI-PEG20 treatment in ASS1-deficient cells. Significantly, we show that ADI-PEG20 treatment can modulate polyamine levels both in vitro and in vivo, thus suggesting that polyamine disruption may be an additional mechanism of action of this therapeutic.
Figure 3.
Reduced Putrescine Levels in Plasma from ASS1-Deficient Mesothelioma Patients with SD/PR after ADI-PEG20 Treatment
(A and B) Untargeted metabolic analysis was performed on plasma isolated from ASS1-deficient mesothelioma patients before and after ADI-PEG20 treatment and best supportive care. Arginine metabolites (A) and citrulline metabolites (B) were identified in plasma from all patients and were significantly decreased and increased after ADI-PEG20 treatment, respectively. The graphs represent the detected concentration (in micromolars) of arginine (A) and citrulline (B). ∗∗∗p < 0.0001.
(C) Untargeted metabolic analysis was performed on plasma isolated from control ASS1-deficient mesothelioma patients before and after best supportive care alone (n = 6) and ASS1-deficient mesothelioma patients with either SD/PR (n = 17) or MR/PD (n = 11) after ADI-PEG20 treatment and best supportive care. The polyamine putrescine was identified in plasma from all patients and was only significantly decreased in the ASS1-deficient patients with SD/PR after ADI-PEG20 treatment and best supportive care. The graph represents the mean centered putrescine peak area. ∗∗∗p < 0.0002.
See also Figure S2B.
Increased Expression of Polyamine Synthesis Genes in ASS1-Deficient Cells
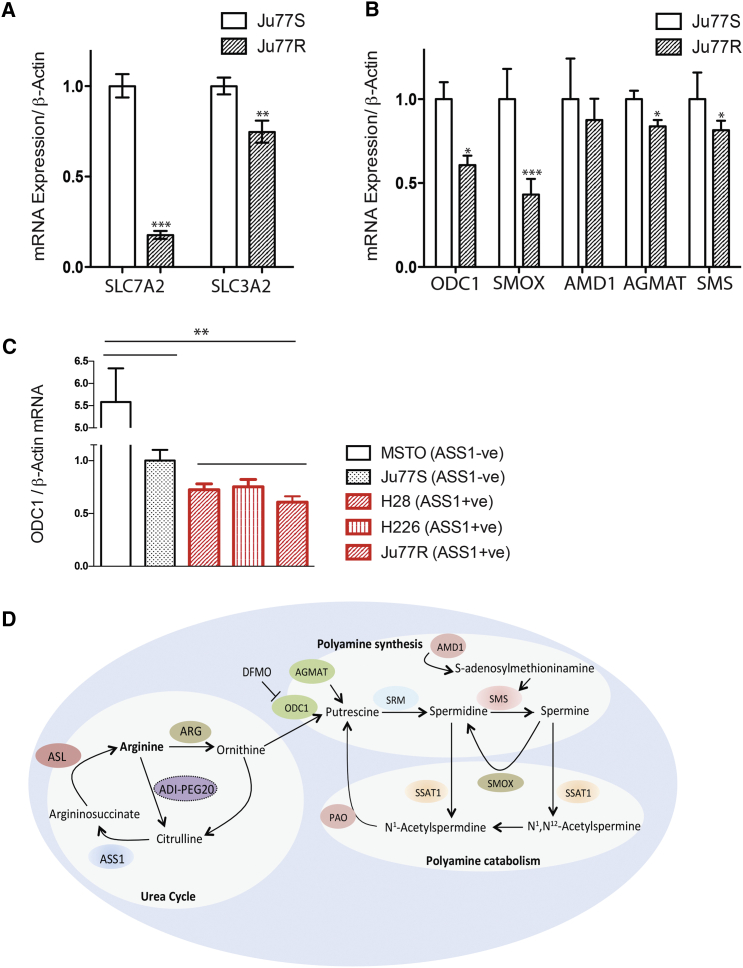
Our results suggest that in the absence of ASS1 and upon arginine deprivation, MPM cells are unable to maintain their levels of polyamine metabolites. To better understand this metabolic reprogramming, we analyzed the global transcriptomic changes that accompany these metabolic alterations. Comparative gene expression analysis of the Ju77S and Ju77R cells was performed using microarrays (Figure S3). This identified 234 differentially expressed genes with a greater than 2-fold difference (adjusted p < 0.05). Many of these differentially expressed genes have critical roles in nutrient uptake, amino acid metabolism, one-carbon metabolism, glycolysis, and nucleotide biosynthesis. As expected, ASS1 was identified as significantly upregulated in Ju77R cells, serving as an internal control for the microarray analysis. In addition, the microarray data highlighted several features that allow the ASS1-negative Ju77S cells to sustain arginine auxotrophy. Ju77S cells have increased expression of the arginine transporters SLC7A2 and SLC3A2, indicating an increased reliance on arginine transport from the extracellular environment (Figure 4A). This is consistent with the dependence on extracellular arginine for survival in the absence of ASS1 expression.
Figure 4.
