Abstract
The highly pathogenic avian influenza subtype H5N1 (HPAI H5N1) is a worldwide zoonotic infectious disease, threatening humans, poultry and wild birds. The role of wild birds in the spread of HPAI H5N1 has previously been investigated by comparing disease spread patterns with bird migration routes. However, the different roles that the southward autumn and northward spring migration might play in virus transmission have hardly been explored. Using direction analysis, we analyze HPAI H5N1 transmission directions and angular concentration of currently circulating viral clades, and compare these with waterfowl seasonal migration directions along major waterfowl flyways. Out of 22 HPAI H5N1 transmission directions, 18 had both a southward direction and a relatively high concentration. Differences between disease transmission and waterfowl migration directions were significantly smaller for autumn than for spring migration. The four northward transmission directions were found along Asian flyways, where the initial epicenter of the virus was located. We suggest waterfowl first picked up the virus from East Asia, then brought it to the north via spring migration, and then spread it to other parts of world mainly by autumn migration. We emphasize waterfowl autumn migration plays a relatively important role in HPAI H5N1 transmission compared to spring migration.
Wild birds are considered to be the natural reservoir for avian influenza viruses, and a particularly high prevalence rate has been observed in waterfowl, including Anseriformes and Charadriiformes1. The highly pathogenic avian influenza subtype H5N1 (HPAI H5N1) is a worldwide zoonotic disease, threatening humans, poultry and wildlife. Since the first detection of HPAI H5N1 in farm geese in China in 1996, up to March 2015 HPAI H5N1 has been found in 69 countries in Asia, Europe, Africa, and the Americas (http://empres-i.fao.org/ empres-i/home). While poultry are the primary reservoir of highly pathogenic avian influenza viruses, wild waterfowl can act as secondary transmitters2,3. Hence, the HPAI H5N1 virus can spread over long distances, not only via trade in poultry and wild-caught birds, but also via the natural movements of wild birds4. Initially, wild birds were most likely infected with HPAI H5N1 via contact with infected poultry in regions where poultry density is high5 (see Fig. 1). Subsequently, asymptomatic infection, as reported in some duck species3,6, and the ability of the virus to survive in wild bird habitats without a host for over a month7, would make it possible for wild birds to disperse HPAI H5N1 across large distances and time spans, without the direct involvement of poultry8.
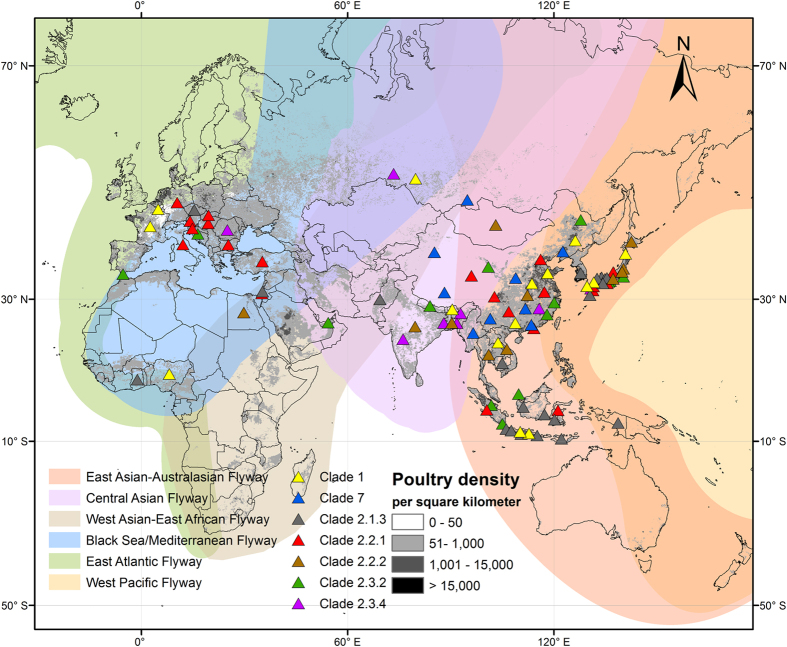
Figure 1. Distribution of HPAI H5N1 genetic clades and poultry density.
One clade sampled along the Mississippi Americas Flyway was omitted from the map due to the small sample size. The map was produced using ArcGIS Desktop 10.3 (www.esri.com).
The first considerable HPAI H5N1 outbreak in migratory waterfowl was recorded at Qinghai Lake in April 2005 (clade 2.2 according to the WHO naming system9) and resulted in mass die-offs for bar-headed geese10,11. Since then, HPAI H5N1 viruses closely related to the Qinghai-like clade 2.2 continued to be isolated along the Eurasian migration flyways and resulted in mortality of millions of wild and domestic birds in Asia, Europe, and Africa4,12,13. Another novel clade, clade 2.3.2, was recorded in Qinghai Lake in 2009 and has caused the death of over two hundred wild birds5,14. Clade 2.3.2.1 was firstly recorded in whooper swan (Cygnus cygnus) in Japan in 20085,14 and has been found in wild waterfowl in Mongolia, Russia, Korea, and Japan during 2009–201113,15,16, and more recently in China in 201517. Other outbreaks of the HPAI H5 lineages H5N8 and H5N2, are considered to have been spread from Asia to Europe and North America via waterfowl migration18,19. These studies highlight that wild birds could carry and spread avian influenza viruses along their flyways.
The role of migratory birds in the spread of HPAI H5N1 has been further investigated by comparing disease spread patterns with bird migration patterns. Six HPAI H5N1 outbreak patterns were found to be spatio-temporally associated with the seasonal migration of waterfowl using a space-time clustering analysis20. By combining space-time and phylogenetic approaches, Liang et al.21 reported possible spread routes for HPAI H5N1 that coincided well with four major flyways. Gaidet et al.22 analyzed a large-scale dataset of wildfowl movements over HPAI H5N1 infected regions in Eurasia and Africa and suggested potential long-distance spread of HPAI H5N1 by migratory birds could occur. According to the relationship between viral transmission and waterfowl migration networks along the East Asian-Australasian and Central Asian Flyway, waterfowl migration was considered as the most likely factor explaining the geographic transmission of HPAI H5N1 viruses (clade 2.3.2)23. Siberia, which intersects with multiple flyways and is considered as one the most important breeding sites for migratory waterfowl, played a vital role in the global HPAI H5N1 transmission, with the highest emigration and second highest immigration rate24.
