Abstract
Purpose
Recommendations for treating patients who carry a BRCA1/2 gene are mainly based on cumulative lifetime risks (CLTRs) of breast cancer determined from retrospective cohorts. These risks vary widely (27% to 88%), and it is important to understand why. We analyzed the effects of methods of risk estimation and bias correction and of population factors on CLTRs in this retrospective clinical cohort of BRCA1/2 carriers.
Patients and Methods
The following methods to estimate the breast cancer risk of BRCA1/2 carriers were identified from the literature: Kaplan-Meier, frailty, and modified segregation analyses with bias correction consisting of including or excluding index patients combined with including or excluding first-degree relatives (FDRs) or different conditional likelihoods. These were applied to clinical data of BRCA1/2 families derived from our family cancer clinic for whom a simulation was also performed to evaluate the methods. CLTRs and 95% CIs were estimated and compared with the reference CLTRs.
Results
CLTRs ranged from 35% to 83% for BRCA1 and 41% to 86% for BRCA2 carriers at age 70 years width of 95% CIs: 10% to 35% and 13% to 46%, respectively). Relative bias varied from −38% to +16%. Bias correction with inclusion of index patients and untested FDRs gave the smallest bias: +2% (SD, 2%) in BRCA1 and +0.9% (SD, 3.6%) in BRCA2.
Conclusion
Much of the variation in breast cancer CLTRs in retrospective clinical BRCA1/2 cohorts is due to the bias-correction method, whereas a smaller part is due to population differences. Kaplan-Meier analyses with bias correction that includes index patients and a proportion of untested FDRs provide suitable CLTRs for carriers counseled in the clinic.
INTRODUCTION
Since the discovery of the BRCA genes 20 years ago, numerous retrospective studies have been performed to estimate the cumulative lifetime risk (CLTR) of breast cancer for pathogenic BRCA1/2 gene mutation carriers.1–44 However, results of these studies show considerable variation: CLTRs by the age of 70 years vary from 27% to 88%, and the width of the 95% CI estimates range from 6% to 97%. Recently, estimates from prospectively collected cohorts were obtained. These were, for BRCA1, 55% to 60% (95% CI, 37% to 76%) and, for BRCA2, 55% to 72% (95% CI, 41% to 88%).45,46 However, because prospective data are limited and available estimates vary, recommendations for managing BRCA1/2 carriers are still primarily based on retrospective risk estimates. Therefore, it is important to identify the source of the large variation in these retrospective estimates. The current lack of clarity can be troublesome for BRCA1/2 carriers and their physicians, particularly in the context of considering preventive treatment options.
The wide range of risk estimates in the retrospective cohorts of BRCA carriers may be attributable to a combination of two main factors: population differences, such as genetic, demographic, and lifestyle factors, and methodologic differences, such as population ascertainment and referral criteria, methods of risk estimation, and correction for selection bias.47–49 Although the observed variation in risk has often been attributed to population differences, it is unclear if some analytic approaches generate systematically higher or lower breast cancer risk estimates and which methods yield more precise estimates than others.
Our objective was to assess the effects of systematically identified risk estimation and bias-correction methods and population factors on CLTR point estimates and 95% CIs. We specifically surveyed published methods, applied them to the Family Cancer Clinic database at our university medical center, and compared our results with a reference using simulated datasets on the basis of this clinical data as well as to published prospective and retrospective data.
PATIENTS AND METHODS
Methods to Estimate the Risk of Breast Cancer in BRCA1/2 Carriers
We searched the literature using the keywords breast cancer, BRCA, and risk in the subject heading and/or title and abstract fields in three databases (PubMed, Embase, and Web of Science) to systematically identify the different risk estimation and bias-correction methods applicable to a clinically ascertained cohort of carriers with a pathogenic BRCA1/2 gene mutation. The search was restricted to studies published in English through July 2014 that included a study population of a clinical cohort of female mutation carriers. A flow diagram of the search is presented in the Data Supplement. The selection procedure and an additional search of the selected articles' reference lists yielded 201 reports of potential relevance, which were then reviewed in detail for their risk-estimation methods. Of these, 184 studies were excluded because the method was not applicable to a retrospective clinical cohort or because only risk ratios were presented.
