Abstract
We have previously described a forward genetic screen in mice for abnormalities of brain development. Characterization of two hydrocephalus mutants by whole-exome sequencing after whole-genome SNP mapping revealed novel recessive mutations in Dnaaf1 and Lrrc48. Mouse mutants of these two genes have not been previously reported. The Dnaaf1 mutant carries a mutation at the splice donor site of exon 4, which results in abnormal transcripts. The Lrrc48 mutation is a missense mutation at a highly conserved leucine residue, which is also associated with a decrease in Lrrc48 transcription. Both Dnaaf1 and Lrrc48 belong to a leucine-rich repeat-containing protein family and are components of the ciliary axoneme. Their Chlamydomonas orthologs are known to be required for normal ciliary beat frequency or flagellar waveform, respectively. Some Dnaaf1 or Lrrc48 homozygote mutants displayed laterality defects, suggesting a motile cilia defect in the embryonic node. Mucus accumulation and neutrophil infiltration in the maxillary sinuses suggested sinusitis. Dnaaf1 mutants showed postnatal lethality, and none survived to weaning age. Lrrc48 mutants survive to adulthood, but had male infertility. ARL13B immunostaining showed the presence of motile cilia in the mutants, and the distal distribution of DNAH9 in the axoneme of upper airway motile cilia appeared normal. The phenotypic abnormalities suggest that mutations in Dnaaf1 and Lrrc48 cause defects in motile cilia function.
Keywords: ENU mutagenesis, motile cilia, Dnaaf1/Lrrc50/Oda7, Lrrc48/FAP134/Drc3, primary ciliary dyskinesia
Motile cilia are important cellular structures for generating fluid flow. Their motility is important for airway mucus clearance, cerebrospinal fluid circulation, leftward extraembryonic fluid flow in the embryonic node, and movement of the sperm and fertilized ovum. Defects in cilia motility can result in human disorders, including primary ciliary dyskinesia (PCD), chronic respiratory tract infection, situs inversus, hydrocephalus, and infertility (Badano et al. 2006; Fliegauf et al. 2007; Brown and Witman 2014; Kurkowiak et al. 2015). While many genes have been already associated with human motile cilia defects, genetic diagnostic tests using only currently known causal genes do not explain all cases. For example, causal genes for one third of the PCD cases still remain to be discovered (Kurkowiak et al. 2015). Mutations causing motile cilia defects identified in various model organisms are important resources for the discovery of causal genes in human patients, and for understanding their biology.
Hydrocephalus results from an accumulation of the cerebrospinal fluid (CSF) due to obstruction of the ventricular system, abnormal production, circulation, or absorption of CSF. Human patients with hydrocephalus and mouse models share similar characteristics, including enlarged head, ventricular dilation, and damage to the ventricular lining and white matter (McAllister 2012; Lee 2013). Mouse models often display postnatal lethality. A cilial defect is one of the causes for the communicating form of hydrocephalus, and the association of hydrocephalus and PCD has been well known (McAllister 2012; Lee 2013). Ependymal cells lining the ventricles and choroid plexus epithelial cells (CPECs) are the multiciliated cells in the brain (Lee 2013; Narita and Takeda 2015). In mice, motile cilia on ependymal cells form postnatally and generate directional CSF flow. CPECs undergo ciliogenesis during midgestation in mice and produce CSF. Cilia on CPECs are immotile but transiently motile during the perinatal period. An ependymal cell cilia defect resulting in abnormal CSF circulation is considered the primary mechanism for cilia-related hydrocephalus, but the association of a CPEC cilia defect with neonatal hydrocephaly in mice has been also suggested (Banizs et al. 2005; Lee 2013; Narita and Takeda 2015).
Abnormal ultrastructure of the ciliary axoneme is a hallmark of a motile cilia defect. The axoneme of motile cilia has a 9 + 2 structure, with an exception of nodal cilia that have 9 + 0 organization (Fliegauf et al. 2007). The 9 + 2 axoneme has nine peripheral microtubule doublets (A and B tubules) and two central single microtubules (central pair), while the 9 + 0 axoneme lacks the central pair. The inner and outer dynein arms (IDA and ODA) extend from the A-tubules toward the B-tubules of neighboring doublets, and are responsible for ciliary movement generation. The A-tubules and adjacent B-tubule of neighboring doublets are linked by the nexin-dynein regulatory complex (N-DRC), which is a conserved structure from algae to humans. It has been proposed that the N-DRC is important for the coordination of microtubule sliding by maintaining microtubule doublet alignment during flagella bending (Porter and Sale 2000; Nicastro et al. 2006; Heuser et al. 2009; Bower et al. 2013). The peripheral microtubules are connected to the central pair by the radial spokes. Mutations in many axonemal structural component genes, including ODA, IDA, and N-DRC, are linked to human motile cilia disorders (Knowles et al. 2013; Kurkowiak et al. 2015).
We have reported previously the identification and mapping of two hydrocephalus mutant lines, described as line 42 and 67, which we ascertained in a forward genetic screen for abnormal brain phenotypes (Ha et al. 2015). In this paper, we report that these lines carry mutations in Dnaaf1 (Dynein axonemal assembly factor 1) and Lrrc48 (Leucine-rich repeat-containing protein 48), respectively. Both of these genes are previously known to encode structural components of the motile cilial axoneme. Dnaaf1, also known as Lrrc50 (Leucine-rich repeat-containing protein 50) or Oda7 (Outer dynein arm 7), is important for ciliary beat frequency in Chlamydomonas reinhardtii and a causal gene for primary ciliary dyskinesia (PCD) in humans (Kamiya 1988; Freshour et al. 2007; Duquesnoy et al. 2009; Loges et al. 2009). The Chlamydomonas ortholog of Lrrc48 is FAP134 (Flagellar associated protein 134) or Drc3 (Dynein regulating complex 3), which is a component of the N-DRC and required for normal flagellar waveform (Pazour et al. 2005; Lechtreck et al. 2009; Lin et al. 2011; Bower et al. 2013; Awata et al. 2015). Of note, mouse mutants of Dnaaf1 and Lrrc48 have not been previously reported. In addition to hydrocephalus, both lines demonstrate laterality defects and sinusitis, which is consistent with the presence of a presumptive motile cilia defect.
Materials and Methods
Animals and genotyping
Generation of Dnaaf1m4Bei and Lrrc48m6Bei mutants was previously described (Ha et al. 2015). Dnaaf1m4Bei was previously named line 42, and is currently listed as m4Bei (MGI:5505439) in the Mouse Genome Informatics (MGI) database. Lrrc48m6Bei was previously named line 67 and is listed as m6Bei (MGI:5505443) in MGI. The mutant lines originated from mutagenized A/J (Jackson Laboratory) and were maintained by serial backcrosses to C57BL6/N (Taconic) females. Genotyping was performed using microsatellite markers and Metaphor agarose (Lonza) gel electrophoresis to identify an A/J region derived from the original mutagenized parent, or using Sanger sequencing to detect the mutations directly. The primer sequences for genotyping are listed in Supplemental Material, Table S1. Animals were maintained in accordance with the guidelines of the National Institutes of Health and the Seattle Children’s Research Institute’s Institutional Animal Care and Use Committee.
