Abstract
The protein product of the Homo sapiens TP53 gene is a transcription factor (p53) that regulates the expression of genes critical for the response to DNA damage and tumor suppression, including genes involved in cell cycle arrest, apoptosis, DNA repair, metabolism, and a number of other tumorigenesis-related pathways. Differential transcriptional regulation of these genes is believed to alter the balance between two p53-dependent cell fates: cell cycle arrest or apoptosis. A number of previously identified p53 cofactors covalently modify and alter the function of both the p53 protein and histone proteins. Both gain- and loss-of-function mutations in chromatin modifiers have been strongly implicated in cancer development; thus, we sought to identify novel chromatin regulatory proteins that affect p53-dependent transcription and the balance between the expression of pro-cell cycle arrest and proapoptotic genes. We utilized an siRNA library designed against predicted chromatin regulatory proteins, and identified known and novel chromatin-related factors that affect both global p53-dependent transcription and gene-specific regulators of p53 transcriptional activation. The results from this screen will serve as a comprehensive resource for those interested in further characterizing chromatin and epigenetic factors that regulate p53 transcription.
Keywords: chromatin, p53, siRNA screen, epigenetics, H. sapiens
The p53 protein (encoded by the TP53 gene in Homo sapiens) is a sequence-specific transcription factor and a master tumor suppressor. Inactivating TP53 mutations are observed in over 50% of human cancers, and the loss of p53 activity leads to genome instability and metabolic dysfunction, ultimately promoting tumor formation (Lawrence et al. 2014). p53 is activated in response to a number of diverse cellular stress signals, including DNA damage, oncogene activation, loss of normal metabolic homeostasis, and telomere attrition, and mediates the expression of cell- and organismal-protective genes involved in processes such as DNA repair, cell cycle arrest, and apoptosis (Kruiswijk et al. 2015).
p53 is negatively regulated through direct interaction and ubiquitination by the E3-ubiquitin ligase MDM2, which mediates proteosomal degradation of p53 during nonstress conditions (Momand et al. 1992; Oliner et al. 1992; Wu et al. 1993). Upon stress, the p53:MDM2 complex is disrupted by kinases like ATM, which directly phosphorylate p53 in the MDM2 interaction domain, leading to stabilization and upregulation of the p53 protein level (Siliciano et al. 1997; Shieh et al. 1997). The transcriptional activity of p53 can be modulated by other cofactors, including a number of other enzymes that directly catalyze posttranslational modifications such as acetylation and methylation on various amino acids (Berger 2010; Dai and Gu 2010). Such modifications may act to fine-tune the p53 response, either through altering the DNA binding specificity of p53 (Luo et al. 2004) or through recruitment of p53 interacting proteins, like 53BP1 (Tong et al. 2015a,b; Barlev et al. 2001).
Many p53 modifying enzymes, such as PCAF/GCN5, SETD8/PR-Set7, and SMYD2, also play critical roles in transcriptional regulation through direct modification of histone proteins (Liu et al. 1999; Huang et al. 2006; Shi et al. 2007). A number of these chromatin regulatory factors and pathways have also been implicated in the maintenance of normal homeostasis, with mutated or hyperactive chromatin modifying enzymes and pathways contributing to disease development, including many cancers (Bannister and Kouzarides 2011; Kouzarides 2007). Given the importance of chromatin regulatory factors in regulating normal and disease-associated transcriptional responses, many small molecule inhibitors are being developed as putative therapeutics for various diseases (Dawson and Kouzarides 2012; Dawson et al. 2012).
Importantly, in response to cellular damage, p53 coordinates the transcription of factors involved in potential oncogenic transformation. The ultimate transcriptional output of p53 leads to a modulation of cell fate; a p53-activated cell can undergo transient cell-cycle arrest until the damage/stress is alleviated, permanent cell cycle arrest (also known as senescence) (Rufini et al. 2013), or cell death via apoptosis (Zilfou and Lowe 2009). The molecular mechanisms that modulate these alternative transcriptional programs have been an area of intense investigation (Andrysik et al. 2013). Of particular interest are chromatin and protein modifications that may underlie the differential transcription. For example, the lysine acetyltransferase KAT5/TIP60 influences transcription of the proapoptotic p53 target gene BBC3/puma without affecting the transcription of prosurvival p53 targets like CDKN1A/p21. KAT5/TIP60 also directly acetylates p53 at lysine 120, suggesting that this modification affects p53 target gene discrimination (Sykes et al. 2006; Tang et al. 2006). These and other results strongly implicate p53 modification and chromatin pathways in the regulation of key pathways underlying the fate of damaged cells; however, the specific mechanisms that result in this fate choice remain elusive.
As discussed above, a number of enzymes capable of directly modifying the p53 protein have been implicated in general regulation of p53-dependent transcription. In addition, we and others observed that p53 binds directly to DNA in a varied chromatin and cis-regulatory element context (Lidor Nili et al. 2010; Sammons et al. 2015; Su et al. 2015), suggesting that chromatin structure and modifications might directly influence p53 activity. We hypothesized that chromatin and epigenetic regulatory mechanisms might modulate p53-dependent transcription and tumor suppression. Thus, we designed the siRNA screen to: 1) identify chromatin and epigenetic regulatory proteins that modulate p53-dependent transcription; and 2) identify new trans-acting factors that could influence the ability of p53 to enact a prosurvival (CDKN1A/p21) or proapoptotic (BBC3/puma) transcriptional program. The results uncovered from this screen provide a strong basis for future studies focused on characterizing key mechanisms underlying p53-mediated cell fate regulation.