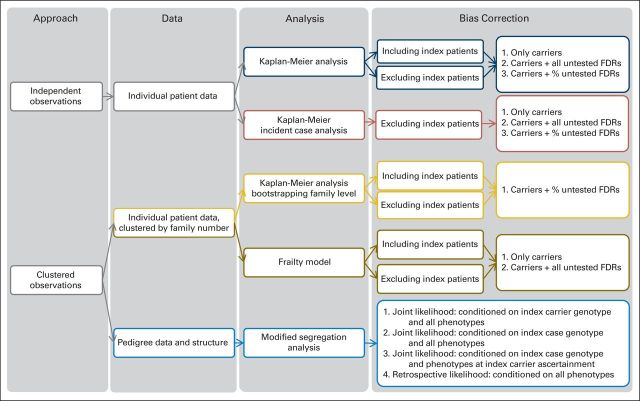
In total, 19 methods for risk estimation were identified and applied to our data (Fig 1 and Data Supplement). Eleven were Kaplan-Meier analyses (including three analyses of incident cases and two analyses with bootstrapping),6,14,19,21,28,37,40,44,50–53 four were frailty models, and four were modified segregation analyses.24,25,27,29,41,54
Fig 1.
Overview of risk estimation and bias correction methods applied to estimate cancer risk. FDR, first-degree relative.
Kaplan-Meier analysis.
Kaplan-Meier analysis allows estimation of survival over time. This nonparametric model can only incorporate independent observations; therefore, familial clustering and subject ascertainment are not taken into account. Bias correction was performed by one or a combination of the following: excluding index patients, including all untested female first-degree relatives (FDRs) who were treated as carriers, including only incident breast cancer cases, or including a proportion of untested female FDRs. The proportion of FDRs was estimated as the ratio of positive and negative DNA tests per age group defined by a 10-year interval from our data.
Kaplan-Meier analysis with bootstrapping at the family level.
Kaplan-Meier analysis with bootstrapping at the family level is a nonparametric analysis in which the 95% CI was corrected for familial clustering by bootstrapping with families as sampling units. Bias correction was performed by including untested FDRs that were weighted on the basis of the calculated posterior probabilities of untested FDRs carrying the mutation given their phenotypes and mutational frequency.
Frailty model.
The frailty model is a semiparametric model in which the familial clustering was accounted for by a hypothetical frailty for shared risk among family members. The frailty term has a multiplicative effect on the baseline hazard and provides a family-specific cancer risk. The marginal or population-averaged CLTR was calculated by integrating out the frailty term.55,56 In this analysis, a semiparametric frailty model with a gamma frailty distribution was used,57 and included only carriers, or carriers and untested relatives.
Modified segregation analysis.
The modified segregation analysis is a semiparametric analysis in which the familial clustering was accounted for by polygenic effects. All members in the pedigree, that is, FDRs and beyond, were included. CLTRs were calculated on the basis of estimated age group–specific hazard ratios and the cancer incidence of the general population.20 Correction for genetic testing and ascertainment bias was performed by maximizing the conditional likelihood of observing the genotypes and phenotypes in the pedigree, given the genotype and phenotype of the index patient or index carrier and the phenotypes of other family members in the pedigree or given all phenotypes only.
Box 1. Definitions.
Index carrier: The first family member (male or female) who tested positive for the mutation, irrespective of their cancer status at the time of the DNA test.
Index patient: If the index carrier was affected by breast and/or ovarian cancer at the time of the DNA test, he/she becomes the index patient. The index patients are a subgroup of the index carriers.
Untested FDRs: Women who did not undergo genetic testing and who are FDRs of a male or female carrier, and therefore have a 50% a priori chance of being a carrier.
Proportion of untested FDRs: The estimated proportion of assumed carriers among the untested FDRs. The proportion of FDRs assumed to be carriers were included in our analyses and treated as carriers.
Incident breast cancer cases: Cases that have arisen after the first positive DNA test in the family, that is, after the date of the index carrier's test. Only years at risk and events from this date forward were included in our analyses.
Application Dataset
To assess the effect of the different methods for statistical analyses and bias correction, we applied them to a well-defined, retrospective clinical cohort consisting of 192 extended BRCA mutation-positive families (112 BRCA1 families and 80 BRCA2 families) from our family cancer clinic.51,52 We also simulated data on the basis of our clinic database and applied all methods to these simulated data. As the true estimates are known in simulation, this helped us to assess the bias of overestimation or underestimation of the CLTR of these methods.
This family cancer clinic at the University Medical Center Groningen is the sole provider of genetic counseling in the northern region of the Netherlands. Information on breast and ovarian cancer and prophylactic surgery was available for 395 female BRCA1 and 232 female BRCA2 carriers and their untested female FDRs (349 in BRCA1 and 176 in BRCA2 families) ≥ 18 years old (Table 1). Pedigree information was available for 2,255 BRCA1 and 1,359 BRCA2 family members, including FDRs and beyond (Table 2). Only one proven carrier was present in 27 (14%) of the 192 families.