Next to the general match between HPAI H5N1 outbreaks and waterfowl migration patterns, several researchers have detected different levels of disease outbreak risk during the distinct waterfowl spring and autumn migration periods. Anatidae were found likely to spread the HPAI H5N1 virus along their autumn migration routes from Russia and Kazakhstan to the Black Sea region25. Bourouiba et al.26 found that repeated outbreaks of HPAI H5N1 at stopovers caused more mortalities during autumn migration than during spring migration. Analysis of the distance between disease outbreak sites in poultry and the closest migratory waterfowl sites in Romania indicated that HPAI H5N1 infections of poultry might occur via exposure to migratory waterfowl during autumn27. Yet, core flyway corridors of wild ducks tracked along the East Asian Flyway during spring migration were not spatio-temporally connected to sites of reported outbreaks28. While these studies suggest potential seasonal variation in waterfowl migration in the spread of HPAI H5N1, whether there is a difference in importance between autumn and spring migration still has to be investigated. Yet, such information is pertinent for understanding the disease transmission mechanisms and is crucial for targeted prevention efforts.
Direction analysis is an efficient way to accurately quantify disease transmission direction and angular concentration. Direction analysis can be used to compare disease spread directions with the distinct seasonal migration directions of waterfowl. However, only few studies adopted direction analysis for determining the spread of HPAI H5N1 and they mostly were restricted to a local or a regional scale. For example, the directions of HPAI H5N1 transmission in Bangladesh29,30 and Romania31 were quantified and compared with local movements of wild birds and poultry. To get a further insight into the spatial-temporal relationships between continental waterfowl migration and HPAI H5N1 transmission, it is necessary to carry out direction analysis at the continental scale.
In this paper we use HPAI H5N1 viral clades as input for direction analysis. The HPAI H5N1 transmission directions and angular concentrations are then compared with waterfowl seasonal migration directions along major waterfowl flyways. We are particularly interested in testing how waterfowl seasonal migration facilitates the transmission of HPAI H5N1. Considering latitudinal migration of waterfowl during their annual life cycle (travelling to the high-latitude breeding grounds in spring and returning to the low-latitude wintering sites in autumn), if disease transmission directions along each flyway point in a southward direction, we postulate that waterfowl autumn migration facilitates transmission, whereas if directions point in a northward direction we postulate that spring migration facilitates transmission. This study is the first to systematically quantify the transmission directions and concentration of HPAI H5N1 viruses along the major flyways and compare these to waterfowl seasonal migration directions. Our findings contribute to global avian influenza surveillance and control.
Results
Spatio-temporal dimensions of HPAI H5N1 viral clades
The sample sites of different clades are shown in Fig. 1. A total of 433 sample sites for clades 1 (n = 34), 2.1.3 (n = 54), 2.2.1 (n = 96), 2.2.2 (n = 20), 2.3.2 (n = 130), 2.3.4 (n = 78), and 7 (n = 21) were obtained for subsequent analysis. All these clades were intensively sampled in East China, Japan, Southeast Asian countries (Indonesia, Malaysia, Cambodia, Thailand, Laos, Vietnam, and Myanmar), Southern Asian countries (India, Bangladesh, Bhutan, Nepal, and Pakistan) and Egypt. All clades except clade 2.2.2 were recovered in Russia. Only clades 2.2.1 and 2.2.2 were found in Mongolia. Clades 1, 2.1.3, and 2.2.1 were found in Africa in Nigeria and Ghana. In Europe, clades 1, 2.1.3, 2.2.1, 2.3.2, and 2.3.4 were sampled from Italy, Bulgaria, Romania, Croatia, Slovenia, Austria, Hungary, Slovakia, Czech Republic, France, Germany, and Belgium.
All seven clades were sampled along the East Asian-Australasian, Central Asian, West Asian-East African, Black Sea/Mediterranean, and West Pacific Flyways. Along the East Atlantic Flyway, clades 1, 2.2.1, 7 (2 sites), 2.1.3 (2 sites), 2.3.2 (2 sites) were sampled. Along the West Asian-East African and Black Sea/Mediterranean Flyways, clade 2.2.2 was only sampled in a single site. Along the Black Sea/Mediterranean Flyway clade 7 was only sampled in a single site. Along the Mississippi Americas Flyway only one clade (2.3.2) was sampled in Washington in 2014; the sample size for this flyway is too small to be included in the direction analysis.
In regards to the temporal dimension, all the seven clades were sampled since 2004, and most of them were firstly found along the East Asian-Australasian Flyway (Table 1). The longevity of each clade is: clade 1 (2004–2012), clade 7 (2004–2015), clade 2.1.3 (2004–2015), clade 2.2.1 (2004–2015), clade 2.2.2 (2004–2013), clade 2.3.2 (2004–2015), and clade 2.3.4 (2004–2015). The number of years each clade was sampled along different flyways is: clade 1 (2–9 years), clade 7 (0–7 years), clade 2.1.3 (1–9 years), clade 2.2.1 (2–11 years), clade 2.2.2 (0–5 years), clade 2.3.2 (1–11 years), and clade 2.3.4 (0–11 years).
Table 1. Number of sequences sampled per HPAI H5N1 clade in different years along main waterfowl flyways.