Materials and Methods
siRNA screen design
The human osteosarcoma cell line U2OS (HTB-96, ATCC) was grown in McCoy’s 5A medium (supplemented with 10% fetal bovine serum and penicillin/streptomycin) at 37 in a standard CO2 incubator. A custom Thermo SmartPool siRNA library targeting chromatin regulatory genes (Supplemental Material, Table S1) was arrayed on a 384-well plate and resuspended in 1 × siRNA Buffer (Dharmacon) at 1 μM. Chromatin regulator targets were manually curated based on a previously published list of putative chromatin regulatory factors (Zuber et al. 2011). Human genes containing domains with previously characterized chromatin regulatory activity (i.e., SET, PHD, Bromo domains, Chromo domains, etc.) were identified using PFAM (EMBL-EBI). All human genes containing these domains were included, even if previously not implicated directly in chromatin regulatory function. Kinases and phosphatases with previously demonstrated direct chromatin regulatory activity were included, and those without were manually removed from the curation.
siRNA was aliquoted into single use 96-well plates with internal control siRNA against TP53, MDM2, and a nonspecific targeting siRNA. Each siRNA was delivered to 11,000 cells via reverse transfection using RNAiMax (Life Technologies) in a 96-well plate to a final concentration of 10 μM, media was changed after 24 hr, and cells were incubated for an additional 48 hr before addition of either DMSO or 100 μM (final) etoposide (Sigma-Aldrich) for 8 hr. PolyA+ RNA was isolated using mRNA Catcher (Life Technologies) and cDNA synthesis was performed on-plate using random hexamer priming. Triplicate qPCR reactions containing cDNA, PowerSybr qPCR Mastermix (Life Technologies), and gene-specific primer pairs were loaded into 384-well plates using an Eppendorf EpMotion 5070, and target gene expression was measured using the relative standard curve method on an ABI 7900HT PCR instrument (Applied Biosystems).
Data analysis and scoring of hits
Target-specific gene expression values for each siRNA knockdown were normalized to mRNA expression of LMNA/lamin A/C. Standard scores (z-scores) were calculated using the equation , where X = gene expression value for an individual target, μ = mean gene expression value across all siRNA experiments, and θ = standard deviation of gene expression values across all siRNA experiments. Values with a z-score representing ± 2 deviations from the standard score were called as hits. Clustering was performed using a 5 × 5 self-organizing map (SOM) with 100 training iterations and implemented using the kohonen package in R (Wehrens and Buydens 2007). Table S1 contains a complete list of gene targets, normalized expression values, and SOM clusters.
Rescreening of a subpool of siRNA and analysis of potential false positives
A random selection of 81 siRNAs were rescreened for their ability to modulate CDKN1A/p21 and BBC3/puma using the same methodology as above with the following changes. First, we screened only using the DMSO/basal condition. Second, we reduced the cut-off for calling a hit in the secondary screen to 1 standard deviation from the mean. Hits present in both the primary and secondary screen are marked with an ampersand in Table 2, Table 3, and Table 4. Putative false positives were then called if they were present but not called as hits in the rescreening experiment. Rescreen expression values can be found in Table S1 under the Secondary Screen tab. Additionally, putative false positives were called if the corresponding expression value for that gene had a value of 0 (not expressed) from a published RNA-seq gene expression analysis (Klijn et al. 2015). Expression values for each gene in the siRNA screen can be found in Table S1. Putative false positives identified using these two methods are now marked with asterisks in Table 2, Table 3, Table 4, and Table 5.
Table 2. Regulators of CDKN1A/p21 and BBC3/puma.
| Downregulated by siRNA | Upregulated by siRNA | ||
|---|---|---|---|
| (Positive Regulator) | (Negative Regulator) | ||
| DMSO | Etoposide | DMSO | Etoposide |
| TP53 | PLK1 | CTCF& | JHDM1D |
| SETD3 | TP53 | MLL2 | |
| NCAPG | PRMT6 | YEATS4 | |
| MBD3L2* | TRERF1 | ||
| TOP1 | EP300& | ||
| SETDB1 | SRCAP& | ||
| PHC1 | USP27X | ||
| HIRIP3& | |||
| KIAA1267& | |||
Genes are listed in order of strongest to weakest phenotypic effect. Full phenotypic values can be found in Table S1. * and & denote putative false positives and secondary screen hits, respectively. Information about identification of putative false positives and secondary screen hits can be found in Materials and Methods. siRNA, small interfering RNA; DMSO, dimethyl sulfoxide.
Table 3. Specific regulators of CDKN1A/p21.
| Downregulated by siRNA | Upregulated by siRNA | ||
|---|---|---|---|
| (Positive Regulator) | (Negative Regulator) | ||
| DMSO | Etoposide | DMSO | Etoposide |
| TBL1XR1 | PCGF2& | EIF2S3 | CHMP4A |
| CBX7& | PRMT2 | SNAPC4 | FKBP1A |
| RCOR2& | SMC2 | CHEK1& | |
| MBD6* | CBX7 | HMG20B& | |
| PCGF2& | TNP1* | CDY1B* | |
| SMC1B | NSD1 | SETD8 | |
| FBXW7 | PCMT1 | KDM4A | |
| TBL1XR1 | YWHAE | ||
| SMC1A | CDC5L& | ||
| SMC1B | SLBP& | ||
| SATB1 | FOXA2 | ||
| HMGA2 | |||
| SNW1 | |||
| TCF7L2 | |||
| MECP2 | |||
Genes are listed in order of strongest to weakest phenotypic effect. Full phenotypic values can be found in Table S1. * and & denote putative false positives and secondary screen hits, respectively. Information about identification of putative false positives and secondary screen hits can be found in Materials and Methods. siRNA, small interfering RNA; DMSO, dimethyl sulfoxide.