Table 1.
Characteristics of Individual Female BRCA1/2 Mutation Carriers (including index patients) and Their Untested First-Degree Female Relatives
| Characteristics |
BRCA1 |
BRCA2 |
||
|---|---|---|---|---|
| Carriers (n = 395) | FDRs (n = 349) | Carriers (n = 232) | FDRs (n = 176) | |
| Genetic test | ||||
| Age of index patient at testing, years, mean (SD) | 48.7 (9.8) | NA | 50.6 (10.5) | NA |
| Age at index carriers' test, years, mean (SD) | 47.1 (19.3) | 63.1 (29.7) | 48.7 (17.9) | 64.2 (27.3) |
| Breast cancer in index patients | ||||
| No. (%) | 78 (80.4) | NA | 56 (90.3) | NA |
| Age, years, mean (SD) | 40.1 (9.0) | NA | 44.9 (9.3) | NA |
| Breast cancer | ||||
| No. (%) | 182 (46.1) | 59 (16.9) | 105 (45.3) | 43 (24.4) |
| Age, years, mean (SD) | 42.5 (9.8) | 45.7 (13.0) | 46.7 (10.3) | 51.1 (12.0) |
| Ovarian cancer in index patients | ||||
| No. (%) | 34 (35.1) | NA | 14 (22.6) | NA |
| Age, years, mean (SD) | 48.4 (7.6) | NA | 54.5 (12.1) | NA |
| Ovarian cancer | ||||
| No. (%) | 89 (22.5) | 41 (11.7) | 25 (10.8) | 11 (6.3) |
| Age, years, mean (SD) | 51.0 (10.1) | 51.4 (10.1) | 55.7 (11.9) | 62.9 (11.8) |
| RRM | ||||
| No. (%) | 84 (21.3) | 1 (0.3) | 48 (20.7) | 0 (0) |
| Age, years, mean (SD) | 41.5 (9.9) | 36.5 (NA) | 43.1 (8.0) | NA |
| RRSO | ||||
| No. (%) | 155 (39.2) | 3 (0.9) | 100 (43.1) | 1 (0.6) |
| Age, mean (SD) | 45.6 (9.0) | 45.8 (17.7) | 47.9 (9.4) | 41.5 (NA) |
Abbreviations: FDR, first-degree relative; NA, not applicable; RRM, risk-reducing mastectomy; RRSO, risk-reducing salpingo-oophorectomy.
Table 2.
Characteristics of BRCA1/2 Mutation Families
| Characteristic |
BRCA1 Families (n = 112) |
BRCA2 Families (n = 80) |
||
|---|---|---|---|---|
| Overall No. (%) | Per Family* Median No. (IQR) | Overall No. (%) | Per Family* Median No. (IQR) | |
| Family members | 2,255 (100) | 15 (10-28) | 1,359 (100) | 13 (10-19) |
| Females | 1,171 (51.9) | 7 (4-14) | 677 (49.8) | 7 (4-9) |
| Index patients | 97 (4.3) | 1 (1-1) | 66 (4.9) | 1 (1-1) |
| Females | 97 (100) | 1 (1-1) | 62 (93.9) | 1 (1-1) |
| Index carriers | 112 (5.0) | 1 (1-1) | 80 (5.9) | 1 (1-1) |
| Females | 111 (99.1) | 1 (1-1) | 73 (91.3) | 1 (1-1) |
| Mutation carriers | 511 (22.7) | 3 (2-6) | 318 (23.4) | 3 (2-5) |
| Untested relatives | 1,105 (49.0) | 7 (5-14) | 615 (45.3) | 6 (4-8) |
| Noncarriers | 639 (28.3) | 4 (2-8) | 426 (31.3) | 4 (2-7) |
| Female cancer† | ||||
| Female breast cancer | 257 (21.9) | 2 (1-3) | 158 (23.3) | 2 (1-2) |
| Ovarian cancer | 138 (11.8) | 2 (1-2) | 40 (5.9) | 0 (0-1) |
| Male breast cancer† | 2 (0.2) | 0 (0-0) | 7 (1.0) | 0 (0-0) |
Abbreviations: IQR, interquartile range; NA, not applicable.
Data are presented irrespective of the family size (ie, not weighted by family size).
Numbers and percentages per sex.
