Abstract
Summary: SBtab is a table-based data format for Systems Biology, designed to support automated data integration and model building. It uses the structure of spreadsheets and defines conventions for table structure, controlled vocabularies and semantic annotations. The format comes with predefined table types for experimental data and SBML-compliant model structures and can easily be customized to cover new types of data.
Availability and Implementation: SBtab documents can be created and edited with any text editor or spreadsheet tool. The website www.sbtab.net provides online tools for syntax validation and conversion to SBML and HTML, as well as software for using SBtab in MS Excel, MATLAB and R. The stand-alone Python code contains functions for file parsing, validation, conversion to SBML and HTML and an interface to SQLite databases, to be integrated into Systems Biology workflows. A detailed specification of SBtab, including examples and descriptions of table types and available tools, can be found at www.sbtab.net.
Contact: wolfram.liebermeister@gmail.com
1 Introduction
Data exchange between experimental and computational biologists plays a central role in Systems Biology. The scientific community has developed a range of standard formats for data and models that facilitate this exchange and increase software interoperability. Prominent examples are the Systems Biology Markup Language (SBML; Hucka et al., 2003) for the exchange of mathematical models, the Systems Biology Graphical Notation (SBGN; Le Novère et al., 2009) for standardized drawings of biological networks, and the Investigation/Study/Assay format (ISA-TAB), a standard spreadsheet format for experimental data and metadata (Sansone et al., 2008). Standards facilitate automated file parsing and guarantee that files contain complete and unambiguous information, which makes research results better comprehensible and reproducible. Standardized formats are often not designed to be human-readable, but rather written and read by software. This makes it harder for users without particular technical knowledge or software skills to employ them efficiently. An alternative approach to fostering data exchange is to provide guidelines for minimal information requirements, as demonstrated by the MIRIAM rules for published models of Systems Biology (Le Novère et al., 2005): These rules specify minimal pieces of model information that must be provided to ensure that model simulations can be reliably reproduced by other researchers. Such rules ensure complete and unambigous information while placing less restrictions on modellers than the aforementioned standardized file formats.
Despite these efforts, the usage of standard formats has not been fully established at the interface of experimental and computational Systems Biology. At this interface, complex models need to be constructed from heterogeneous data types (descriptions of network topologies as well as omics, thermodynamic and kinetic data) and using innovative methods. A number of formats are employed by different subcommunities to approach these tasks: While SBML is popular among modellers, experimentalists typically publish their data, including network structures, as spreadsheets or delimiter-separated text files. In contrast to standardized formats—which also exist for many types of experimental data—spreadsheets provide high flexibility, but file structure and naming conventions vary widely. Integrating these diverse and heterogeneous data into computational models can be tedious, since data often need to be reformatted, associated with unique identifiers and annotated. Even without moving from spreadsheets to other formats, simple conventions regarding the usage of names, annotations and syntax, as well as automatic tools for file validation and conversion, could facilitate this process, help avoid errors and experiment repetitions.
2 Results and implementation
Combining the advantages of standardized formats with the flexibility of spreadsheet files, we developed SBtab as a set of conventions for spreadsheets and delimited text files. SBtab defines table structures and naming conventions that make tables easy to parse and support precise and complete information in data files. A simple example of an SBtab table is shown in Figure 1: The attributes in the first line provide general information about the dataset, followed by a line with defined column headers (marked by the ! character). Additional data can be placed into ‘uncontrolled’ columns, to which no restrictions apply.
Fig. 1.
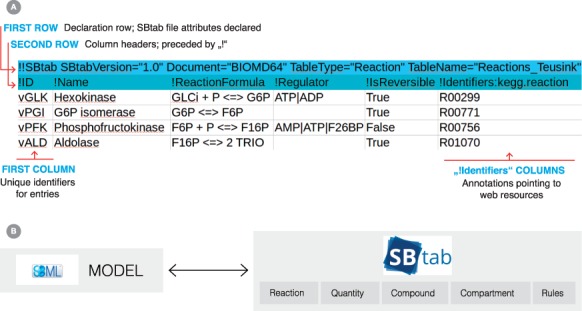
(A) The depicted SBtab table type ‘Reaction’ describes biochemical reactions in upper glycolysis. The table comprises information about reaction modifiers and reversibility, and annotates reactions with identifiers from the KEGG database. (B) Structural information about a biochemical network model can be converted between SBML and SBtab formats. The SBtab tables refer to different types of SBML elements (e.g. Reaction, Compound, Compartment, Quantity, Rules, Events)
In designing the format, we tried to adopt useful standards from other places. Identifiers.org, for example, provides a simple and safe mechanism to refer to the entries of databases and web repositories (Juty et al., 2012). Instead of defining our own format for database references, we simply rely on this mechanism. This makes SBtab files easy to process and may contribute to promoting this useful existing standard. SBtab offers predefined table types that represent diverse kinds of data, e.g. experimental time series, biochemical model parameters (e.g. kinetic constants), or descriptions of SBML-compliant network models. Beyond these predefined table types, SBtab can be tailored to particular types of data to be exchanged. A detailed description of SBtab, together with many examples, is given in the SBtab specification document (http://arxiv.org/abs/1502.01463, Liebermeister et al., 2015).
