Abstract
Summary: We previously reported the multi-modal Dynamic Cross Correlation (mDCC) method for analyzing molecular dynamics trajectories. This method quantifies the correlation coefficients of atomic motions with complex multi-modal behaviors by using a Bayesian-based pattern recognition technique that can effectively capture transiently formed, unstable interactions. Here, we present an open source toolkit for performing the mDCC analysis, including pattern recognitions, complex network analyses and visualizations. We include a tutorial document that thoroughly explains how to apply this toolkit for an analysis, using the example trajectory of the 100 ns simulation of an engineered endothelin-1 peptide dimer.
Availability and implementation: The source code is available for free at http://www.protein.osaka-u.ac.jp/rcsfp/pi/mdcctools/, implemented in C ++ and Python, and supported on Linux.
Contact: kota.kasahara@protein.osaka-u.ac.jp
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Molecular dynamics (MD) simulations are a promising method to investigate the dynamical behaviors of various molecular systems with atomic details. Although recent advances in the computer technologies have realized the long-term simulations of large systems, the huge amount of trajectory data thus generated is not easily interpreted. In order to tackle this problem, analyses toolkits have been extensively developed, such as MDAnalysis (Michaud-Agrawal et al., 2011), Wordom (Seeber et al., 2011) and VMD (Humphrey et al., 1996). Long-term trajectories reflect complex behaviors of the local and global conformational changes of molecules. The distributions of atomic coordinates may be unimodal, i.e. adequately described by a cluster that is approximated by a single mean and standard deviation, or multi-modal, with several spatially distinct clusters, often slowly interchanging with relatively rapid fluctuations within each cluster. For example, a common multi-modal local motion in proteins results from the rearrangements of hydrogen bonds associated with transient flipping of side chains. The analysis of multi-modal motions, which are not each describable by a single Gaussian distribution, is not straightforward.
We previously proposed a new analysis method, named ‘multi-modal Dynamic Cross Correlation (mDCC)’ (Kasahara et al., 2014), as a variant of the conventional Dynamic Cross Correlation (DCC) method (McCammon, 1984). Because DCC calculates the correlations between atomic motions, based on deviations from the averaged coordinate of each atom, it does not make sense when atoms undergo multi-modal motions. To characterize such multi-modal motions, the mDCC method takes advantage of a Bayesian statistics-based pattern recognition technique (Attias, 1999), and classifies the distributions of atomic coordinates into some clusters, or modes. We applied this method to analyze transcription factor-DNA interactions, and found that many transient, multi-modal interactions are formed at interfaces between proteins and DNA. See Supplementary Materials S1 and S2 for details of the method.
Here, we present an open source, easy-to-use toolkit for the mDCC method. This toolkit performs the full analysis techniques applied in our previous work (Kasahara et al., 2014), and not only covers the correlative coefficients of multi-modal atomic motions, but also enables visualization of the results effectively as a heatmap and a complex network diagram, powered by standard software such as Cytoscape (Shannon et al., 2015) and R (R Core Team, 2003). As the output files are simple tab-separated texts, users can apply their favorite software for visualization. Users can easily learn how to use this toolkit via the attached tutorial document, with the trajectory of the 100 ns simulation of an engineered endothelin-1 peptide dimer as an example.
2 Implementation
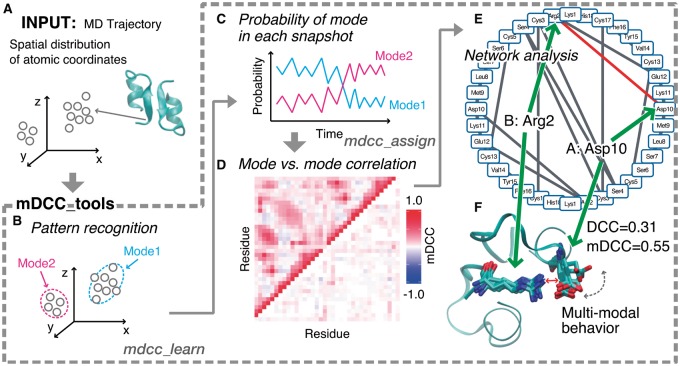
Figure 1 summarizes the mDCC analysis by using mDCC_toolkit, which is composed of two C ++ programs and several scripts. mDCC_tools handles a variety of trajectory file formats, such as Gromacs, AMBER and CHARMM, by taking advantage of MDAnalysis library (Fig. 1A). In addition, files in PRESTO format (Mashimo et al., 2013) and tab-separated text files are also accepted.
Fig. 1.
