Abstract
Background
Many species of bivalves exhibit a unique system of mtDNA transmission named Doubly Uniparental Inheritance (DUI). Under this system, species have two distinct, sex-linked mitochondrial genomes: the M-type mtDNA, which is transmitted by males to male offspring and found in spermatozoa, and the F-type mtDNA, which is transmitted by females to all offspring, and found in all tissues of females and in somatic tissues of males. Bivalves with DUI also have sex-specific mitochondrial ORFan genes, (M-orf in the M mtDNA, F-orf in the F mtDNA), which are open reading frames having no detectable homology and no known function. DUI ORFan proteins have previously been characterized in silico in a taxonomically broad array of bivalves including four mytiloid, one veneroid and one unionoid species. However, the large evolutionary distance among these taxa prevented a meaningful comparison of ORFan properties among these divergent lineages. The present in silico study focuses on a suite of more closely-related Unionoid freshwater mussel species to provide more reliably interpretable information on patterns of conservation and properties of DUI ORFans. Unionoid species typically have separate sexes, but hermaphroditism also occurs, and hermaphroditic species lack the M-type mtDNA and possess a highly mutated version of the F-orf in their maternally transmitted mtDNA (named H-orf in these taxa). In this study, H-orfs and their respective proteins are analysed for the first time.
Results
Despite a rapid rate of evolution, strong structural and functional similarities were found for M-ORF proteins compared among species, and among the F-ORF and H-ORF proteins across the studied species. In silico analyses suggest that M-ORFs have a role in transport and cellular processes such as signalling, cell cycle and division, and cytoskeleton organisation, and that F-ORFs may be involved in cellular traffic and transport, and in immune response. H-ORFs appear to be structural glycoproteins, which may be involved in signalling, transport and transcription. Our results also support either a viral or a mitochondrial origin for the ORFans.
Conclusions
Our findings reveal striking structural and functional similarities among proteins encoded by mitochondrial ORFans in freshwater mussels, and strongly support a role for these genes in the DUI mechanism. Our analyses also support the possibility of DUI systems with elements of different sources/origins and different mechanisms of action in the distantly-related DUI taxa. Parallel situations to the novel mitochondrially-encoded functions of freshwater mussel ORFans present in some other eukaryotes are also discussed.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2986-6) contains supplementary material, which is available to authorized users.
Keywords: Mitochondrial DNA, Mitochondrial ORFans, Mitochondrial inheritance, Doubly uniparental inheritance of mitochondria, Bivalvia, Unionoida
Background
Metazoan mitochondrial genomes (mtDNAs) are typically small, circular genomes without introns that encode two ribosomal RNAs, 22 transfer RNAs, and 13 proteins involved in ATP production [1, 2]. Strict maternal inheritance (SMI) of mtDNA is predominant among animals with limited or no paternal contribution [3]. There are, however, many exceptions to these characteristics (e.g. [4–6]). Anomalous gene contents have been found among metazoan mtDNAs, particularly in invertebrates (reviewed in [6]). For example, duplications of typical protein-coding genes have been reported in several mollusc species, including cephalopods, aplacophorans, and bivalves. Additional ‘atypical’ protein-coding genes with non-OXPHOS functions have been reported in cnidarians, sponges, and placozoans (e.g. dnaB, tatC); and mitochondrial ORFans, i.e. genes with unknown function, have been identified in cnidarians, and also in bivalves with doubly uniparental inheritance of mtDNA (DUI), which is the only known exception to SMI in animals [6].
DUI has been reported in marine and freshwater bivalves, specifically the orders Mytiloida, Nuculanoida, Unionoida, and Veneroida [7–10]. Species with DUI possess mitochondrial genomes that are transmitted in a sex-specific manner (known as a female-transmitted F-type and a male-transmitted M-type mtDNA, respectively). Haploid eggs typically contain mitochondria with only F-type mtDNA (but see [11, 12]), while sperm mitochondria, which enter the egg when fertilization occurs, only contain the M-type [10]. If the embryo develops as a female, sperm mitochondria are dispersed and/or destroyed, leading to homoplasmic females (similar to what happens under SMI) [10]. However, if the embryo develops as a male, sperm mitochondria remain grouped together, and are eventually sequestered in the germ line, which becomes homoplasmic for the M mtDNA [13, 14]. Males are therefore heteroplasmic individuals, with mitochondria inherited from their mother containing the F-type mtDNA present throughout their soma, and mitochondria inherited from their father containing the M-type mtDNA in their germ line cells (in males M mtDNA can also be found in variable proportions in somatic tissues [9, 10]). DNA divergence between conspecific M- vs. F-type mitochondrial genomes over 40 % has been found in many species [10].
The mitochondrial genomes of bivalve species with DUI also contain additional, sex-specific protein-coding genes known as mitochondrial ORFans - F-orfs and M-orfs in the F- and M-type mtDNAs, respectively - whose products are exported from the organelle and may be involved in functions other than energy production [15–20]. For example, in freshwater mussels, species typically have separate sexes (gonochorism or dioecy), but hermaphroditic species also occur rarely [21, 22]. In gonochoric species, an absolute correlation has been observed between the presence of DUI and novel sex-specific proteins encoded by the F- and M-type mtDNAs (F-ORF and M-ORF), whereas hermaphroditic species lack the M-type altogether [16]. Hermaphroditic species appear to follow the SMI rule of mitochondrial transmission and individual mussels have only one type of mtDNA, called H-type [16]. The H-type is remarkably similar to (and evolutionarily derived from) the F-type mtDNA of closely-related gonochoric species except for the novel ORFan gene (named H-orf in these species), which is a highly mutated version of the F-orf in their sister taxa [16] (Fig. 1). For these reasons, Breton et al. [16] proposed a connection between DUI and the maintenance of separate sexes in freshwater mussels. However, the link between DUI and sex determination, and the cause of deviation from the “SMI rule” in bivalves remain open questions.
Fig. 1.
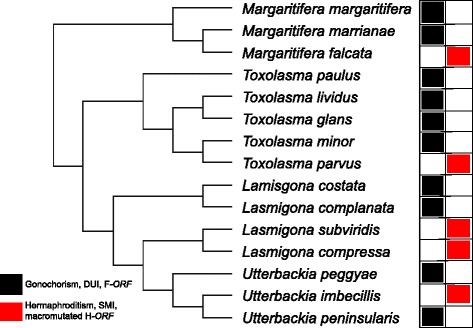
Simplified phylogeny of some gonochoric and hermaphroditic unionoid bivalves redrawn after Breton et al. [16]. The presence of DUI (Doubly Uniparental Inheritance) with F- and M-type mtDNAs in gonochoric species is indicated in black, whereas hermaphroditism with SMI (Strict Maternal Inheritance) is indicated in red. Species in black have F-ORFs containing one predicted transmembrane (TM) helix in their N-terminal portion, whereas hermaphroditic species have macromutated H-ORFs with repeat units and sometimes more than one predicted TM helix [16]
The first in-depth bioinformatic analysis of the structures and potential functions of F-ORF and M-ORF proteins was performed by Milani et al. [18] on the following DUI bivalve species: the marine mussels Musculista senhousia, Mytilus edulis, Mytilus galloprovincialis, Mytilus trossulus and Mytilus californianus (Mytiloida), the marine clam Ruditapes philippinarum (Veneroida), and the freshwater mussel Venustaconcha ellipsiformis (Unionoida). M-orf and F-orf nucleotide sequences were found to be highly variable, with mostly non-synonymous mutations, indicating rapid evolution and supporting previous claims that these protein-coding genes are the fastest-evolving mitochondrial genes in bivalves with DUI [16–18]. Despite this fast rate of evolution, structural similarities in their translated amino acid sequences were observed among species and ORFan proteins were predicted to share similar functions. For example, F-ORFs were largely predicted to bind and interact with nucleic acids, associate with membranes for cell adhesion and/or signalling, or play a role in immune response. M-ORFs were also predicted to be membrane-associated and interact with nucleic acids, primarily for signalling, cell differentiation and development, and also for cytoskeleton formation and dynamics, ubiquitination, apoptosis, and immune response [18]. Even if hit probabilities in the proteins were sometimes low and the regions of similarity were of short lengths, several clues suggested that the respective novel ORFans originated from endogenization of viral DNA [18, 19]. However, obtaining satisfactory alignments including F-ORFs and M-ORFs from all species was impossible due to the highly divergent nature of these proteins [18]. This indicated either that their fast rate of evolution erased any evidence of ORFan sequence similarities (homology) among species or that the ORFans originated from independent virus endogenization events [18]. It is also conceivable that the ORFans originated from different sources/processes but evolved similar function(s) in these distantly related DUI species, particularly if DUI evolved independently more than once [18]. Other than a viral origin, there are at least three other possibilities for the source of these mitochondrial ORFans: they may have originated from (i) a duplicated and diverged mitochondrial gene, (ii) a gene composed from previously non-coding mitochondrial sequences, or (iii) a gene transferred from the nucleus to the mitochondrion (e.g. [17]).
Unfortunately, it is not currently possible to confirm whether or not the mitochondrial ORFans in phylogenetically unrelated DUI species are homologous because of their high divergence and our incomplete knowledge regarding their distribution in bivalves. One option to better understand the origin(s) and function(s) of a subset of these ORFans is to compare a suite of more closely related sequences within a single order of bivalves. Freshwater mussels (Unionoida) offer an excellent opportunity for this for at least two reasons: (1) they are an evolutionarily old group of bivalves, suggesting that their ORFans have an ancient origin and that DUI in this group might be one of the first examples of this phenomenon in bivalves [23], and (2) complete F and M genomes or F-orf, M-orf and H-orf sequences are available for several gonochoric species and five independently evolved hermaphroditic species (e.g. [16, 23, 24]). All of these taxa belong to the family Unionidae (except for Margaritifera falcata [Margaritiferidae]), but recently we have sequenced the F and M mtDNAs from Cumberlandia monodonta (Margaritiferidae) and Hyridella menziesii (Hyriidae) (these genomes have been sequenced at the sequencing platform of McGill University [Montreal, Canada] using the genome sequencer FLX sequencing service), and these genomes possess an F-orf and an M-orf, suggesting that these unique genes have been present and functioning continuously for >200 million years in this group ([16, 23]; Guerra et al. unpublished).
The present study aims to predict the origin, structure, and function of the F-ORF and M-ORF protein sequences in Unionoida, and to analyze the H-ORFs for the first time. Our results confirm that they are encoded by the fastest evolving genes in unionoid mitochondrial genomes, that they share structural and functional similarities, and that their respective ORFans could have a viral or a mitochondrial origin, leading us to revisit the evolutionary scenario of multiple origins of DUI [18, 19].
Methods
Sequences used in the analyses
ORFan, cox1, and atp8 nucleotide sequences of unionoid bivalve species were either obtained from the National Center for Biotechnology Information (NCBI) or from newly sequenced mitochondrial genomes (i.e. H. menziesii and C. monodonta; Guerra et al. unpublished). All species and GenBank entries used in this study are listed in Table 1 (note that M-orf sequences for Lasmigona complanata, Margaritifera margaritifera and Toxolasma lividus have not been obtained; Additional file 1: Table S1). The sequences were translated with ORF Finder [25] using the invertebrate mitochondrial genetic code and analyzed at the nucleotide and/or amino acid level (see below). Because M-ORF and F-ORF protein sequences vary little within a species, only one sequence was used for each gonochoric species. H-ORF sequences are highly variable within species [16], and so multiple sequences were analyzed per species to provide a more complete picture of intraspecific H-ORF evolution and potential functionality.
Table 1.