Exome sequencing
Genomic DNAs were prepared using a DNeasy kit (Qiagen) from the liver tissue of hydrocephalus mutants. Whole-exome sequencing was performed to analyze the entire coding and splice-junction sequence at the Broad Institute (Cambridge, MA) as a part of the Mouse Mutant Resequencing project (https://www.broadinstitute.org/mouse-mutant-resequencing). Reads were mapped to the mouse genome mm10 using BWA-MEM (Li and Durbin 2009), and duplicate reads marked using picard (http://broadinstitute.github.io/picard). Subsequent processing and variant calling was completed using the Genome Analysis Toolkit (GATK) (McKenna et al. 2010). The average coverages were 65X and 72X of the exome for Dnaaf1m4Bei and Lrrc48m6Bei, respectively, with > 91.4% of the exome covered by 10 or more reads. To identify the causal mutations, we considered all nonsynonymous, nonsense, or splicing mutations within recombinant intervals defined by SNP analysis. The Dnaaf1m4Bei and Lrrc48m6Bei lines each had only one homozygous variant that fitted these criteria.
RT-PCR and qRT-PCR
Total RNA was isolated from the brains of wild-type and mutant mice using Trizol reagent (Sigma). A Superscript III First-Strand RT-PCR kit (ThermoFisher Scientific) was used for cDNA synthesis following the manufacturer’s instructions, using the same amount of RNA. RT-PCR was performed using the cDNA and primers indicated in Table S2, designed using Primer3plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). Primer pairs for qRT-PCR were designed using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast) and tested to determine their specificity, optimal reaction temperature, melt curve/peak, and efficiency. qRT-PCR was performed using KAPA SYBR FAST qPCR Kits (KAPA Biosystems) and the CFX96 Real-Time System with a C1000 Thermal Cycler (BioRad). cDNAs from four mice for each genotype (wild-type and homozygote mutant) were used, and the reactions for each sample were triplicated. Mean Cq values for each sample were calculated from the triplicate measurement, Lrrc48 expression was normalized to Actb, relative expression levels compared with the wild-type were calculated, and statistical analysis was performed using REST (Relative Expression Software Tool, http://rest.gene-quantification.info). Power analysis was performed using PS (Power and Sample size calculation software, http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize) to determine sample size, and four samples were enough to reject the null hypothesis with probability 0.9.
Analysis of Dnaaf1 splicing defect
RT-PCR products from the wild-type and Dnaaf1m4Bei mutant were cloned into the pCR2.1 vector using a TOPO TA cloning kit (Invitrogen). Plasmid DNAs were isolated from single colonies, and wild-type and mutant clones were screened by PCR and sequenced using M13 forward and reverse primers. Selected clones with insert sizes similar to the major RT-PCR bands were amplified using the same primers (Dnaaf1 e3-6, Table S2), and the sizes of PCR products were compared to the RT-PCR products by electrophoresis.
Histology
For Nissl staining, the brains were fixed in Bouin’s fixative for 24 hr and then in 10% phosphate-buffered formalin overnight. The fixed brains were embedded in paraffin and sectioned using a Leica microtome. Sections 10-μm thick were stained using FD Thionin solution (FD NeuroTechnologies) following the manufacturer’s instruction.
For hematoxylin and eosin staining of the maxillary sinuses, the whole heads were fixed in Bouin’s fixative and decalcified using Poly-NoCal solution (Polysciences). After 4 hr (P7 mice) or 8 hr (16 week-old mice) incubation, endpoint of decalcification was determined using a Poly-NoCal endpoint determination kit (Polysciences). The decalcified heads were processed, embedded in paraffin, sectioned 10 μm in thickness, and then hematoxylin and eosin staining was performed. Imaging of histology slides was done using a Leica DM4000B upright microscope and DFC310FX camera.
Immunohistochemistry
The tissues were fixed in 4% PFA in PBS overnight, incubated in 30% sucrose until they sank, and then embedded in OCT compound. Sections 10-μm thick were mounted on the glass slides and immunostaining was performed. The brain sections were incubated with blocking solution (3% goat serum, 3% bovine serum albumin, 0.3% Triton X-100 in PBS) for 2 hr, with mouse anti-ARL13B (NeuroMab, clone N295B/66, 1:200) and rabbit anti-dynein heavy chain (DNAH9) antibody (Abcam, ab133968, 1:200) overnight at 4°, with the secondary antibodies (Alexa Fluor 488 goat anti-mouse IgG2a and Alexa Fluor 568 goat anti-rabbit IgG) for 2 hr at room temperature, and then with Hoechst for 5 min. The immunostained sections were imaged using Leica TCS SP5 or Zeiss LSM710 confocal microscopes.
Data availability
DNA sequence data is available in the SRA sequence archive using the accession number: SRP076681. Otherwise, all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Identification and analysis of Dnaaf1 and Lrrc48 mutations
The originally reported recombination intervals based on mouse genome assembly version mm9 were chr8:105–130 Mb for line 42 and chr11:47–69 Mb for line 67 (Ha et al. 2015). Mb positions used in this paper are from mm10 unless indicated otherwise, and SNPs flanking the original recombinant interval and updated map positions are as follows: rs3705275 (chr8:103,336,314 bp) and rs6400423 (chr8:126,582,425 bp) in line 42, and rs13481014 (chr11:48,117,382 bp) and rs13481084 (chr11:68,770,517 bp) in line 67. Whole-exome sequencing was performed to identify recessive mutations within the homozygous regions previously defined using whole-genome SNP panels (Ha et al. 2015). Mutations in Dnaaf1 and Lrrc48 were the only nonsynonymous or splice-site homozygous variants found in the recombinant intervals defined by SNP analysis.
We determined that the mutant mice used for exome sequencing included recombinants within the original interval; therefore, additional analysis of the exome sequencing data was done to identify a minimal region carrying homozygous A/J strain-specific SNPs. We have previously described a detailed method of mutant mapping utilizing exome sequencing data (Gallego-Llamas et al. 2015). This further narrowed the interval to chr8:119.4–129.4 Mb for line 42 and chr11:54.1–63.8 Mb for line 67. This increased resolution excluded some candidate genes; for example, Dnah2 at chr11:69,420,809–69,549,108 bp. Lists of genes in the newly defined interval and exome sequencing coverage are shown in Figure S1. Importantly, the histologically-confirmed hydrocephalus phenotype has remained 100% concordant with homozygosity for the mutations in Dnaaf1 (N = 12) and Lrrc48 (N = 16), and we have never observed hydrocephalus in mice that are not homozygous at these loci (N > 100 for both lines).