Expression of Polyamine Metabolic Enzymes Are Increased in ASS1-Deficient Cells
(A and B) Real-time qPCR analysis of RNA extracted from Ju77S and Ju77R cells. (A) mRNA expression was measured for the arginine transporter genes using SLC7A2, SLC3A2, and β-actin-specific TaqMan probes. (B) mRNA expression was measured for the polyamine metabolic enzymes using ODC1, SMOX, AMD1, AGMAT, SMS, and β-actin-specific TaqMan probes. β-actin was used as a control. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005.
(C) Real-time qPCR analysis of RNA extracted from a panel of ASS1-deficient (MSTO and Ju77S) and ASS1-proficient (H28, H226, and Ju77R) MPM cell lines. mRNA expression was measured for the polyamine metabolic enzyme, ODC1, and β-actin using gene-specific TaqMan probes. β-actin was used as a control. ∗∗p < 0.005 upon comparison of the ASS1−ve cell lines (MSTO and Ju77S) versus the ASS1+ve cell lines (H28, H226, and Ju77R).
(D) Schematic representation of key enzymes and their roles in the urea cycle, polyamine synthesis, and polyamine catabolism. These enzymes are ASS1, ASL, arginine (ARG), ODC1, AGMAT, spermidine synthase (SRM), SMS, AMD1, SSAT1, SMOX, and polyamine oxidase (PAO).
For (A)–(C), experiments were carried out in triplicate and error bars represent SEM. See also Figure S3.
Several enzymes involved in polyamine metabolism had a significant increase in expression in the ASS1-deficient Ju77S cells (Figures 4B and 4C). These include the rate-limiting enzyme in polyamine biosynthesis ornithine decarboxylase (ODC1), adenosylmethionine decarboxylase 1 (AMD1), spermine synthase (SMS), spermine oxidase (SMOX), and agmatinase (AGMAT; Figure 4D). Real-time qPCR validation experiments confirmed consistent increased expression of genes involved in polyamine biosynthesis in ASS1-deficient cells by real-time qPCR (Figure 4B). ODC1 was observed to be significantly increased in the panel of ASS1-negative cells in comparison to the ASS1-positive MPM cells (Figure 4C), suggesting that the increase in expression of polyamine metabolic enzymes may be a general feature of ASS1 loss rather than a specific effect in Ju77 cells. Altogether, our results suggest that ASS1-deficient cells maintain their polyamine pools by decreased catabolism and increased synthesis through upregulation of polyamine biosynthetic enzymes.
ASS1-Deficient Cells Are Synthetically Lethal with Polyamine Inhibition
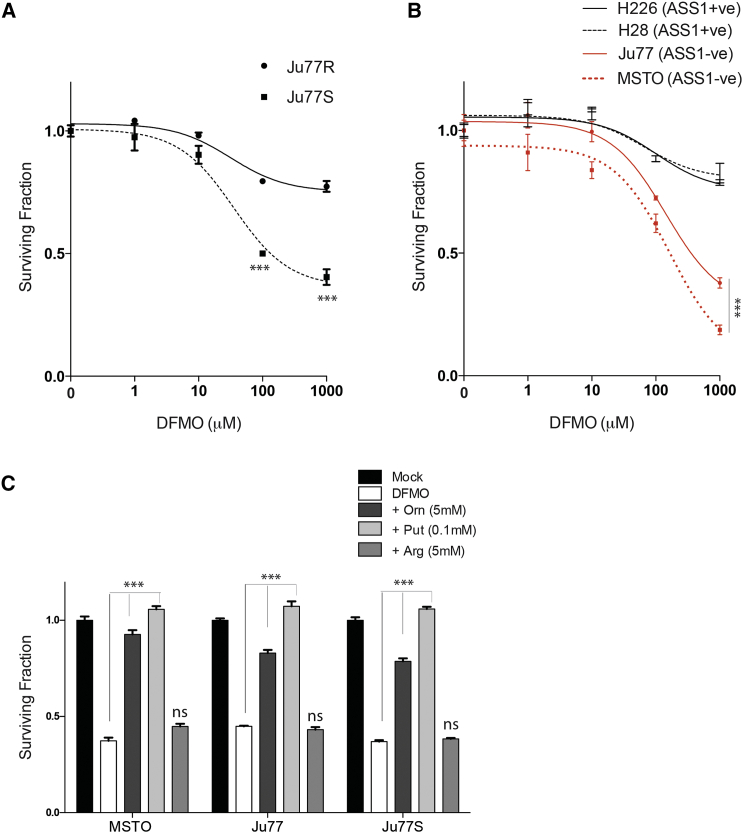
Our data suggest that the inability of ASS1-deficient cells to synthesize arginine results in a compensatory increase in polyamine biosynthetic enzymes and a decrease in polyamine catabolism in an effort to maintain polyamine pools. Furthermore, these pools are susceptible to perturbation by arginine deprivation. We therefore hypothesized that ASS1-deficient cells may be reliant on expression of these metabolic enzymes for survival. To investigate this, we treated Ju77S and Ju77R cells with the irreversible ODC1 inhibitor D,L-α-difluoromethylornithine (DFMO; Figure 4D) to block polyamine biosynthesis and analyzed cell viability after 4 days (Figure 5A). Significantly, we observed an increased sensitivity in the ASS1-deficient cells to DFMO treatment (p < 0.0005) in comparison to the ASS1-proficient cells, suggesting a synthetic lethal interaction between ASS1 loss and inhibition of polyamine synthesis. To further investigate this, we treated a panel of ASS1-deficient and ASS1-proficient MPM cells with increasing concentrations of DFMO and analyzed cell viability (Figure 5B). Significantly, we observed increased sensitivity in the ASS1-deficient cells in the panel of MPM cell lines (p < 0.0005), suggesting that ASS1-deficient cells are reliant on the increase in polyamine metabolic gene expression for survival. To determine whether this sensitivity was due to polyamine depletion, rather than an off-target effect on arginine levels, we treated Ju77S, Ju77, and MSTO cells with DFMO in addition to ornithine, putrescine, or arginine (Figure 5C). Our data show that addition of polyamines, but not