During the normal course of genetic risk counseling, patients were asked to provide information on their family history, and family pedigrees were drawn. In a previous study that included 185 of the current families, pedigrees were drawn and data on family members were collected.51 Data from this previous study were recorded in a database and updated through September 2011.52 The database contained information on the BRCA mutation, pedigree structure, date of birth, date of death or last contact, date of breast and/or ovarian cancer diagnosis, date of prophylactic surgery, and carrier status of family members. Missing were 2% to 3% of the dates of death or dates of breast and/or ovarian cancer and 10% of the dates of birth. These missing values were imputed by using national tumor-, period-, and/or age-specific incidence and/or survival rates.54
This clinical dataset was used as basis for generating 50 datasets with 100,000 three-generation families consisting of 18 relatives. For each individual, we generated a mutational status for the BRCA genes, a polygenic component that represents other familial risk factors, follow-up time, breast cancer status, and censoring events (Appendix, online only).
Statistical Analysis
Population-averaged CLTRs and 95% CIs (floating 95% CIs for modified segregation analysis20) were estimated by using the different risk-estimation and bias-correction methods we had identified. In all analyses, primary breast cancer cases were counted as events, and the censoring time was defined as the first date of the following events: diagnosis of ovarian cancer, risk-reducing mastectomy, risk-reducing salpingo-oophorectomy, or death or last contact.
First, the effects of the risk-estimation and bias-correction methods were assessed by comparing the CLTR estimates by age 70 and the width of the corresponding 95% CI in our real clinic data. Second, the CLTR estimates and 95% CIs were compared with the reference in our simulated data. We specifically calculated mean CLTRs and 95% CIs widths for the 50 simulated datasets. Third, we assessed the effect of study-population factors by comparing the CLTR estimates and 95% CIs from our clinic data to the published CLTR estimates that had been obtained by the same method.
Box 2. Statistical Terms.
Right censoring: By the time that a censoring event occurs, a woman has not developed breast cancer. Years at risk and events after the right censoring are not counted in the analyses.
Bootstrapping at family level: Randomly drawing samples (with the same sample size) from the original dataset to estimate the CI. The samples are of the same size as the original dataset; therefore, one family can be included multiple times in the same data sample.
Width of 95%CI: Indicator of the uncertainty around the CLTRs. It is calculated by subtracting the lower CI from the upper CI.
Relative bias: Measure of underestimation and overestimation of the reference CLTR, calculated as: (estimated CLTR − reference CLTR)/reference CLTR.
Kaplan-Meier and frailty model analyses were performed with a statistical program (version 22; SPSS, Chicago, IL) and with R software (R Foundation for Statistical Computing, Vienna, Austria).57,58 Modified segregation analyses were performed in MENDEL (Department of Human Genetics, University of California, Los Angeles, CA) using additional subroutines.20,59,60
RESULTS
Comparison of Breast Cancer Risk Estimates and CIs Across Analytical Methods
Table 3 shows the CLTR estimate and 95% CI for each method on the basis of the real data. Kaplan-Meier analyses (including all carriers with bias correction by excluding index patients, including all untested FDRs, or including a proportion of untested FDRs) yielded estimates by age 70 years of 35% to 66% (width of the 95% CI, 10% to 19%) for BRCA1 carriers and 41% to 73% (width of the 95% CI, 13% to 26%) for BRCA2 carriers. Overall, analyses that excluded the index patients and included all untested female FDRs yielded the lowest CLTRs (for BRCA1, 35%; 95% CI, 30% to 40%; and for BRCA2, 41%; 95% CI, 34% to 49%).
Table 3.