Sequences analyzed in the present study for gonochoric species with DUI and hermaphroditic species with SMI
| Species | mtDNA type | Accession number | ORF names |
|---|---|---|---|
| Subfamiliy Ambleminae | |||
| Quadrula quadrula | M | FJ809751.1 | Qqu-Morf |
| M | FJ809751.1 | Qqu-Mcox1 | |
| M | FJ809751.1 | Qqu-Matp8 | |
| F | FJ809750.1 | Qqu-Forf | |
| F | FJ809750.1 | Qqu-Fcox1 | |
| F | FJ809750.1 | Qqu-Fatp8 | |
| Toxolasma lividus | F | HM849457.1 | Tli-Forf |
| Toxolasma parvum | H | KU728097 | Tpa-Horf |
| Venustaconcha ellipsiformis | M | FJ809752.1 | Vel-Morf |
| M | FJ809752.1 | Vel-Mcox1 | |
| M | FJ809752.1 | Vel-Matp8 | |
| F | FJ809753.1 | Vel-Forf | |
| F | FJ809753.1 | Vel-Fcox1 | |
| F | FJ809753.1 | Vel-Fatp8 | |
| Subfamiliy Anodontinae | |||
| Anodonta anatina | M | KF030962.1 | Aan-Morf |
| F | KF030964.1 | Aan-Forf | |
| Subfamiliy Gonideinae | |||
| Inversidens japanensis | M | AB055624.1 | Ija-Morf |
| M | AB055624.1 | Ija-Mcox1 | |
| M | AB055624.1 | Ija-Matp8 | |
| F | AB055625.1 | Ija-Forf | |
| F | AB055625.1 | Ija-Fcox1 | |
| F | AB055625.1 | Ija-Fatp8 | |
| Solenaia carinatus | M | KC848655.1 | Sca-Morf |
| M | KC848655.1 | Sca-Mcox1 | |
| M | KC848655.1 | Sca-Matp8 | |
| F | KC848654.1 | Sca-Forf | |
| F | KC848654.1 | Sca-Fcox1 | |
| F | KC848654.1 | Sca-Fatp8 | |
| Subfamiliy Hyriidae | |||
| Hyridella menziesii | M | KU728093 | Hme-Morf |
| M | KU728094 | Hme-Mcox1 | |
| F | KU728092 | Hme-Forf | |
| F | AY785394.1 | Hme-Fcox1 | |
| Subfamiliy Margaritiferinae | |||
| Cumberlandia monodonta | M | KU728095 | Cmo-Morf |
| M | KU728096 | Cmo-Mcox1 | |
| F | HM849375.1 | Cmo-Forf | |
| F | KF647374.1 | Cmo-Fcox1 | |
| Margaritifera falcata | H | HM849545.1 | Mfa-Horf (top- bottom 1–4) |
| H | HM856634.1 | ||
| H | HM849547.1 | ||
| H | HM849548.1 | ||
| H | HM856634.1 | Mfa-Hcox1 (top-bottom 1–2) | |
| H | NC_015476.1 | ||
| Margaritifera margaritifera | F | HM849399.1 | Mma-Forf |
| F | HM849095.1 | Mma-Fcox1 | |
| Subfamiliy Unioninae | |||
| Lasmigona complanata | F | HM849393.1 | Lco-Forf |
| Lasmigona compressa | H | HM849534.1 | Lco-Horf (top-bottom 1–2) |
| H | HM849535.1 | ||
| H | HM856638.1 | Lco-Hcox1 (top-bottom 1–2) | |
| H | NC_015481.1 | ||
| Lasmigona subviridis | H | HM849542.1 | Lsu-Horf (top-bottom 1–2) |
| H | HM849543.1 | ||
| Pyganodon grandis | M | FJ809755.1 | Pgr-Morf |
| M | FJ809755.1 | Pgr-Mcox1 | |
| M | FJ809755.1 | Pgr-Matp8 | |
| F | FJ809754.1 | Pgr-Forf | |
| F | FJ809754.1 | Pgr-Fcox1 | |
| F | FJ809754.1 | Pgr-Fatp8 | |
| Utterbackia imbecillis | H | HM849591.1 | Uim-Horf (top-bottom 1–7) |
| H | HM849595.1 | ||
| H | HM849594.1 | ||
| H | HM849601.1 | ||
| H | HM849606.1 | ||
| H | HM849597.1 | ||
| H | HM849584.1 | ||
| H | NC_015479 | Uim-Hcox1 (top-bottom 1–2) | |
| H | HM856637.1 | ||
| Utterbackia peninsularis | M | HM856635.1 | Upe-Morf |
| M | HM856635.1 | Upe-Mcox1 | |
| M | HM856635.1 | Upe-Matp8 | |
| F | HM856636.1 | Upe-Forf | |
| F | HM856636.1 | Upe-Fcox1 | |
| F | HM856636.1 | Upe-Fatp8 | |
Note: M M mtDNA in a DUI gonochoric breeding system, F F mtDNA in a DUI gonochoric breeding system, H H mtDNA in a non-DUI hermaphroditic breeding system
Analyses of ORFan sequences and protein secondary structures
Alignments of ORFan, cox1, and atp8 nucleotide and translated protein sequences were performed with M-COFFEE (DNA) and PSI-COFFEE (proteins) [26]. Nucleotide and amino acid p-distances, as well as a codon-based test of positive selection using the Nei-Gojobori method [27], were calculated using MEGA6 [28] with variance estimated using 500 bootstrap repetitions. The program VISTA [29] was used to display the level of sequence conservation between M vs. M, F vs. F, and F vs. H complete mitochondrial genomes. M- and F-type mtDNAs were not compared due to their previous characterization that showed extreme intraspecific sequence divergences [16, 23]. Hydropathy profiles of each amino acid sequence were calculated with the ProtScale tool at ExPASy [30] using the method of Kyte and Doolittle [31]. Putative transmembrane (TM) helices were identified using a variety of protein signature recognition methods implemented by the following programs: Phobius [32], InterProScan (TMHMM) [33], TMPred [34], TOPCONS [35], and Predict Protein [36].
Functional analyses of ORFan proteins
Evidence of signal peptides (SPs) was sought using Phobius [26], InterProScan [33], PrediSi [37], and SignalP [38]. Motif Scan [39] and HHpred [40] were used to search for known functional sequence motifs and domains. TPRpred [41] was used to search for potential tetratricopeptide repeat (TPR) or pentatricopeptide repeat (PPR) motifs. The following procedures were used to predict the function of ORFan proteins: (1) we performed BLASTp, tBLASTx, and PSI-BLAST searches against NCBI entire non-redundant protein database (NRDB) and against mitochondrial proteins only (last accessed July, 2015) with default parameters [42], as well as FASTA and PSI-BLAST searches against UniProt (release 2015_05) with default parameters, at the EBI websites [43] and [42], respectively; (2) we used hmmbuild (v3.1b2; downloaded from http://hmmer.janelia.org) [44] to generate two HMM profiles from both the F-ORF and M-ORF protein alignments (four profiles in total; see below) (H-ORFs were not considered given their scattered phylogenetic distribution and independent evolutionary histories) using default and custom parameters (for the latter procedure, the options --fast --symfrac 0 --fragthresh 0 --wnone --enone were used), and performed profile HMM – sequence comparison against UniProtKB, Swissprot, PDB, QfO, and Pfamseq databases using HMMER hmmsearch [44] with default parameters (E-value cutoff = 0.001); (3) for profile HMM – profile HMM comparisons, we used HHpred, which compares HMM profiles with databases of HMMs representing proteins with known structure (e.g. PDB, SCOP) or annotated protein families (e.g. PFAM, SMART, CDD, COGs, KOGs); and (4) the following programs were also used to predict the function of ORFan proteins: @tome2, which predicts tertiary structure and searches for similarity to proteins with structures solved [45]; I-TASSER, which uses a hierarchical protein structure modeling approach that is based on the secondary-structure enhanced profile–profile threading alignment [46]; and PredictProtein, which predicts aspects of protein structure (secondary structure, solvent accessibility, transmembrane helices [TMSEG] and strands, coiled-coil regions, disulfide bonds and disordered regions) and function (identification of functional regions, homology-based inference of Gene Ontology terms, comprehensive subcellular localization prediction, protein-protein binding sites, protein-polynucleotide binding sites and predictions of the effect of point mutations [non-synonymous SNPs] on protein function) [36]. For BLASTp and PredictProtein all matches with E-values <1.0 were kept, while for position-specific iterative or PSI-BLAST all matches with E-values <0.01 were kept as recommended by the program (except for PSI-BLAST analyses against NCBI mitochondrial genes only, where E-values <1.0 were kept, see below). For I-TASSER, all top templates and structural analogs were recorded. All @tome2 results were kept. Motif Scan results not marked as “questionable” or “weak” were kept. Hits described as “uncharacterized,” “putative,” “unknown,” or “predicted” were not kept.
Results
Rate of evolution of ORFan genes and proteins
The amino acid sequences of ORFans were generally not well conserved among unionoid species. As seen in Fig. 2, a good comprehensive alignment including all M-ORF sequences was not possible due to their high divergence, however, sequences from the same subfamily produced good alignments (Fig. 2b–d). A common feature of M-ORFs is that they are all lysine-rich proteins frequently with poly-K strings, a characteristic that is apparently absent in F-ORF and H-ORF amino acid sequences. Similar to M-ORF sequences, F-ORF sequences from the same subfamily or family produced better alignments than for all species (Fig. 3). Finally, because phylogenetic analysis indicates that the H-ORFs were formed by five independent evolutionary events [15], interspecific alignment is not possible for hermaphrodite ORFans, and alignments between hermaphrodite H-ORFs and closely-related gonochoric species F-ORFs were mainly of low quality (Additional file 2: Figure S1). In instances where multiple H-ORFs were available for a given species of hermaphrodite, these protein sequences were only aligned intraspecifically.
Fig. 2.
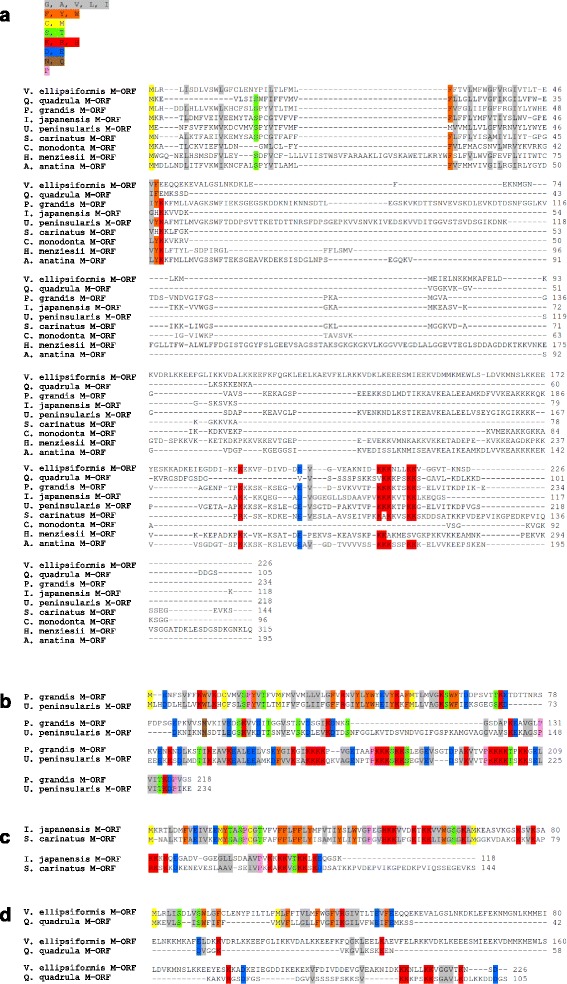
Alignments of M-ORF protein sequences. Global alignments and alignments for each subfamily are shown. a All M-ORF sequences, b M-ORFs from the subfamily Unioninae, c M-ORFs from the subfamily Gonideinae, d M-ORFs from the subfamily Ambleminae. Colour coding is applied to amino acid groups conserved in ≥70 % of sequences. Grey, aliphatic amino acids; orange, aromatic amino acids; yellow, sulfur amino acids; green, amino acids bearing a hydroxyl group; red, basic amino acids; blue, acidic amino acids; brown, amino acids with an amide group; pink, cyclic amino acids
Fig. 3.