arginine, can rescue the synthetic lethal effect with DFMO treatment, confirming that this sensitivity is specifically due to polyamine depletion (p < 0.0005). To further investigate this synthetic lethal relationship, we next determined whether addition of DFMO to ADI-PEG20 treatment could have a synergistic effect by performing combination index (CI) experiments. Combination studies were carried out over the range of drug concentrations, and the CI values were calculated using CalcuSyn software (Table 1). CI = 1 indicates an additive effect between two drugs, whereas CI < 1 or CI > 1 indicates synergism or antagonism, respectively. We observed that a combination of DFMO+ADI-PEG20 is synergistic upon treatment with these drugs at specific concentrations (10 μg/ml ADI-PEG20 + 10 nM DFMO and 100 μg/ml ADI-PEG20 + 100 nM DFMO).
Figure 5.
Inhibition of Polyamine Synthesis Is Synthetically Lethal in ASS1-Deficient MPM Cells
(A and B) Ju77S and Ju77R MPM cells (A) and a panel of ASS1-deficient (MSTO and Ju77) and ASS1-proficient (H28 and H226) MPM cell lines (B) were treated with increasing concentrations of the polyamine inhibitor DFMO (0, 1, 10, 100, and 1,000 μM). After 4 days of treatment, cell viability was measured using an ATP-based luminescence assay.
(C) The Ju77, Ju77S, and MSTO cells were treated with mock (DMSO; 0.01%), DFMO (1,000 μM), DFMO and ornithine (Orn; 5 mM), DFMO and arginine (Arg; 5 mM), or DFMO and putrescine (Put; 0.1 mM). After 4 days of treatment, cell viability was measured using an ATP-based luminescence assay.
∗∗∗p < 0.0005. Experiments were carried out in triplicate, and error bars represent SEM.
Table 1.
CI Values of Combined Treatment of ADI-PEG20 and DFMO at Specified Concentrations, Calculated with CalcuSyn Software, for Ju77S and Ju77R Cells
| Cell Line | ADI-PEG20 (μg/ml) | DFMO (nM) | CI |
|---|---|---|---|
| Ju77S | 10 | 10 | 0.368 |
| 100 | 100 | 0.499 | |
| Ju77R | 10 | 10 | 0.418 |
| 100 | 100 | 0.581 |
Altogether, our data suggest that in ASS1-deficient cells, polyamine levels are maintained by a compensatory increase in expression of polyamine biosynthetic enzymes and a decrease in polyamine catabolism. The increased reliance of ASS1-deficient cells on these pathways has uncovered a vulnerability that can be exploited therapeutically by arginine and/or polyamine depletion.
Discussion
MPM is a highly chemoresistant cancer associated with poor prognosis. Approximately 50% of MPMs do not express ASS1, making them sensitive to arginine-depriving agents such as ADI-PEG20. This personalized therapeutic strategy has shown activity in a randomized phase II trial (Szlosarek et al., 2013). However, resistance to ADI-PEG20 remains a significant clinical problem, and its precise mechanism has yet to be thoroughly investigated. We have generated a model of resistance to arginine deprivation in MPM cells that is characterized by demethylation of the ASS1 promoter and re-expression of ASS1. Metabolomic and transcriptomic profiling of these cells uncovered a previously unappreciated connection between ASS1 loss and polyamine metabolism, a finding we were able to confirm in MPM patients undergoing arginine deprivation therapy. Our data suggest that targeting polyamine synthesis using the clinically available polyamine inhibitor DFMO may provide a therapeutic strategy for the treatment of ASS1-negative MPMs. DFMO is clinically approved for the treatment of African trypanosomiasis and hirsutism and is in clinical trials as both a chemotherapeutic and a chemopreventative agent in a number of malignancies. Future studies will determine whether combinatorial treatment with ADI-PEG20 and DFMO can overcome the generation of resistance to arginine deprivation by simultaneously targeting two compensatory metabolic pathways. Significantly, ASS1 loss not only occurs in MPM tumors but also is a feature of numerous other tumor types, suggesting this therapeutic combination may extend beyond treatment of MPM. Additional studies are required to elucidate the full potential of this therapeutic strategy.