Overview of the mDCC analysis. (A) Input data for the analysis. (B) The pattern recognition on the atomic coordinates. (C) Assessing probabilities for each mode. (D) Visualization of all-against-all correlation coefficients. Each column and row indicates each residue. The color gradation from blue to red corresponds to negative and positive correlations. The upper- and lower-triangle depict the mDCC and mDCC-DCC values, respectively. (E) A network diagram. The edges indicate the contacting residue pairs with positive correlation. The interaction including multi-modal behavior is shown as the red edge. (F) An example of multi-modal behavior in engineered endothelin-1 peptide dimer
The analysis is performed by the following programs:
mdcc_learn (Fig. 1B) recognizes the multi-modal motions of each atom from a MD trajectory. By parameter fitting of the Gaussian mixture model, the spatial distribution of the atomic coordinates is classified into some Gaussian functions, each referred to as a ‘mode’.
mdcc_assign (Fig. 1C) calculates the probability of the event that an observed atomic coordinate ri(t) belongs to a mode k (Gaussian element) for all i (atom) and for all k over the total time t.
cal_mdcc.py (Fig. 1D) calculates the mDCC values between modes. The correlation map shows the maximum mDCC value of each pair of residues (upper triangle). The map also indicates the difference from the conventional DCC values in the lower-triangle, where the residue pairs with large differences from the DCC show multi-modal behaviors. The R-script for drawing this heatmap is included in this package.
The network diagram (Fig. 1E) provides a bird’s-eye view of the interactions in the molecular system. Each node indicates each residue, and each edge indicates a pair of residues with highly positive mDCC values (≥0.5 is used in this example) and atomic contacts (the minimum distance ≤5 Å). The toolkit generates files readable by Cytoscape, one of the standard programs for complex network analyses. In addition, the importance of each residue can be quantified by the betweenness values, which are calculated by nx_centrality.py. The betweenness quantifies the centrality of each node in the network. High betweenness values imply that the node plays an important role in the network (see the Supplementary Material S3).
The simulation trajectory of the engineered endothelin-1 peptide dimer (PDB: 1t7h) in a 150 mM NaCl solution is included in this package, as a tutorial example. The 100 ns simulation in the NPT ensemble was performed by using Gromacs (Pronk et al., 2013). Users can readily trace our analyses starting from the trajectory file, in a step-by-step manner. The analysis revealed the transient interactions between Asp10 in chain A and Arg2 in chain B (the red edge in Fig 1E), which had mDCC and DCC values of 0.55 and 0.31, respectively. A transient flipping motion of Asp10 side-chain resulted in breaking the salt bridge with Arg2 (Fig. 1F). See our previous publication for more details regarding the theory and application to a more complex molecular assembly, consisting of two transcription factors on a double-stranded DNA (Kasahara et al., 2014).
Although this method has been tailored for analyses of MD trajectories, it can be applied to any multi-dimensional distribution (see the software documentation).
Supplementary Material
Acknowledgements
The MD simulation was performed on the TSUBAME2.5 supercomputer at the Tokyo Institute of Technology, provided through the HPCI System Research Projects (Project IDs: hp140032 and hp150015). The supercomputer resource for the post-simulation analyses was provided by the National Institute of Genetics, Research Organization of Information and Systems, Japan.
Funding
This work was supported by the Japan Society for the Promotion of Science KAKENHI, Grant-in-Aid for Scientific Research on Innovative Areas, Grant Number 24118001.
Conflict of Interest: none declared.
References
- Attias H. (1999) Inferring parameters and structure of latent variable models by variational Bayes. UAI’99 Proceedings of the Fifteenth Conference on Uncertainty in Artificial Intelligence, Stockholm, Sweden, 30 July—1, 21–30 August.
- Humphrey W. et al. (1996) VMD: visual molecular dynamics. J. Mol. Graphics, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Kasahara K. et al. (2014) A novel approach of dynamic cross correlation analysis on molecular dynamics simulations and its application to Ets1 dimer-DNA complex. PLoS One, 9, e112419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T. et al. (2013) Molecular dynamics simulations accelerated by GPU for biological macromolecules with a non-ewald scheme for electrostatic interactions. J. Chem. Theory Comput., 9, 5599–5609. [DOI] [PubMed] [Google Scholar]
- McCammon J.A. (1984) Protein dynamics. Rep. Progr. Phys., 47, 1–46. [Google Scholar]
- Michaud-Agrawal N. et al. (2011) MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem., 32, 2319–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk S. et al. (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics, 29, 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2015) R: The R Project for Statistical Computing. http://r-project.org. https://cran.r-project.org/doc/FAQ/R-FAQ.html#Citing-R
- Seeber M. et al. (2011) Wordom: A userfriendly program for the analysis of molecular structures, trajectories, and free energy surfaces. J. Comput. Chem., 32, 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.