| Clade | Flyway | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | AAF | 60 | 79 | 4 | 15 | 23 | 1 | 11 | 4 | 2 | |||
| CAF | 1 | 2 | |||||||||||
| WEF | 1 | 6 | 6 | 1 | 1 | ||||||||
| BMF | 1 | 13 | 11 | 1 | 1 | ||||||||
| EAF | 7 | 5 | |||||||||||
| WPF | 2 | 1 | 2 | ||||||||||
| 7 | AAF | 5 | 3 | 5 | 20 | 1 | 1 | 4 | |||||
| CAF | 2 | 2 | 4 | 1 | |||||||||
| WEF | 2 | 3 | 1 | 1 | |||||||||
| BMF | 3 | 1 | 1 | ||||||||||
| WPF | 1 | 1 | |||||||||||
| 2.1.3 | AAF | 4 | 9 | 22 | 75 | 15 | 18 | 20 | 9 | 1 | 1 | ||
| CAF | 3 | 3 | 5 | 1 | 1 | 1 | |||||||
| WEF | 6 | 3 | 5 | ||||||||||
| BMF | 7 | 1 | |||||||||||
| EAF | 2 | 1 | |||||||||||
| WPF | 3 | 6 | 7 | ||||||||||
| 2.2.1 | AAF | 3 | 4 | 6 | 23 | 4 | 28 | 22 | 31 | 8 | 18 | 4 | |
| CAF | 1 | 2 | 2 | 5 | 11 | 7 | 1 | 17 | 2 | 2 | |||
| WEF | 31 | 70 | 58 | 30 | 30 | 54 | 7 | 1 | 2 | ||||
| BMF | 2 | 55 | 70 | 55 | 30 | 30 | 54 | 7 | 1 | 7 | |||
| EAF | 4 | 1 | 3 | ||||||||||
| WPF | 2 | 1 | 1 | 1 | 2 | 7 | 31 | ||||||
| 2.2.2 | AAF | 1 | 7 | 5 | 3 | 2 | |||||||
| CAF | 3 | 6 | 7 | 2 | 4 | ||||||||
| WEF | 6 | 1 | 1 | ||||||||||
| BMF | 6 | 1 | 1 | ||||||||||
| WPF | 5 | ||||||||||||
| 2.3.2 | AAF | 7 | 19 | 14 | 43 | 21 | 49 | 34 | 122 | 90 | 41 | 40 | |
| CAF | 1 | 1 | 2 | 6 | 22 | 5 | 23 | 55 | 22 | 2 | 1 | ||
| WEF | 2 | 4 | 23 | 5 | 22 | 22 | 2 | 5 | 7 | 15 | |||
| BMF | 1 | 3 | 4 | 26 | 5 | 22 | 22 | 2 | 5 | 6 | 14 | ||
| EAF | 4 | ||||||||||||
| WPF | 2 | 2 | 4 | 12 | 7 | 42 | 1 | 6 | 1 | ||||
| 2.3.4 | AAF | 3 | 20 | 29 | 110 | 57 | 32 | 15 | 11 | 10 | 1 | 1 | |
| CAF | 2 | 4 | 1 | 7 | 6 | 7 | 4 | 2 | 2 | ||||
| WEF | 2 | 2 | 1 | 8 | 8 | 5 | 2 | 16 | 1 | ||||
| BMF | 1 | 1 | 1 | 8 | 8 | 5 | 2 | 16 | 3 | ||||
| WPF | 1 | 2 | 2 | 1 | 10 | 1 |
AAF, CAF, WEF, BMF, EAF, and WPF represent the East Asian-Australasian Flyway, Central Asian Flyway, West Asian-East African Flyway, Black Sea/Mediterranean Flyway, East Atlantic Flyway, and West Pacific Flyway, respectively. For each clade, flyways with no sequence data sampled are not shown.
Directions of HPAI H5N1 transmission and waterfowl seasonal migration
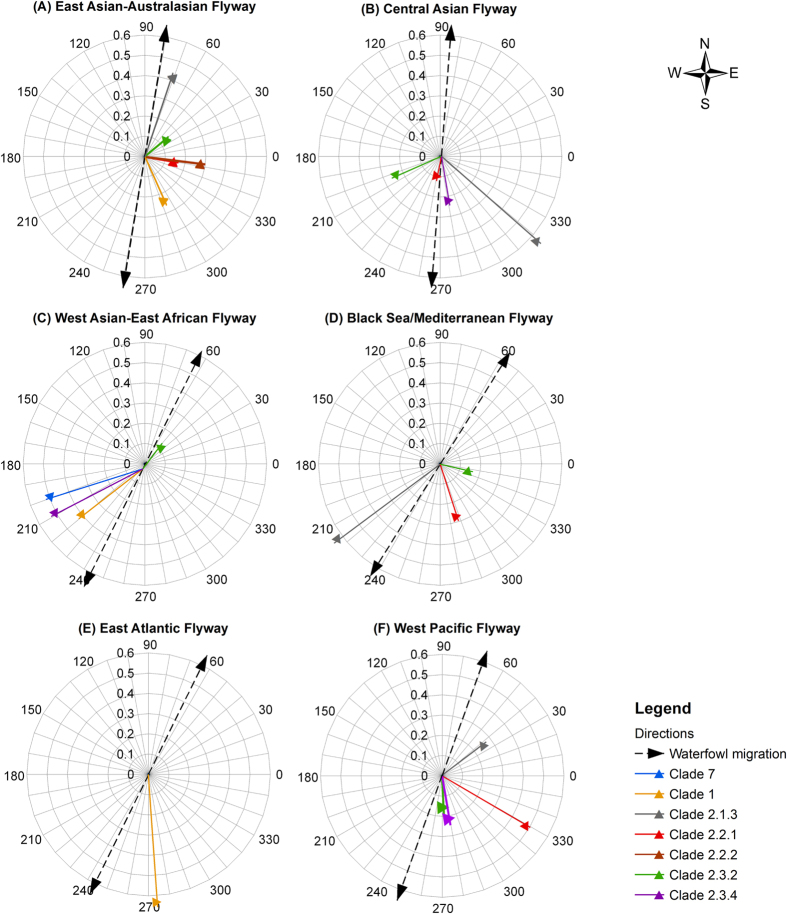
Table 2 show the transmission direction and angular concentration of HPAI H5N1 genetic clades. Table 3 shows the waterfowl seasonal migration directions and concentrations in the four infected flyways. For all directions, 90 degree is due north and 0 degree is due east. We define directions within 0–180 degree as northward directions and 180–360 degree as southward directions. In total 22 directions with significant angular concentrations (P ≤ 0.01) were found along the East Asian-Australasian (2 northward and 3 southward), Central Asian (5 southward), West Asian-East African (1 northward and 3 southward), Black Sea/Mediterranean (3 southward), East Atlantic (1 southward) and West Pacific (1 northward and 3 southward) Flyways (Tables 2 and 3). Three flyways with either large or small sample size yielded both southward and northward directions, indicating there was no correlation between sample size and the detection of bi-directional patterns. The mean concentration for southward disease transmission directions (0.41) was higher than for northward directions (0.25).