The concept of synthetic lethality has provided a means to exploit specific genetic vulnerabilities in cancer cells that can be targeted for the development of personalized therapeutics. In this study, we describe a synthetic lethal relationship between loss of ASS1 and inhibition of polyamine synthesis. Previously, others have shown that synthetic lethal interactions can occur between different compensating metabolic pathways. Inhibition of heme oxygenation is synthetically lethal in fumarate hydratase-deficient hereditary leiomyomatosis and renal-cell cancer (HLRCC) cells (Frezza et al., 2011a). In addition, a nicotinamide adenine dinucleotide (NAD+) synthesis inhibitor induces synthetic lethality in lactose dehydrogenase-A-deficient tumor cells (Le et al., 2010). Studies have also shown that dual inhibition of two energy-producing pathways, using metformin and the glycolysis inhibitor 2-deoxyglucose, results in a synthetic lethal interaction in in vivo tumor mouse models (Cheong et al., 2011). Advances in high-throughput genomic technologies have greatly increased our understanding of the genetics of tumor cells and have identified numerous mutations in metabolic enzymes, including isocitrate dehydrogenase 1, fumarate hydratase, and succinate dehydrogenase, in a range of tumor types (Frezza et al., 2011b, King et al., 2006, Muller et al., 2012, Tomlinson et al., 2002, Yan et al., 2009). Elucidation of these metabolic deficiencies highlights compensatory metabolic pathways that may be targeted through a synthetic lethal strategy.
Loss of ASS1 has been described for numerous tumor types, including MPM; consequently, ASS1 has been confirmed as a bona fide tumor suppressor gene. However, despite increasing interest in the role of ASS1, the overall functional significance of ASS1 loss in carcinogenesis has yet to be fully elucidated. Rabinovich et al. (2015) described diversion of aspartate for thymidine synthesis as a major consequence of ASS1 loss leading to enhanced tumor cell proliferation. Polyamines affect numerous processes in carcinogenesis, including increasing cell proliferation, modulating apoptosis, and increasing expression of genes affecting tumor invasion and metastasis (Gerner and Meyskens, 2004). Increased polyamine levels have been reported in many tumor types (Gerner and Meyskens, 2004), and overexpression of ODC1 was identified as a potential biomarker of colorectal cancer risk and progression (Brabender et al., 2001, McGarrity et al., 1990, Wang et al., 1996). In addition, numerous studies have shown that polyamines have the ability to regulate expression of a number of oncogenes (Bachrach et al., 2001, Tabib and Bachrach, 1999). All these observations suggest that the deregulated polyamine metabolism resulting from ASS1 loss described herein may be partly driving the tumorigenic phenotype of these cells and contributing to the tumor suppressive function of ASS1.
In addition to uncovering a dependence on polyamine synthesis in ASS1-deficient tumor cells, our results point to a potential mechanism of resistance to ADI-PEG20, with demethylation driving ASS1 upregulation. Studies of tumor tissue and/or plasma from trials of patients with MPM undergoing arginine deprivation will be critical to assessing the relevance of these findings in the clinic. Based on our data, putrescine levels may be used as an early response marker, predicting those patients likely to respond to ADI-PEG20 treatment. Moreover, ASS1 re-expression is likely to modulate sensitivity to alternative drugs and drug combinations, the study of which is possible by the availability of the Ju77S and Ju77R cell lines.
Altogether, we have elucidated a compensatory metabolic network in tumor cells such that in the absence of the arginine biosynthetic enzyme ASS1, the polyamine metabolic pathway is upregulated. Collectively, our data suggest that this increase in polyamine metabolic gene expression drives cellular proliferation in ASS1-deficient cells and can be exploited clinically through the use of polyamine inhibitors.
Experimental Procedures
Cell Culture
MSTO, Ju77, H28, and H226 cell lines were purchased from ATCC and routinely grown in RPMI-1640 media (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C/5% CO2. Cell lines were authenticated by short-tandem repeat (STR) profiling (DNA Diagnostics Centre). The compounds ADI-PEG20 (Polaris Pharmaceuticals) and DFMO (Santa Cruz) were obtained commercially. To generate Ju77R, 1 × 106 Ju77 cells were incubated with 10 μg/ml ADI-PEG20 in RPMI-1640 media supplemented with 2% FBS and re-treated every week. After approximately 2 months, colonies were picked and seeded in a 96-well plate with 10 μg/ml ADI-PEG20. A surviving clone (Ju77R) was gradually expanded and maintained in 1 μg/ml ADI-PEG20.
ADI-PEG20 Cell Viability Assays
Cells were seeded in 96-well plates (1 × 103 to 2 × 103 cells/well) 24 hr before treatment with ADI-PEG20 serially diluted in RPMI-1640 media containing 2% dialyzed FBS. After 4 days, cell viability was assessed with the ATP-based, luminescence assay CellTiter-Glo (Promega).