To simplify the work with SBtab documents, we provide free online tools and software for using SBtab in MS Excel, Python, MATLAB and R. The online tools on www.sbtab.net comprise an automatic syntax validator for SBtab files and a tool for converting models from SBtab to SBML and vice versa. The same functions are also provided as Python code, as an R interface and as an add-in for MS Excel. The latter has been implemented in C# and enables the manipulation of SBML files via the SBtab interface from within Excel. Using the Python-based SBtab parser and object classes, support for SBtab can be easily integrated into Systems Biology workflows. The potential applications are numerous: The structure of SBtab resembles those of relational databases, and we also provide Python code for the import and export of SBtab to and from an SQLite database. Using such a database, model or data elements can be queried via SQL statements, which are supported by virtually all programming languages. This can be used in workflows comprising data storage, manipulation or creation of models (e.g. for the systematic construction of kinetic models; Stanford et al., 2013), conversion of SBML files into human-readable formats, or incorporation of experimental data into models (e.g. SBtab as an input format for parameter balancing; Lubitz et al., 2010).
3 Conclusion
SBtab is a flexible, table-based format for data exchange in Systems Biology that comes with tools for diverse groups of users. The use of SBtab can be beneficial both for programmers and for end users with little technical background: Programmers can use the open-source code to easily integrate the functionality into their own software. Excel users can use the MS Excel add-in to conveniently edit SBtab and SBML files in their normal working environment. Unlike specialized formats such as SBML, SBtab is designed to be human-readable: It relies on existing spreadsheets and defines rules that ensure complete and unambiguous information. Adapting existing experimental data files to the SBtab format is simple (since it does not enforce complex syntax restrictions) and rewarding, because the SBtab format is oriented towards established and accepted standards. Hence, SBtab has the potential to bridge the gap between the vast amounts of empirical data, typically stored in spreadsheets, and software that requires structured input formats. Currently, it is used for data storage (e.g. source data of the eQuilibrator web tool; Flamholz et al., 2012) and modelling workflows (e.g. Stanford et al., 2013), and is supported by the Data Repository for Kinetic Models of Biological Systems (KiMoSys; Costa et al., 2014). We envisage that SBtab will foster an easy exchange of data for applications in which no specialized data formats have been established, or where these formats would be too restrictive to use. We also encourage users to customize SBtab for their needs and to tell us about interesting use cases, to support the further development of the format.
Acknowledgements
The authors wish to thank Jannis Uhlendorf, Marvin Schulz, Dagmar Waltemath, Dirk Wiesenthal, Anne Lubitz and the many colleagues listed in the SBtab specification for their various contributions to the development of SBtab.
Funding
This work was supported by the German Research Foundation (grant number Ll 1676/2-1 to WL) and by the German Ministry for Education and Research (BMBF [grant number 0316184B to EK]). EN is supported by a SystemsX.ch Transition Postdoc Fellowship (TPdF).
Conflict of Interest: none declared.
References
- Costa R.S. et al. (2014) KiMoSys: a web-based repository of experimental data for KInetic MOdels of biological SYStems. BMC Syst. Biol., 8, 85.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamholz A. et al. (2012) eQuilibrator – the biochemical thermodynamics calculator. Nucleic Acids Res., D1, D770–D775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M. et al. (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics, 19, 524–531. [DOI] [PubMed] [Google Scholar]
- Juty N. et al. (2012) Identifiers.org and MIRIAM Registry: community resources to provide persistent identification. Nucleic Acids Res., 40, D580–D586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novère N. et al. (2005) Minimum information requested in the annotation of biochemical models (MIRIAM). Nat. Biotechnol., 23, 1509–1515. [DOI] [PubMed] [Google Scholar]
- Le Novère N. et al. (2009) The systems biology graphical notation. Nat. Biotechnol., 27, 735–741. [DOI] [PubMed] [Google Scholar]
- Liebermeister W. et al. (2015). SBtab – conventions for structured data tables in systems biology. ArXiv e-print service. [Google Scholar]
- Lubitz T. et al. (2010) Parameter balancing in kinetic models of cell metabolism. J. Phys. Chem. B, 114, 16298–16303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone S.A. et al. (2008) The first RSBI (ISA-TAB) workshop: can a simple format work for complex studies? OMICS J. Integr. Biol., 12, 143–149. [DOI] [PubMed] [Google Scholar]
- Stanford N. et al. (2013) Systematic construction of kinetic models from genome-scale metabolic networks. PLoS One, 8, e79195. [DOI] [PMC free article] [PubMed] [Google Scholar]


