Abstract
Summary: We present a practical computational pipeline to readily perform data analyses of protein–protein interaction networks by using genetic and functional information mapped onto protein structures. We provide a 3D representation of the available protein structure and its regions (surface, interface, core and disordered) for the selected genetic variants and/or SNPs, and a prediction of the mutants’ impact on the protein as measured by a range of methods. We have mapped in total 2587 genetic disorder-related SNPs from OMIM, 587 873 cancer-related variants from COSMIC, and 1 484 045 SNPs from dbSNP. All result data can be downloaded by the user together with an R-script to compute the enrichment of SNPs/variants in selected structural regions.
Availability and Implementation: PinSnps is available as open-access service at http://fraternalilab.kcl.ac.uk/PinSnps/
Contact: franca.fraternali@kcl.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
High-throughput experiments are routinely performed to decipher genetic, metabolic and protein-protein interaction networks (PPINs) and bioinformaticians are compelled to develop efficient and accurate tools to assist decision-making based on available data from multiple sources (Chung et al., 2015; Fernandes et al., 2010; Lu et al., 2013). Bioinformatics applications, which merge available genomic, interaction and structural data, can be broadly classified into exploratory or predictive tools. The former comprises of tools which map and visualize the merged data (Kelley et al., 2015; Lees et al., 2010; Mosca et al., 2015; Niknafs et al., 2013; Pappalardo and Wass, 2014; Ryan et al., 2009; Vazquez et al., 2015), while predictive tools are quantitative estimators of the potential impact of SNPs/variants and offer an assessment in terms of scores or pseudo free-energy metrics (Adzhubei et al., 2010; Betts et al., 2015; Li et al., 2014; Ng and Henikoff, 2003; Pires et al., 2014a; Pires et al., 2014b; Pires et al., 2016; Yates et al., 2014).
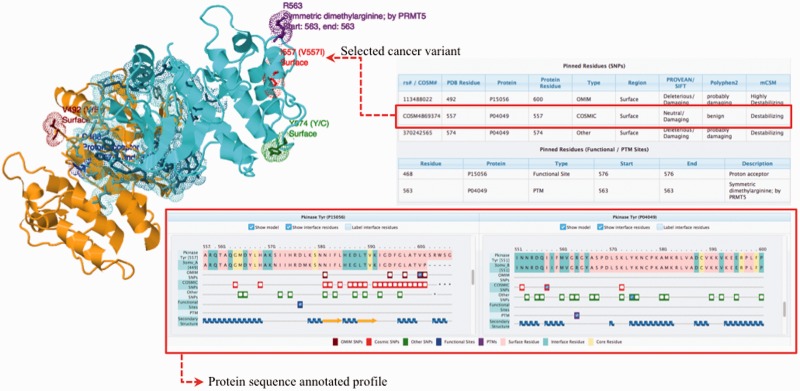
In this application, we use 3D interactome networks and their homologs to highlight how human variants and disease-causing mutations may affect protein function and complex stability. Recent studies have used the structural information of PPINs to understand the molecular mechanisms of binding partner selection (Fornili et al., 2013). These reliable methods only consider the interactions that have a representative 3D structure or a close homolog with a 3D structure to add weight to the existence of the observed protein interactions (or network links) in a given PPIN (Hooda and Kim, 2012; Kim et al., 2006; Lees et al., 2011; Meyer et al., 2013; Mosca et al., 2013; Wang et al., 2012). Multiple studies have pointed out that the interfaces of protein complexes harbours mutations associated with diseases (Espinosa et al., 2014; Gao et al., 2015; Kamburov et al., 2015; Nishi et al., 2013; Studer et al., 2013; Wang et al., 2012; Yates and Sternberg, 2013a,b). The evaluation of the impact of genomic variation on coding regions can be enhanced by mapping SNPs to distinct regions of protein structure, i.e. surface, interface or core. To generate a comprehensive mapping of available SNPs onto PPINs, the automatic pipeline PinSnps has been developed (for details see Supplementary Fig. S2); this extracts structure-integrated human PPINs, enriched with information from homologous protein domains with sequence identity higher than 30%. The main strengths and differences to previous approaches lie in (i) the use of homologous structures of human protein sequences in the PPINs to map the studied variants, which more than doubles the available positional 3D information; (ii) the mapping onto predefined protein regions (surface, core, interface) along with the mapping of functional sites and Post-Translational Modifications (PTMs) (obtained from UniProt (UniProt Consortium, 2015)). This information, together with precompiled predictions of the SNP/variant’s impact from multiple predictors, can help users to quantitatively assess and evaluate the functional implications of their studied variants. The annotation of both intra- and inter-domain disordered regions as predicted by DISOPRED2 (Ward et al., 2004) has also been included in the pipeline, as recent studies imply the importance of these regions in regulating biological functions (Cline and Karchin, 2011; Gibbs and Showalter, 2015; Wright and Dyson, 2015); (iii) allowing the users to download the query data in various file formats (Fig. 1).