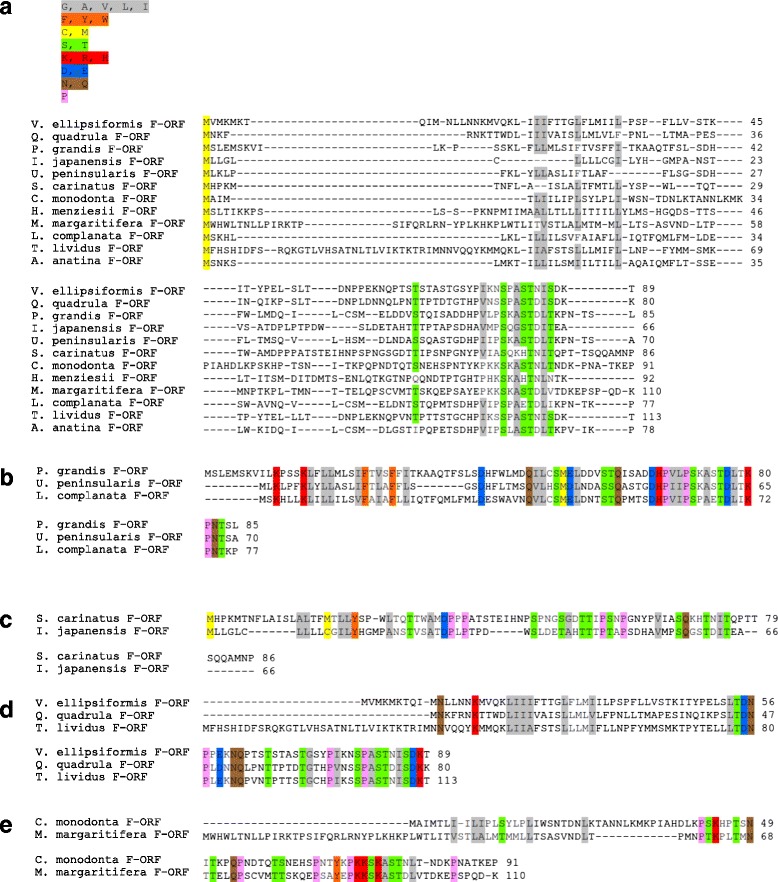
Alignments of F-ORF protein sequences. Global alignments and alignments for each subfamily are shown. a all F-ORF sequences, b F-ORFs from the subfamily Unioninae, c F-ORFs from the subfamily Gonideinae, d F-ORFs from the subfamily Ambleminae (e) F-ORFs from the subfamily Margaritiferidae. Colour coding is applied to amino acid groups conserved in ≥70 % of sequences. Grey, aliphatic amino acids; orange, aromatic amino acids; yellow, sulfur amino acids; green, amino acids bearing a hydroxyl group; red, basic amino acids; blue, acidic amino acids; brown, amino acids with an amide group; pink, cyclic amino acids
The p-distances for nucleotide and amino acid ORFan sequences as well as the outcome of the test of positive selection are reported in Table 2 (M-ORFs and F-ORFs) and Table 3 (H-ORFs), along with the values for cox1 and atp8 sequences taken from the same sex-specific mtDNAs. Table 4 shows the p-distances for within-genus comparisons of F-ORFs versus H-ORFs. In all cases, the novel ORFs have interspecific p-distances several times higher than cox1 and higher than atp8, which typically represent the slowest- and fastest- evolving mitochondrial protein-coding genes, respectively, in both freshwater mussels and in animals in general [16, 47]. For all groups of sequences, we observed no significant probability of rejecting the null hypothesis of neutral selection in favor of the alternative hypothesis of positive selection. The level of sequence conservation between M vs. M, F vs. F, and F vs. H complete mitochondrial genomes also confirmed that mitochondrial ORFans are the fastest evolving genes in the mtDNA of freshwater mussels with DUI (Additional file 3: Figure S2).
Table 2.
p-distances (p-D) and standard error (SE) values for mitochondrial M-orfs, F-orfs, cox1 and atp8 in freshwater mussel subfamilies
| Subfamily | Gene (N) | Nucleotide | Amino acid | p | ||
|---|---|---|---|---|---|---|
| p-D | SE | p-D | SE | |||
| Unioninae | F-orf (3) | 0.355 | 0.023 | 0.467 | 0.047 | 1.000 |
| F-cox1 (2) | 0.103 | 0.007 | 0.014 | 0.005 | 1.000 | |
| F-atp8 (2) | 0.300 | 0.011 | 0.333 | 0.015 | 1.000 | |
| M-orf (2) | 0.350 | 0.018 | 0.502 | 0.034 | 1.000 | |
| M-cox1 (2) | 0.165 | 0.010 | 0.094 | 0.012 | 1.000 | |
| M-atp8 (2) | 0.250 | 0.010 | 0.267 | 0.013 | 1.000 | |
| Gonideinae | F-orf (2) | 0.469 | 0.033 | 0.692 | 0.058 | 1.000 |
| F-cox1 (2) | 0.132 | 0.008 | 0.033 | 0.008 | 1.000 | |
| F-atp8 (2) | 0.400 | 0.025 | 0.222 | 0.010 | 1.000 | |
| M-orf (2) | 0.384 | 0.025 | 0.552 | 0.044 | 1.000 | |
| M-cox1 (2) | 0.175 | 0.009 | 0.130 | 0.015 | 1.000 | |
| M-atp8 (2) | 0.301 | 0.019 | 0.421 | 0.039 | 1.000 | |
| Ambleminae | F-orf (3) | 0.351 | 0.024 | 0.508 | 0.041 | 1.000 |
| F-cox1 (2) | 0.128 | 0.009 | 0.033 | 0.007 | 1.000 | |
| F-atp8 (2) | 0.278 | 0.018 | 0.370 | 0.031 | 1.000 | |
| M-orf (2) | 0.421 | 0.027 | 0.687 | 0.047 | 1.000 | |
| M-cox1 (2) | 0.179 | 0.010 | 0.145 | 0.015 | 1.000 | |
| M-atp8 (2) | 0.211 | 0.012 | 0.233 | 0.017 | 1.000 | |
| Margaritiferinae | F-orf (2) | 0.393 | 0.029 | 0.705 | 0.050 | 1.000 |
| F-cox1 (2) | 0.164 | 0.009 | 0.068 | 0.009 | 1.000 | |
Note: N number of sequences used. The probability of rejecting the null hypothesis of strict-neutrality (d N = d S) in favor of the alternative hypothesis (d N > d S) (in the p column) is shown. d S and d N are the numbers of synonymous and nonsynonymous substitutions per site, respectively
Table 3.
p-distances (p-D) and standard error (SE) values of mitochondrial H-orfs and cox1 in hermaphroditic freshwater mussels
| Species | Gene (N) | Nucleotide | Amino acid | p | ||
|---|---|---|---|---|---|---|
| p-D | SE | p-D | SE | |||
| Utterbackia imbecillis | H-orf (7) | 0.070 | 0.008 | 0.181 | 0.022 | 1.000 |
| cox1 (2) | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 | |
| Margaritifera falcata | H-orf (4) | 0.003 | 0.002 | 0.004 | 0.004 | 1.000 |
| cox1 (2) | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 | |
| Lasmigona compressa | H-orf (2) | 0.029 | 0.007 | 0.065 | 0.017 | 1.000 |
| cox1 (2) | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 | |
| Lasmigona subviridis | H-orf (2) | 0.016 | 0.005 | 0.021 | 0.010 | 1.000 |
Note: N number of sequences used. Multiple cox1 sequences were not available for L. subviridis. The probability of rejecting the null hypothesis of strict-neutrality (d N = d S) in favor of the alternative hypothesis (d N > d S) (in the p column) is shown. d S and d N are the numbers of synonymous and nonsynonymous substitutions per site, respectively
Table 4.
p-distances (p-D) and standard error (SE) values of mitochondrial F-orfs vs H-orfs and Fcox1 vs Hcox1 in comparisons between gonochoric vs. closely related hermaphroditic freshwater mussel species
| Species | Genes | Nucleotide | Amino acid | ||
|---|---|---|---|---|---|
| p-D | SE | p-D | SE | ||
| Utterbackia peninsularis vs U. imbecillis | |||||
| F-ORF vs. H-ORF1 | 0.338 | 0.034 | 0.691 | 0.055 | |
| F-ORF vs. H-ORF2 | 0.310 | 0.032 | 0.721 | 0.054 | |
| F-ORF vs. H-ORF3 | 0.343 | 0.031 | 0.743 | 0.051 | |
| F-ORF vs. H-ORF4 | 0.335 | 0.034 | 0.729 | 0.054 | |
| F-ORF vs. H-ORF5 | 0.333 | 0.031 | 0.714 | 0.052 | |
| F-ORF vs. H-ORF6 | 0.333 | 0.031 | 0.714 | 0.052 | |
| F-ORF vs. H-ORF7 | 0.310 | 0.030 | 0.739 | 0.055 | |
| Mean | 0.329 | 0.030 | 0.722 | 0.052 | |
| F-COX1 vs. H-COX1-1 | 0.547 | 0.012 | 0.020 | 0.006 | |
| F-COX1 vs. H-COX1-2 | 0.547 | 0.012 | 0.020 | 0.006 | |
| Mean | 0.547 | 0.0012 | 0.020 | 0.006 | |
| Margaritifera margaritifera vs M. falcata | |||||
| F-ORF vs. H-ORF1 | 0.339 | 0.025 | 0.491 | 0.048 | |
| F-ORF vs. H-ORF2 | 0.336 | 0.026 | 0.491 | 0.049 | |
| F-ORF vs. H-ORF3 | 0.358 | 0.024 | 0.491 | 0.049 | |
| F-ORF vs. H-ORF4 | 0.336 | 0.026 | 0.491 | 0.049 | |
| Mean | 0.342 | 0.025 | 0.491 | 0.049 | |
| F-COX1 vs. H-COX1-1 | 0.469 | 0.022 | 0.000 | 0.000 | |
| F-COX1 vs. H-COX1-2 | 0.469 | 0.021 | 0.000 | 0.000 | |
| Mean | 0.469 | 0.021 | 0.000 | 0.000 | |
| Lasmigona complanata vs L. compressa | |||||
| F-ORF vs. H-ORF1 | 0.218 | 0.028 | 0.394 | 0.059 | |
| F-ORF vs. H-ORF2 | 0.255 | 0.027 | 0.395 | 0.055 | |
| Mean | 0.237 | 0.027 | 0.395 | 0.057 | |
| Lasmigona complanata vs L. subviridis | |||||
| F-ORF vs. H-ORF1 | 0.269 | 0.029 | 0.429 | 0.054 | |
| F-ORF vs. H-ORF2 | 0.295 | 0.029 | 0.442 | 0.055 | |
| Mean | 0.282 | 0.029 | 0.436 | 0.054 | |
| Toxolasma lividus vs T. parvum | F-ORF vs. H-ORF | 0.443 | 0.027 | 0.736 | 0.044 |
Note: Bold numbers indicate mean values
Conserved structures in ORFan protein sequences
One TM helix was predicted near the N-terminus of all M-ORFs (Fig. 4 and Additional file 1: Table S2), except for H. menziesii M-ORF sequence, for which one N-terminal and two additional TM helices were predicted. PrediSi and SignalP both returned predicted SPs for all M-ORF sequences, however, the programs rarely agreed about the length of the predicted SP (Additional file 1: Table S3). One TM helix near the N-terminus was also predicted in all F-ORF sequences, with an SP predicted to overlap with this TM structure, except in the case of the T. lividus F-ORF for which the location of the SP was uncertain (Fig. 5 and Additional file 1: Tables S2 and S3). All H-ORFs contained one predicted TM helix near the N-terminus as well, except for U. imbecillis H-ORFs that contained multiple predicted TM helices, but only the location of the first TM helix (closest to the N-terminus) was predicted with high confidence (Fig. 5 and Additional file 1: Table S4). U. imbecillis H-ORFs also returned variable SP predictions, whereas all other H-ORF sequences contain one predicted SP overlapping with the N-terminal TM helix (Additional file 1: Table S5). Although they could not be confidently aligned (see Additional file 3: Figure S2), F-ORFs and H-ORFs of closely related species showed some structural similarities in the localization of the TM helices and SPs (Fig. 5). Importantly, all H-ORFs contain tandem repeats (L. compressa possesses between 3 to 7 tandemly repeated sequence motifs of 20 or 21aa; L. subviridis 7 to 9 repeats of 17aa; T. parvum 2 to 3 repeats of 47aa; M. falcata 2 to 3 repeats of 11aa; and U. imbecillis 2 to 4 repeats of 11 or 21aa), which are not found in F-ORFs and account for most of the difference in length between F-ORFs and H-ORFs of closely related species (Additional file 3: Figure S2).
Fig. 4.