Table 2. HPAI H5N1 transmission directions of genetic clades.
| Clade | Flyway | Number of sample sites | Direction | Concentration | P-value |
|---|---|---|---|---|---|
| 1 | East Asian-Australasian Flyway | 23 | 293.94 | 0.2586 | 0.001 |
| Central Asian Flyway | 3 | 277.76 | 0.9904 | 0.002 | |
| West Asian-East African Flyway | 5 | 217.89 | 0.4000 | 0.001 | |
| Black Sea/Mediterranean Flyway | 7 | 215.29 | 0.1054 | 0.388 | |
| East Atlantic Flyway | 3 | 274.00 | 0.6667 | 0.007 | |
| 2.1.3 | East Asian-Australasian Flyway | 42 | 71.49 | 0.4319 | 0.001 |
| Central Asian Flyway | 7 | 318.69 | 0.6147 | 0.001 | |
| West Asian-East African Flyway | 6 | 331.50 | 0.4322 | 0.019 | |
| Black Sea/Mediterranean Flyway | 6 | 216.65 | 0.9422 | 0.001 | |
| West Pacific Flyway | 12 | 37.28 | 0.2711 | 0.002 | |
| 2.2.1 | East Asian-Australasian Flyway | 59 | 350.34 | 0.1655 | 0.001 |
| Central Asian Flyway | 19 | 258.57 | 0.1242 | 0.005 | |
| West Asian-East African Flyway | 14 | 237.58 | 0.0814 | 0.171 | |
| Black Sea/Mediterranean Flyway | 23 | 287.90 | 0.2934 | 0.001 | |
| West Pacific Flyway | 21 | 329.68 | 0.4999 | 0.001 | |
| 2.2.2 | East Asian-Australasian Flyway | 10 | 352.79 | 0.2973 | 0.007 |
| Central Asian Flyway | 8 | 20.58 | 0.1163 | 0.264 | |
| 2.3.2 | East Asian-Australasian Flyway | 91 | 39.89 | 0.1505 | 0.001 |
| Central Asian Flyway | 27 | 203.83 | 0.2671 | 0.001 | |
| West Asian-East African Flyway | 17 | 52.82 | 0.1577 | 0.001 | |
| Black Sea/Mediterranean Flyway | 20 | 347.13 | 0.1649 | 0.001 | |
| West Pacific Flyway | 25 | 272.64 | 0.1891 | 0.001 | |
| 2.3.4 | East Asian-Australasian Flyway | 51 | 18.93 | 0.0370 | 0.049 |
| Central Asian Flyway | 20 | 279.67 | 0.2447 | 0.001 | |
| West Asian-East African Flyway | 12 | 207.38 | 0.5083 | 0.001 | |
| Black Sea/Mediterranean Flyway | 13 | 191.97 | 0.1112 | 0.062 | |
| West Pacific Flyway | 20 | 279.67 | 0.2447 | 0.001 | |
| 7 | East Asian-Australasian Flyway | 14 | 105.94 | 0.0945 | 0.238 |
| Central Asian Flyway | 6 | 234.46 | 0.3037 | 0.066 | |
| West Asian-East African Flyway | 4 | 197.55 | 0.5000 | 0.002 |
Table 3. Waterfowl seasonal migration directions.
| Flyway | Spring migration | P-value | Autumn migration | |||||
|---|---|---|---|---|---|---|---|---|
| Number of sites | Direction | Concentration | Number of sites | Direction | Concentration | P-value | ||
| East Asian-Australasian Flyway | 431 | 80.56 | 0.7458 | 0.001 | 431 | 260.56 | 0.7458 | 0.001 |
| Central Asian Flyway | 564 | 85.67 | 0.7184 | 0.001 | 564 | 265.67 | 0.7184 | 0.001 |
| West Asian-East African Flyway | 382 | 63.44 | 0.8272 | 0.001 | 382 | 243.44 | 0.8272 | 0.001 |
| Black Sea/Mediterranean Flyway | 462 | 58.04 | 0.7444 | 0.001 | 462 | 238.04 | 0.7444 | 0.001 |
| East Atlantic Flyway | 380 | 63.78 | 0.6928 | 0.001 | 380 | 243.78 | 0.6928 | 0.001 |
| West Pacific Flyway | 188 | 70.21 | 0.8210 | 0.001 | 188 | 250.21 | 0.8210 | 0.001 |
Most (18 out of 22) of the calculated HPAI H5N1 transmission directions overlapping with the major flyways were oriented southward and matched better with autumn than with spring waterfowl migration direction (Fig. 2). The differences between the disease transmission direction and waterfowl migration direction along the six flyways were normally distributed (Kolmogorov-Smirnov Test, N = 44, P = 0.16). The HPAI H5N1 transmission directions showed a significantly smaller difference with autumn migration directions than with spring migration directions (Paired-Samples T Test, t = −2.21, N1 = N2 = 22, P = 0.04) along these flyways (Fig. 3A). The concentrations of significant transmission directions were not normally distributed (Kolmogorov-Smirnov Test, N = 22, P = 0.01). Although the mean concentration for southward transmission directions was higher than for northward ones, they did not significantly differ from each other (Mann-Whitney U Test, Z = 1.28, N1 = 18, N2 = 4, P = 0.20) (Fig. 3B).
Figure 2. Directions of HPAI H5N1 transmission and waterfowl seasonal migration.
The graphs represents calculated direction (degree) for HPAI H5N1 genetic clades for the six main infected flyways; the length of the arrow (see axis) represents the angular concentration of each direction; all the directions displayed are statistically significant (P ≤ 0.01) in direction tests; the concentrations for waterfowl migration directions are all greater than 0.6.
Figure 3. Differences between HPAI H5N1 transmission and waterfowl seasonal migration.