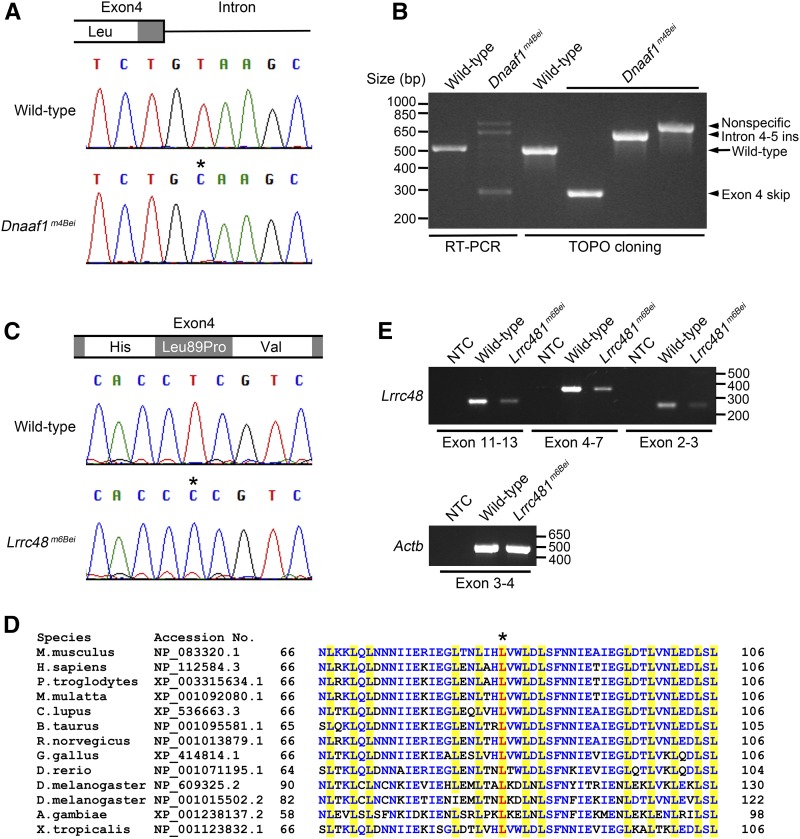
Line 42 had a mutation in Dnaaf1 (NM_026648) at chr8:119582732 bp (exon 4: c.556 + 2T > C). This is in the splice donor site of exon 4, two bases distal from the exon (Figure S2A). The mutant line is referred to Dnaaf1m4Bei in this paper, as listed in the MGI database. Sanger sequencing analysis was performed to confirm the mutation (Figure 1A). The mutation at the splice donor site suggested the likelihood of a splicing defect, therefore the Dnaaf1 transcript was analyzed by RT-PCR using primers designed to amplify a fragment spanning exon 3 and exon 6. The wild-type mice showed a band at expected size 501 bp, while multiple bands with incorrect sizes were detected in the Dnaaf1m4Bei mutant (Figure 1B). This result confirmed that the Dnaaf1m4Bei mutation causes abnormal splicing. To examine abnormal transcripts, RT-PCR products were cloned into a plasmid vector, and individual clones were analyzed using PCR and sequencing (Figure 1B). No abnormal Dnaaf1 transcripts were obtained from the wild type. In contrast, 48 clones from a Dnaaf1m4Bei mutant were analyzed and no normal transcript of 501 bp was found. Eleven mutant clones had 222 bp of the entire exon 4 skipped (Figure 1B, c.335_556del). Five mutant clones retained the entire intron 4–5 between exon 4 and 5 (Figure 1B, [c.556 + 2T > C + c.556_557ins556 + 1_556 + 116]). The remainder of clones had sequences unrelated to Dnaaf1.
Figure 1.
Mutations in Dnaaf1 and Lrrc48. (A) A chromatogram from Sanger sequencing showing a T > C mutation (asterisk) at the junction of exon 4 and intron. (B) RT-PCR result showing amplification of a fragment corresponding to exon 3–6 of Dnaaf1 (lanes 1 and 2). Multiple bands with incorrect sizes were detected in Dnaaf1m4Bei, indicating a splicing defect. Cloned RT-PCR products revealed abnormal transcripts with exon 4 skipping (lane 4, insert size 279 bp) or intron 4–5 insertion (lane 5, insert size 617 bp). The largest band (nonspecific) was not a Dnaaf1 transcript. (C) A chromatogram from Sanger sequencing showing a T > C mutation (asterisk) causing an amino acid change (Leu89Pro). (D) Multiple protein sequence alignment adapted from NCBI HomoloGene search result, including conserved leucine-rich repeat domains near the mutation. Leucine residues in the domain are highlighted in yellow. The leucine residue substituted in Lrrc48m6Bei mutants is shown in red (asterisk). Other conserved residues are marked in blue. (E) RT-PCR results using three different primer sets spanning the Lrrc48 transcript. All amplified weaker bands from the mutant. Actb (Beta-actin) was used as a control. NCBI, National Center for Biotechnology Information; NTC, no template control; RT-PCR, reverse transcription polymerase chain reaction.
Transcripts with exon 4 skipped would encode a DNAAF1 protein with an in-frame deletion of 74 amino acids (p.G112_S186delinsA). Dnaaf1 is a leucine-rich repeat-containing protein (Duquesnoy et al. 2009), and its predicted protein domain structures are shown in Figure S3A. Exon 4 deletion will remove three out of six leucine-rich repeats. Intron 4–5 insertion introduces a S186C substitution at the junction with the addition of 43 amino acids followed by a premature stop codon (p.S186CfsX43), which would result in a large truncation of 448 C-terminal amino acids.
A mutation in Lrrc48 (NM_029044) at Chr11:60363569 bp (exon4: c.T266C: p.L89P) was found in line 67 by the whole-exome sequencing analysis (Figure S2B). This is a missense mutation that substitutes proline for leucine. The mutant line is referred to as Lrrc48m6Bei in this paper, as listed in the MGI database. A chromatogram from Sanger sequencing analysis is shown in Figure 1C. Lrrc48 is also a leucine-rich repeat-containing protein (Figure S3B), and the mutation alters a highly-conserved leucine residue (Figure 1D) located within the leucine-rich repeat domain (Bella et al. 2008). Importantly, the PolyPhen score for this missense mutation is 1.0 (Adzhubei et al. 2010). In addition, analysis of the Lrrc48 transcript using three independent primer sets suggested that the mutant mice have decreased expression (Figure 1E). qRT-RCR analysis using primers for exon 8–9 confirmed a reduction of Lrrc48 expression in the mutant to 34% of wild-type levels (P < 0.001, Figure S4).