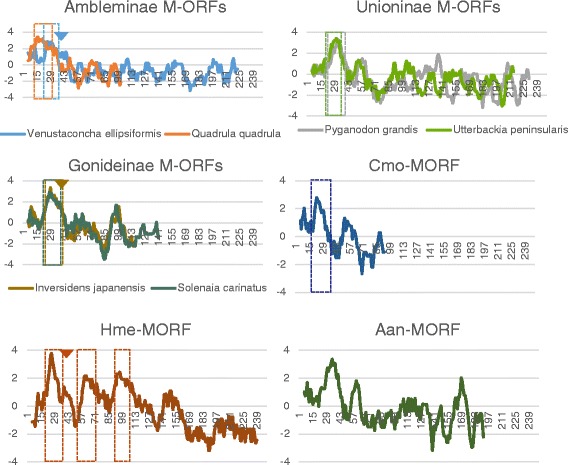
Hydrophobicity profiles of M-ORFs. Boxes indicate predicted TM helices, arrowheads indicate the end of predicted SPs. X-axis is amino acid position, Y-axis is hydrophobicity. Aan, Anodonta anatina; Cmo, Cumberlandia monodonta; Hme, Hyridella menziesii
Fig. 5.
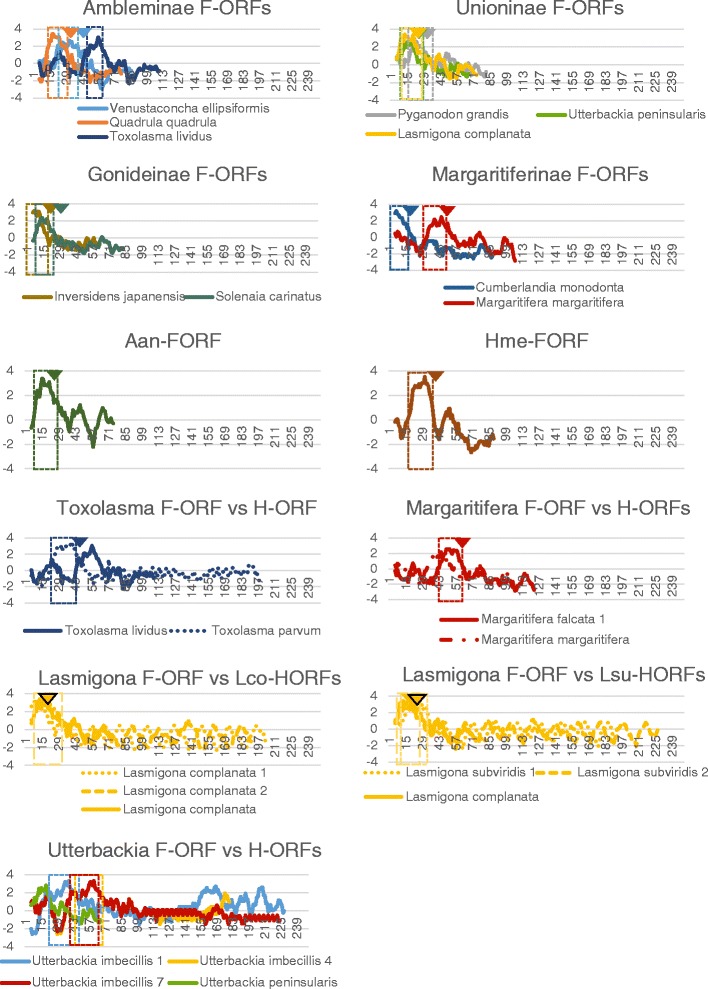
Hydrophobicity profiles of F-ORFs (top) and H-ORFs vs. F-ORFs (bottom). Boxes indicate predicted TM helices, arrowheads indicate the end of predicted SPs. X-axis is amino acid position, Y-axis is hydrophobicity. Aan, Anodonta anatina; Cmo, Cumberlandia monodonta; Hme, Hyridella menziesii; Lco-HORFs, Lasmigona compressa H-ORFs; Lsu-HORFs, Lasmigona subviridis H-ORFs. For hermaphroditic species, only sequences with different hydrophobicity profiles are shown
Motif and functional domain scans: frequently recurring HHpred hits and potential ligand-binding sites
Six HHpred hits consistently appeared highly ranked in the results of M-ORFs, F-ORFs and H-ORFs: (1) prepilin-type processing-associated H-X9-DG domain, (2) outer membrane insertion C-terminal signal, (3) LPXTG cell wall anchor domain, (4) X-X-X-Leu-X-X-Gly heptad repeats, (5) GlyGly-CTERM domain, and (6) a pentatricopeptide repeat (PPR) domain. Probabilities were all >92 % (which the developers state can be interpreted literally [40]), and ranks were typically 1–6 in variable order, with very few of these hits falling outside of the top 10 (Additional file 1: Tables S6 and S7). Fig. 6 shows the position of these six hits in the protein sequences analyzed. Other less recurring motifs and domains are presented in detail in Additional file 1: Table S8 and S9.
Fig. 6.
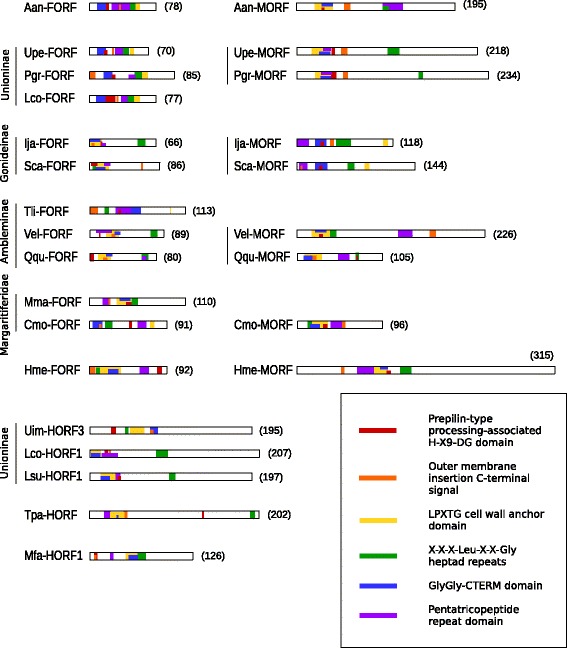
Position of motifs frequently recurring in HHpred hits. Protein length in amino acids is indicated in parentheses. One representative sequence was chosen for each hermaphroditic species
Inferred homologies and prediction of binding sites both indicated that ORFan proteins may bind several ligands (Table 5). All M-ORFs returned hits to protein-binding, DNA-binding and RNA-binding proteins, all F-ORFs returned hits to protein-binding and RNA-binding proteins, and all H-ORF sequences returned hits to protein-binding, DNA-binding, RNA-binding and carbohydrates-binding proteins.
Table 5.
Summary of hits to ligand-binding sites in M-ORFs, F-ORFs and H-ORFs
| Protein | DNA | RNA | Protein | Carbohydrate | Ion | Lipid | ATP |
|---|---|---|---|---|---|---|---|
| Vel-MORF | X | X | X | X | X | X | |
| Qqu-MORF | X | X | X | X | |||
| Pgr-MORF | X | X | X | X | X | X | |
| Ija-MORF | X | X | X | X | X | ||
| Upe-MORF | X | X | X | X | X | ||
| Sca-MORF | X | X | X | X | X | ||
| Cmo-MORF | X | X | X | X | X | ||
| Hme-MORF | X | X | X | X | X | X | |
| Aan-MORF | X | X | X | X | X | ||
| Total | 9 | 9 | 9 | 3 | 8 | 3 | 6 |
| Vel-FORF | X | X | X | X | X | X | |
| Qqu-FORF | X | X | X | X | X | X | X |
| Pgr-FORF | X | X | X | X | X | X | |
| Ija-FORF | X | X | X | X | X | ||
| Upe-FORF | X | X | X | X | X | ||
| Sca-FORF | X | X | X | X | X | X | |
| Cmo-FORF | X | X | X | ||||
| Hme-FORF | X | X | X | X | X | ||
| Lco-FORF | X | X | X | X | X | X | |
| Tli-FORF | X | X | X | X | X | X | |
| Mma-FORF | X | X | X | X | |||
| Aan-FORF | X | X | X | X | X | ||
| Total | 11 | 12 | 12 | 7 | 10 | 4 | 8 |
| Uim-HORF1 - 3 | X | X | X | X | X | X | |
| Uim-HORF4 - 7 | X | X | X | X | X | ||
| Mma-HORF1, 2, 4 | X | X | X | X | X | ||
| Mma-HORF3 | X | X | X | X | |||
| Tpa-HORF | X | X | X | X | X | X | X |
| Lco-HORF1 | X | X | X | X | |||
| Lco-HORF2 | X | X | X | X | X | X | |
| Lsu-HORF1 - 2 | X | X | X | X | X | ||
| Total | 14 | 14 | 14 | 10 | 14 | 2 | 6 |
Note: Bold numbers indicate mean values
Prediction of molecular function: hits to viral proteins
As mentioned above, a recent study proposed a viral origin for the mitochondrial ORFans in DUI bivalves [18]. Therefore, we first scanned our results obtained with all programs for protein function prediction, i.e. BLAST, HMMER, HHpred, @tome2, I-TASSER, and PredictProtein, for supported hits to viral proteins (Table 6). For H-ORFs, M. falcata primarily returned envelope proteins, L. subviridis returned capsid and envelope proteins, L. compressa returned proteins that interact with receptors, T. parvum returned a protein that regulates the degradation of a receptor, and U. imbecillis returned capsid proteins and other structural proteins. M-ORFs returned nucleoproteins (A. anatina and H. menziesii), membrane proteins (I. japanensis and S. carinatus), and proteins with a role in replication, life cycle, and apoptosis (A. anatina, U. peninsularis, I. japanensis and V. ellipsiformis). F-ORF hits were mostly parts of the viral capsid and viral envelope (S. carinatus, T. lividus and M. margaritifera), receptors/fibre proteins (M. margaritifera and C. monodonta), or proteins involved in cell cycle and translation (P. grandis and I. japanensis).
Table 6.