Cumulative Lifetime Risk of (in %) and 95% CI of Breast Cancer in BRCA1/2 Carriers by Age 70 Years by Method of Analysis
| Bias Correction Method* |
BRCA1 |
BRCA2 |
||||
|---|---|---|---|---|---|---|
| n/N | CLTR | 95% CI | n/N | CLTR | 95% CI | |
| Kaplan-Meier analysis | ||||||
| Including index patients | 161/395 | 66.4 | 58.7 to 74.0 | 101/232 | 72.9 | 63.2 to 81.8 |
| Including index patients and including proportion of untested FDRs | 212/590 | 54.5 | 49.0 to 60.3 | 139/332 | 63.6 | 56.2 to 70.9 |
| Including index patients and including all untested FDRs | 218/744 | 43.6 | 38.9 to 48.5 | 144/408 | 51.9 | 45.5 to 58.6 |
| Excluding index patients | 87/298 | 54.3 | 45.2 to 64.0 | 46/170 | 55.7 | 43.0 to 69.1 |
| Excluding index patients and including proportion of untested FDRs | 138/493 | 45.4 | 39.4 to 52.0 | 84/270 | 52.1 | 43.6 to 61.2 |
| Excluding index patients and including all untested FDRs | 144/647 | 34.9 | 31.1 to 40.2 | 89/346 | 40.5 | 33.7 to 48.1 |
| Kaplan-Meier incident cases analysis† | ||||||
| Excluding index patients | 23/167 | 83.4 | 62.5 to 96.2 | 10/114 | 86.0 | 56.9 to 99.0 |
| Excluding index patients and including proportion of untested FDRs | 25/232 | 75.6 | 56.3 to 90.9 | 10/139 | 77.6 | 51.5 to 95.5 |
| Excluding index patients and including all untested FDRs | 26/289 | 67.2 | 49.1 to 84.2 | 10/137 | 72.7 | 47.6 to 92.7 |
| Kaplan-Meier analysis with bootstrapping at family level | ||||||
| Including index patients and including proportion of untested FDRs | 208/495 | 72.8 | 65.4 to 80.2 | 139/332 | 80.4 | 71.6 to 89.3 |
| Excluding index patients and including proportion of untested FDRs | 136/403 | 66.0 | 57.5 to 74.4 | 84/270 | 70.5 | 54.3 to 86.6 |
| Frailty model analysis | ||||||
| Including index patients | 161/395 | 67.4 | 59.6 to 75.1 | 101/232 | 73.9 | 64.2 to 83.7 |
| Including index patients and including all untested FDRs | 218/744 | 44.7 | 39.4 to 49.9 | 144/408 | 53.3 | 46.5 to 60.2 |
| Excluding index patients | 87/298 | 54.4 | 45.0 to 63.8 | 46/170 | 56.2 | 41.7 to 70.7 |
| Excluding index patients and including all untested FDRs | 144/647 | 35.1 | 29.8 to 40.4 | 89/346 | 41.3 | 33.9 to 48.8 |
| Modified segregation analysis‡ | ||||||
| Joint likelihood conditioned on genotype of index carriers and all phenotypes | 156/1,060 | 36.6 | 18.8 to 50.4 | 96/604 | 42.4 | 14.9 to 61.0 |
| Joint likelihood conditioned on genotype of index patients and all phenotypes | 158/1,074 | 40.7 | 25.6 to 52.8 | 98/615 | 49.4 | 30.5 to 63.1 |
| Joint likelihood conditioned on genotype of index patients and phenotypes at time of index patients' DNA test | 158/1,074 | 57.1 | 43.7 to 67.3 | 98/615 | 53.2 | 34.8 to 66.4 |
| Retrospective likelihood conditioned only on all phenotypes | 230/1,171 | 52.8 | 43.2 to 60.8 | 151/677 | 67.4 | 55.8 to 75.9 |
Abbreviations: CLTR, cumulative lifetime risk; FDRs, first-degree relatives; n, total number of events (ie, female breast cancer); N, total number of women at risk in the analysis.
Right censoring at date of first event (which might be diagnosis of breast cancer, ovarian cancer, risk-reducing mastectomy, risk-reducing salpingo-oophorectomy, or last contact or death).
Incident case analysis includes only years at risk and events after the date of the first positive DNA test in the family.
Modeling the probability of breast cancer conditioned on the genotype and phenotype of the index patients or index carriers, and/or the phenotype of relatives.
Including only incident cases yielded the highest CLTRs and the widest 95% CIs, with values of 67% to 83% (width of the 95% CI, 34% to 35%) for BRCA1 carriers and 73% to 86% (width of the 95% CI, 42% to 45%) for BRCA2 carriers. Estimates from the incident cases analyses including all carriers and FDRs were similar to the estimated CLTRs without any correction. For BRCA1, the result was 66% (95% CI, 59% to 74%) and for BRCA2, 73% (95% CI, 63% to 82%).
The bootstrap approach (including all carriers, with bias corrected by excluding index patients and including a proportion of FDRs) yielded estimates by age 70 years of 66% to 73% (width of the 95% CIs, 15% to 17%) for BRCA1 carriers and 70% to 80% (width of the 95% CIs, 18% to 32%) for BRCA2 carriers. Their point estimates were higher and their 95% CIs were wider than those of the Kaplan-Meier analyses that included a proportion of FDRs.
The frailty model, including all carriers, with bias corrected by excluding index patients and/or including all untested FDRs, produced point estimates similar to those of the Kaplan-Meier analyses. However, the range of the 95% CIs was somewhat wider. The CLTRs were 35% to 67% (width of the 95% CI, 11% to 19%) for BRCA1 carriers and 41% to 74% (width of the 95% CI, 14% to 29%) for BRCA2 carriers.