Hydrocephalus
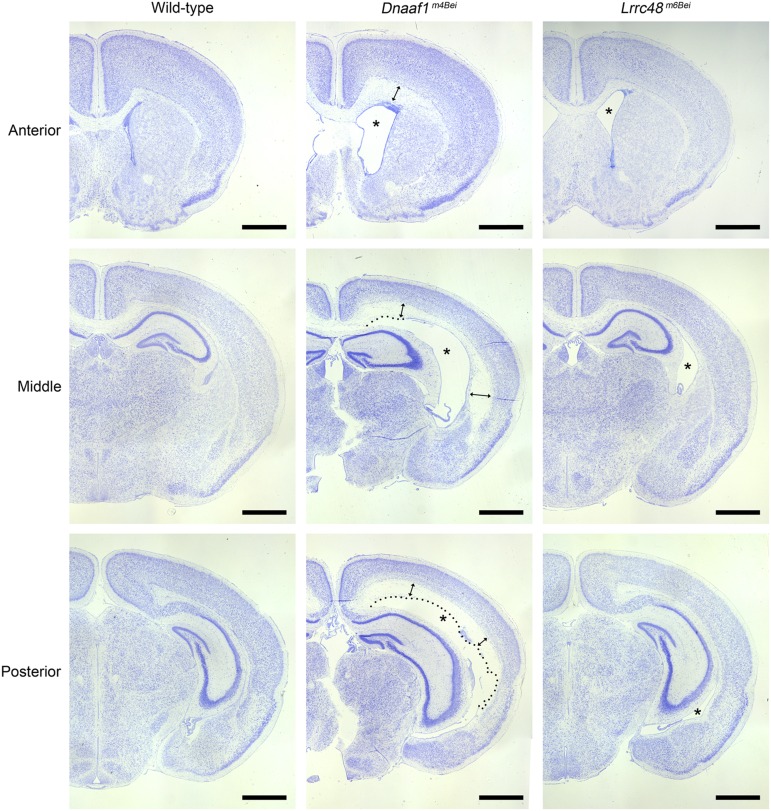
Both mutant lines were originally ascertained as having hydrocephalus prior to P21 (Ha et al. 2015). At birth, both Dnaaf1m4Bei and Lrrc48m6Bei mutant brains appeared grossly normal. The lateral ventricle size, aqueduct opening, and choroid plexus morphology were comparable to wild-type littermates. The lack of aqueduct stenosis at birth suggests that the mutants have a communicating form of hydrocephalus or ventriculomegaly, although we did not exclude the possibility that obstructive hydrocephalus develops secondarily afterward. At postnatal day 10 (P10), changes in the brain structure are already evident (Figure 2). The lateral ventricles and third ventricle were significantly enlarged. In mutants with severe hydrocephalus, the ependymal lining was disrupted, and transependymal edema of the white matter was noticeable. Dnaaf1m4Bei mutants usually displayed more severe hydrocephalus compared to Lrrc48m6Bei.
Figure 2.
Hydrocephalus phenotype. Nissl-stained images of coronal sections are shown. Enlarged lateral ventricles are marked with asterisks. Enlargement of the third ventricles is also evident in both mutants. Dotted lines in Dnaaf1m4Bei mutant images indicate disruption in ependymal lining. Double-headed arrows indicate transependymal edema. Scale bars, 1 mm.
Postnatal lethality of Dnaaf1m4Bei
Most of the Dnaaf1m4Bei mutants were severely runted compared with littermates (Figure S5) and frequently died during the second postnatal week. Postnatal lethality is described in Table 1. Many homozygotes (17.1% of total pups and 68.3% of expected homozygote pups) survived at P1, while only 8.6% of total pups and 34.3% of expected homozygote pups survived at P10. Furthermore, no homozygote was recovered at weaning (P21) when 33 mice from six litters born from heterozygote mating pairs were genotyped (Chi-square test, P = 0.0037). Although similarly severe hydrocephalus or runting was occasionally observed in litters born from Lrrc48m6Bei heterozygote mating pairs, most Lrrc48m6Bei homozygotes were able to survive to adulthood. A total of 71 mice from 11 litters were genotyped, and 18 (25.4%) were homozygotes.
Table 1. Postnatal lethality and laterality defect.
| Age | P0–P1a | P10 | P21 | |||
|---|---|---|---|---|---|---|
| Dnaaf1m4Bei | Observedb | Expected | Observedc | Expected | Observedd | Expected |
| Wild-type | 29 | 20.5 | 21 | 17.5 | 12 | 8.25 |
| Heterozygote | 39 | 41 | 43 | 35 | 21 | 16.5 |
| Homozygote | 14 | 20.5 | 6 | 17.5 | 0 | 8.25 |
| Laterality defect | 8 (57.1%) | 2 (33.3%) | N/Ae | |||
| Situs inversus | 3 | 0 | ||||
| Heterotaxy | 5 | 2 | ||||
| Lrrc48m6Bei | Observed | Expected | Observed | Expected | Observed | Expected |
| Wild-type | 21 | 18 | 24 | 19.75 | 22 | 17.75 |
| Heterozygote | 33 | 36 | 36 | 39.5 | 31 | 35.5 |
| Homozygote | 18 | 18 | 19 | 19.75 | 18 | 17.75 |
| Laterality defect | 2 (11.1%) | 1 (5.3%) | N/Ae | |||
| Situs inversus | 2 | 0 | ||||
| Heterotaxy | 0 | 1 | ||||
N/A, not applicable.
Only one litter was P0 out of total 11 litters.
Chi-square test, P = 0.0583 (two-tailed).
Chi-square test, P = 0.0065 (two-tailed).
Chi-square test, P = 0.0037 (two-tailed).
Laterality defect was not examined at P21.
Male infertility of Lrrc48m6Bei
We observed during mouse husbandry that male Lrrc48m6Bei homozygotes never generated pups, while homozygote females were fertile. To confirm the male infertility phenotype, six homozygote males between the age of 8–14 wk were set up with four different wild-type females. All six males were able to plug at least one female, and a total of nine out of 24 females were plugged. However, none of the females became pregnant.