Hits to viral proteins from structural prediction analyses
| Gene | Hit | Function | Position |
|---|---|---|---|
| Aan-MORF | Nucleoprotein, Andes virus [Atome 2; 41.16] | Nucleoprotein | NA |
| Regulatory protein MNT, Enterobacteria phage P22 [Atome 2; 21.14] | Gene regulation | NA | |
| Upe-MORF | Uncharacterized protein 56B, Sulfolobus islandicus [Atome 2; 27.96] | Transcription repressor | NA |
| Pgr-MORF | Matrix protein 1, Influenza A virus [Atome 2; 39.16] | Matrix protein | NA |
| Helix-destabilizing protein, Enterobacteria phage T7 [Atome 2; 18.55] | DNA binding protein | NA | |
| Ija-MORF | Nonstructural protein 5A, Bovine viral diarrhea virus 1-CP7 [Atome 2; 33.37] | Membrane protein | NA |
| Functional anti-apoptotic factor vBCL-2 homolog, Human herpesvirus 8 [Atome 2; 27.14] | Apoptosis | NA | |
| Sca-MORF | Nonstructural protein 5A, Bovine viral diarrhea virus 1-CP7 [Atome 2; 22.35] | Membrane protein | NA |
| Vel-MORF | Macrophage galactose N-acetyl-galactosamine specific lectin 2 [Hhpred; 93.40] | C-type lectin | 20–171 |
| RhUL123, Macacine herpesvirus 3 [I-TASSER; TM score 0.671] | Viral life cycle | NA | |
| Phosphoprotein, Measels virus [Atome 2; 49.33] | Unknown function | NA | |
| Tail needle protein gp26, Enterobacteria phage P22 [Atome 2; 48.96] | Fibrous protein | NA | |
| Qqu-MORF | Virion RNA polymerase, Bacteriophage n4 [I-TASSER; TM score 0.542] | Transferase | NA |
| Cmo-MORF | No hits to viral proteins | ||
| Hme-MORF | Nucleoprotein, Andes virus [Atome 2; 63.91] | Nucleoprotein | NA |
| Aan-FORF | No hits to viral proteins | ||
| Upe-FORF | BM2 protein, Influenza B virus (B/Taiwan/70061/2006) [Atome 2; 42.29] | Transport protein | NA |
| Pgr-FORF | V-cyclin, Human herpesvirus 8 [I-TASSER; norm. TM score 0.517] | Cell cycle | NA |
| Lco-FORF | Herpes simplex virus protein ICP47, Herpes simplex virus (type 1/strain 17) [Atome 2; 46.61] | Membrane protein | NA |
| Ija-FORF | Non-structural RNA-binding protein 34, Simian rotavirus A/SA11 (2) [Atome 2; 48.04, 28.60] | Translation | NA |
| Sca-FORF | Major capsid protein (protein P3), Enterobacteria phage PRD1 [Atome 2; 80.01] | Capsid protein | NA |
| Tli-FORF | Envelope protein E, Dengue virus 2 Thailand/16681/84 [Atome 2; 46.45] | Envelope protein | NA |
| Vel-FORF | V1V2 region of HIV-1 on 1FD6 scaffold, Human immunodeficiency virus 1 [Atome 2; 57.65] | Immune system | NA |
| Qqu-FORF | HIV-1 matrix protein, Human immunodeficiency virus 1 (2) [Atome 2; 83.13, 72.79] | Matrix protein | NA |
| Mma-FORF | ODV-E18: Occlusion-derived virus envelope protein ODV-E18 (2) [Hhpred; 72.05, 62.79] | Envelope protein | 21–62 |
| Adenovirus fibre, Human adenovirus 2 [Atome 2; 27.29] | Fibre protein | 23–55 | |
| Fibre protein 2 (receptor-binding domain), Human adenovirus 41 [I-TASSER; 18.06] | Fibre protein, receptor binding | NA | |
| NA | |||
| Cmo-FORF | Virus attachment protein globular domain (49835) SCOP seed sequence: d1h7za [Hhpred; 21.78] | Viral attachment, entry into host cell | 50–68 |
| Adenovirus fibre protein; cell receptor recognition, receptor, Human adenovirus type 3 [Hhpred; 21.71] | Fibre protein, Cell receptor recognition | 44–68 | |
| Fibre protein, Human adenovirus 37 [Atome 2; 31.21] | NA | ||
| Fibre protein, Human adenovirus 2 [Atome 2; 30.90] | NA | ||
| Type 5 fibre protein, Human adenovirus 5 [Atome 2; 30.46] | NA | ||
| Fibre protein, Human adenovirus 41 [Atome 2; 24.60] | NA | ||
| Hme-FORF | Nucleoprotein, Influenza A virus [Atome 2; 80.49] | RNA binding protein | NA |
| Uim-HORFs | HIV-1 capsid, Human immunodeficiency virus 1 [I-TASSER; TM score 0.513] | Capsid protein | NA |
| Gag Polyprotein, Human immunodeficiency virus 1 [I-TASSER; TM score 0.510] | Precursor protein | NA | |
| Capsid protein P24, Human immunodeficiency virus type 2 [I-TASSER; TM score 0.504] | Capsid protein | NA | |
| Nucleoprotein, Andes virus [Atome 2; 44.18] | Nucleoprotein | NA | |
| Protein ICP47, Herpes simplex virus [Atome 2; 37.48] | Membrane protein | NA | |
| LdOrf-129 peptide, Lymantria dispar multiple nucleopolyhedrovirus (2) [BLASTP, PSIBLAST; 2e-06, 7e-10] | Structual protein | 74–144 | |
| ORF-132 protein, Lymantria dispar multiple nucleopolyhedrovirus (2) [BLASTP, PSIBLAST; 4e-06, 2e-09] | Unknown | 74–131 | |
| orf-126 protein, Lymantria dispar multiple nucleopolyhedrovirus [PSIBLAST; 4e-08] | Unknown | 72–140 | |
| Central variable region protein, African swine fever virus [PSIBLAST; 6e-08, 7e-07] | Unknown | 60–154 | |
| Central variable region protein, African swine fever virus [PSIBLAST; 7e-08] | Unknown | 60–130 | |
| pB602L, African swine fever virus tick/South Africa/Pretoriuskop Pr4/1996 [PSIBLAST; 8e-08] | Structural capsid protein, chaperone in capsid assembly (several hits) | 65–153 | |
| U1, Hyposoter didymator ichnovirus [PSIBLAST; 3e-07] | Spliceosomal RNA | 65–137 | |
| gp7, Salmonella phage epsilon15 [I-TASSER; norm. Z-score 1.32] | DNA transfer protein | NA | |
| Long tail fibre protein p37, Enterobacteria phage T4 [I-TASSER; norm. Z-score 1.30] | Fibre protein | 88–166 | |
| RhUL123, Macacine herpesvirus 3 [I-TASSER; TM score 0.617] | Viral life cycle | NA | |
| Nucleoprotein, Andes virus [Atome 2; 39.59] | Nucleoporin (several hits) | NA | |
| LdOrf-129 peptide, Lymantria dispar multiple nucleopolyhedrovirus [PSIBLAST; 8e-10] | Structual protein | NA | |
| ORF-132 protein, Lymantria dispar multiple nucleopolyhedrovirus [PSIBLAST; 5e-09] | Unknown | NA | |
| DNA stabilization protein, Salmonella phage HK620 [I-TASSER; Z-score 1.09] | DNA binding & stabilization | 87–188 | |
| Hexon protein, Human adenovirus 5 [I-TASSER; Z-score 1.01] | Major coat protein | 139–223 | |
| Human T-cell leukemia virus type II matrix protein, Human T-lymphotropic virus 2 [I-TASSER; Z-score 1.00] | Matrix protein | NA | |
| Herpes simplex virus protein ICP47, Herpes simplex virus (type 1/strain 17) [Atome 2; 1.72] | Blocks the major histocompatibility complex class I antigen presentation pathway | NA | |
| Lco-HORFs | Long tail fiber protein P37, Enterobacteria phage T4 [I-TASSER; Z-score 1.01] | Receptor binding viral protein | NA |
| Capsid protein, Rubella virus strain M33 [Atome 2; 83.05] | Capsid component | NA | |
| VPU protein, Human immunodeficiency virus 1 [Atome 2; 43.79] | Regulates degradation of receptor molecule CD4 (several hits) | NA | |
| Lsu-HORFs | Major capsid protein, Synechococcus phage Syn5 [I-TASSER; Z-score 1.66] | Capsid component | NA |
| RhUL123, Macacine herpesvirus 3 [I-TASSER; TM score 0.547] | Viral life cycle | 69–195 | |
| Herpes virus major outer envelope glycoprotein (BLLF1) [BLASTP/PSIBLAST; 2.73e-03] | Envelope protein | NA | |
| Short tail fiber protein, Enterobacteria phage T4 [I-TASSER; Z-score 2.14] | Structural protein | NA | |
| Major capsid protein, Synechococcus phage Syn5 [I-TASSER; Z-score 2.19] | Capsid component (several hits) | NA | |
| Coat protein, Enterobacteria phage P22 [I-TASSER; TM score 0.520] | Coat component | NA | |
| Herpes virus major outer envelope glycoprotein (BLLF1) [BLASTP/PSIBLAST; 4.85e-04] | Envelope protein | NA | |
| Tpa-HORF | VPU protein (Trans-membrane domain), Human immunodeficiency virus 1 [Atome 2; 33.16] | Regulates degradation of receptor molecule CD4 (several hits) | NA |
| Mfa-HORFs | ODV-E18: Occlusion-derived virus envelope protein ODV-E18 [Hhpred; 74.97] | Envelope protein (several hits) | 33–73 |
| Herpes_TK_C: Thymidine kinase from Herpesvirus C-terminal, Herpesvirus (2) [Hhpred; 48.70, 48.13] | ATP binding, thymidine kinase (several hits) | 33–73 | |
| Adenovirus fibre, Human adenovirus 2 [Atome 2; 34.11] | Fibre protein, receptor binding (several hits) | NA |
Note: I-TASSER: Norm. Z-score > 1 indicates a good alignment; TM-score > 0.5 indicates a similar fold with query [46]; position = amino acid position in the query sequence; NA not applicable
Prediction of molecular function: hits to mitochondrial proteins
Besides viral hits, most of the sequences analyzed also returned hits to proteins involved in energy production, including proteins of the mitochondrial electron transport system, so we tested the similarity of the ORFan proteins to standard mtDNA-encoded ones with BLAST searches. Our analyses predicted M-ORFs mostly as subunit 5 of the NADH-Ubiquinone Oxidoreductase complex I of the mitochondrial electron transport chain (NAD5) for 5 species out of 9, and/or ATP8 of the ATP synthase complex V for 5 species, but only with very low support (i.e. E-values ranged between 6e-04 and <1.0, the limit chosen for this analysis) (see Table 7). This latter result was also supported by a moderately significant domain hit identified in C. monodonta, i.e. pfam02326 or Mt_ATP-synt_B, a superfamily that corresponds to the subunit 8 of the F0 complex of plants (E-value 4e-03). Specifically, C. monodonta M-ORF shares similarities in its N-terminal amino-acid sequence with ATP8 sequences from plant but also from non-plant species (Additional file 4: Figure S3). However, similar results were not found for other M-ORF protein sequences (data not shown).
Table 7.
List of BLAST hits for mitochondrial ORFans in freshwater mussels searched against NCBI NRDB mitochondrial proteins
| Species Name | M-ORFs | F-ORFs | H-ORFs |
|---|---|---|---|
| Anodonta anatina | NAD7 (0.61) | --- | |
| --- | atp9 (0.19) | ||
| Cumberlandia monodonta | ATP8 (0.81) | --- | |
| --- | nad2 (6e-08) | ||
| Hyridella menziesi | ATP8 (0.61) | NAD2 (0.33) | |
| nad4 (6e-04) | nad2 (0.022) | ||
| Lasmigona complanata | --- | ||
| nad2 (0.094) | |||
| Lasmigona compressa | F-ORF (4e-05) | ||
| f-orf (2e-05) | |||
| Lasmigona subviridis | F-ORF (6e-09) | ||
| f-orf (2e-05) | |||
| nad1 (0.64) | |||
| Inversidens japanensis | ATP8 (0.62) | --- | |
| nad5 (0.001) | nad2 (0.22) | ||
| atp8 (0.048) | |||
| cox1 (0.15) | |||
| Margaritifera falcata | COX1 (0.94) | ||
| --- | |||
| Margaritifera margaritifera | NAD5 (0.093) | ||
| NAD2 (0.23) | |||
| nad2 (0.15) | |||
| Pyganodon grandis | NAD5 (0.046) | --- | |
| atp9 (0.30) | cytb (0.13) | ||
| Quadrula quadrula | NAD5 (0.026) | NAD5 (0.31) | |
| ATP8 (0.070) | nad2 (0.56) | ||
| atp9 (0.30) | |||
| Solenaia carinatus | COX1 (0.41) | --- | |
| NAD5 (0.99) | nad2 (0.018) | ||
| nad5 (0.33) | |||
| Toxolasma lividus | --- | ||
| --- | |||
| Toxolasma parvum | F-ORF (0.020) | ||
| --- | |||
| Utterbackia imbecillis | --- | ||
| nad2 (0.061) | |||
| Utterbackia peninsularis | NAD5 (0.38) | --- | |
| nad2 (0.31) | cox1 (0.056) | ||
| Venustaconcha ellipsiformis | NAD4 (0.19) | NAD4 (0.55) | |
| CYTB (0.21) | nad2 (0.14) | ||
| ATP8 (0.94) | |||
| nad4 (0.15) |
Note: Protein name and (e-values <1.0) identified using PSI-BLAST and tBLASTx are indicated above in capital letters and below in italics, respectively. Hits to freshwater mussel mitochondrial ORF homologs are not presented, except for the highly divergent H-ORFs
For F-ORFs, the most recurring hit (8 species out of 12) was subunit 2 of the mitochondrial complex I (NAD2), again with quite low support (E-values ranged between 6e-08 and <1.0). The lowest E-value was obtained with the F-ORF sequence of C. monodonta, but only for a short fragment of 20 amino acids sharing similarities with the NAD2 protein of the trematode Fasciola sp. The alignment of C. monodonta F-ORF and NAD2 protein sequences revealed poor similarities (Additional file 5: Figure S4), and identical results were also obtained in other studied gonochoric species (data not shown). Finally, BLAST searches of H-ORFs principally identified F-ORFs (3 species out of 5), with moderate E-values (Table 7).