Modified segregation analyses with a conditional joint likelihood yielded lower CLTRs by age 70 years. Results were 37% to 57% (width of the 95% CI, 24% to 32%) for BRCA1 carriers and 42% to 53% (width of the 95% CI, 32% to 46%) for BRCA2 carriers. When the likelihood was conditional solely on the basis of phenotypes, the CLTR of 53% (95% CI, 43% to 61%) for BRCA1 carriers was still relatively low. However, the CLTR of 67% (95% CI, 56% to 76%) for BRCA2 carriers was relatively high. The analyses with conditioning of the genotype on the basis of index carriers or index patients were most comparable with the Kaplan-Meier analyses that included all FDRs, while excluding or including index patients.
Relative Bias of Breast Cancer Risk Estimates in Simulated Data Across Analytical Methods
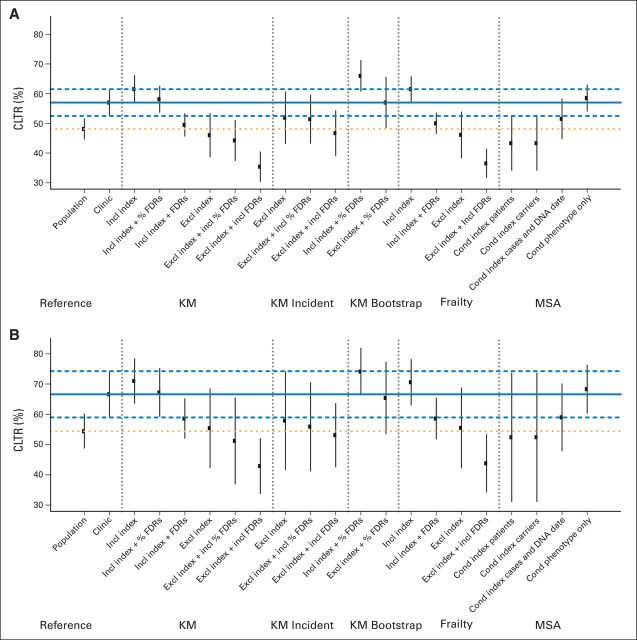
The CLTR of all methods varied from 35% to 66% in BRCA1 mutation carriers and from 43% to 74% in BRCA2 mutation carriers (Fig 2). Compared with the reference, this translated into a variation in the relative bias of −38% to +16% and −36% to +11%, respectively (Appendix Table 2, online only).
Fig 2.
Comparison of each method's cumulative lifetime risks (CLTRs; 95% CI) by age 70 years with the reference estimate in (A) BRCA1 and (B) BRCA2 mutation carriers on the basis of the simulated data. The solid and dashed horizontal lines represent the CLTR and 95% CI of the reference for clinic-based cohorts, and the dotted line represents the reference CLTR for population-based cohorts. Cond, conditioned on; Excl, excluding; FDR, first-degree relative; Incl, including; KM, Kaplan-Meier; MSA, modified segregation analysis.
Bias-correction methods that yielded the smallest bias and uncertainty were Kaplan-Meier analysis with inclusion of index patients and untested FDRs (+2.0%; SD, 2.1, in BRCA1 carriers and +0.9%; SD, 3.6, in BRCA2 carriers) and the modified segregation analysis conditioned on phenotype only (+2.7%; SD, 2.2, in BRCA1 carriers and +2.5%; SD, 3.4, in BRCA2 carriers). Kaplan-Meier analysis with bootstrapping at the family level was, on average, the least biased, but its uncertainty was relatively higher because relative bias differed for all datasets, with +0.0%, (SD, 5.7) in BRCA1 carriers and −1.8% (SD, 8.7) in BRCA2 carriers.
Kaplan-Meier analyses with exclusion of the index patient and inclusion of FDRs, as well as the modified segregation analyses conditioned on all phenotypes and genotypes of the index patient or carrier, produced the most underestimated risk with relative biases greater than 20%. However, these methods yielded risk estimates that approximated the risk of a carrier in the general population.
Comparison With Published Results from Retrospective Studies Using the Same Methods
The risk difference between our CLTRs and estimates from the identified publications with the same method varied from 1% to 35% for BRCA1 mutation carriers and from 1% to 37% for BRCA2 mutation carriers (Table 4). Median risk variation in the CLTRs from all Kaplan-Meier analyses was 6% for BRCA1 and 8% for BRCA2 carriers. Median variation was 14% and 25%, respectively for the modified segregation analyses. For some methods, complete comparison was not possible because the published estimates were for the combined event of breast or ovarian cancer,6,14,21,44 or the estimates were only for BRCA1 carriers.25,37
Table 4.