Laterality defect
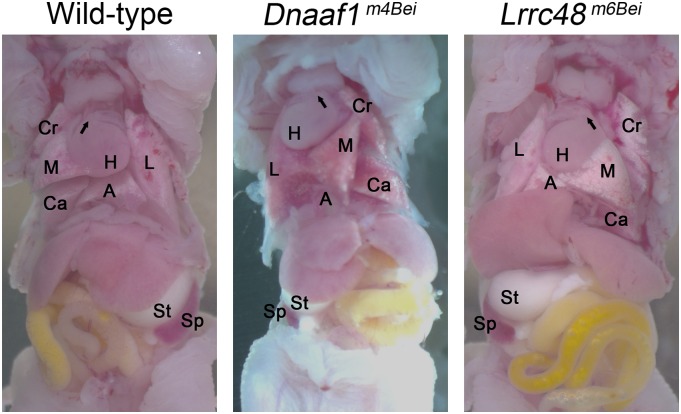
To assess if the mutants have a laterality defect, the visceral organs were examined at P1 and P10. Mutants with situs inversus were found from both Dnaaf1m4Bei and Lrrc48m6Bei mutant lines (Figure 3). The numbers of mutants with laterality defects, including situs inversus and heterotaxy, are shown in Table 1. Dnaaf1m4Bei mutants express laterality defects more frequently than Lrrc48m6Bei mutants. Combining observations at P1 and P10, 52.6% and 8.1% of homozygotes from Dnaaf1m4Bei and Lrrc48m6Bei mutant lines, respectively, showed laterality defects.
Figure 3.
Situs inversus phenotype. The position of the heart (H), lung lobes (L, Cr, M, Ca, and A), stomach (St), and spleen (Sp) shows a laterality defect. Arrows indicate the angle of pulmonary trunk. A, accessary lobe; Ca, right caudal lobe; Cr, right cranial lobe; L, left lobe; M, right middle lobe.
Sinusitis
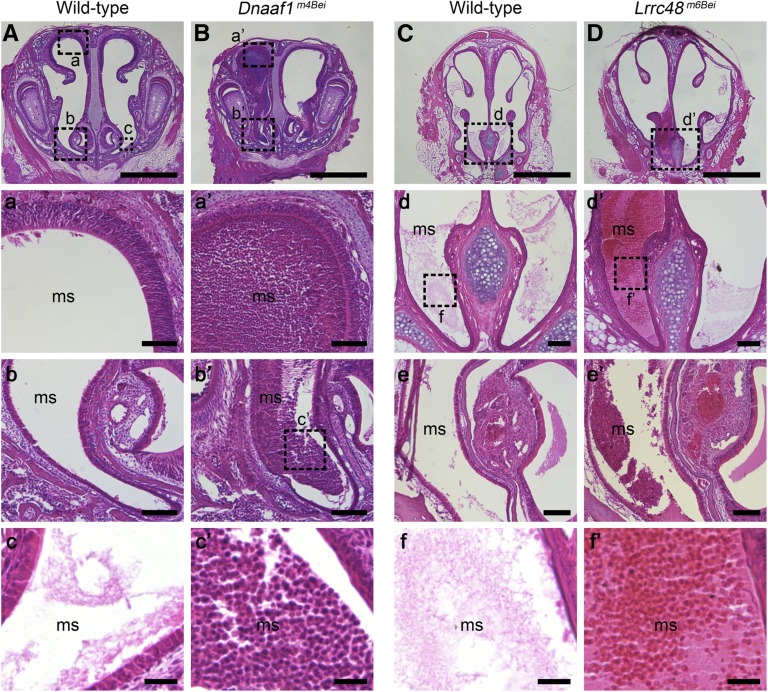
As sinusitis is a common feature of human PCD, histological analysis of maxillary sinuses was performed (Figure 4). Both Dnaaf1m4Bei and Lrrc48m6Bei mutants showed similar phenotypes to those reported for other mouse PCD models (Ibanez-Tallon et al. 2002; Kobayashi et al. 2002; Sironen et al. 2011; Lucas et al. 2012); this included mucus accumulation and infiltration of neutrophils. Dnaaf1m4Bei mutants displayed a severe phenotype at P7, while Lrrc48m6Bei mutants displayed a mild phenotype at older age (16 wk).
Figure 4.
Mucus accumulation in the sinuses. (A–D) Coronal sections of the maxillary sinuses (ms) from mutant and littermate wild-type mice are stained using hematoxylin and eosin. Higher magnification images of the boxed region (a–f for wild-type and a’–f’ for mutants) are shown in lower panels. e and e’ are from more posterior paranasal cavity sections. Severe mucus accumulation and infiltration of neutrophils (c’) were obvious in a Dnaaf1m4Bei mutant at P7. Lrrc48m6Bei mutants show milder phenotype that is undetectable until later (16 week-old). Scale bars: A–D, 1 mm; a, a’, b, b’, d’, e, and e’, 100 μm; c, c’, f, and f’, 25 μm.
Examination of the motile cilia
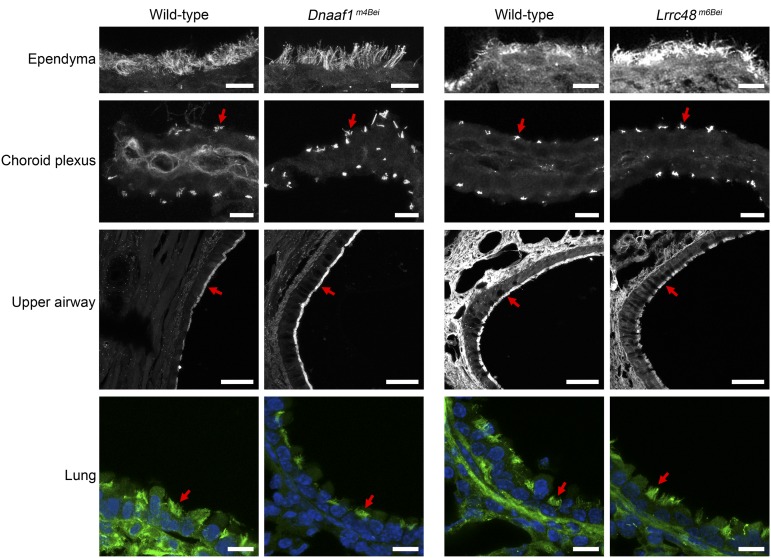
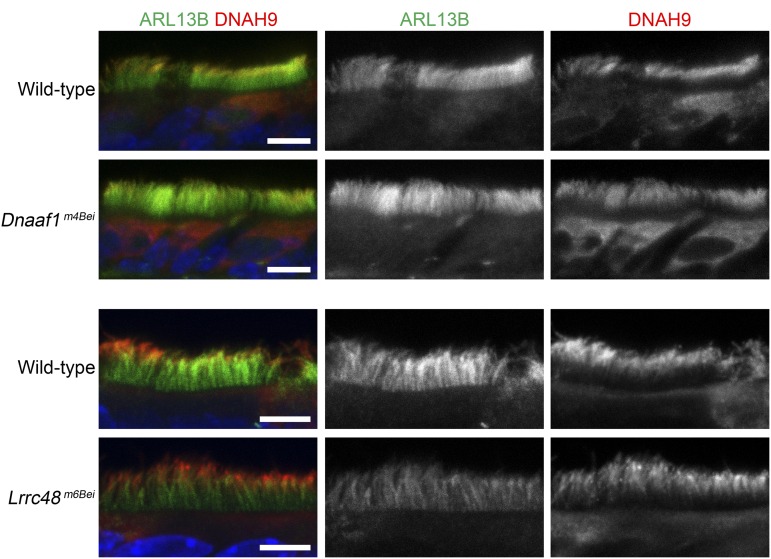
We examined the motile cilia using ARL13B as a marker and did not find any obvious abnormalities; the cilia were present in the ependyma and choroid plexus of the lateral ventricles, upper airway, and bronchi of the lung (Figure 5). It has been previously shown that outer dynein arm heavy chain components, including DNAH9, are absent in the motile cilia of primary respiratory cells from human patients carrying DNAAF1/LRRC50 mutations (Loges et al. 2009). We performed coimmunostaining of ARL13B and DNAH9 on the upper airway sections and found that DNAH9 was present and normally localized in the motile cilia of mutant mice (Figure 6).