Table 4. Specific regulators of BBC3/puma.
| Downregulated by siRNA | Upregulated by siRNA | ||
|---|---|---|---|
| (Positive Regulator) | (Negative Regulator) | ||
| DMSO | Etoposide | DMSO | Etoposide |
| MCM2 | SUDS3 | DLX2* | BAZ1B |
| SFMBT1 | SETD8 | HMGN2 | DACH1 |
| MBD3L1* | TADA2A | ZNF24& | MBD2 |
| SETD1B | KAT5 | NUP62& | DLX2* |
| CEBPB | TCF7L1 | RARA | DMPK |
| PRMT3 | NAP1LF | WNT5A | |
| FGF19* | SMARCA5 | YWHAB | |
| NDEL1 | PAM | SOX12 | |
| PRMT5 | MCM10 | ||
| SMC4 | |||
Genes are listed in order of strongest to weakest phenotypic effect. Full phenotypic values can be found in Table S1. * and & denote putative false positives and secondary screen hits, respectively. Information about identification of putative false positives and secondary screen hits can be found in Materials and Methods. siRNA, small interfering RNA; DMSO, dimethyl sulfoxide.
Table 5. Specific regulators of TP53/p53.
| Downregulated by siRNA | Upregulated by siRNA | ||
|---|---|---|---|
| (Positive Regulator) | (Negative Regulator) | ||
| DMSO | Etoposide | DMSO | Etoposide |
| LMNB1 | SMYD3 | ERCC6 | UBE2I |
| ERCC6 | PRMT8 | RNF20 | DIDO1 |
| HMGN1 | JUN | PHF5A | CDHD9 |
| FKBP2 | PRDM1 | MLL5 | DFFB |
| HDAC9 | NFKB1 | SMARCD2 | |
| SIN3A | KAT2A | HMGN4 | |
| ATM | NCOA3 | HMGN1 | |
| CREB1 | HMGN1 | ||
| KDM5D | CXXC1 | ||
| FOXC2 | SIN3A | ||
| HDAC6 | HDAC1 | ||
| SFMBT2 | |||
| ESR2 | |||
Genes are listed in order of strongest to weakest phenotypic effect. Full phenotypic values can be found in Table S1. * and & denote putative false positives and secondary screen hits, respectively. Information about identification of putative false positives and secondary screen hits can be found in Materials and Methods. siRNA, small interfering RNA; DMSO, dimethyl sulfoxide.
Data availability
Table S1 contains all normalized gene expression data across all screening conditions. Screen data are also available at the GenomeRNAi repository under accession number GR00389-S.
Results and Discussion
Chromatin-focused siRNA screen performance
Based on previous observations of overlap between p53 and chromatin regulatory pathways, we used an siRNA-based approach to identify epigenetic or chromatin regulators of p53-dependent transcriptional activity. We designed a targeted “epigenetic bookcase” of siRNA molecules against 589 genes involved in chromatin regulatory mechanisms (Table S1). These genes include known and putative histone modifying enzymes, chromatin remodelers, chaperones, and cofactors. We reasoned that the reduced complexity of this targeted bookcase compared to a full genome library would facilitate the identification of direct p53 regulators, as opposed to chromatin factors that affect pathways far upstream of p53 activity at DNA.
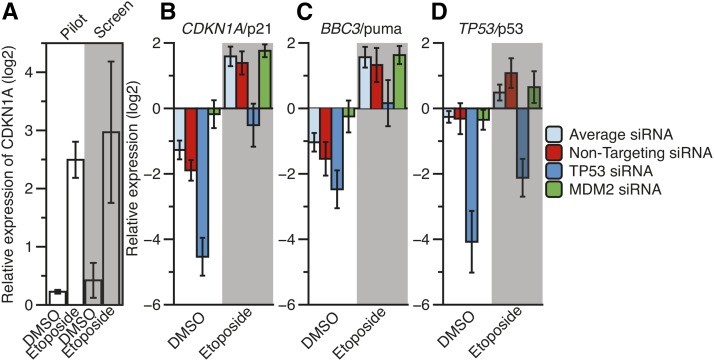
We further developed and optimized an automated RT-qPCR-based readout to maximize efficiency and reduce variability. We utilized U2OS osteosarcoma cell lines, which contain wild-type p53 alleles. The workflow measured expression of CDKN1A/p21 in response to p53 activation following 8 hr of treatment with etoposide, a potent DNA damaging agent and p53 activator. Pilot experiments measuring CDKN1A/p21 expression in control DMSO or etoposide-treated U2OS cells across 80 biological replicates demonstrated a z-score of 0.54, suggesting that this RT-qPCR-based assay would robustly identify putative p53 regulators (Birmingham et al. 2009). We screened 589 gene-specific siRNA pools for the ability to modulate the expression of three different p53 target genes (CDKN1A/p21, BBC3/puma, and TP53/p53) following 8 hr of 100 μM etoposide treatment or DMSO control in U2OS cells (Figure 1). We also measured the expression of a control gene (LMNA/lamin A). This experimental rationale allowed for the identification of genes that regulate p53-dependent transcription at both the basal level (DMSO) and under activated conditions (etoposide), and would allow for the elimination of those factors whose knockdown resulted in global transcriptional changes in genes unrelated to p53 (lamin A). The targeted siRNA screen measuring CDKN1A/p21 transcription mirrored the pilot experiment, although we observed more variation in the siRNA screen, as expected (Figure 2A). Importantly, nontargeting siRNA and control siRNA treatments targeting TP53 or MDM2 produced expected results, indicating that the siRNAs approach was feasible and successful. Nontargeting siRNA displayed comparable expression values as the average of all experimental targets, as expected (Figure 2, B–D). In contrast, treatment with control TP53 siRNAs led to severely reduced CDKN1A/p21, BBC3/puma, and TP53/p53 expression (Figure 2, B–D). MDM2 siRNA controls resulted in increased basal CDKN1A and BBC3 expression, but not TP53, as expected of the role of MDM2 in posttranscriptional regulation of p53 protein activity (Figure 2B). Results of statistical tests for pairwise comparison of control siRNA experiments can be found in Table 1.
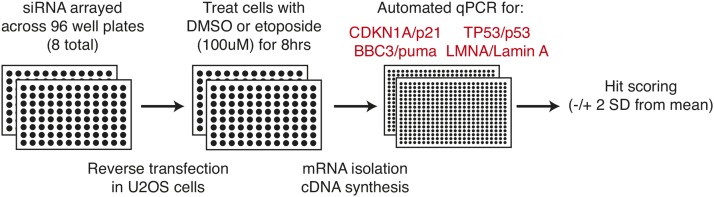
Figure 1.