Fig. 1.
PinSnps user interface overview. The complex between Raf1 (P04049, coloured in cyan) and Braf (P15056, coloured in orange) is shown. The protein sequence annotated profile of the complex shows the sequence alignment of the query protein sequence and the available PDB structure sequences. A more detailed description of the platform interactive output is given in the Supplementary Figure S1
2 Implementation and features
The PPIN used in this study has been derived as a non-redundant set of protein interactions from the list of human PPIs given in Supplementary Table S1. The current release includes data of 16 603 proteins, of which 4673 have a resolved structure and 4962 have a homologous structure (Supplementary Fig. S3).
PinSnps is, to our knowledge, one of the largest collections of variants mapped onto 3D coordinates. SNPs from dbSNP (Sherry et al., 2001), consisting of common and germ-line disease variants (the later originally from OMIM (Hamosh et al., 2005)), together with somatic cancer mutations from COSMIC (Forbes et al., 2015) have been mapped onto cognate 3D structures and, when not available, to their homologous structures. The use of homologous structures expands significantly the number of SNPs/variants mapped onto 3D positions within folded domains. The enrichment of disease-associated variants in specific regions of proteins can be quantified using Formula S1 and the R script which is provided on the PinSnps ‘Downloads’ webpage (see example in Supplementary Fig. S4).
We present a number of case studies and more detailed instructions on the web server’s ‘Help’ page and in the Supplementary Materials.
2.1 Protein sequence annotated profiles
Each protein in the PPIN is transformed into a sequence-annotated string (we refer to this as ‘profile’) that represents the fingerprint of the user-selected information. These profiles were generated based on information obtained from sequence alignments, available structural information, human genetic data (from dbSNP, OMIM and COSMIC) and UniProt protein functional site and PTM annotations. PSI-BLAST (Altschul et al., 1997) was used to identify resolved and homologous structures of human proteins by searching against sequences of the Protein Data Bank (Berman et al., 2000). Homologous structures with more than 80% coverage of the human protein domain sequence and with more than 30% sequence identity were selected. Each protein was annotated with domain boundaries according to Pfam (Finn et al., 2014). Alignments between sequences of query protein domains and available protein structure sequences were performed using T-Coffee (Notredame et al., 2000). The classification of structural regions, i.e. the definition of surface, interface and core regions, was based on the surface area analysis of POPSCOMP (Kleinjung and Fraternali, 2005).
Conflict of Interest: none declared.