Figure 5.
The presence of cilia in the mutants. Cilia in ependyma and choroid plexus of the brain, upper airway, and lung were visualized by anti-ARL13B immunostaining (gray or green). The nuclei were stained using Hoechst (blue). Red arrows indicate representative cilia. Scale bars: ependyma, choroid plexus, and lung, 10 μm; upper airway, 50 μm.
Figure 6.
Normal localization of DNAH9 in the mutants. Anti-ARL13B (green, middle) and anti-DNAH9 (red, right) coimmunostaining was performed. DNAH9 is distally localized in the ciliary axoneme of the motile cilia in the upper airway. The nuclei were stained using Hoechst (blue). Scale bars, 5 μm.
Discussion
Murine forward genetic screens for developmental phenotypes have generated numerous mutants with cilial defects. Interestingly, genes encoding cilial proteins have been identified relatively frequently from multiple independent developmental screens, regardless of whether the screen was specifically targeting laterality defects, embryonic lethality, organogenesis defects, or discrete phenotypes such as congenital heart defects or brain patterning phenotypes, indicating a crucial role of cilia during the development of multiple organ systems (Kasarskis et al. 1998; Tran et al. 2008; Ermakov et al. 2009; Rao Damerla et al. 2014; Ha et al. 2015; Li et al. 2015). Proteomic analyses of cilia have revealed the presence of a surprisingly large number of proteins despite the small size of this organelle (Ostrowski et al. 2002; Pazour et al. 2005; Ishikawa et al. 2012), and new genes required for cilial function are still being discovered. Of particular note is a recent comprehensive screen for congenital heart disease, which revealed an enrichment of mutations associated with cilial function and laterality defects (Ermakov et al. 2009; Li et al. 2015),
In this paper, we report the characterization of two novel mouse models of cilial defects, Dnaaf1m4Bei and Lrrc48m6Bei, which both display hydrocephalus and laterality defects. The Dnaaf1m4Bei mutant carries a splice site mutation that affects transcription with high penetrance; the abnormal transcripts that are generated would result in either a large internal deletion or premature protein truncation. The Lrrc48m6Bei mutant carries a missense mutation in a highly conserved leucine-rich repeat. This mutant also shows a reduction in expression; however, as mice that are heterozygous for null mutations of genes required for motile ciliary function appear normal (e.g., Tan et al. 2007), it seems unlikely that the reduced gene expression accounts for the mutant phenotype.
Both genes are known to encode proteins required for motile cilia and flagellar function; however, mouse mutants of Dnaaf1 and Lrrc48 have not been previously reported. Despite the presence of motile cilia and the normal localization of DNAH9 in these mutants, their phenotypes suggest defects in motile cilia function. Considering what is already known about the Chlamydomonas orthologs of these genes, as discussed below, it is possible that mutations in mice may cause abnormalities in axonemal ultrastructure and cilia motility, rather than a ciliogenesis defect. Further analysis of the axonemal ultrastructure and ciliary motility might illuminate the nature of motile cilia dysfunction in Dnaaf1m4Bei and Lrrc48m6Bei mutants.
Lrrc48 has been previously described as an ortholog of the C. reinhardtii gene Flagella Associated Protein (FAP) 134 (Pazour et al. 2005; Lechtreck et al. 2009; Lin et al. 2011; Bower et al. 2013). FAP134 protein was isolated from the flagellar proteome and its localization in the flagellar axoneme has been confirmed (Pazour et al. 2005; Lechtreck et al. 2009). Another peptide sequencing analysis identified FAP134 to be DRC3 (Lin et al. 2011). DRC3 protein is one of the eleven known components of N-DRC. DRC3 was originally identified by a proteomics analysis of flagellar axonemal polypeptides, using a series of flagella motility mutants (Huang et al. 1982; Piperno et al. 1992; Lin et al. 2011; Bower et al. 2013).
More specifically, FAP134/DRC3 was found to be absent or reduced in the flagellar axoneme of Chlamydomonas mutants pf2 and sup-pf-3 (Bower et al. 2013). These mutant strains are suppressor strains that reverse flagellar immotility in paralysis mutant strains lacking the central microtubule pairs (Huang et al. 1982). However, pf2 and sup-pf-3 do not carry mutations in the FAP134/DRC3 gene, but in the DRC4 and ODA2 genes, respectively (Rupp et al. 1996; Rupp and Porter 2003). They display an overall structural defect of N-DRC involving multiple N-DRC components; as such, the flagellar motility defect observed in pf2 and sup-pf-3 is not direct evidence to prove the requirement of FAP134/DRC3. More recently, the drc3 mutant was reported to display slower swimming speed due to an abnormal flagellar waveform (Awata et al. 2015). This mutant, generated by an insertional mutagenesis, has a large deletion including the first seven exons of FAP134/DRC3. All previously known N-DRC components were found present in the axoneme of the drc3 mutant, providing direct evidence for the importance of FAP134/DRC3 in flagellar motility (Awata et al. 2015).
Our Lrrc48m6Bei mutant phenotype, as the first reported mouse mutant of the FAP134/DRC3 ortholog, demonstrates a presumptive requirement for this gene in mammalian motile cilial function. Consistent with the observation that Chlamydomonas N-DRC component mutants have no change in flagellar length (Piperno et al. 1992; Awata et al. 2015), ciliogenesis was normal in our mouse mutants. We did not pursue further analysis of axonemal ultrastructure as it requires high-resolution in situ cryo-electron tomography. A study using this technique has recently visualized DRC3 localization to the L1 protrusion of the nexin linker region of N-DRC and its direct interaction with the dynein g motor domain of an inner dynein arm, IA5 (Heuser et al. 2009; Awata et al. 2015; Song et al. 2015). The drc3 mutant displayed a mild alteration of N-DRC structure restricted to a small portion near DRC3, unlike other N-DRC component mutants (Awata et al. 2015). This type of ultrastructural defect would not be easily detectable using conventional transmission electron microscopy.