Profile HMM – sequence comparisons for F-ORFs and M-ORFs
The hmmsearch analyses with HMM profiles for F-ORF and M-ORF alignments gave different numbers of hits for default vs. custom profiles. In general, the custom profiles were more “stringent” in terms of hit yield among all databases analysed, giving fewer total results than the default ones. Except for one hit for the M-ORF profiles, freshwater mussel ORFan sequences were the only significant hits (E-value <0.001) returned for all profiles, and they will not be considered. Therefore, we will describe all the hits other than unionoids ORFans (even those with E-values higher than the cutoff) in terms of functional recurrence. Results are presented in Additional file 1: Table S10 and S11.
Overall, F-ORF hits for both profiles are related to membrane association, virus life cycle, and interaction with nucleic acids (Additional file 1: Table S10 and S11). The M-ORF default profile frequently returned hits associated with membranes, related to energy production in bacteria or eukaryotes, transport or movement, or other functions related to membranes (Additional file 1: Table S10 and S11). The Excalibur domain protein, predicted two times with borderline significance (E-values 0.0011 and 0.0018), also has functions in DNA binding and repair and transcription regulation. Other recurring predicted functions are interaction with RNA (pre-rRNA processing, translation initiation, tRNA modification, poly-(A) RNA binding for nuclear import, posttranscriptional expression regulation) and with amino acids and proteins (protein transport, protein modification, or involvement in cytoskeleton rearrangements). Some hits suggest the possible insertion of DNA from foreign sources such as viruses (e.g. hits to viral delta antigens of hepatitis delta virus that are related to viral life cycle, i.e. invasion in host cell and nucleus, replication) and bacteria (a transposition protein gene from E. coli Tn7 transposon). The M-ORF custom profile returned four additional results, all involved in protein and/or membrane interactions.
Prediction of molecular function (all sequences, all programs except hmmsearch)
Finally, we compiled the results obtained for all ORFans with all other programs for protein function prediction (i.e. BLAST, HHpred, @tome2, I-TASSER, and PredictProtein). Fig. 7 summarizes the most frequent categories of hits for biological processes or molecular functions for freshwater mussel mitochondrial ORFans (i.e. those returned for over 75 % of all analyzed species for each ‘sex’) and Additional file 6: Figure S5 and Additional file 1: Table S12-S37 contain detailed hits and recurring functions (i.e. biological processes, cellular components/subcellular localizations and molecular functions). Overall, the most common hits for all M-ORFs, F-ORFs and H-ORFs were transmembrane proteins, proteins involved in nucleic acid binding and transcription, protein binding proteins, and proteins involved in cellular signalling and transport (Fig. 7). In particular, all M-ORFs returned hits to proteins involved in cell adhesion, migration and proliferation, and the predicted subcellular localizations for M-ORFs were membranes and mostly organelles (endoplasmic reticulum, mitochondria, Golgi and nucleus). Other hits for M-ORFs included proteins related to developmental processes (e.g. embryonic development) and structural activity (Figs. 6 and 7 and Additional file 1: Table S10-S37).
Fig. 7.
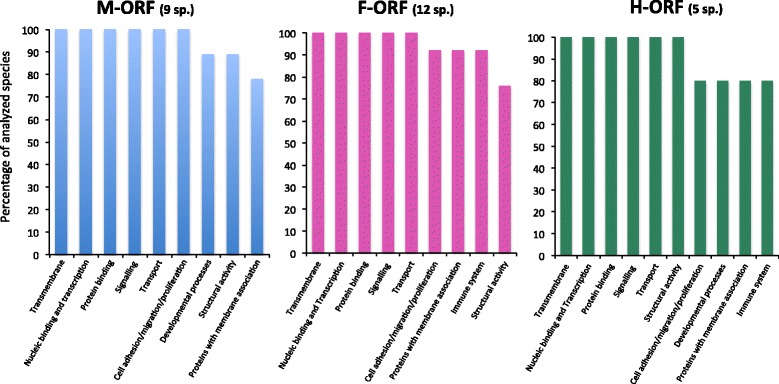
Most frequent categories of hits for biological processes or molecular functions for freshwater mussel mitochondrial ORFans. Categories presented are those returned for over 75 % of all analyzed species for each ‘sex’ (the number of analyzed species for each sex is indicated in parentheses). Blue, M-ORF; pink, F-ORF; green, H-ORF
The most common hits for F-ORFs included proteins with membrane association (e.g. proteins involved in trafficking and transport functions such as SNAP receptors and kinases). Many hits also pointed to a role in immune response. The mitochondria, Golgi, and ER were predicted subcellular localizations for F-ORFs (an extracellular localization was also suggested) (Fig. 7 and Additional file 1: Tables S10-S37). For H-ORFs, structural proteins, particularly collagen and collagen-like proteins were the most common categories, closely followed by membrane-associated proteins, proteins involved in developmental processes and immune response. An extracellular localisation was also suggested for H-ORFs (Fig. 7 and Additional file 1: Tables S10-S37).
Discussion
Evolution of freshwater mussel ORFan sequences and protein structures
One general feature observed in mitochondrial ORFan sequences of marine [18] and freshwater bivalves with DUI (present study) is their higher p-distance values at the amino acid level compared to their own nucleotide sequences, suggesting a rapid rate of evolution. However, the null hypothesis of strict-neutrality (dN = dS) was not rejected in favor of the alternative hypothesis of positive selection (dN > dS) (although dN > dS is an extremely conservative test that may miss instances in which positive selection is happening [48]). Despite low sequence conservation, M-ORF and F-ORF proteins appear structurally conserved among species, suggesting that their biological functions might be conserved as well.
Compared to F-ORFs from gonochoric species, H-ORFs from hermaphroditic unionoids contain repeat units and sometimes different hydropathy profiles (e.g. U. imbecillis vs. U. peninsularis). One possible mechanism for the duplication of repeats independently in the H-orf sequences is slippage due to DNA hairpins, a common mechanism implicated in the creation of short protein repeats [49, 50]. These distinctive features of the five H-ORFs could indicate changes of function from that of the homologous F-ORFs in gonochoric species. The high level of amino acid sequence and structural similarities of the H-ORF protein within species, as well as its recent detection in the transcriptome of the hermaphroditic species U. imbecillis (Capt et al. unpublished) suggest that it is functional.
Proteins that contain tandem repeats frequently interact with other proteins or ligands such as DNA or RNA (e.g. [50, 51]). A classic example in organelles is the pentatricopeptide repeat (PPR) protein family, and PPR hits were found in all ORFan protein sequences using HHpred. PPR proteins contain variable numbers of tandem repeats and function in transcription, RNA processing, splicing, stability, editing, and translation [51]. Interestingly, PPR proteins are key elements of the only known sex determination system in which the mitochondrial DNA is involved, i.e. in hermaphroditic plants exhibiting cytoplasmic male sterility (CMS) [51]. PPR proteins appear to function as nuclear-encoded restorers of fertility in CMS plants, which suppress mtDNA-encoded factors that inhibit the production of viable pollen [51]. It has been hypothesized that in unionids with DUI the loss of the M mitochondrial genome and macromutations in the F-orf gene (i.e. acquisition of tandem repeats) could enable an individual to produce both sperm and eggs leading to hermaphroditism [16].
Conserved motifs and domains: mitochondrial export of ORFan proteins
In this unionoid-specific study, we found the same pattern of homology detection hits for M-ORFs and F-ORFs as presented in Milani et al. [18], i.e. motifs and domains involved in cell membrane/surface anchoring, transcription and post-transcriptional processes. Two notable differences involved hits involved in cleavage/methylation and protein transport.
So far, the protein products of the F-orf and M-orf genes in unionoids have been studied only in the species Venustaconcha ellipsiformis [16]. Using immunoelectron microscopy, the F-ORF protein has been localized not only to egg mitochondria, but also to the nuclear envelope and the egg nucleoplasm [16]. Interestingly, the F-ORF protein was also found on the inner mitochondrial membrane of some sperm mitochondria [52], which are thought to contain only M mtDNA [53]. Because small proteins may diffuse into the nucleus without a specific targeting signal, the nuclear localization in eggs may not be specific, however, mitochondrial localization depends on an N-terminus signal peptide [54, 55]. Because the F mtDNA is not present in DUI bivalve sperm mitochondria [53], either there is a version of the F-orf gene in the nuclear genome (or another nuclear-encoded gene product is capable of reacting with the antibody), or the mtDNA-encoded F-ORF protein is exported from F-type mitochondria and imported via an N-terminal signal peptide into sperm mitochondria. Examination of a freshwater mussel nuclear genome (currently underway in our laboratory) will help test these hypotheses.
Subcellular localization of the M-ORF protein has not yet been studied, but our in silico detection of nuclear localization signals in several M-ORF sequences, and of hits related to protein movement, are consistent with the hypothesis that this protein is exported from the organelle. Such results have been observed in the venerid clam Ruditapes philippinarum, in which the M-ORF protein was immunolocalized in both mitochondria and the nucleus of sperm [19]. Hence, mitochondrial ORFan proteins in DUI bivalves likely have multiple roles in different cellular compartments ([16, 18, 19], present study), explaining the existence of functional domains for interacting with diverse cellular elements.
The process for mitochondrial exporting of F-ORF or M-ORF proteins remains unexplained. In fact, while mitochondrial import of proteins is well-studied in eukaryotes [56], the process of mitochondrial export is still obscure (e.g. [57]). The export of cell death effectors [58], retrograde signals humanin and MOTS-c [59], and small peptides to trigger retrograde nuclear signalling in mitochondrial unfolded protein response in mammals are all partially characterized, but mitochondrial protein export of larger molecules is relatively unstudied (e.g. [57, 60]). Further work is needed to better understand mitochondrial export in animals.
Putative origin for freshwater mussel mitochondrial ORFans
As mentioned, prior in silico analyses pointed to a possible viral origin of bivalve mitochondrial ORFans, although the probability of some hits were low and the regions of similarity were short [18]. Except for the M-ORF of C. monodonta and the F-ORF of A. anatina, our results revealed the presence of at least one viral hit for each sequence analyzed (consistent with the viral hypothesis), but with low probability values and short regions of similarity. We also consistently obtained hits with stronger probability values for bacterial or metazoan genes (Table 6 and Additional file 1: Tables S12-S37). Consequently, we cannot exclude other organisms or other processes [61–63] as the source of these ORFan genes. For example, gene duplication is thought to be the mechanism underlying the origin of most novel genes, and thus represents one of the most important processes for functional innovation during evolution [62]. Interestingly, several sequences returned hits to proteins involved in mitochondrial energy production, including proteins of the electron transport system, suggesting that duplication and neofunctionalization of a mitochondrial gene could be the source of freshwater mussel mitochondrial ORFans. Several M-ORF sequences returned hits to the subunit ATP8 of the mitochondrial ATP synthase complex V (Table 7), and M-ORF profiles to subunit b of bacterial ATP synthase. These results are interesting for two reasons. First, the atp8 and M-orf genes occur beside one another in a region corresponding to one of the three gene order rearrangements observed between female and male mtDNAs of freshwater mussels [16]. Second, the atp8 gene is highly modified or reported as missing in other bivalve species with DUI due to its short length and rapid evolution causing difficulties in annotation (e.g. [64–66]). It is conceivable that a duplication event (as described in several other animal mtDNAs [67]) of the region containing the atp8 gene occurred in an ancestral freshwater mussel species with DUI. One of the two duplicate atp8 copies could have evolved new male-specific functional properties, giving rise to the M-orf gene. The identification of a conserved domain of the Mt_ATP-synt_B superfamily in the M-ORF protein sequence of C. monodonta, i.e. a domain found at the N terminus of subunit 8 of the F0 complex of mitochondrial ATP synthases from plants and algae, also provides further support for the above scenario (Additional file 4: Figure S3). In a variety of plant species, this N-terminal conserved domain is not only found in ATP8 but also in CMS proteins (coupled to novel C-terminus domains as a result of mt genome rearrangements) that are associated with reduction in ATPase activity in male-sterile lines (e.g. [68, 69]). Considering this, both mitochondrial ATP8 and bacterial subunit b hits for M-ORF protein sequences may indicate a mitochondrial localization for M-ORF in the F0 subunit of complex V, the region of ATP synthase where protons pass through the inner membrane from the intermembrane space to the matrix. Examples of mtDNA-encoded non-canonical subunits of the F0 complex are already known from studies on protists [70] and plants [68, 69], and unionoid M-ORFs might be a metazoan version of this scenario. Questions for future studies include whether (1) the M-ORF in these species forms part of complex V thereby altering mitochondrial membrane potential, and (2) whether sperm mitochondrial inheritance could be effected by such a mechanism (as proposed by [71]).