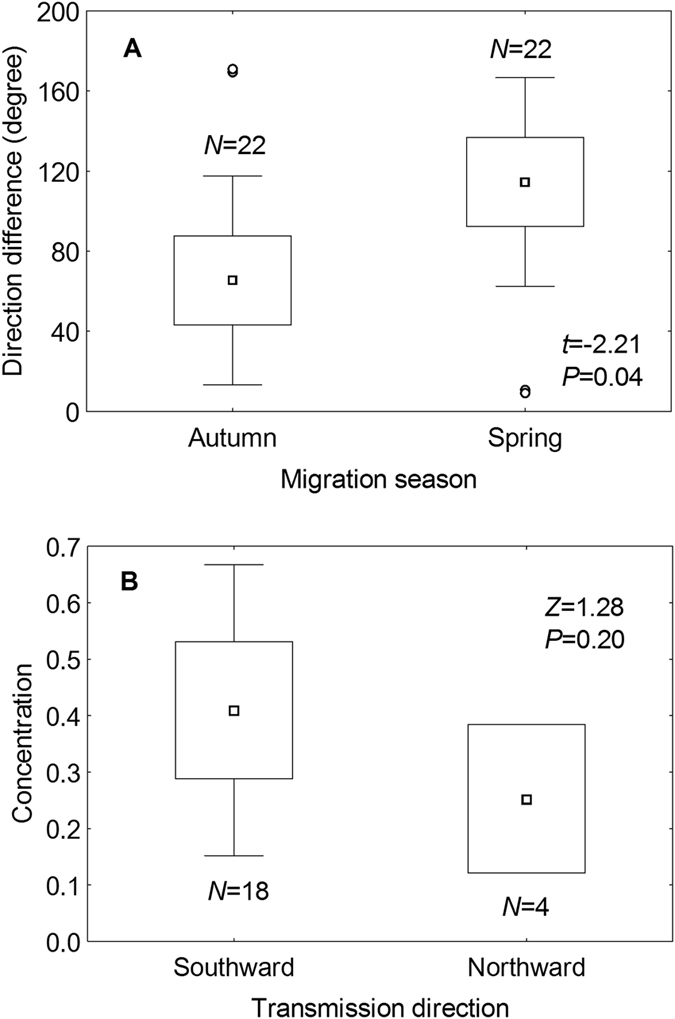
The boxplot compares differences in (A) direction differences with waterfowl autumn and spring migration directions; (B) the concentrations of southward and northward disease transmission directions. The boxes represent the standard error range; the bars cover the standard deviation range; the midpoint is the mean of each group; and the circles show outliers.
Discussion
This study quantified the HPAI H5N1 spread directions derived from genetic clades and compared them with waterfowl seasonal migration directions. Eighteen out of 22 HPAI H5N1 transmission directions were oriented southwards, in close proximity to waterfowl autumn migration directions. Moreover, the concentrations for southward disease transmission directions were higher than that for northward directions. Our findings emphasize a relatively important role of waterfowl autumn migration in facilitating HPAI H5N1 cross-continental transmission compared to spring migration.
Possible explanations for the close proximity of HPAI H5N1 transmission directions to waterfowl southward autumn migration are as follows. Firstly, southward autumn migration includes juveniles born the previous breeding season, meaning the population size is larger in autumn than in spring. Due to the positive relationship between virus prevalence and migratory waterfowl density32, a higher migratory waterfowl density in autumn may facilitate an increased avian influenza prevalence33. Secondly, the population structure with more vulnerable juveniles leads to a higher disease infection rate during autumn migration. Olsen et al.34 indicated a higher low pathogenic avian influenza (LPAI) virus prevalence in juveniles in early autumn. Waterfowl with prior exposure to LPAI show immunity to HPAI H5N135. Juveniles are immunologically naive in comparison to adults and are therefore more prone to being infected by and shedding avian influenza viruses33,36. Thirdly, autumn migration is less synchronized and more flexible than spring migration. Various species migrate north in spring and gather in their northern breeding grounds creating a virus pool21,24. After breeding, they migrate southwards to their wintering sites using different migration routes, which could cause a redistribution of avian influenza viruses and contribute to the survival of the virus across a wide geographic range. This process would also help transmit the virus to the resident avian population in the wintering sites of waterfowl37.
HPAI H5N1 southward transmission has been reported in previous studies using phylogenetic analysis in combination with trajectory analysis of cross-continent viral movements. Liang et al.21 found that the HPAI H5N1 virus spread in a southward direction along the West Asian-East African and Black Sea/Mediterranean Flyways using Siberian breeding land as a hinge area. Li et al.38 observed a “high-to-low latitude” transmission pattern coinciding with the autumn migration of waterfowl along the Central Asian and East Asian-Australasian Flyways. As indicated by Newman et al.39, along the Central Asian Flyway, the spatial pattern of viral evolution derived from phylogeographic mapping shows southward movements, coinciding with the autumn migration of waterfowl. However, previous studies36,37 that linked outbreak data with individual bird satellite tracking data also reported northward transmission directions. Disease directions identified from outbreak data tend to overlook the genetic similarity of these outbreaks and might lead to less rigorous conclusions.
We found both northward and southward HPAI H5N1 spreads along the East Asian-Australasian, West Asian-East African, and West Pacific Flyways. This reflects a unique role of East Asia in HPAI H5N1 transmission. Southern China has been hypothesized as the initial epicenter of the epidemic as the HPAI H5N1 virus was first discovered there in poultry40. Clade 2.3.2 was suggested to be transmitted from southern Asia to Mongolia by waterfowl on their way to their northern breeding grounds15, which is consistent with the northward transmission directions of clade 2.3.2 in our results. This suggests that migratory birds initially got infected with the HPAI H5N1 virus in East Asia, via contact with poultry, and then spread it northwards during spring migration21,24. The higher number of southwards disease spread directions suggest that after the virus was brought to north, waterfowl circulated the virus to other parts of world mainly via southward autumn migration. Although poultry transportation cannot be ruled out as a vector in the identified patterns of virus transmission, it would not be expected to leave nearly unidirectional transmission patterns. Furthermore, a similar pattern of disease spread has been reported in the newly emerged HPAI H5N8 virus17,41,42. The H5N8 virus was initially found in China in 2013, was hypothesized to next have been brought to Siberia by waterfowl via northwards spring migration in 2014, and subsequently in the same year spread to Europe, North America and East Asia along different flyways via southwards autumn migration17,41,42.
The continental relay transmission of avian influenza is complex, involving successively infected birds, not necessarily of the same species, but particularly those that share the same habitat22. HPAI H5N1 transmission might be aggravated by a large density and diversity of waterfowl with asynchronous timing of arrival and departure41. The avian influenza virus can survive longer in cold environments. For example, it can retain infectivity in lake water for more than 30 days at 0 °C, and in faeces for more than 30 days at 4 °C7. High HPAI H5N1 infection rates in wild birds might be related to waterfowl movements and congregation along the 0 °C isotherm42. This facilitates disease transmission via a “relay race”, i.e., the virus in water body is taken up by different migratory bird populations at different times. The pathobiology of the HPAI H5N1 virus infection varies between different waterfowl species, from fatal to asymptomatic43. Such characteristic can cause a mismatch between the timing of HPAI H5N1 outbreaks and waterfowl presence. The interaction between waterfowl and domestic poultry in shared habitat also facilitates HPAI H5N1 transmission8,44,45. This makes it possible for wildfowl to spread HPAI H5N1 over more than one annual migration cycle. Previous studies directly comparing disease outbreak patterns with individual bird migration patterns28,46,47,48,49 tended to overlook such relay and delay effects.