Western Blot
Cells were lysed in 20 mM Tris (pH 8), 200 mM NaCl, 1 mM EDTA, 0.5% (v/v) tergitol-type NP-40 (NP40), and 10% glycerol supplemented with protease inhibitors (Roche). Equivalent amounts of protein were electrophoresed on 4%–12% Novex precast gels (Invitrogen) and transferred to nitrocellulose membrane. After blocking for 1 hr in 1 × Tris-buffered saline/5% non-fat dried milk, membranes were incubated overnight at 4°C in primary antibody, including anti-ASS1 (HPA020934, Sigma-Aldrich), anti-ASL (HPA016646, Sigma-Aldrich), anti-c-Myc (#5650, Cell Signaling Technology [CST]), anti-vinculin (#4650, CST), and anti-β-actin-HRP (#4970, CST). Membranes were incubated with anti-immunoglobulin G-horseradish peroxidase and visualized by chemiluminescent detection (Supersignal West Pico Chemiluminescent Substrate, Pierce). Immunoblotting for β-actin or vinculin was performed as a loading control.
siRNA Transfections
For siRNA transfections, cells were transfected with individual siRNA oligonucleotides (QIAGEN) using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer’s instructions. As a control for each experiment, cells were left untransfected or transfected with a non-targeting control siRNA and were concurrently analyzed.
Methylation Analysis
Genomic DNA was isolated from 1 × 106 cells using a DNeasy kit (QIAGEN). Then, 500 ng was bisulfite converted using the EZ DNA methylation kit (Zymo Research). Pyrosequencing was performed using the PyroMark ID System (Biotage). The primers used for amplification of bisulfite-treated DNAs were designed to cover a region including seven CpG sites of the ASS1 promoter. The primers were 5′-biotin-TGTGTTTATAATTTGGGATGG-3′ (forward primer) and 5′-CCTCCTCCTCTAAACCC-3′ (reverse primer), which amplified a 145 bp region. PCR was performed in a final volume of 30 μl containing 0.5 μM of each primer, 200 μmol/L of each dinucleotide triphosphate, 0.05 units of AmpliTaq Gold 360 DNA polymerase in buffer containing 1.5 mmol/l MgCl2, 3 μl of 360 GC Enhancer (Applied Biosystems), and 2 μl of bisulfite-treated DNA as a template. The initial denaturation step (95°C, 10 min) was followed by 35 cycles of 30 s at 95°C, 30 s at 52°C, and 40 s at 72°C and a final extension step at 72°C for 10 min. Then, 3 μl of the PCR products were visualized by gel electrophoresis and 25 μl were subjected to pyrosequencing using the reverse sequencing primer at a final concentration of 0.5 μM. Purification and subsequent processing of the biotinylated single-strand DNA was done according to the manufacturer’s instructions using the Pyro Gold reagents kit (Biotage). Resulting data were analyzed and quantified with the PyroMark CpG Software (Biotage). Positive (commercial methylated DNA) and negative (placenta DNA) controls were included and treated, as well as samples. Pyrograms of the control DNA were analyzed to confirm complete bisulfite conversion.
Real-Time qPCR
RNA was extracted from cells using either the TRIzol Reagent (Life Technologies) or the RNeasy kit (QIAGEN) and quantified using a Nanodrop spectrophotometer (Thermo Scientific). Then, 500 ng of RNA was reverse transcribed using the Omniscript cDNA synthesis kit (QIAGEN) and 1μl of cDNA was used in each qPCR reaction. Multiplex PCR was performed on an ABI Prism 7500 Real-Time PCR Instrument (Applied Biosystems), and data were analyzed using the ΔΔCt method. TaqMan probes were purchased from Applied Biosystems.
Gene Expression Microarray Analysis
The 2 × 105 cells were plated in a 6-well plate, and RNA was extracted the following day using the RNeasy kit. Quantification and quality control of RNA were performed on the Nanodrop spectrophotometer and an Agilent Bioanalyser (Agilent Technologies), respectively. Global gene expression profiling was performed on an Illumina HT12v4 array at the Genome Centre (Queen Mary University of London). Raw image files were processed and analyzed in Illumina BeadStudio. This was followed by quality control and normalization of array data using the upgraded version of the O-miner transcriptomics analysis platform (Cutts et al., 2012) adapting the Lumi R Bioconductor package (https://www.bioconductor.org/packages/release/bioc/html/lumi.html). Differential expression analysis was performed using Limma (Ritchie et al., 2015), and the p values were further adjusted using the Benjamini and Hocheberg procedure (Benjamini and Hochberg, 1995). Significantly differentially expressed genes were further identified using a double threshold of the adjusted value of p < 0.05 and the absolute value of fold change > 2. Gene expression data have been deposited at GEO: GSE73970.