Supplementary Material
References
- Adzhubei I.A. et al. (2010) A method and server for predicting damaging missense mutations. Nat. Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F. et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M. et al. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts M.J. et al. (2015) Mechismo: predicting the mechanistic impact of mutations and modifications on molecular interactions. Nucleic Acids Res., 43, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.S. et al. (2015) Bridging topological and functional information in protein interaction networks by short loops profiling. Sci. Rep., 5, 8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M.S., Karchin R. (2011) Using bioinformatics to predict the functional impact of SNVs. Bioinformatics, 27, 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa O. et al. (2014) Deriving a mutation index of carcinogenicity using protein structure and protein interfaces. PLoS One, 9, e84598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes L.P. et al. (2010) Protein networks reveal detection bias and species consistency when analysed by information-theoretic methods. PLoS One, 5, e12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D. et al. (2014) Pfam: the protein families database. Nuclei. Acids Re., 42, D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes S.A. et al. (2015) COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res., 43, D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornili A. et al. (2013) Specialized dynamical properties of promiscuous residues revealed by simulated conformational ensembles. J. Chem. Theory Comput., 9, 5127–5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M. et al. (2015) Insights into disease-associated mutations in the human proteome through protein structural analysis. Structure, 23, 1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs E.B., Showalter S.A. (2015) Quantitative biophysical characterization of intrinsically disordered proteins. Biochemistry, 54, 1314–1326. [DOI] [PubMed] [Google Scholar]
- Hamosh A. et al. (2005) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res., 33, D514–D517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooda Y., Kim P.M. (2012) Computational structural analysis of protein interactions and networks. Proteomics, 12, 1697–1705. [DOI] [PubMed] [Google Scholar]
- Kamburov A. et al. (2015) Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc. Natl. Acad. Sci. USA, 112, E5486–E5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A. et al. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc., 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.M. et al. (2006) Relating three-dimensional structures to protein networks provides evolutionary insights. Science, 314, 1938–1941. [DOI] [PubMed] [Google Scholar]
- Kleinjung J., Fraternali F. (2005) POPSCOMP: an automated interaction analysis of biomolecular complexes. Nucleic Acids Res., 33, W342–W346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees J. et al. (2010) Gene3D: merging structure and function for a Thousand genomes. Nucleic Acids Res., 38, D296–D300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees J.G. et al. (2011) Systematic computational prediction of protein interaction networks. Phys. Biol., 8, 035008. [DOI] [PubMed] [Google Scholar]
- Li M. et al. (2014) Predicting the impact of missense mutations on protein–protein binding affinity. J. Chem. Theory Comput., 10, 1770–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H.C. et al. (2013) Protein–protein interaction networks studies and importance of 3D structure knowledge. Expert. Rev. Proteomics, 10, 511–520. [DOI] [PubMed] [Google Scholar]
- Meyer M.J. et al. (2013) INstruct: a database of high-quality 3D structurally resolved protein interactome networks. Bioinformatics, 29, 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca R. et al. (2013) Interactome3D: adding structural details to protein networks. Nat. Methods, 10, 47–53. [DOI] [PubMed] [Google Scholar]
- Mosca R. et al. (2015) dSysMap: exploring the edgetic role of disease mutations. Nat. Methods, 12, 167–168. [DOI] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. (2003) SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res., 31, 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niknafs N. et al. (2013) MuPIT interactive: webserver for mapping variant positions to annotated, interactive 3D structures. Hum. Genet., 132, 1235–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi H. et al. (2013) Cancer missense mutations alter binding properties of proteins and their interaction networks. PLoS One, 8, e66273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C. et al. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol., 302, 205–217. [DOI] [PubMed] [Google Scholar]
- Pappalardo M., Wass M.N. (2014) VarMod: modelling the functional effects of non-synonymous variants. Nucleic Acids Res., 42, W331–W336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D.E. et al. (2014a) DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res., 42, W314–W319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D.E. et al. (2014b) mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics, 30, 335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D.E. et al. (2016) In silico functional dissection of saturation mutagenesis: Interpreting the relationship between phenotypes and changes in protein stability, interactions and activity. Sci. Rep., 6, 19848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. et al. (2009) LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics, 25, 1431–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S.T. et al. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res., 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer R.A. et al. (2013) Residue mutations and their impact on protein structure and function: detecting beneficial and pathogenic changes. Biochem. J., 449, 581–594. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium. (2015) UniProt: a hub for protein information. Nucleic Acids Res., 43, D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez M. et al. (2015) Structure-PPi: a module for the annotation of cancer-related single-nucleotide variants at protein-protein interfaces. Bioinformatics, 31, 2397–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. (2012) Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat. Biotechnol., 30, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J.J. et al. (2004) The DISOPRED server for the prediction of protein disorder. Bioinformatics, 20, 2138–2139. [DOI] [PubMed] [Google Scholar]
- Wright P.E., Dyson H.J. (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol., 16, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates C.M., Sternberg M.J. (2013a) The effects of non-synonymous single nucleotide polymorphisms (nsSNPs) on protein-protein interactions. J. Mol. Biol., 425, 3949–3963. [DOI] [PubMed] [Google Scholar]
- Yates C.M., Sternberg M.J. (2013b) Proteins and domains vary in their tolerance of non-synonymous single nucleotide polymorphisms (nsSNPs). J. Mol. Biol., 425, 1274–1286. [DOI] [PubMed] [Google Scholar]
- Yates C.M. et al. (2014) SuSPect: enhanced prediction of single amino acid variant (SAV) phenotype using network features. J. Mol. Biol., 426, 2692–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.