As a way forward, we analyzed disease transmission directions based on viral clades and calculated waterfowl spring and autumn migration directions separately for each flyway, based on the actual distribution of wetlands. The migration directions generated in this way are considered to encompass the total waterfowl population migrating along this flyway, in contrast to the migration pattern derived from satellite tracking data of just a handful of individuals. However, our findings should be further validated by using a large number of individual bird tracks. In this respect, birds that conduct exploratory movements that cross over flyways50 should be considered. Waterfowl tracking data recording information on the age of the birds involved can be used to integrate the age effect on HPAI H5N1 transmission.
In conclusion, we highlight a relatively important role of southward waterfowl migration in cross-continental HPAI H5N1 transmission compared to spring migration, especially after the virus had been introduced to the north. This knowledge facilitates further epidemiological investigations in terms of explaining and predicting the avian influenza transmission in an ecological context. On top of the routine monitoring of waterfowl seasonal migration, we suggest to strengthen the surveillance and control at waterfowl breeding sites, stopover sites during autumn migration, and wintering sites. Siberia is not only the major hub in the global HPAI H5N1 transmission network24, but also the common starting point of autumn migration for the different flyways, so we recommend future investigation into disease transmission at the northern breeding sites. Inadequate understanding of the migration ecology of most avian species may limit our ability to model and predict disease transmission51,52. Therefore, detailed bird migration data are needed to further validate the findings of this study.
Methods
Data
We obtained a total of 2867 hemagglutinin (HA) sequences with at least 1600 nucleotides, sampled from avian hosts during 2004–2015, from the Epiflu Database in GISAID (www.gisaid.org). We used the NIAID Influenza Research Database (IRD)53 at http://www.fludb.org, which uses phylogenetic analysis, to classify HA sequences to clade according to the WHO classification scheme9. The information of each HPAI H5N1 sequence includes sequence name, clade name, host species, location and year of sampling. The locations of the sequences are generally available at a country level, except for several countries such as China and Russia for which province- or city-level data are available. The geographic centroid of the given country or province was used to describe the geographic location of each sequence. Sequences sampled in the same location and year were merged into a single sample site. We excluded clades that have not been detected since 2008 and focused on those seven clades marked as currently circulating by Smith et al.54, namely clade 1, clade 2.1.3, clade 2.2.1, clade 2.2.2, clade 2.3.2, clade 2.3.4 and clade 7.
The major waterfowl flyways identified in the global monitoring program of Wetland International and the Level 1 Global Lakes and Wetlands Database (GLWD-1)55 were used for further direction analysis of waterfowl seasonal migration. Gridded poultry density was obtained from the Global Poultry Density (2005) Dataset in FAO GeoNetwork (www.fao.org/geonetwork/).
Direction analysis for HPAI H5N1 transmission
The direction method56 was applied to calculate the direction and concentration of HPAI H5N1 transmission. Whether the spread of disease tended to be in a particular direction, and the concentration of the direction, were tested with the direction statistic. The null hypothesis for the direction statistic is that the transmission directions across time are dissimilar to each other. The test statistic is a vector and consists of both directions and angular concentrations. The direction is the mean direction of the chains of HPAI H5N1 transmission connected from one HPAI H5N1 clade sample site to another. We used the relative time-connection matrix, which assumed each sample site is related to all sites after it. The concentration is the reciprocal of angular variance of a HPAI H5N1 transmission chain, ranging from 0 to 1; the larger the concentration is, the more the subsequent clade sample sites point to the same direction. The significance of the test statistic was estimated by 999 Monte Carlo simulations. An angular concentration is significant at a 0.01 level when it ranks in the top ten among the random simulated concentrations.
HPAI H5N1 viral clades overlapping with a specific flyway were utilized as the input for the direction statistics. The geographic coordinates of viral clades were transformed into projected x, y coordinates under Mercator (world) projection, which minimizes the distortion on angels. We estimated the transmission directions and angular concentrations of HPAI H5N1 viral clades across each flyway. The direction statistic was calculated with ClusterSeer version 2.5 (www.biomedware.com).
Waterfowl seasonal migration directions
Waterfowl mostly undertake latitudinal seasonal migration, using wetlands as their stopover sites. We therefore consider the wetlands inside the range of major flyways as potential stopover sites for waterfowl. The GLWD-1 database consists of 3,067 large lakes with more than 50 km2 area and 654 large reservoirs with more than 0.5 km3 storage capacity from all over the world55. The geometric centers of each wetland site were transformed into projected x, y coordinates under world Mercator projection. According to HPAI H5N1 outbreak regions, a total of 1,416 wetlands within a latitude from −10 to 90 and a longitude from −12 to 180 were selected for direction statistics. The temporal information of each wetland was defined based on their latitudes. Generally, waterfowl migrate south during autumn migration, so the lower the latitude of a wetland is, the later waterfowl might encounter it during their migration. For spring migration the reverse applies. Waterfowl seasonal migration directions and concentrations were calculated by direction statistics across different flyways.
Comparison between HPAI H5N1 transmission and waterfowl seasonal migration directions
HPAI H5N1 transmission directions and waterfowl migration directions with a significant level of P ≤ 0.01 were maintained for further analysis. The difference between disease transmission and waterfowl migration directions was the absolute value of each disease transmission direction in a flyway minus the waterfowl migration direction in the same flyway. Therefore, each disease transmission direction generated two differences, one for waterfowl autumn migration direction (dA) and another for waterfowl spring migration direction (dS). We then evaluated the normality of these differences with the Kolmogorov-Smirnov test. If they were normally distributed, a Paired-Samples T Test would be conducted to test the difference between dA and dS. If not, a Wilcoxon Matched Pairs Test would be conducted.
We also tested whether there is a significant difference between the concentration of northward (0–180 degree) and southward directions (180–360 degree). The normality of concentrations was tested with a Kolmogorov-Smirnov test; we applied Independent-Samples T Test for normally distributed data and otherwise a Mann Whitney U Test. All statistical analyses were run in Statistica 7 (http://www.statsoft.com/).
Additional Information
How to cite this article: Xu, Y. et al. Southward autumn migration of waterfowl facilitates cross-continental transmission of the highly pathogenic avian influenza H5N1 virus. Sci. Rep. 6, 30262; doi: 10.1038/srep30262 (2016).