Metabolomic Analysis
For the untargeted metabolomics, 100 μl of plasma were extracted with 300 μl of methanol containing 0.1% formic acid. Intracellular metabolites were extracted from 5 × 106 cells using 80% methanol containing 1% formic acid. After vortexing, samples were incubated on ice for 30 min and then centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was transferred and evaporated under SpeedVac (Savant, Thermo Scientific). Dried extracts were reconstituted in 50 μl of 10% acetonitrile (containing 0.1% formic acid), and 10 μl were injected into the ultra-high performance liquid chromatography (UPLC)-mass spectrometry (MS) system. UPLC-MS was used to quantify the intracellular metabolites as described (Pandher et al., 2009). For the targeted metabolomics, metabolites were extracted from cells (1 × 106) using an 80% methanol solution. Samples were mixed and incubated on ice for 30 min. After centrifuging, supernatant was separated and evaporated in a SpeedVac. Dried extracts were reconstituted in 50 μl of 85% acetonitrile (containing 0.5% acetic acid), and 10 μl were injected into the UPLC-tandem mass spectrometry (MS/MS) system. The method used to quantify putrescine, spermine, and spermidine was according to Shin et al. (2011).
Patient Plasma Samples
Plasma samples were collected from patients with mesothelioma after written informed consent at St. Bartholomew’s Hospital. The protocol was approved by the East London and City Research Ethics Committee. All studies comply with the rules of the review board and by the revised Helsinki protocol. The samples were collected at baseline and after ADI-PEG20 treatment and best supportive care or best supportive care alone weekly over 9 weeks of treatment.
Analysis of Drug Combination
Drug synergy was determined by the CI method using CalcuSyn software (Biosoft). Data obtained from the growth inhibitory experiments were used to perform these analyses. The CI method is a mathematical and quantitative representation of a pharmacologic interaction between two drugs. Using growth inhibitory data and computerized software, CI values were generated over a range of 0.05–0.90 (reflecting 5%–90% growth inhibition). CI = 1 indicates an additive effect between two drugs, whereas CI < 1 or CI > 1 indicates synergism or antagonism, respectively.
Statistical Analysis
Unless stated otherwise, data represent SEM of at least three independent experiments. The two-tailed paired Student’s t test was used to determine statistical significance, with p < 0.05 regarded as significant.
Author Contributions
Experimental design and execution were conducted by M.L., E.G., M.O.F., and M.M. Data interpretation was performed by M.L., E.G., P.S., and S.A.M. Microarray analysis was performed by A.N., J.W., and C.C. Methylation analysis was performed by L.L. and C.L.N. The manuscript was written by M.L. and S.A.M. and edited by E.G., M.O.F., M.M., L.L., C.L.N., A.N., J.W., C.C., and P.S.
Acknowledgments
We thank all members of the S.A.M. lab for helpful discussions. We thank Dr. P. Ribeiro and Dr. T.V. Sharp for critical reading of the manuscript. This work was supported by funding from the British Lung Foundation (APG12-10 and MESO15-12), the June Hancock Mesothelioma Research Fund, and Cancer Research UK (C16420/A18066). E.G. acknowledges research funding from the Barry Reed Cancer Research Fund.
Published: July 21, 2016
Footnotes
Supplemental Information includes three figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.06.097.
Accession Numbers
The accession number for the gene expression data reported in this paper is GEO: GSE73970.
Supplemental Information
References
- Allen M.D., Luong P., Hudson C., Leyton J., Delage B., Ghazaly E., Cutts R., Yuan M., Syed N., Lo Nigro C. Prognostic and therapeutic impact of argininosuccinate synthetase 1 control in bladder cancer as monitored longitudinally by PET imaging. Cancer Res. 2014;74:896–907. doi: 10.1158/0008-5472.CAN-13-1702. [DOI] [PubMed] [Google Scholar]
- Amelio I., Cutruzzolá F., Antonov A., Agostini M., Melino G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014;39:191–198. doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrach U., Wang Y.C., Tabib A. Polyamines: new cues in cellular signal transduction. News Physiol. Sci. 2001;16:106–109. doi: 10.1152/physiologyonline.2001.16.3.106. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., B. 1995;57:289–300. [Google Scholar]
- Brabender J., Lord R.V., Danenberg K.D., Metzger R., Schneider P.M., Uetake H., Kawakami K., Park J.M., Salonga D., Peters J.H. Upregulation of ornithine decarboxylase mRNA expression in Barrett’s esophagus and Barrett’s-associated adenocarcinoma. J. Gastrointest. Surg. 2001;5:174–181, discussion 182. doi: 10.1016/s1091-255x(01)80031-7. [DOI] [PubMed] [Google Scholar]