Schematic of steps for reverse transcription-quantitative PCR (RT-qPCR) methodology for screening a chromatin-focused siRNA library. qPCR, quantitative polymerase chain reaction; DMSO, dimethyl sulfoxide; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SD, standard deviation; siRNA, small interfering RNA.
Figure 2.
The normalized, relative expression of CDKN1A/p21 after DMSO or 8 hr of etoposide treatment (100 μM final) in a pilot RT-qPCR screen and in the experimental siRNA screen (A). Average (B) CDKN1A/p21, (C) BBC3/puma, and (D) TP53/p53 expression values under DMSO and etoposide conditions across each experimental RT-qPCR plate for nontargeting, TP53, and MDM2 siRNA. Results from T-tests of pairwise comparisons can be found in Table 1. DMSO, dimethyl sulfoxide; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; siRNA, small interfering RNA.
Table 1. Results of T-tests for screen-average siRNA related to Figure 2.
| CDKN1A DMSO | CDKN1A Etoposide | BBC3 DMSO | BBC3 Etoposide | TP53 DMSO | TP53 Etoposide | |
|---|---|---|---|---|---|---|
| Nontargeting | 9.96E-4 | 0.2407 | 0.0287 | 0.2875 | 0.7663 | 5.57E-3 |
| TP53 | 9.54E-10 | 9.73E-7 | 1.93E-5 | 1.51E-4 | 2.12E-8 | 1.20E-8 |
| MDM2 | 3.12E-5 | 0.1946 | 1.43E-3 | 0.6630 | 0.4927 | 0.4037 |
DMSO, dimethyl sulfoxide.
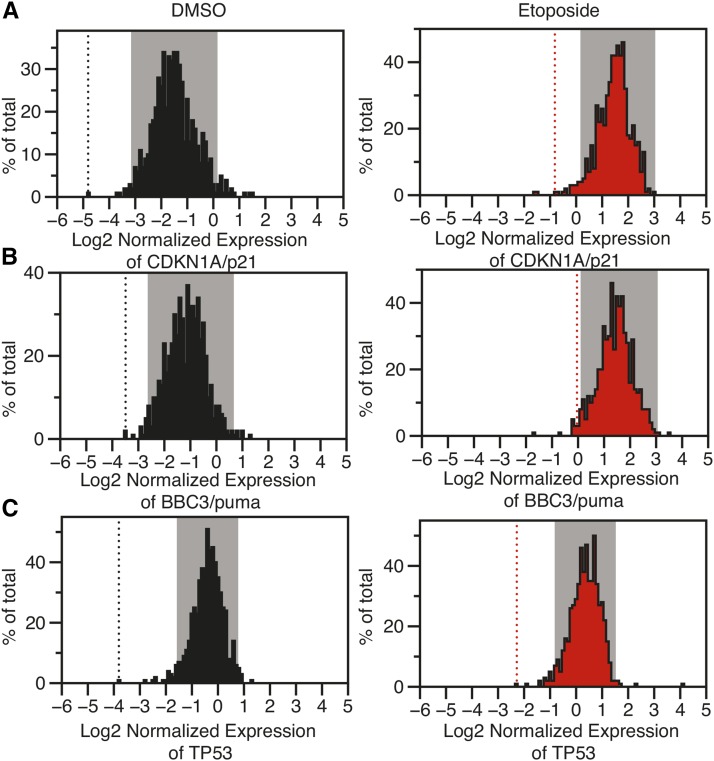
Normalized gene expression values for each of the target mRNA molecules were normally distributed (Figure 3). Therefore, we used a z-score-based cut-off to call hits as siRNA treatments that altered expression ± 2 standard deviations from the mean of all expression values for that target gene (Birmingham et al. 2009). Gray boxes in Figure 3 depict gene expression values that fall within 2 standard deviations from the mean, and are thus not called as hits. As an important control, we note that TP53 siRNA, included in our bookcase (Table S1), scored as a hit for all six conditions tested using these criteria, and are presented as dashed lines in Figure 3.
Figure 3.
The distribution of relative RNA expression for (A) CDKN1A/p21, (B) BBC3/puma, and (C) TP53/p53. Gray windows represent data with z-scores between –2 and 2. Dotted lines represent the expression value for TP53 knockdown siRNA used as an internal control within the screen. siRNA, small interfering RNA.
Positive regulators of CDKN1A/p21 and BBC3/puma
We identified 10 genes that behave as putative positive regulators of both CDKN1A/p21 and BBC3/puma transcription (Table 2). Based on the scoring strategy described above, these genes have z-scores < −2 for both CDKN1A/p21 and BBC3/puma, either in the basal (DMSO-treated) condition or after etoposide treatment. As expected, knockdown of TP53 itself led to the most dramatic decrease in basal CDKN1A and BBC3 expression of all genes tested (Figure 3, A and B, DMSO). PLK1 knockdown resulted in a significant reduction in CDKN1A, BBC3, and TP53 mRNA, suggesting potentially indirect effects on p53 target gene transcription through reduced p53 protein expression. siRNA-mediated knockdown of SETD3 and NCAPG reduced CDKN1A and BBC3 mRNA expression under basal conditions (DMSO), but not upon etoposide treatment. Interestingly, TOP1 (topoisomerase 1) scored as a positive regulator of p53 by reducing p53 target gene transcription after treatment with etoposide. This is of particular interest, since etoposide is a potent topoisomerase II poison that acts to inhibit repair of TopoII-induced dsDNA breaks, which, in turn, is a potent activator of p53-dependent signaling (Fortune and Osheroff 2000; Pommier et al. 2010). Both TOP1 and TOP2 have been implicated directly in positive gene regulation through their ability to relax DNA coiling (King et al. 2013; Madabhushi et al. 2015). Therefore, alternative p53-activating stimuli like the MDM2 inhibitor nutlin 3A (Vassilev et al. 2004) may prove to be useful in further investigating the roles for TOP1 and TOP2 in regulating p53-dependent transcription.