Acknowledgments
This research was supported by the National Natural Science Foundation of China (No. 41471347) and Tsinghua University (No. 2012Z02287). We thank Fred de Boer (Wageningen University, the Netherlands) for his comments on an earlier version of the manuscript. We are grateful to Tao Zhang (Tsinghua University, China) for his assistance with phylogenetic analyses and Zezhong Wang (Peking University, China) for his assistance with spatial data processing.
Footnotes
Author Contributions Y.S. conceived and designed the analyses. Y.X. analyzed performed the Analyses. Y.X., P.G., B.W. and Y.S. contributed materials and analysis tools. Y.X., P.G., B.W. and Y.S. wrote the paper.
References
- Munster V. J. et al. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3, e61, 10.1371/journal.ppat.0030061 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M. et al. Flying over an infected landscape: distribution of highly pathogenic avian influenza H5N1 risk in South Asia and satellite tracking of wild waterfowl. EcoHealth 7, 448–458, 10.1007/s10393-010-0672-8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keawcharoen J. et al. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 14, 600–607, 10.3201/eid1404.071016 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. et al. H5N1 avian influenza re-emergence of Lake Communication Qinghai: phylogenetic and antigenic analyses of the newly isolated viruses and roles of migratory birds in virus circulation. J. Gen. Virol. 89, 697–702, 10.1099/vir.0.83419-0 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X. et al. Clade 2.3.2 Avian Influenza Virus (H5N1), Qinghai Lake Region, China, 2009-2010. Emerg. Infect. Dis. 17, 560–562, 10.3201/eid1703.100948 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J. Virol. 80, 5976–5983, 10.1128/JVI.00110-06 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R. G., Yakhno M., Hinshaw V. S., Bean W. J. & Murti K. G. Intestinal influenza - replication and characterization of influenza-viruses in ducks. Virology 84, 268–278, 10.1016/0042-6822(78)90247-7 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y., de Boer W. F. & Gong P. Different Environmental Drivers of Highly Pathogenic Avian Influenza H5N1 Outbreaks in Poultry and Wild Birds. PLoS One 8, e53362, 10.1371/journal.pone.0053362 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Resp. 8, 384–388, 10.1111/irv.12230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436, 191–192, 10.1038/nature03974 (2005). [DOI] [PubMed] [Google Scholar]
- Liu J. et al. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 309, 1206–1206, 10.1126/science.1115273 (2005). [DOI] [PubMed] [Google Scholar]
- Saad M. D. et al. Possible avian influenza (H5N1) from migratory bird, Egypt. Emerg. Infect. Dis. 13, 1120–1121, 10.3201/eid1307.061222 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H.-i. et al. Genetic characterization and pathogenicity assessment of highly pathogenic H5N1 avian influenza viruses isolated from migratory wild birds in 2011, South Korea. Virus Res. 160, 305–315, 10.1016/j.virusres.2011.07.003 (2011). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. New avian influenza virus (H5N1) in wild birds, Qinghai, China. Emerg. Infect. Dis. 17, 265, 10.3201/eid1702.100732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoda Y. et al. Characterization of H5N1 highly pathogenic avian influenza virus strains isolated from migratory waterfowl in Mongolia on the way back from the southern Asia to their northern territory. Virology 406, 88–94, 10.1016/j.virol.2010.07.007 (2010). [DOI] [PubMed] [Google Scholar]
- Sakoda Y. et al. Reintroduction of H5N1 highly pathogenic avian influenza virus by migratory water birds, causing poultry outbreaks in the 2010-2011 winter season in Japan. J. Gen. Virol. 93, 541–550, 10.1099/vir.0.037572-0 (2012). [DOI] [PubMed] [Google Scholar]
- Bi Y. et al. Highly Pathogenic Avian Influenza A(H5N1) Virus Struck Migratory Birds in China in 2015. Sci. Rep. 5, 10.1038/srep12986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen J. H., Herfst S. & Fouchier R. A. M. How a virus travels the world. Science 347, 616–617, 10.1126/science.aaa6724 (2015). [DOI] [PubMed] [Google Scholar]
- Pasick J. et al. Reassortant Highly Pathogenic Influenza A H5N2 Virus Containing Gene Segments Related to Eurasian H5N8 in British Columbia, Canada, 2014. Sci. Rep. 5, 10.1038/srep09484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y. et al. Spatio-temporal dynamics of global H5N1 outbreaks match bird migration patterns. Geospat. Health 4, 65–78, 10.4081/gh.2009.211 (2009). [DOI] [PubMed] [Google Scholar]
- Liang L. et al. Combining spatial-temporal and phylogenetic analysis approaches for improved understanding on global H5N1 transmission. PLoS One 5, e13575, 10.1371/journal.pone.0013575 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidet N. et al. Potential spread of highly pathogenic avian influenza H5N1 by wildfowl: dispersal ranges and rates determined from large-scale satellite telemetry. J. Appl. Ecol. 47, 1147–1157, 10.1111/j.1365-2664.2010.01845.x (2010). [DOI] [Google Scholar]
- Tian H. et al. Avian influenza H5N1 viral and bird migration networks in Asia. Proc. Natl. Acad. Sci. USA 112, 172–177, 10.1073/pnas.1405216112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. et al. Global and Local Persistence of Influenza A(H5N1) Virus. Emerg. Infect. Dis. 20, 1287–1295, 10.3201/eid2008.130910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M. et al. Anatidae migration in the Western Palearctic and spread of highly pathogenic avian influenza H5N1 virus. Emerg. Infect. Dis. 12, 1650–1656, 10.3201/eid1211.060223 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourouiba L. et al. Spatial dynamics of bar-headed geese migration in the context of H5N1. J. R. Soc. Interface 7, 1627–1639, 10.1098/rsif.2010.0126 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M. P., Maftei D. N., Apostu C. L. & Suru A. R. Association Between Outbreaks of Highly Pathogenic Avian Influenza Subtype H5N1 and Migratory Waterfowl (Family Anatidae) Populations. Zoonoses Public Hlth. 56, 1–9, 10.1111/j.1863-2378.2008.01150.x (2009). [DOI] [PubMed] [Google Scholar]