Dnaaf1 is also called Lrrc50 (human and zebrafish) or Oda7 (Chlamydomonas reinhardtii), and its function is relatively well characterized. In Chlamydomonas, a genomic deletion that included exon 1 of the Oda7 gene was identified in an arg7, oda7 mutant strain (Freshour et al. 2007). This strain was ascertained in a screen for slow-swimming phenotypes with reduced flagellar beat frequency, and the entire outer dynein arm (ODA) was absent in many of these mutants (Kamiya and Okamoto 1985). Analysis of axonemal ultrastructure revealed that the Oda7 mutant lacks any observable pool of outer row dynein heavy chain α (Fowkes and Mitchell 1998). Freshour et al. (2007) found that ODA7 interacts with both outer row and I1 inner row dyneins, and that the mutation in ODA7 prevents axonemal outer row dynein assembly by blocking the cytoplasmic assembly of heavy chains and intermediate chains.
In zebrafish, the lrrc50hu255H mutant carrying a nonsense mutation at a conserved leucine residue was identified from ENU mutagenesis (van Rooijen et al. 2008). This mutant displayed an ultrastructural defect of the ciliary axoneme, including the absence of ODA and misalignments of outer microtubule doublets. A range of motile cilia defects in multiple organs were observed: the absence of motile cilia in the nose and neural tube, irregular cilia distribution in Kupffer’s vesicle, abnormal cilia morphology and distribution in the anterior and posterior pronephric duct, reduced kinocilium number and length in the lateral line organ, and reduced cilia motility measured by dye-excretion test. As a result, the mutant has a laterality defect and develops pronephric tubular cysts.
Importantly, both Dnaaf1 and Lrrc48 mutations have a clinical relevance. DNAAF1 mutations have been found in PCD patients with or without situs inversus (Duquesnoy et al. 2009; Loges et al. 2009) and seminoma patients (Basten et al. 2013). Analysis of the axonemal ultrastructure in human PCD patients revealed the absence of both dynein arms (Duquesnoy et al. 2009; Loges et al. 2009). It is likely that Dnaaf1m4Bei mutants have a similar axonemal ultrastructure defect, given that ODA structure is conserved from algae to zebrafish to human. DNAAF1 is included in genetic diagnostic testing of PCD (Knowles et al. 2013; Kurkowiak et al. 2015; Lobo et al. 2015; Marshall et al. 2015). In contrast, the direct clinical relevance of LRRC48 mutation has not yet been reported. However, orthologs of multiple N-DRC component genes have been implicated in human PCD, including CCDC164/DRC1 and CCDC65/DRC2 (Austin-Tse et al. 2013; Horani et al. 2013; Wirschell et al. 2013). It was found that LRRC48 protein, which is normally localized in the axoneme, was reduced in PCD patients with CCDC164/DRC1 mutation (Wirschell et al. 2013).
In summary, we report the phenotypic and genomic characterization of mice carrying mutations in Dnaaf1 and Lrrc48, which are presumptively required for the function of primary motile cilia. For unknown reasons, mouse models of PCD frequently display hydrocephalus, which is less common in humans than other PCD symptoms such as respiratory infection and situs inversus. In addition to differences in shape and size, human and mouse brains have structural and physiological differences that may influence their susceptibility to PCD-associated hydrocephalus (reviewed in Lee 2013). Despite this, it is likely that the underlying physiology of motile cilial function is conserved across species; as such, the Dnaaf1m4Bei and Lrrc48m6Bei mutants can serve as valuable models of human PCD.
Supplementary Material
Acknowledgments
We thank Mingyue Lun, Haiyan Qiu, and George C. Talbott for technical assistance. This work was supported by National Institutes of Health grants 5R01MH081187 and 5R01HD36404.
Footnotes
Supplemental material is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.030791/-/DC1
Communicating editor: A. S. McCallion
Literature Cited
- Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., et al. , 2010. A method and server for predicting damaging missense mutations. Nat. Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin-Tse C., Halbritter J., Zariwala M. A., Gilberti R. M., Gee H. Y., et al. , 2013. Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 defects as causing primary ciliary dyskinesia. Am. J. Hum. Genet 93: 672–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awata J., Song K., Lin J., King S. M., Sanderson M. J., et al. , 2015. DRC3 connects the N-DRC to dynein g to regulate flagellar waveform. Mol. Biol. Cell 26: 2788–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N., 2006. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7: 125–148. [DOI] [PubMed] [Google Scholar]
- Banizs B., Pike M. M., Millican C. L., Ferguson W. B., Komlosi P., et al. , 2005. Dysfunctional cilia lead to altered ependyma and choroid plexus function, and result in the formation of hydrocephalus. Development 132: 5329–5339. [DOI] [PubMed] [Google Scholar]
- Basten S. G., Davis E. E., Gillis A. J., van Rooijen E., Stoop H., et al. , 2013. Mutations in LRRC50 predispose zebrafish and humans to seminomas. PLoS Genet. 9: e1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bella J., Hindle K. L., McEwan P. A., Lovell S. C., 2008. The leucine-rich repeat structure. Cell. Mol. Life Sci. 65: 2307–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R., Tritschler D., Vanderwaal K., Perrone C. A., Mueller J., et al. , 2013. The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol. Biol. Cell 24: 1134–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. M., Witman G. B., 2014. Cilia and Diseases. Bioscience 64: 1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquesnoy P., Escudier E., Vincensini L., Freshour J., Bridoux A. M., et al. , 2009. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am. J. Hum. Genet. 85: 890–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakov A., Stevens J. L., Whitehill E., Robson J. E., Pieles G., et al. , 2009. Mouse mutagenesis identifies novel roles for left-right patterning genes in pulmonary, craniofacial, ocular, and limb development. Dev. Dyn. 238: 581–594. [DOI] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., Omran H., 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8: 880–893. [DOI] [PubMed] [Google Scholar]
- Fowkes M. E., Mitchell D. R., 1998. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol. Biol. Cell 9: 2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour J., Yokoyama R., Mitchell D. R., 2007. Chlamydomonas flagellar outer row dynein assembly protein ODA7 interacts with both outer row and I1 inner row dyneins. J. Biol. Chem. 282: 5404–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Llamas J., Timms A. E., Geister K. A., Lindsay A., Beier D. R., 2015. Variant mapping and mutation discovery in inbred mice using next-generation sequencing. BMC Genomics 16: 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S., Stottmann R. W., Furley A. J., Beier D. R., 2015. A forward genetic screen in mice identifies mutants with abnormal cortical patterning. Cereb. Cortex 25: 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T., Raytchev M., Krell J., Porter M. E., Nicastro D., 2009. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J. Cell Biol. 187: 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horani A., Brody S. L., Ferkol T. W., Shoseyov D., Wasserman M. G., et al. , 2013. CCDC65 mutation causes primary ciliary dyskinesia with normal ultrastructure and hyperkinetic cilia. PLoS One 8: e72299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Ramanis Z., Luck D. J., 1982. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell 28: 115–124. [DOI] [PubMed] [Google Scholar]