Negative regulators of CDKN1A/p21 and BBC3/puma
We identified 10 genes that are predicted to act as negative regulators of both CDKN1A/p21 and BBC3/puma expression, with mRNA expression z-scores > 2 for both genes (Table 2). Nine of these 10 genes increased CDKN1A and BBC3 mRNA expression under basal conditions; only JHDM1D/KDM7A further increased activation of CDKN1A/p21 and BBC3/puma after etoposide treatment. JHDM1D catalyzes the removal of dimethylation from lysine 9 of histone H3 (H3K9me2) (Horton et al. 2010), a canonically repressive histone modification catalyzed by SETDB1 (Schultz et al. 2002), which was identified above as a positive regulator of p53 transcription. SETDB1 and JHDM1D regulate transcription through respective deposition and removal of heterochromatin-associated histone modifications, an activity that is predicted to have an opposite effect on p53-dependent transcription than what is observed in this screen. Further investigation is required to determine whether SETDB1 and JHDM1D control p53 through transcription-associated histone modifications or through direct interaction/modification of p53.
Knockdown of the boundary element and chromatin looping factor CTCF has been previously shown to derepress BBC3/puma transcription in the basal state (Gomes and Espinosa 2010). We observed that siRNA-mediated knockdown of CTCF led to upregulation of BBC3/puma, as well as derepression of CDKN1A/p21, further suggesting that chromatin looping may function in the regulation of p53 targets (Merkenschlager and Odom 2013; Kim et al. 2015). The SRCAP complex, which catalyzes exchange of H2A.Z for H2A in nucleosomes, contains both the catalytic protein SRCAP and YEATS4/GAS41, a previously identified repressor of p53 activity (Park and Roeder 2006; Wong et al. 2007; Pikor et al. 2013). In our screen, knockdown of either SRCAP or YEATS4/GAS41 led to increased mRNA expression of p53 target genes. SRCAP represses ΔNp63 target genes through its H2A.Z deposition activity (Gallant-Behm et al. 2012) and appears to behave similarly with p53 transcriptional targets in our screen. This suggests a potential common repressive mechanism for p53 family transcription factors through SRCAP complex-mediated H2A.Z deposition.
Specific regulators of CDKN1A/p21, BBC3/puma, or TP53/p53
We next examined genes that behaved as gene-specific regulators of CDKN1A/p21, BBC3/puma, or TP53/p53 (Table 3, Table 4, and Table 5). Strikingly, there were many siRNA targets that were specific for either CDKN1A/p21 or BBC3/puma. For example, KAT5/TIP60 is a lysine acetyltransferase that catalyzes the acetylation of histone H4 lysine 16 (H4K16ac) and is critical for DNA repair (Tang et al. 2013). Beyond the canonical role in histone acetylation and DNA damage, KAT5/TIP60 directly interacts with and acetylates p53 at lysine 120. This activity modulates the ability of p53 to activate BBC3/puma, but not CDKN1A/p21 (Sykes et al. 2006; Tang et al. 2006). Consistent with these data, KAT5 knockdown in our screen specifically inhibited etoposide-induced BBC3/puma expression but not expression of CDKN1A/p21. Similarly, knockdown of SNW1/SKIP1 affected only CDKN1A/p21 mRNA expression, consistent with reports of SNW1 binding to the CDKN1A gene to regulate cotranscriptional splicing (Chen et al. 2011).
Treatment of U2OS cells with siRNA directed against the product of the ERCC6 gene, a protein commonly known as CSB, led to a specific reduction of TP53 expression in both the DMSO and etoposide conditions without affecting downstream p53 targets (Table 5). ERCC6/CSB mutations lead to Cockayne Syndrome, which is characterized by nervous system and DNA damage, and premature aging phenotypes (Mallery et al. 1998). Reduction in TP53 expression after treatment with siRNA targeting ERCC6/CSB is consistent with observations that CSB partially regulates the DNA damage response (Proietti-De-Santis et al. 2006) and directly interacts with the p53 protein in vivo (Latini et al. 2011; Lake et al. 2011) We observed that siRNA treatment against SMYD2/SMYD2 led to an upregulation of TP53 mRNA after DMSO treatment (Table 5). SMYD2 catalyzes the methylation of the p53 protein at lysine 370, which represses p53-dependent transcription (Huang et al. 2006). Our data suggests that a number of known and unknown chromatin regulatory proteins, such as ERCC6/CSB and SMYD2, may influence TP53/p53 mRNA levels without a concomitant regulation of downstream p53 target genes.
CBX7 and PCGF2 are both members of the polycomb group family complex PRC1, and their reduction by siRNA treatment strongly diminished CDKN1A/p21 mRNA expression without affecting BBC3/puma (Table 3). Canonically, polycomb group complexes regulate facultative heterochromatin and mediate gene repression (Di Croce and Helin 2013), and CBX7 is a tumor suppressor in the hematopoeitic lineage (Fortune and Osheroff 2000; Klauke et al. 2013). Our results suggest a novel role for PRC1 complexes as potential direct coactivators for p53-mediated transcription of CDKN1A; however, we cannot rule out an indirect role of PCGF2 and CBX7 knockdown leading to derepression of a CDKN1A/p21 repressor. Further investigation is required to fully characterize the role of PRC1-mediated p53-dependent transcription regulation.