Individual F-ORF sequences also returned many hits pointing to mitochondrial membrane proteins, often NAD2, although with relatively low E-values. Nonetheless, this is interesting because nad2 and the F-orf genes are also typically localized beside one another in a region corresponding to the only gene order rearrangement observed among F mtDNAs in freshwater mussels with DUI [15]. It is plausible that this region was duplicated with subsequent adaptation of one of the two copies of nad2. The nad2 gene is also localized beside the F-orf gene in the marine clam Ruditapes philippinarum [66] (but this is not the case for all species with DUI). Finally, and not surprisingly, all H-ORF sequences returned hits to F-ORF sequences (Table 7), and many hits for F-ORF profiles are annotated H-ORFs, supporting previous results that H-orf genes are derived from F-orf genes [16]. With a rapid rate of evolution, the mitochondrial ORFans would have rapidly lost their resemblance to the highly conserved mitochondrial genes from which they evolved. Our results do not refute the hypothesis that these ORFans originated from viral sequences, but they open up the possibility of a mitochondrial origin for these genes, specifically ATP8 and NAD2 for the M-ORF and F-ORF in unionoids, respectively.
Predicted functions for freshwater mussel mitochondrial ORFans
The absolute linkage of a hermaphroditic breeding system, the absence of an M genome and highly modified F-ORFs (i.e. H-ORFs in hermaphrodites) has led to the hypothesis that the F-ORF and M-ORF proteins likely have coordinated roles in maintaining gonochorism in freshwater mussels [16]. Furthermore, these roles must be concordantly modified to produce a hermaphroditic individual [16]. Milani et al. [18, 19] suggested that the M-ORF protein might play a role in aggregating sperm-derived mitochondria in early-stage male embryos. Our analysis of M-ORF sequences indicated connections with cytoskeleton proteins involved in microtubule-binding and actin-binding (e.g. ankyrin). With their predicted SPs and TM helices, M-ORFs may target sites outside sperm mitochondria and be responsible for their cellular positioning in developing embryos. It has been suggested that mitochondrial dynamics (e.g., motility, fusion, etc.) must include “signalling” from the respective individual mitochondrion [72]. Although no protein of the dynamics machinery has been identified in bivalves yet, the mtDNA-encoded M-ORF in bivalves with DUI is an ideal candidate for direct control of sperm mitochondria. As hypothesized by Milani et al. [18], the M-ORF protein could be a masculinizing factor and sperm from males with high amounts of transcript and/or protein would shift embryo development toward maleness. Yusa et al. [73], in their DUI sex-determination model, predicted the existence of such secondary or minor sex-determining mitochondrial factors. Like the M-ORF, if the F-ORF is a feminizing factor, and because macromutational modifications to the F-orf gene are always associated with hermaphroditism, it is tempting to speculate that the F-ORF protein could participate in the inhibition of testicular development in embryos that will become females, and the extreme modifications seen in H-ORFs could explain why development of some testicular tissue is not completely inhibited in hermaphrodites.
Conclusions
Because the evolutionary distance among mytilids, venerids, and unionids did not allow for a meaningful comparison of mitochondrial ORFans [18], we decided to perform in silico analyses on more closely related ORFan sequences within the order Unionoida. Our findings, in agreement with previous data by Milani et al. [18, 19], reveal high levels of sequence divergence among ORFans, yet with conserved predicted structures, motifs and domains. These ORFans might have originated either from viral horizontal gene transfers or mitochondrial gene duplications but they have evolved rapidly to the point that a clear signature of their origin is not easily recognizable. Our study, which also strongly supports a role for these ORFans in the DUI mechanism, is in line with the growing body of literature extending our understanding of metazoan mitochondrial genome function beyond exclusively OXPHOS related roles (e.g. [18, 59, 74, 75]. DUI as well as other intriguing systems like the recently discovered maternally transmitted sex distortion in booklice that is associated with extremely divergent mitochondria [76], represent interesting cases to look for and better understand antagonistic interactions between distorting mitochondria and nuclear suppressors similar to CMS in plants. If the F-ORF and M-ORF proteins in bivalves with DUI are indeed antagonistic molecules, i.e. with the F-ORF participating in the inhibition of testicular development in female developing embryos and the M-ORF participating in the inhibition of ovarian development in male developing embryos, this could explain why macromutations in the F-ORF protein (that turns it into a H-ORF) would allow for testis development in otherwise female gonads (i.e. hermaphroditism). However, the precise mechanisms underlying DUI and sex determination in bivalves remain to be elucidated.
Abbreviations
ATP, adenosine triphosphate; atp8, ATP synthase subunit 8; CMS, cytoplasmic male sterility; cox1, cytochrome c oxidase subunit 1; CTERM, C-terminal; DNA, deoxyribonucleic acid; dnaB, DNA helicase; DUI, doubly uniparental inheritance; ER, endoplasmic reticulum; HMM, hidden Markov model; mtDNA, mitochondrial DNA; MY, million years; nad2, NADH dehydrogenase subunit 2; nad5, NADH dehydrogenase subunit 5; NADH, nicotiamide adenine dinucleotide, reduced form; NCBI, national Center for Biotechnology Information; NRDB, non-redundant protein database; ORF, open reading frame; ORFan, open reading frame without homology to a known protein; PPR, pentatricopeptide repeat; RNA, ribonucleic Acid; SMI, strict Maternal Inheritance; SP, signal peptide; tatC, twin-arginine translocase, subunit C; TM, transmembrane; TPR, tetratricopeptide repeat
Acknowledgements
We thank France Dufresne, two anonymous reviewers and the editor for critical reading of the manuscript. This work was supported by funding from the Natural Sciences and Engineering Research Council of Canada (grant no., RGPIN/435656-2013 to S.B. and grant no., RGPIN/217175-2013 to D.T.S.). A.M. was financially supported by a Natural Sciences and Engineering Research Council of Canada scholarship.
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its Additional files].
Authors’ contributions
SB conceived the study, participated in its design and coordination and drafted the manuscript. AM performed the analyses and drafted the manuscript. DG performed part of the analyses and drafted the manuscript. DTS assisted with analytical recommendations and drafting the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional files
Results of in silico analyses. (DOCX 368 kb)
Alignments of F-ORFs and H-ORFs of closely related species. Colour coding is applied to amino acid groups conserved in ≥70 % of sequences. Grey, aliphatic amino acids; orange, aromatic amino acids; yellow, sulfur amino acids; green, amino acids bearing a hydroxyl group; red, basic amino acids; blue, acidic amino acids; brown, amino acids with an amide group; pink, cyclic amino acids. Green box: conserved C-terminal domain identified in [16]; blue underlining: repetitive sequences. UpeFORF, U. peninsularis F-ORF; UimHORF, U. imbecillis H-ORF; TliFORF, T. lividus F-ORF; TpaHORF, T. parvum H-ORF; MmaFORF, M. margaritifera F-ORF; MfaHORF, M. falcata H-ORF; LcoFORF, L. complanata F-ORF; LcoHORF, L. compressa H-ORF; LsuHORF, L. subviridus H-ORF. (PDF 1185 kb)
Percentage of similarity between complete mitochondrial genomes of freshwater mussels with DUI. Each graph shows the percent of conservation between genomes at any given coordinate. The top and bottom percentage bounds are shown to the right of every row. The pink regions are conserved non-protein-coding sequences, the dark blue regions are protein-coding genes, the white regions are non-coding sequences. (A) M vs. M genome comparison between two closely related species (Utterbackia peninsularis and Pyganodon grandis, GenBank accession numbers HM856635 and FJ809754, respectively) showing that the M-ORF gene shows low level of sequence conservation compared to other protein-coding genes. (B) F vs. F genome comparison between two closely related species (U. peninsularis and P. grandis, GenBank accession numbers HM856636 and FJ809755, respectively) showing that the F-ORF gene shows low level of sequence conservation compared to other protein-coding genes. (C) F vs. H genome comparison between two closely related species (Utterbackia peninsularis and U. imbecillis, GenBank accession numbers HM856636 and HM856637, respectively) showing that the F-ORF/H-ORF gene region shows low level of sequence conservation compared to other protein-coding genes. (PDF 160 kb)
Protein sequence alignment of Cumberlandia monodonta M-ORF and ATP8, along with ATP8 from the most diverse members of the Mt_ATP-synt_B superfamily (pfam02326). Homo sapiens ATP8 has also been included for comparison. The alignment was generated using T-COFFEE. The most conserved N-terminal domain, i.e. the best aligned portion, is in red; the rest of the sequences are rather badly aligned (in green). Consensus is shown and indicates good (red), intermediate (yellow), and bad alignment (green), and insertion/deletion (in blue). Cumberland, Cumberlandia monodonta; H_sapiens, Homo sapiens; Malawimonas, Malawimonas sp. (Excavate); Thraustoch, Thraustochytrium sp. (Stramenopiles); Mesostigma, Mesostigma sp. (Streptophyta); Reclinomon, Reclinomonas sp. (Protozoa); Porphyra, Porphyra sp. (Rhodophyta); Cyanidiosc, Cyanidioschyzon sp. (Rhodophyta); Pseudendoc, Pseudendoclonium sp. (Chlorophyta); Acanthamoe, Acanthamoeba sp. (Amoebozoa); Nephroselm, Nephroselmis sp. (Streptophyta). (PDF 236 kb)
Protein sequence alignment of Cumberlandia monodonta F-ORF and NAD2. The alignment was generated using T-COFFEE. Consensus is shown and indicates identical (*) and similar (: and.) amino acids. Description of the data: Protein sequence alignment of Cumberlandia monodonta F-ORF and NAD2. (PDF 141 kb)
Position of frequently recurring functions in HHpred and BLAST hits for (a) M-ORFs, (b) F-ORFs, and (c) and (d) H-ORFs. Hits with positions were grouped into categories and traced together, showing hot spots of functionality. Protein length in amino acids is indicated in parentheses. (PDF 301 kb)
Contributor Information
Alyssa Mitchell, Email: alyssa.mitchell@umontreal.ca.
Davide Guerra, Email: davide.guerra@umontreal.ca.
Donald Stewart, Email: don.stewart@acadiau.ca.
Sophie Breton, Email: s.breton@umontreal.ca.