- Cheong J.H., Park E.S., Liang J., Dennison J.B., Tsavachidou D., Nguyen-Charles C., Wa Cheng K., Hall H., Zhang D., Lu Y. Dual inhibition of tumor energy pathway by 2-deoxyglucose and metformin is effective against a broad spectrum of preclinical cancer models. Mol. Cancer Ther. 2011;10:2350–2362. doi: 10.1158/1535-7163.MCT-11-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutts R.J., Dayem Ullah A.Z., Sangaralingam A., Gadaleta E., Lemoine N.R., Chelala C. O-miner: an integrative platform for automated analysis and mining of -omics data. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gks432. W560-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delage B., Fennell D.A., Nicholson L., McNeish I., Lemoine N.R., Crook T., Szlosarek P.W. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int. J. Cancer. 2010;126:2762–2772. doi: 10.1002/ijc.25202. [DOI] [PubMed] [Google Scholar]
- Delage B., Luong P., Maharaj L., O’Riain C., Syed N., Crook T., Hatzimichael E., Papoudou-Bai A., Mitchell T.J., Whittaker S.J. Promoter methylation of argininosuccinate synthetase-1 sensitises lymphomas to arginine deiminase treatment, autophagy and caspase-dependent apoptosis. Cell Death Dis. 2012;3:e342. doi: 10.1038/cddis.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feun L., You M., Wu C.J., Kuo M.T., Wangpaichitr M., Spector S., Savaraj N. Arginine deprivation as a targeted therapy for cancer. Curr. Pharm. Des. 2008;14:1049–1057. doi: 10.2174/138161208784246199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C., Zheng L., Folger O., Rajagopalan K.N., MacKenzie E.D., Jerby L., Micaroni M., Chaneton B., Adam J., Hedley A. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477:225–228. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- Frezza C., Zheng L., Tennant D.A., Papkovsky D.B., Hedley B.A., Kalna G., Watson D.G., Gottlieb E. Metabolic profiling of hypoxic cells revealed a catabolic signature required for cell survival. PLoS ONE. 2011;6:e24411. doi: 10.1371/journal.pone.0024411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner E.W., Meyskens F.L., Jr. Polyamines and cancer: old molecules, new understanding. Nat. Rev. Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- Huang H.Y., Wu W.R., Wang Y.H., Wang J.W., Fang F.M., Tsai J.W., Li S.H., Hung H.C., Yu S.C., Lan J. ASS1 as a novel tumor suppressor gene in myxofibrosarcomas: aberrant loss via epigenetic DNA methylation confers aggressive phenotypes, negative prognostic impact, and therapeutic relevance. Clin. Cancer Res. 2013;19:2861–2872. doi: 10.1158/1078-0432.CCR-12-2641. [DOI] [PubMed] [Google Scholar]
- Jobgen W.S., Fried S.K., Fu W.J., Meininger C.J., Wu G. Regulatory role for the arginine-nitric oxide pathway in metabolism of energy substrates. J. Nutr. Biochem. 2006;17:571–588. doi: 10.1016/j.jnutbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- King A., Selak M.A., Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–4682. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- Kramer D.L., Diegelman P., Jell J., Vujcic S., Merali S., Porter C.W. Polyamine acetylation modulates polyamine metabolic flux, a prelude to broader metabolic consequences. J. Biol. Chem. 2008;283:4241–4251. doi: 10.1074/jbc.M706806200. [DOI] [PubMed] [Google Scholar]
- Lan J., Tai H.C., Lee S.W., Chen T.J., Huang H.Y., Li C.F. Deficiency in expression and epigenetic DNA Methylation of ASS1 gene in nasopharyngeal carcinoma: negative prognostic impact and therapeutic relevance. Tumour Biol. 2014;35:161–169. doi: 10.1007/s13277-013-1020-8. [DOI] [PubMed] [Google Scholar]
- Le A., Cooper C.R., Gouw A.M., Dinavahi R., Maitra A., Deck L.M., Royer R.E., Vander Jagt D.L., Semenza G.L., Dang C.V. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl. Acad. Sci. USA. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuzzi V., Alessandrì M.G., Casarano M., Battini R., Cioni G. Arginine and glycine stimulate creatine synthesis in creatine transporter 1-deficient lymphoblasts. Anal. Biochem. 2008;375:153–155. doi: 10.1016/j.ab.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Lind D.S. Arginine and cancer. J. Nutr. 2004;134:2837S–2841S. doi: 10.1093/jn/134.10.2837S. discussion 2853S. [DOI] [PubMed] [Google Scholar]
- Liu R., Li Q., Ma R., Lin X., Xu H., Bi K. Determination of polyamine metabolome in plasma and urine by ultrahigh performance liquid chromatography-tandem mass spectrometry method: application to identify potential markers for human hepatic cancer. Anal. Chim. Acta. 2013;791:36–45. doi: 10.1016/j.aca.2013.06.044. [DOI] [PubMed] [Google Scholar]