Self-organizing map clustering to identify genes with similar p53-dependent transcriptional profiles
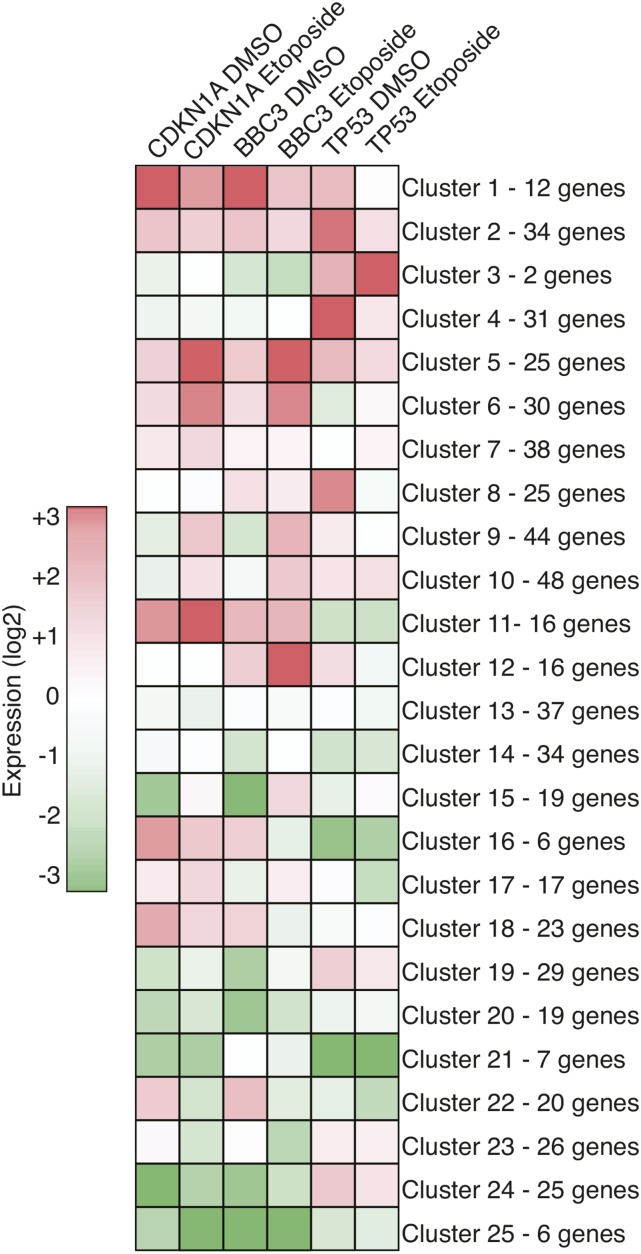
Following review of the screen results, we reasoned that using a stringent z-score cut-off to call screen hits might preclude identification of potential p53 regulators that behave similarly to known regulators, or other novel hits within the screen, but fall outside of the z-score cut-off. Therefore, we used normalized expression values of CDKN1A/p21, BBC3/puma, and TP53/p53 under both DMSO and etoposide treatment conditions as the input for self-organizing map (SOM) analysis (Kohonen 1987; Wehrens and Buydens 2007) to define groups of genes that similarly regulate p53 target genes (Figure 4 and Table S1).
Figure 4.
A self-organizing map (SOM) analysis was used to create 25 clusters that behave similarly across the six gene expression values. All clustering data can be found in Table S1. DMSO, dimethyl sulfoxide.
TP53 siRNA was grouped into Cluster 21 with ATM, ERCC6, HMGA2, LMNB1, PRMT2, and SIN3A siRNA. This cluster is characterized by the lowest levels of TP53 expression across all tested siRNA and reduced CDKN1A/p21 and BBC3/puma levels. Thus, these genes may function as positive regulators of TP53 transcription and act upstream of direct regulation of CDKN1A/p21 or BBC3/puma. ATM was identified as a hit using our initial hit criteria and clusters here with TP53. ERCC6, a chromatin remodeling protein also known as CSB, has previously been implicated in positive regulation of p53 transcriptional activity (Lake et al. 2011). In contrast, Cluster 25 contained six genes (MBD3L2, PLK1, PRMT6, SETD1B, SMC1A, and TOP1) that displayed reduced CDKN1A/p21 and BBC3/puma expression and also likely act as positive regulators of TP53-dependent transcription, but without reduced TP53 expression.
Cluster 1 contained 12 genes that are characterized by a dramatic increase in basal expression of CDKN1A and BBC3, yet with only moderate effects on TP53 expression. CTCF and the SRCAP chromatin remodeling complex members (YEATS4 and SRCAP; discussed above) are grouped into this cluster, along with eight genes that were identified as hits using the z-score-based cut-off. Interestingly, SOM analysis places ATG7 into Cluster 1, despite failing to be called a hit using the strict z-score cut-off criteria. ATG7 was previously described as a direct p53 binding protein, and a loss of ATG7 led to increased BBC3/puma expression, similar to the results of our primary screen (Lee et al. 2012).
In contrast, Cluster 5 is specific for genes that negatively regulate etoposide-induced p53 transcription of CDKN1A and BBC3, as siRNA-mediated knockdown of these genes leads to increased expression of CDKN1A and BBC3. Clusters 11 and 12 can be characterized as negative regulators of etoposide-induced CDKN1A or BBC3 transcription, respectively. Overall, these SOM analysis-derived clusters can serve as an alternative reference for selecting putative regulators of gene-specific p53 transcription for further study.
We performed limited validation by rescreening 81 random siRNA targets for CDKN1A/p21 and BBC3/puma expression after DMSO treatment. We reasoned that random selection of siRNA targets would not skew the distribution of expression values and would allow us to use z-score-based hit selection, similar to our original scoring system. A total of 19 putative CDKN1A/p21 or BBC3/puma regulators were present in the rescreen. Overall, 17 out of 19 primary hits were called as hits in the secondary screening (marked with ampersands in Table 2, Table 3, and Table 4), with MBD6 and MBD3L1 being called as putative false positives. Consistent with the utility of the approach, MBD3L1 was already listed as a potential false positive due to its low mRNA expression in U2OS cells. DLX2, which was also called as a false positive based on RNA expression, was again scored as a hit in the secondary screen, suggesting strong off-target effects mediated by DLX2-targeting siRNA. Interestingly, although we call DLX2 a false positive based on mRNA expression analysis of U2OS cells, the results of our primary and secondary screen are consistent with the recent discovery that DLX2 interferes with ATM-p53 signaling and functions normally as a negative regulator of p53 activity (Wang et al. 2016). It should be noted that, because the same siRNA complexes were used in both the primary and secondary screening approach, orthogonal knockdown approaches and/or siRNA pool deconvolution should be performed in order to truly validate any siRNA target identified in these screens.