Comparison Between This Study and Published Studies of the Cumulative Lifetime Risk of Breast Cancer by Age 70 and 95% CIs of Right-Censored Analyses by Method of Analysis
| Analysis |
BRCA1 |
BRCA2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Present Study |
Published Study* |
Present Study |
Published Study* |
|||||||
| N | CLTR (95% CI) | N | CLTR (95% CI) | Study | N | CLTR (95% CI) | N | CLTR (95% CI) | Study | |
| Kaplan-Meier analysis | ||||||||||
| Including index patients | 395 | 66 (59 to 74) | 40 | 64 (39 to 78) | Beristain et al50 | 232 | 73 (63 to 82) | 50 | 69 (40 to 84) | Beristain et al50 |
| 308 | 71 (67 to 82) | Van der Kolk et al51 | 433 | 72 (64 to 78) | Vos et al52 | |||||
| 656 | 72 (66 to 78) | Vos et al52 | 394 | 78 (69 to 85) | Vos et al52 | |||||
| 483 | 73 (68 to 78) | Brose et al19 | 178 | 88 (82 to 93) | Van der Kolk et al51 | |||||
| 1,580 | 76 (71 to 79) | Vos et al52 | 220 | 88 (81 to 95) | Tea et al53 | |||||
| 264 | 85 (75 to 97) | Kroiss et al28 | ||||||||
| Including index patients and including proportion of untested FDRs | 590 | 55 (49 to 60) | 839 | 68 (65 to 71) | Evans et al40 | 332 | 64 (56 to 71) | 603 | 75 (72 to 78) | Evans et al40 |
| Excluding index patients | 167 | 54 (45 to 64) | 24 | 36 (5 to 57) | Beristain et al50 | 114 | 56 (43 to 69) | 34 | 38 (12 to 56) | Beristain et al50 |
| 16 fam | ±52 (NA) | Dorum et al6 | 305 | 61 (50 to 69) | Vos et al52 | |||||
| 77 | 53 (35 to 75) | Vogl et al37 | 269 | 64 (50 to 75) | Vos et al52 | |||||
| 14 fam | ±57 (NA) | Dorum et al6 | 120 | 78 (69 to 88) | Van der Kolk et al51 | |||||
| 462 | 58 (51 to 66) | Heimdal et al21 | ||||||||
| 467 | 58 (50 to 66) | Vos et al52 | ||||||||
| 214 | 60 (55 to 66) | Van der Kolk et al51 | ||||||||
| 1,091 | 68 (62 to 73) | Vos et al52 | ||||||||
| Joint likelihood conditioned on genotype of index carriers and all phenotypes | 112 fam | 37 (19 to 50) | 582 fam | 45 (36 to 52) | Brohet et al54 | 80 fam | 42 (15 to 61) | 176 fam | 27 (14 to 38) | Brohet et al54 |
| 155 fam | 52 (26 to 69) | Milne et al41 | 164 fam | 47 (29 to 60) | Milne et al41 | |||||
| 2 fam | 64 (28 to 96) | Tesoriero et al29 | 27 fam | 75 (0 to 97) | Antoniou et al30 | |||||
| 25 fam | 72 (0 to 93) | 6 fam | 79 (48 to 98) | Tesoriero et al29 | ||||||
| Modified segregation analysis | ||||||||||
| Joint likelihood conditioned on genotype of index patients and all phenotypes | 112 fam | 41 (26 to 53) | 28 fam | 48 (22 to 82) | Scott et al24 | 80 fam | 49 (31 to 63) | 23 fam | 74 (50 to 93) | Scott et al24 |
| Joint likelihood conditioned on all phenotypes | 112 fam | 53 (43 to 61) | 1 fam | 49 (13 to 96) | Southey et al25 | 80 fam | 67 (56 to 76) | NA | NA | NA |
| 1 fam | 39 (29 to 49) | Vogl et al37 | ||||||||
Abbreviations: CLTR, cumulative lifetime risk; fam, families; FDRs, first-degree relatives; NA, not applicable.
Estimates from studies identified in our literature search.
DISCUSSION
Published CLTRs of breast cancer in BRCA1/2 carriers vary widely, most likely because of a combination of differences in the study populations and applied methods. We aimed, first, to assess the effects of different methods of risk estimation and bias correction on the CLTRs and 95% CIs generated in a large, homogeneous, retrospective clinic-based cohort of BRCA1/2 carriers and, second, to assess the effect of differences in study populations. We applied 19 methods that resulted in CLTRs between 35% and 83% for BRCA1 carriers and between 41% and 86% for BRCA2 carriers; widths of the 95% CIs varied between 10% and 35% and between 11% and 46%, respectively. Bias correction by including index patients and a proportion of untested FDRs or by conditioning the likelihood function only on phenotypic data yielded rather accurate CLTRs for the context of our family cancer clinic. Comparison of our CLTRs with retrospective CLTRs estimated by applying the same method showed risk variations between 1% and 37%.
