Abstract
Planococcus maritimus MKU009 is a Gram positive cocci and a moderate halophilic bacterium isolated from marine water of Pichavaram, South East Coast of India. Here we report the draft genome of Planococcus maritimus MKU009 with a total size of 3,251,644 bp with N50 value of 1681571 bp. The overall G+C content of the genome was 47.27%. The carotenoid producing crtN, crtB, crtP and crtI genes were located within the first contig of the genome assembly. This genome source will provide insights into functional genomics of carotenoid production and metabolic engineering.
Keywords: Carotenoids, Genome sequence, Planococcus maritimus.
Introduction
Carotenoids are naturally occurring pigments which are widely distributed in bacteria, algae and other eukaryotic organisms. These pigments are safe alternative to synthetic derived pigments due to their toxic, carcinogenic and tetratogenic properties 1,2. Planococcus maritimus have been isolated from sea water 3 and industrial effluent environments 4. It produces diverse secondary metabolite such as osmotic stabilizing agents 5, anticancer agents 6 and methyl glucosyl-3,4-dehydro-apo-8-lycopenoate, a novel carotenoid with antioxidant activity 7. Yellow pigmented Planococcus maritimus MKU009 was isolated from surface marine waters of Pichavaram (11.45N, 79.79E), South East Coast, Tamil Nadu, India, by spread plate method using Zobell marine agar (Hi-media, ZMB 2216). Phylogentic analysis of 16S rRNA sequence of this strain showed 99% similarity to Planococcus maritimus strain TF-9 3.
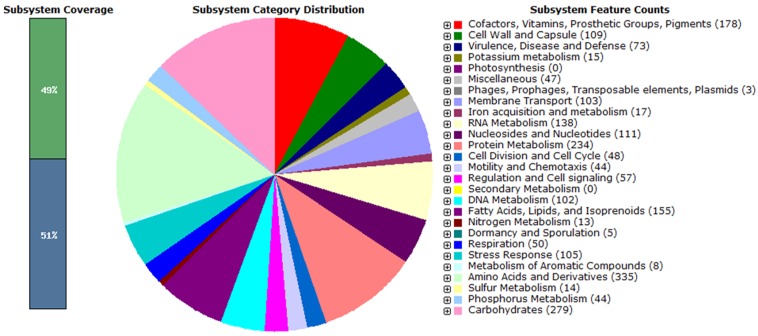
The genomic DNA of Planococcus maritimus MKU009 was extracted using the DNeasy kit (Qiagen, Hilden, Germany) and the whole genome was sequenced using the Illumina MiSeq sequencing platform with a paired-end library, which was performed by Genotypic Technology, Bangalore, India. A total of 3,251,644 Illumina reads were assembled using SPAdes genome assembler version 3.6.2 8, which resulted in 69 contigs (above the length of 200 bp) with an N50 value of 16,81,571bp. The largest contig assembled in this study has the length of 16,81,571 bp. The cumulative G+C content of the genome assembly was 47.27%. Gene annotation was performed using the Rapid Annotations using Subsystems Technology (RAST) 9 and 3,217 protein-coding sequences (CDS's) were predicted in the draft genome. There are 64 tRNAs in the genome assembly predicted using ARAGORN 10.
The DNA sequences of the predicted 3,217 CDS's were obtained from RAST in multi-fasta format and analyzed with KAAS automated annotation server. It was resulted with the KO assignments for 1,655 CDS's out of the 3,217 entries and the remaining 1,562 CDS were left blank by KAAS 11. The 1,655 CDS's with KO assignments were analyzed for their metabolic roles using KEGG Pathway Reconstruct tool (http://www.genome.jp/kegg/tool/map_pathway.html) 11. It has identified four genes involved in carotenoid biosynthesis (Pathway No: 00906). Genes encoding for enzymes involved in carotenoid biosynthetic pathway such as diapophytoene desaturase (crtN), Phytoene synthase (crtB), Phytoene desaturase (crtP) and Phytoene dehydrogenase (crtI) were present on the genome of Planococcus maritimus MKU009. In addition, KEGG pathway analysis also revealed specific genes involved in the degradation of benzoate, aminobenzoate, fluorobenzoate, chloroalkane, chloroalkene, chlorocyclohexane, chlorobenzene, toluene, xylene, dioxin, naphthalene and metabolism of xenobiotics by cytochrome p450. This genome source can provide insights into the functional genomics of carotenoid production with industrial applications and biodegradation of hydrocarbons.
Nucleotide sequence accession numbers
The whole-genome shotgun assembly has been deposited at GenBank 12 under the accession no LTZG00000000. Details about the Planococcus maritimus MKU009 genome sequencing bioproject, biosample and raw reads submitted to Sequence Read Archive (SRA) were deposited with the following accession numbers: PRJNA313006, SAMN04511041, SRX1598158.
Figure .
Graphical abstract of Planococcus maritimus MKU009.
Acknowledgments
This project was financially supported by the Department of Biotechnology, Government of India under Rapid Grant for Young Investigator, India (Grant No. BT/PR15135/GBD/27/331/20110). We also acknowledge the DST-Purse program of the School of Biotechnology, Madurai Kamaraj University, Tamil Nadu, India.
References
- 1.Kirti K, Amita S, Priti S, Mukesh Kumar A, Jyoti S. Colorful world of microbes:carotenoids and their applications. Adv Biol. 2014;2014:1–13. [Google Scholar]
- 2.Soliev AB, Hosokawa K, Enomoto K. Bioactive pigments from marine bacteria: applications and physiological roles. Evid Based Complement Alternat Med. 2011;2011:1–17. doi: 10.1155/2011/670349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon JH, Weiss N, Kang KH, Oh TK, Park YH. Planococcus maritimus sp.nov, isolated from seawater of a tidal flat in Korea. Int.J.Syst.Evol.Micro-biol. 2003;53:2013–2017. doi: 10.1099/ijs.0.02557-0. [DOI] [PubMed] [Google Scholar]
- 4.Varsha C, Arpana J. Screening of significant Nutrient Parameters for Pigment Production from newly isolated organism Planococcus maritimus AHJ_2 Using Plackett Burman Design. J. Microbiol. Biotech. Res. 2013;3(1):79–83. [Google Scholar]
- 5.Joghee NN, Gurunathan J. Planococcus maritimus VITP21synthesizes (2-acetamido-2-deoxy-a-D-glucopyranosyl)-(1-2)-b-D-fructofuranose under osmotic stress: a novel protein stabilizing sugar osmolyte. Carbohydr Res. 2014;383:76–81. doi: 10.1016/j.carres.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Krishnaveni M, Jayachandran S. Inhibition of MAP kinases and down regulation of TNF-⍺, IL-β and COX-2 genes by the crude extracts from marine bacteria. Biomed Pharmacother. 2009;63(7):469–476. doi: 10.1016/j.biopha.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Shindo K, Endo M, Miyake Y, Wakasugi K, Morritt D, Bramley PM, Fraser PD, Kasai H, Misawa N. Methyl Glucosyl-3,4-dehydro-apo-8-lycopenoate, a Novel Antioxidative Glyco-C30-carotenoic Acid Produced by a Marine Bacterium Planococcus maritimus. J. Antibiot. 2008;61(12):729–735. doi: 10.1038/ja.2008.86. [DOI] [PubMed] [Google Scholar]
- 8.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST server: Rapid Annotations using Subsystems Technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:182–185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. Genbank. Nucleic Acids Res. 2012;40:48–53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]