- Long Y., Tsai W.B., Wangpaichitr M., Tsukamoto T., Savaraj N., Feun L.G., Kuo M.T. Arginine deiminase resistance in melanoma cells is associated with metabolic reprogramming, glucose dependence, and glutamine addiction. Mol. Cancer Ther. 2013;12:2581–2590. doi: 10.1158/1535-7163.MCT-13-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnes C., Fauland A., Gander E., Narath S., Ratzer M., Eisenberg T., Madeo F., Pieber T., Sinner F. Polyamines in biological samples: rapid and robust quantification by solid-phase extraction online-coupled to liquid chromatography-tandem mass spectrometry. J. Chromatogr. A. 2014;1331:44–51. doi: 10.1016/j.chroma.2013.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S., Mandal A., Johansson H.E., Orjalo A.V., Park M.H. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc. Natl. Acad. Sci. USA. 2013;110:2169–2174. doi: 10.1073/pnas.1219002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlpine J.A., Lu H.T., Wu K.C., Knowles S.K., Thomson J.A. Down-regulation of argininosuccinate synthetase is associated with cisplatin resistance in hepatocellular carcinoma cell lines: implications for PEGylated arginine deiminase combination therapy. BMC Cancer. 2014;14:621. doi: 10.1186/1471-2407-14-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarrity T.J., Peiffer L.P., Bartholomew M.J., Pegg A.E. Colonic polyamine content and ornithine decarboxylase activity as markers for adenomas. Cancer. 1990;66:1539–1543. doi: 10.1002/1097-0142(19901001)66:7<1539::aid-cncr2820660718>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Muller F.L., Colla S., Aquilanti E., Manzo V.E., Genovese G., Lee J., Eisenson D., Narurkar R., Deng P., Nezi L. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488:337–342. doi: 10.1038/nature11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott P.A., Carvajal R.D., Pandit-Taskar N., Jungbluth A.A., Hoffman E.W., Wu B.W., Bomalaski J.S., Venhaus R., Pan L., Old L.J. Phase I/II study of pegylated arginine deiminase (ADI-PEG 20) in patients with advanced melanoma. Invest. New Drugs. 2013;31:425–434. doi: 10.1007/s10637-012-9862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandher R., Ducruix C., Eccles S.A., Raynaud F.I. Cross-platform Q-TOF validation of global exo-metabolomic analysis: application to human glioblastoma cells treated with the standard PI 3-Kinase inhibitor LY294002. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009;877:1352–1358. doi: 10.1016/j.jchromb.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Rabinovich S., Adler L., Yizhak K., Sarver A., Silberman A., Agron S., Stettner N., Sun Q., Brandis A., Helbling D. Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature. 2015;527:379–383. doi: 10.1038/nature15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Fung S.M., Mohan S., Fung H.L. Simultaneous bioanalysis of L-arginine, L-citrulline, and dimethylarginines by LC-MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011;879:467–474. doi: 10.1016/j.jchromb.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synakiewicz A., Stachowicz-Stencel T., Adamkiewicz-Drozynska E. The role of arginine and the modified arginine deiminase enzyme ADI-PEG 20 in cancer therapy with special emphasis on Phase I/II clinical trials. Expert Opin. Investig. Drugs. 2014;23:1517–1529. doi: 10.1517/13543784.2014.934808. [DOI] [PubMed] [Google Scholar]
- Szlosarek P.W., Klabatsa A., Pallaska A., Sheaff M., Smith P., Crook T., Grimshaw M.J., Steele J.P., Rudd R.M., Balkwill F.R., Fennell D.A. In vivo loss of expression of argininosuccinate synthetase in malignant pleural mesothelioma is a biomarker for susceptibility to arginine depletion. Clin. Cancer Res. 2006;12:7126–7131. doi: 10.1158/1078-0432.CCR-06-1101. [DOI] [PubMed] [Google Scholar]
- Szlosarek P.W., Luong P., Phillips M.M., Baccarini M., Stephen E., Szyszko T., Sheaff M.T., Avril N. Metabolic response to pegylated arginine deiminase in mesothelioma with promoter methylation of argininosuccinate synthetase. J. Clin. Oncol. 2013;31:e111–e113. doi: 10.1200/JCO.2012.42.1784. [DOI] [PubMed] [Google Scholar]
- Tabib A., Bachrach U. Role of polyamines in mediating malignant transformation and oncogene expression. Int. J. Biochem. Cell Biol. 1999;31:1289–1295. doi: 10.1016/s1357-2725(99)00098-9. [DOI] [PubMed] [Google Scholar]
- Tomlinson I.P., Alam N.A., Rowan A.J., Barclay E., Jaeger E.E., Kelsell D., Leigh I., Gorman P., Lamlum H., Rahman S., Multiple Leiomyoma Consortium Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- Tsai W.B., Aiba I., Lee S.Y., Feun L., Savaraj N., Kuo M.T. Resistance to arginine deiminase treatment in melanoma cells is associated with induced argininosuccinate synthetase expression involving c-Myc/HIF-1alpha/Sp4. Mol. Cancer Ther. 2009;8:3223–3233. doi: 10.1158/1535-7163.MCT-09-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun Z.Y., Possemato R. Amino acid management in cancer. Semin. Cell Dev. Biol. 2015;43:22–32. doi: 10.1016/j.semcdb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelzang N.J., Rusthoven J.J., Symanowski J., Denham C., Kaukel E., Ruffie P., Gatzemeier U., Boyer M., Emri S., Manegold C. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J. Clin. Oncol. 2003;21:2636–2644. doi: 10.1200/JCO.2003.11.136. [DOI] [PubMed] [Google Scholar]
- Wang W., Liu L.Q., Higuchi C.M. Mucosal polyamine measurements and colorectal cancer risk. J. Cell. Biochem. 1996;63:252–257. doi: 10.1002/(sici)1097-4644(19961101)63:2<252::aid-jcb12>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Wise D.R., Thompson C.B. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.