In summary, our primary chromatin-focused siRNA screen identified both previously known and putative regulators of p53-dependent transcription. Additional investigation will be required to characterize the specific mechanisms of these enzymes in either directly modulating p53 activity or in gene-specific regulation of the local chromatin environment at CDKN1A, BBC3, or TP53.We note that measurement of mRNA expression as the screen readout does not preclude the possibility that our identified p53 regulatory genes operate at the level of mRNA stability; however, we minimized the influence of other posttranscriptional mechanisms like mRNA translation or protein stability that are possible through measurement of CDKN1A/p21 or BBC3/puma protein levels. The results of this targeted chromatin siRNA screen will provide a useful foundation and resource for future investigation into the molecular mechanisms regulating p53-dependent transcription and the balance between prosurvival and proapoptotic transcriptional programs.
Supplementary Material
Acknowledgments
We thank Parisha Shah for editing assistance and other members of the Berger lab for helpful comments and discussions. Work in this manuscript was supported by an American Cancer Society Postdoctoral Fellowship awarded to M.A.S., and by a National Institutes of Health National Cancer Institute grant (CA078831) to S.L.B.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.031534/-/DC1
Communicating editor: B. J. Andrews
Literature Cited
- Andrysik Z., Kim J., Tan A. C., Espinosa J. M., 2013. A genetic screen identifies tcf3/e2a and triap1 as pathway-specific regulators of the cellular response to p53 activation. Cell Reports 3: 1346–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J., Kouzarides T., 2011. Regulation of chromatin by histone modifications. Cell Res. 21: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev N. A., Liu L., Chehab N. H., Mansfield K., Harris K. G., et al. , 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8: 1243–1254. [DOI] [PubMed] [Google Scholar]
- Berger S. L., 2010. Keeping p53 in check: a high-stakes balancing act. Cell 142: 17–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham A., Selfors L. M., Forster T., Wrobel D., Kennedy C. J., et al. , 2009. Statistical methods for analysis of high-throughput rna interference screens. Nat. Methods 6: 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhang L., Jones K. A., 2011. Skip counteracts p53-mediated apoptosis via selective regulation of p21cip1 mrna splicing. Genes Dev. 25: 701–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Gu W., 2010. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol. Med. 16: 528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M. A., Kouzarides T., 2012. Cancer epigenetics: from mechanism to therapy. Cell 150: 12–27. [DOI] [PubMed] [Google Scholar]
- Dawson M. A., Kouzarides T., Huntly B. J., 2012. Targeting epigenetic readers in cancer. N. Engl. J. Med. 367: 647–657. [DOI] [PubMed] [Google Scholar]
- Di Croce L., Helin K., 2013. Transcriptional regulation by polycomb group proteins. Nat. Struct. Mol. Biol. 20: 1147–1155. [DOI] [PubMed] [Google Scholar]
- Fortune J. M., Osheroff N., 2000. Topoisomerase ii as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol. 64: 221–253. [DOI] [PubMed] [Google Scholar]
- Gallant-Behm C. L., Ramsey M. R., Bensard C. L., Nojek I., Tran J., et al. , 2012. Deltanp63alpha represses anti-proliferative genes via h2a.z deposition. Genes Dev. 26: 2325–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes N. P., Espinosa J. M., 2010. Gene-specific repression of the p53 target gene puma via intragenic ctcf-cohesin binding. Genes Dev. 24: 1022–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J. R., Upadhyay A. K., Qi H. H., Zhang X., Shi Y., et al. , 2010. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 17: 38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Perez-Burgos L., Placek B. J., Sengupta R., Richter M., et al. , 2006. Repression of p53 activity by smyd2-mediated methylation. Nature 444: 629–632. [DOI] [PubMed] [Google Scholar]
- Kim S., Yu N. K., Kaang B. K., 2015. Ctcf as a multifunctional protein in genome regulation and gene expression. Exp. Mol. Med. 47: e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I. F., Yandava C. N., Mabb A. M., Hsiao J. S., Huang H. S., et al. , 2013. Topoisomerases facilitate transcription of long genes linked to autism. Nature 501: 58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauke K., Radulovic V., Broekhuis M., Weersing E., Zwart E., et al. , 2013. Polycomb cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat. Cell Biol. 15: 353–362. [DOI] [PubMed] [Google Scholar]
- Klijn C., Durinck S., Stawiski E. W., Haverty P. M., Jiang Z., et al. , 2015. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 33: 306–312. [DOI] [PubMed] [Google Scholar]
- Kohonen T., 1987. Adaptive, associative, and self-organizing functions in neural computing. Appl. Opt. 26: 4910–4918. [DOI] [PubMed] [Google Scholar]
- Kouzarides T., 2007. Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Kruiswijk F., Labuschagne C. F., Vousden K. H., 2015. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 16: 393–405. [DOI] [PubMed] [Google Scholar]