- Ibanez-Tallon I., Gorokhova S., Heintz N., 2002. Loss of function of axonemal dynein Mdnah5 causes primary ciliary dyskinesia and hydrocephalus. Hum. Mol. Genet. 11: 715–721. [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Thompson J., Yates J. R., 3rd, Marshall W. F., 2012. Proteomic analysis of mammalian primary cilia. Curr. Biol. 22: 414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R., 1988. Mutations at twelve independent loci result in absence of outer dynein arms in Chylamydomonas reinhardtii. J. Cell Biol. 107: 2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R., Okamoto M., 1985. A mutant of Chlamydomonas reinhardtii that lacks the flagellar outer dynein arm but can swim. J. Cell Sci. 74: 181–191. [DOI] [PubMed] [Google Scholar]
- Kasarskis A., Manova K., Anderson K. V., 1998. A phenotype-based screen for embryonic lethal mutations in the mouse. Proc. Natl. Acad. Sci. USA 95: 7485–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles M. R., Daniels L. A., Davis S. D., Zariwala M. A., Leigh M. W., 2013. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am. J. Respir. Crit. Care Med. 188: 913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Watanabe M., Okada Y., Sawa H., Takai H., et al. , 2002. Hydrocephalus, situs inversus, chronic sinusitis, and male infertility in DNA polymerase lambda-deficient mice: possible implication for the pathogenesis of immotile cilia syndrome. Mol. Cell. Biol. 22: 2769–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkowiak M., Zietkiewicz E., Witt M., 2015. Recent advances in primary ciliary dyskinesia genetics. J. Med. Genet. 52: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K. F., Luro S., Awata J., Witman G. B., 2009. HA-tagging of putative flagellar proteins in Chlamydomonas reinhardtii identifies a novel protein of intraflagellar transport complex B. Cell Motil. Cytoskeleton 66: 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L., 2013. Riding the wave of ependymal cilia: genetic susceptibility to hydrocephalus in primary ciliary dyskinesia. J. Neurosci. Res. 91: 1117–1132. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Klena N. T., Gabriel G. C., Liu X., Kim A. J., et al. , 2015. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 521: 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Tritschler D., Song K., Barber C. F., Cobb J. S., et al. , 2011. Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J. Biol. Chem. 286: 29175–29191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo J., Zariwala M. A., Noone P. G., 2015. Primary ciliary dyskinesia. Semin. Respir. Crit. Care Med. 36: 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loges N. T., Olbrich H., Becker-Heck A., Haffner K., Heer A., et al. , 2009. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am. J. Hum. Genet. 85: 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas J. S., Adam E. C., Goggin P. M., Jackson C. L., Powles-Glover N., et al. , 2012. Static respiratory cilia associated with mutations in Dnahc11/DNAH11: a mouse model of PCD. Hum. Mutat. 33: 495–503. [DOI] [PubMed] [Google Scholar]
- Marshall C. R., Scherer S. W., Zariwala M. A., Lau L., Paton T. A., et al. , 2015. Whole-exome sequencing and targeted copy number analysis in primary ciliary dyskinesia. G3 (Bethesda) 5: 1775–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister J. P., 2nd, 2012. Pathophysiology of congenital and neonatal hydrocephalus. Semin. Fetal Neonatal Med. 17: 285–294. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita K., Takeda S., 2015. Cilia in the choroid plexus: their roles in hydrocephalus and beyond. Front. Cell. Neurosci. 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D., Schwartz C., Pierson J., Gaudette R., Porter M. E., et al. , 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313: 944–948. [DOI] [PubMed] [Google Scholar]
- Ostrowski L. E., Blackburn K., Radde K. M., Moyer M. B., Schlatzer D. M., et al. , 2002. A proteomic analysis of human cilia: identification of novel components. Mol. Cell. Proteomics 1: 451–465. [DOI] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B., 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170: 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Mead K., Shestak W., 1992. The inner dynein arms I2 interact with a “dynein regulatory complex” in Chlamydomonas flagella. J. Cell Biol. 118: 1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M. E., Sale W. S., 2000. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151: F37–F42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Damerla R., Gabriel G. C., Li Y., Klena N. T., Liu X., et al. , 2014. Role of cilia in structural birth defects: insights from ciliopathy mutant mouse models. Birth Defects Res. C Embryo Today 102: 115–125. [DOI] [PubMed] [Google Scholar]
- Rupp G., Porter M. E., 2003. A subunit of the dynein regulatory complex in Chlamydomonas is a homologue of a growth arrest-specific gene product. J. Cell Biol. 162: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp G., O’Toole E., Gardner L. C., Mitchell B. F., Porter M. E., 1996. The sup-pf-2 mutations of Chlamydomonas alter the activity of the outer dynein arms by modification of the gamma-dynein heavy chain. J. Cell Biol. 135: 1853–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironen A., Kotaja N., Mulhern H., Wyatt T. A., Sisson J. H., et al. , 2011. Loss of SPEF2 function in mice results in spermatogenesis defects and primary ciliary dyskinesia. Biol. Reprod. 85: 690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Awata J., Tritschler D., Bower R., Witman G. B., et al. , 2015. In situ localization of N and C termini of subunits of the flagellar nexin-dynein regulatory complex (N-DRC) using SNAP tag and cryo-electron tomography. J. Biol. Chem. 290: 5341–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. Y., Rosenthal J., Zhao X. Q., Francis R. J., Chatterjee B., et al. , 2007. Heterotaxy and complex structural heart defects in a mutant mouse model of primary ciliary dyskinesia. J. Clin. Invest. 117: 3742–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. V., Haycraft C. J., Besschetnova T. Y., Turbe-Doan A., Stottmann R. W., et al. , 2008. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat. Genet. 40: 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooijen E., Giles R. H., Voest E. E., van Rooijen C., Schulte-Merker S., et al. , 2008. LRRC50, a conserved ciliary protein implicated in polycystic kidney disease. J. Am. Soc. Nephrol. 19: 1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M., Olbrich H., Werner C., Tritschler D., Bower R., et al. , 2013. The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat. Genet. 45: 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequence data is available in the SRA sequence archive using the accession number: SRP076681. Otherwise, all data necessary for confirming the conclusions presented in the article are represented fully within the article.