References
- 1.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–80. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity (Edinb) 2008;101:301–20. doi: 10.1038/hdy.2008.62. [DOI] [PubMed] [Google Scholar]
- 3.Birky CW. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu Rev Genet. 2001;35:125–48. doi: 10.1146/annurev.genet.35.102401.090231. [DOI] [PubMed] [Google Scholar]
- 4.Kayal E, Bentlage B, Collins AG, Kayal M, Pirro S, Lavrov DV. Evolution of linear mitochondrial genomes in medusozoan cnidarians. Genome Biol Evol. 2012;4:1–12. doi: 10.1093/gbe/evr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doublet V, Souty-Grosset C, Bouchon D, Cordaux R, Marcadé I. A thirty million year-old inherited heteroplasmy. PLoS One. 2008;3:e2938. doi: 10.1371/journal.pone.0002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breton S, Milani L, Ghiselli F, Guerra D, Stewart DT, Passamonti M. A resourceful genome: updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends Genet. 2014;30:555–64. doi: 10.1016/j.tig.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Breton S, Beaupré HD, Stewart DT, Hoeh WR, Blier PU. The unusual system of doubly uniparental inheritance of mtDNA: isn’t one enough? Trends Genet. 2007;23:465–74. doi: 10.1016/j.tig.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Boyle EE, Etter RJ. Heteroplasmy in a deep-sea protobranch bivalve suggests an ancient origin of doubly uniparental inheritance of mitochondria in Bivalvia. Mar Biol. 2013;160:413–22. doi: 10.1007/s00227-012-2099-y. [DOI] [Google Scholar]
- 9.Passamonti M, Ghiselli F. Doubly uniparental inheritance: two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA Cell Biol. 2009;28:79–89. doi: 10.1089/dna.2008.0807. [DOI] [PubMed] [Google Scholar]
- 10.Zouros E. Biparental Inheritance Through Uniparental Transmission: The Doubly Uniparental Inheritance (DUI) of Mitochondrial DNA. Evol Biol. 2013;40:1–31. doi: 10.1007/s11692-012-9195-2. [DOI] [Google Scholar]
- 11.Obata M, Sano N, Kawamura K, Komaru A. Inheritance of two M type mitochondrial DNA from sperm and unfertilized eggs to offspring in Mytilus galloprovincialis. Dev Growth Differ. 2007;49:335–44. doi: 10.1111/j.1440-169X.2007.00930.x. [DOI] [PubMed] [Google Scholar]
- 12.Chakrabarti R, Walker JM, Chapman EG, Shepardson SP, Trdan RJ, Curole JP, Watters GT, Stewart DT, Vijayaraghavan S, Hoeh WR. Reproductive function for a C-terminus extended, male-transmitted cytochrome c oxidase subunit II protein expressed in both spermatozoa and eggs. FEBS Lett. 2007;581:5213–9. doi: 10.1016/j.febslet.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao L, Kenchington E, Zouros E. Differential Segregation Patterns of Sperm Mitochondria in Embryos of the Blue Mussel (Mytilus edulis) Genet Soc Am. 2004;894:883–94. doi: 10.1534/genetics.166.2.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milani L, Ghiselli F, Passamonti M. Sex-linked mitochondrial behavior during early embryo development in Ruditapes philippinarum (Bivalvia Veneridae) a species with the Doubly Uniparental Inheritance (DUI) of mitochondria. J Exp Zool B Mol Dev Evol. 2012;318:182–9. doi: 10.1002/jez.b.22004. [DOI] [PubMed] [Google Scholar]
- 15.Breton S, Beaupre HD, Stewart DT, Piontkivska H, Karmakar M, Bogan AE, Blier PU, Hoeh WR. Comparative Mitochondrial Genomics of Freshwater Mussels (Bivalvia: Unionoida) With Doubly Uniparental Inheritance of mtDNA: Gender-Specific Open Reading Frames and Putative Origins of Replication. Genetics. 2009;183:1575–89. doi: 10.1534/genetics.109.110700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breton S, Stewart DT, Shepardson S, Trdan RJ, Bogan AE, Chapman EG, Ruminas AJ, Piontkivska H, Hoeh WR. Novel protein genes in animal mtDNA: A new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol Biol Evol. 2011;28:1645–59. doi: 10.1093/molbev/msq345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breton S, Ghiselli F, Passamonti M, Milani L, Stewart DT, Hoeh WR. Evidence for a fourteenth mtDNA-encoded protein in the female-transmitted mtDNA of marine Mussels (Bivalvia: Mytilidae) PLoS One. 2011;6:e19365. doi: 10.1371/journal.pone.0019365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milani L, Ghiselli F, Guerra D, Breton S, Passamonti M. A comparative analysis of mitochondrial ORFans: New clues on their origin and role in species with Doubly Uniparental Inheritance of mitochondria. Genome Biol Evol. 2013;5:1408–34. doi: 10.1093/gbe/evt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milani L, Ghiselli F, Maurizii MG, Nuzhdin SV, Passamonti M. Paternally transmitted mitochondria express a new gene of potential viral origin. Genome Biol Evol. 2014;6:391–405. doi: 10.1093/gbe/evu021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milani L, Ghiselli F. Mitochondrial activity in gametes and transmission of viable mtDNA. Biol Direct. 2015;10:22. doi: 10.1186/s13062-015-0057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heller J. Hermaphroditism in molluscs. Biol J Linn Soc. 1993;48:19–42. doi: 10.1111/j.1095-8312.1993.tb00874.x. [DOI] [Google Scholar]
- 22.Hoeh WR, Frazer KS, Naranjo-Garcia E, Trdan RJ. A phylogenetic perspective on the evolution of simultaneous hermaphroditism in a freshwater mussel Clade (Bivalvia: Unionidae: Utterbackia) Malacol Rev. 1995;28:25–42. [Google Scholar]
- 23.Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, Hoeh WR. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol Biol. 2010;10:50. doi: 10.1186/1471-2148-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue T, Chen M, Wang G, Han Z, Li J. The complete F-type mitochondrial genome of Chinese freshwater mussel Anodonta euscaphys. Mitochondrial DNA. 2015;26:263–4. doi: 10.3109/19401736.2013.823190. [DOI] [PubMed] [Google Scholar]
- 25.Rombel IT, Sykes KF, Rayner S, Johnston SA. ORF-FINDER: A vector for high-throughput gene identification. Gene. 2002;282:33–41. doi: 10.1016/S0378-1119(01)00819-8. [DOI] [PubMed] [Google Scholar]
- 26.Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang J-M, Taly J-F, Notredame C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–7. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–26. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32(Web Server):W273–9. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. New York: Humana Press; 2005. pp. 571–607. [Google Scholar]
- 31.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–32. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 32.Käll L, Krogh A, Sonnhammer EL. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J Mol Biol. 2004;338:1027–36. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Zdobnov EM, Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–8. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 34.Hofmann K, Stoffel W. TMbase-A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 35.Bernsel A, Viklund H, Falk J, Lindahl E, von Heijne G, Elofsson A. Prediction of membrane-protein topology from first principles. Proc Natl Acad Sci. 2008;105:7177–81. doi: 10.1073/pnas.0711151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32(Web Server issue):W321–6. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen H, Engelbrecht J, Brunak S, Heijne G. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites Cited by me. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 38.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 39.Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38(Database):D161–6. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server):W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karpenahalli MR, Lupas AN, Söding J. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics. 2007;8:2. doi: 10.1186/1471-2105-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson WR. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-V. [DOI] [PubMed] [Google Scholar]
- 44.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pons J-L, Labesse G. @TOME-2: a new pipeline for comparative modeling of protein-ligand complexes. Nucleic Acids Res. 2009;37:W485–91. doi: 10.1093/nar/gkp368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pesole G, Gissi C, De Chirico A, Saccone C. Nucleotide substitution rate of mammalian mitochondrial genomes. J Mol Evol. 1999;48:427–34. doi: 10.1007/PL00006487. [DOI] [PubMed] [Google Scholar]
- 48.Kreitman M. Methods to detect selection in populations with applications to the human. Annu Rev Genom Hum G. 2000;1:539–59. doi: 10.1146/annurev.genom.1.1.539. [DOI] [PubMed] [Google Scholar]
- 49.Djian P. Evolution of simple repeats in DNA and their relation to human disease. Cell. 1998;94:155–60. doi: 10.1016/S0092-8674(00)81415-4. [DOI] [PubMed] [Google Scholar]
- 50.Björklund ÅK, Ekman D, Elofsson A. Expansion of Protein Domain Repeats. PLoS Comput Biol. 2006;2:e114. doi: 10.1371/journal.pcbi.0020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manna S. An overview of pentatricopeptide repeat proteins and their applications. Biochimie. 2015;113:93–9. doi: 10.1016/j.biochi.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Shepardson SP, Heard WH, Breton S, Hoeh WR. Light and Transmission Electron Microscopy of Two Spermatogenic Pathways and Unimorphic Spermatozoa in Venustaconcha ellipsiformis (Conrad, 1836) (Bivalvia: Unionoida) Malacologia. 2012;55:263–84. doi: 10.4002/040.055.0207. [DOI] [Google Scholar]
- 53.Venetis C, Theologidis I, Zouros E, Rodakis GC. No evidence for presence of maternal mitochondrial DNA in the sperm of Mytilus galloprovincialis males. Proc Biol Sci. 2006;273:2483–9. doi: 10.1098/rspb.2006.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terry LJ, Wente SR. Flexible Gates: Dynamic Topologies and Functions for FG Nucleoporins in Nucleocytoplasmic Transport. Eukaryot Cell. 2009;8:1814–27. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu F, Shi J, Zhou J, Gu J, Chen Q, Li J, Cheng W, Mao D, Tian L. ANK6, a mitochondrial ankyrin repeat protein, is required for male–female gamete recognition in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2010;107:22332–7. doi: 10.1073/pnas.1015911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dudek J, Rehling P, van der Laan M. Mitochondrial protein import: Common principles and physiological networks. Biochem Biophys Acta. 1883;2013:274–85. doi: 10.1016/j.bbamcr.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 57.Ng F, Tang BL. Pyruvate dehydrogenase complex (PDC) export from the mitochondrial matrix. Mol Membr Biol. 2014;31:207–10. doi: 10.3109/09687688.2014.987183. [DOI] [PubMed] [Google Scholar]
- 58.Bernardi P. The mitochondrial permeability transition pore: a mystery solved? Front Physiol. 2013;4:1–12. doi: 10.3389/fphys.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim S-J, Mehta H, Hevener AL, de Cabo R, Cohen P. The Mitochondrial-Derived Peptide MOTS-c Promotes Metabolic Homeostasis and Reduces Obesity and Insulin Resistance. Cell Metab. 2015;21:443–54. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang BL. Mitochondrial Protein in the Nucleus. Cell Bio. 2015;4:23–9. [Google Scholar]
- 61.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25:404–13. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 62.Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010;20:1313–26. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- 64.Breton S, Stewart DT, Hoeh WR. Characterization of a mitochondrial ORF from the gender-associated mtDNAs of Mytilus spp. (Bivalvia: Mytilidae): identification of the “missing” ATPase 8 gene. Mar Genomics. 2010;3:11–8. doi: 10.1016/j.margen.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 65.Burzynski A, Wenne R. Comparative Genomics of Marine Mussels (Mytilus spp.) Gender Associated mtDNA : Rapidly Evolving atp8. J Mol Evol. 2010;71:385–400. doi: 10.1007/s00239-010-9393-4. [DOI] [PubMed] [Google Scholar]
- 66.Ghiselli F, Milani L, Guerra D, Chang PL, Breton S, Nuzhdin SV, Passamonti M. Structure, Transcription, and Variability of Metazoan Mitochondrial Genome: Perspectives from an Unusual Mitochondrial Inheritance System. Genome Biol Evol. 2013;5:1535–54. doi: 10.1093/gbe/evt112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boore JL, Brown WM. Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: sequence and gene arrangement comparisons indicate that Pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Mol Biol Evol. 2000;17:87–106. doi: 10.1093/oxfordjournals.molbev.a026241. [DOI] [PubMed] [Google Scholar]
- 68.Schnable PS, Wise RP. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 1998;3:175–80. doi: 10.1016/S1360-1385(98)01235-7. [DOI] [Google Scholar]
- 69.Sabar M, Gagliardi D, Balk J, Leaver CJ. ORFB is a subunit of F1F0-ATP synthase: insight into the basis of cytoplasmic male sterility in sunflower. EMBO Rep. 2003;4:381–6. doi: 10.1038/sj.embor.embor800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burger G, Lang BF, Braun H, Marx S. The enigmatic mitochondrial ORF ymf39 codes for ATP synthase chain b. Nucleic Acids Res. 2003;31:2353–60. doi: 10.1093/nar/gkg326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Milani L. Mitochondrial membrane potential : a trait involved in organelle inheritance? Biol Lett. 2015;11:20150732. doi: 10.1098/rsbl.2015.0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwarzländer M, Finkemeier I. Mitochondrial Energy and Redox Signaling in Plants. Antioxid Redox Signal. 2013;18:2122–44. doi: 10.1089/ars.2012.5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yusa Y, Breton S, Hoeh WR. Population genetics of sex determination in Mytilus mussels: Reanalyses and a model. J Hered. 2013;104:380–5. doi: 10.1093/jhered/est014. [DOI] [PubMed] [Google Scholar]
- 74.Capt C, Passamonti M, Breton S. The human mitochondrial genome may code for more than 13 proteins. Mitochondrial DNA. 2015 doi: 10.3109/19401736.2014.1003924. [DOI] [PubMed] [Google Scholar]
- 75.Cobb LJ, Lee C, Xiao J, Yen K, Wong RG, Nakamura HK, Mehta HH, Gao Q, Ashur C, Huffman DM, Wan J, Muzumdar R, Barzilai N, Cohen P. Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers. Aging. 2016;8:796–809. doi: 10.18632/aging.100943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perlman SJ, Hodson CN, Hamilton PT, Opit GP, Gowen BE. Maternal transmission, sex ratio distortion, and mitochondria. Proc Natl Acad Sci U S A. 2015;112:1–7. doi: 10.1111/nyas.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its Additional files].