- Lake R. J., Basheer A., Fan H. Y., 2011. Reciprocally regulated chromatin association of cockayne syndrome protein b and p53 protein. J. Biol. Chem. 286: 34951–34958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latini P., Frontini M., Caputo M., Gregan J., Cipak L., et al. , 2011. Csa and csb proteins interact with p53 and regulate its mdm2-dependent ubiquitination. Cell Cycle 10: 3719–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M. S., Stojanov P., Mermel C. H., Robinson J. T., Garraway L. A., et al. , 2014. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I. H., Kawai Y., Fergusson M. M., Rovira I., Bishop A. J., et al. , 2012. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science 336: 225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidor Nili E., Field Y., Lubling Y., Widom J., Oren M., et al. , 2010. p53 binds preferentially to genomic regions with high dna-encoded nucleosome occupancy. Genome Res. 20: 1361–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Scolnick D. M., Trievel R. C., Zhang H. B., Marmorstein R., et al. , 1999. p53 sites acetylated in vitro by pcaf and p300 are acetylated in vivo in response to dna damage. Mol. Cell. Biol. 19: 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Li M., Tang Y., Laszkowska M., Roeder R. G., et al. , 2004. Acetylation of p53 augments its site-specific dna binding both in vitro and in vivo. Proc. Natl. Acad. Sci. USA 101: 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R., Gao F., Pfenning A. R., Pan L., Yamakawa S., et al. , 2015. Activity-induced dna breaks govern the expression of neuronal early-response genes. Cell 161: 1592–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallery D. L., Tanganelli B., Colella S., Steingrimsdottir H., van Gool A. J., et al. , 1998. Molecular analysis of mutations in the csb (ercc6) gene in patients with cockayne syndrome. Am. J. Hum. Genet. 62: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M., Odom D. T., 2013. Ctcf and cohesin: linking gene regulatory elements with their targets. Cell 152: 1285–1297. [DOI] [PubMed] [Google Scholar]
- Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J., 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69: 1237–1245. [DOI] [PubMed] [Google Scholar]
- Oliner J. D., Kinzler K. W., Meltzer P. S., George D. L., Vogelstein B., 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358: 80–83. [DOI] [PubMed] [Google Scholar]
- Park J. H., Roeder R. G., 2006. Gas41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation. Mol. Cell. Biol. 26: 4006–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikor L. A., Lockwood W. W., Thu K. L., Vucic E. A., Chari R., et al. , 2013. Yeats4 is a novel oncogene amplified in non-small cell lung cancer that regulates the p53 pathway. Cancer Res. 73: 7301–7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., Leo E., Zhang H., Marchand C., 2010. Dna topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proietti-De-Santis L., Drane P., Egly J. M., 2006. Cockayne syndrome b protein regulates the transcriptional program after uv irradiation. EMBO J. 25: 1915–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufini A., Tucci P., Celardo I., Melino G., 2013. Senescence and aging: the critical roles of p53. Oncogene 32: 5129–5143. [DOI] [PubMed] [Google Scholar]
- Sammons M. A., Zhu J., Drake A. M., Berger S. L., 2015. Tp53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res. 25: 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D. C., Ayyanathan K., Negorev D., Maul G. G., and F. J. Rauscher 3rd, 2002. Setdb1: a novel kap-1-associated histone h3, lysine 9-specific methyltransferase that contributes to hp1-mediated silencing of euchromatic genes by krab zinc-finger proteins. Genes Dev. 16: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Kachirskaia I., Yamaguchi H., West L. E., Wen H., et al. , 2007. Modulation of p53 function by set8-mediated methylation at lysine 382. Mol. Cell 27: 636–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh S. Y., Ikeda M., Taya Y., Prives C., 1997. Dna damage-induced phosphorylation of p53 alleviates inhibition by mdm2. Cell 91: 325–334. [DOI] [PubMed] [Google Scholar]
- Siliciano J. D., Canman C. E., Taya Y., Sakaguchi K., Appella E., et al. , 1997. Dna damage induces phosphorylation of the amino terminus of p53. Genes Dev. 11: 3471–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D., Wang X., Campbell M. R., Song L., Safi A., et al. , 2015. Interactions of chromatin context, binding site sequence content, and sequence evolution in stress-induced p53 occupancy and transactivation. PLoS Genet. 11: e1004885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes S. M., Mellert H. S., Holbert M. A., Li K., Marmorstein R., et al. , 2006. Acetylation of the p53 dna-binding domain regulates apoptosis induction. Mol. Cell 24: 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Cho N. W., Cui G., Manion E. M., Shanbhag N. M., et al. , 2013. Acetylation limits 53bp1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20: 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Luo J., Zhang W., Gu W., 2006. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24: 827–839. [DOI] [PubMed] [Google Scholar]
- Tong Q., Cui G., Botuyan M. V., Rothbart S. B., Hayashi R., et al. , 2015a Structural plasticity of methyllysine recognition by the tandem tudor domain of 53bp1. Structure 23: 312–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Q., Mazur S. J., Rincon-Arano H., Rothbart S. B., Kuznetsov D. M., et al. , 2015b An acetyl-methyl switch drives a conformational change in p53. Structure 23: 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., et al. , 2004. In vivo activation of the p53 pathway by small-molecule antagonists of mdm2. Science 303: 844–848. [DOI] [PubMed] [Google Scholar]
- Wang Y., Xu Q., Sack L., Kang C., Elledge S. J., 2016. A gain-of-function senescence bypass screen identifies the homeobox transcription factor dlx2 as a regulator of atm-p53 signaling. Genes Dev. 30: 293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrens R., Buydens L., 2007. Self- and super-organising maps in r: the kohonen package. J. Stat. Softw. 21: 1–19. [Google Scholar]
- Wong M. M., Cox L. K., Chrivia J. C., 2007. The chromatin remodeling protein, srcap, is critical for deposition of the histone variant h2a.z at promoters. J. Biol. Chem. 282: 26132–26139. [DOI] [PubMed] [Google Scholar]
- Wu X., Bayle J. H., Olson D., Levine A. J., 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7: 1126–1132. [DOI] [PubMed] [Google Scholar]
- Zilfou J. T., Lowe S. W., 2009. Tumor suppressive functions of p53. Cold Spring Harb. Perspect. Biol. 1: a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J., Shi J., Wang E., Rappaport A. R., Herrmann H., et al. , 2011. Rnai screen identifies brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Table S1 contains all normalized gene expression data across all screening conditions. Screen data are also available at the GenomeRNAi repository under accession number GR00389-S.