- Takekawa J. Y. et al. Migration of waterfowl in the East Asian flyway and spatial relationship to HPAI H5N1 outbreaks. Avian Dis. 54, 466–476, 10.1637/8914-043009-Reg.1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S. S., Ersboll A. K., Biswas P. K., Christensen J. P. & Toft N. Spatio-temporal magnitude and direction of highly pathogenic avian influenza (H5N1) outbreaks in Bangladesh. PLoS One 6, e24324, 10.1371/journal.pone.0024324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani M. G., Ward M. P., Giasuddin M., Islam M. R. & Kalam A. The spread of highly pathogenic avian influenza (subtype H5N1) clades in Bangladesh, 2010 and 2011. Prev. Vet. Med. 114, 21–27, 10.1016/j.prevetmed.2014.01.010 (2014). [DOI] [PubMed] [Google Scholar]
- Ward M. P., Maftei D., Apostu C. & Suru A. Geostatistical visualisation and spatial statistics for evaluation of the dispersion of epidemic highly pathogenic avian influenza subtype H5N1. Vet. Res. 39, 22, 10.1051/vetres:2007063 (2008). [DOI] [PubMed] [Google Scholar]
- Gaidet N. et al. Understanding the ecological drivers of avian influenza virus infection in wildfowl: a continental-scale study across Africa. Proc. Biol. Sci. 279, 1131–1141, 10.1098/rspb.2011.1417 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk J. G. B. et al. Juveniles and migrants as drivers for seasonal epizootics of avian influenza virus. J. Anim. Ecol. 83, 266–275, 10.1111/1365-2656.12131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B. et al. Global patterns of influenza a virus in wild birds. Science 312, 384–388, 10.1126/science.1122438 (2006). [DOI] [PubMed] [Google Scholar]
- Fereidouni S. R. et al. Highly Pathogenic Avian Influenza Virus Infection of Mallards with Homo- and Heterosubtypic Immunity Induced by Low Pathogenic Avian Influenza Viruses. Plos One 4, 10.1371/journal.pone.0006706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoye B. J., Fouchier R. A. M. & Klaassen M. Host behaviour and physiology underpin individual variation in avian influenza virus infection in migratory Bewick’s swans. Proc. R. Soc. Lond. B Biol. Sci. 279, 529–534, 10.1098/rspb.2011.0958 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill N. J. et al. Migration strategy affects avian influenza dynamics in mallards (Anas platyrhynchos). Mol. Ecol. 21, 5986–5999, 10.1111/j.1365-294X.2012.05735.x (2012). [DOI] [PubMed] [Google Scholar]
- Li R., Jiang Z. & Xu B. Global spatiotemporal and genetic footprint of the H5N1 avian influenza virus. Int. J. Health Geogr. 13, 10.1186/1476-072x-13-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman S. H. et al. Eco-virological approach for assessing the role of wild birds in the spread of avian influenza H5N1 along the Central Asian Flyway. PLoS One 7, e30636, 10.1371/journal.pone.0030636 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer D. U. et al. Implications of global and regional patterns of highly pathogenic avian influenza virus H5N1 clades for risk management. Vet. J. 190, 309–316, 10.1016/j.tvjl.2010.12.022 (2011). [DOI] [PubMed] [Google Scholar]
- Altizer S., Bartel R. & Han B. A. Animal migration and infectious disease risk. Science 331, 296–302, 10.1126/science.1194694 (2011). [DOI] [PubMed] [Google Scholar]
- Reperant L. A., Fuckar N. S., Osterhaus A. D., Dobson A. P. & Kuiken T. Spatial and temporal association of outbreaks of H5N1 influenza virus infection in wild birds with the 0 degrees C isotherm. PLoS Pathog. 6, e1000854, 10.1371/journal.ppat.1000854 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. K., Thomas C. & Swayne D. E. Variability in pathobiology of South Korean H5N1 high-pathogenicity avian influenza virus infection for 5 species of migratory waterfowl. Vet. Pathol. 47, 495–506, 10.1177/0300985809359602 (2010). [DOI] [PubMed] [Google Scholar]
- Cappelle J. et al. Risks of Avian Influenza Transmission in Areas of Intensive Free-Ranging Duck Production with Wild Waterfowl. EcoHealth 11, 109–119, 10.1007/s10393-014-0914-2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit P. S., Bunn D. A., Pande S. A. & Aly S. S. Modeling highly pathogenic avian influenza transmission in wild birds and poultry in West Bengal, India. Sci. Rep. 3, 10.1038/srep02175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser D. J. et al. Wild bird migration across the Qinghai-Tibetan plateau: a transmission route for highly pathogenic H5N1. PLoS One 6, e17622, 10.1371/journal.pone.0017622 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratanakorn P. et al. Satellite Tracking on the Flyways of Brown-Headed Gulls and Their Potential Role in the Spread of Highly Pathogenic Avian Influenza H5N1 Virus. PLoS One 7, 10.1371/journal.pone.0049939 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman S. H. et al. Migration of whooper swans and outbreaks of highly pathogenic avian influenza H5N1 virus in eastern Asia. PLoS One 4, e5729, 10.1371/journal.pone.0005729 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takekawa J. Y. et al. Movements of wild ruddy shelducks in the Central Asian flyway and their spatial relationship to outbreaks of highly pathogenic avian influenza H5N1. Viruses 5, 2129–2152, 10.3390/v5092129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangeluwe D. The odyssey of the Bewick Swan – another route to Greece – Belgian Ringing Scheme. Available at: http://odnature.naturalsciences.be/bebirds/en/blog_swans/post_174 (Accessed: 1st April 2016) (2015).
- Si Y., Xin Q., Prins H. H., de Boer W. F. & Gong P. Improving the quantification of waterfowl migration with remote sensing and bird tracking. Sci. Bull. 60, 1984–1993, 10.1007/s11434-015-0930-9 (2015). [DOI] [Google Scholar]
- Yasue M., Feare C. J., Bennun L. & Fiedler W. The epidemiology of H5N1 avian influenza in wild birds: Why we need better ecological data. Bioscience 56, 923–929, 10.1641/0006-3568(2006)56[923:teohai]2.0.co;2 (2006). [DOI] [Google Scholar]
- Squires R. B. et al. Influenza Research Database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Resp. 6, 404–416, 10.1111/j.1750-2659.2011.00331.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Continued evolution of highly pathogenic avian influenza A (H5N1): updated nomenclature. Influenza Other Resp. 6, 1–5, 10.1111/j.1750-2659.2011.00298.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B. & Döll P. Development and validation of a global database of lakes, reservoirs and wetlands. J. Hydrol. (Amst.) 296, 1–22, 10.1016/j.jhydrol.2004.03.028 (2004). [DOI] [Google Scholar]
- Jacquez G. et al. ClusterSeer User Guide 2: Software for identifying disease clusters. Ann Arbor, MI: TerraSeer Press, (2002). [Google Scholar]