Without any bias correction, the CLTR in our study population was 67% (95% CI, 59% to 74%) for BRCA1 carriers and 73% (95% CI 63% to 82%) for BRCA2 carriers. Bias correction resulted in a stepwise decreasing effect in the CLTRs compared with the unadjusted CLTR. The only exception was the bootstrap approach for which including a proportion of FDRs resulted in higher risk estimates compared with no inclusion of FDRs. The posterior probability of assumed carriers among the untested FDRs was probably overestimated and led to overestimation of the number of carriers among affected FDRs or to underestimation of carriers among unaffected FDRs. In the simulation, however, this bootstrap approach with exclusion of index patients yielded, on average, the least biased CLTRs: +0% (SD, 5.7) for BRCA1 and −1.8% (SD, 8.7) for BRCA2. Because this uncertainty is high, this method needs further exploration.
Overall, low CLTRs were produced from the analyses that excluded all index patients but included all untested FDRs (for BRCA1, 35%; 95% CI, 31% to 40%; and for BRCA2, 41%; 95% CI, 34% to 48%) and by the modified segregation analysis in which the likelihood was conditioned on the genotype and phenotype of the index carriers and all other phenotypes in the family (for BRCA1, 36%; 95% CI, 19% to 50%; and BRCA2, 42%; 95% CI, 15% to 61%). Kaplan-Meier analyses excluding index patients and including FDRs and modified segregation analyses with conditioning on the basis of the genotype and phenotypes produced estimates approximating the risk for carriers in the general population. Here, no ascertainment bias is present. However, because not all of these carriers will enter the family cancer clinic, these estimates are substantially lower.
High CLTRs were produced by the incident case analyses that included only carriers. The result for BRCA1 was 83% (95% CI, 63% to 96%) and for BRCA2, 86% (95% CI, 57% 99%). This could have been a result of genetic testing bias among relatives and to having more follow-up information about affected relatives than about unaffected relatives. This explanation is likely given the simulation results, for which follow-up was complete and the CLTR was underestimated (≥ −9%). However, because the number at risk and the number of events were low, and because the CIs were large and overlapped with the reference CLTRs, results regarding this method still are uncertain.
The width of the 95% CI depends on two main factors: first, the number of women at risk and the number of events in the analysis and, second, whether familial clustering is taken into account. The Kaplan-Meier analyses excluding index patients, the analyses of incident cases, and the modified segregation analyses all led to relatively large SEs. Accounting for familial clustering in the analyses of individual subjects (ie, frailty model and Kaplan-Meier analysis with bootstrapping at the family level) had only a small positive effect on the 95% CIs: width of the 95% CIs, > 0% to 4.3% for BRCA1 and > 0.5% to 14.7% for BRCA2. This small effect was probably because not all the women in the family were FDRs. The greatest effect on the CI was seen in the bootstrap approach and was probably because the approach used to calculate the proportions for including FDRs and because no structure was imposed for familial clustering.
In general, risk estimates from prospective studies are considered most reliable. CLTRs most similar to those of the largest prospective clinic-based cohort (EMBRACE [Epidemiological Study of Familial Breast Cancer])45 were the Kaplan-Meier analyses with bias correction by either including index patients with a proportion of FDRs (risk difference compared with EMBRACE, −5.5% for BRCA1 and +6.0% for BRCA2) or by solely excluding index patients (risk difference, −5.7% for BRCA1 and +1.3% for BRCA2), and those of the modified segregation analyses with conditioning of the phenotypes restricted to those at the time of the index carrier's DNA test (risk difference, −2.9% for BRCA1 and +3.4% for BRCA2). Although population differences may interfere with the comparison, the good performance of the first method was confirmed in the simulation.
Methods of previous retrospective cohort studies included in this study produced estimated CLTRs of 30% to 85% for BRCA1 carriers and 27% to 88% for BRCA2 carriers.6,19,21,24–26,28–30,37,40,50–54 These risk ranges are broader than the range of estimates on the basis of our current dataset. However, for each method, we still observed considerable variation (as high as 37%) when we compared our estimates with previously published estimates. This demonstrates that there are other factors in addition to the risk- and bias-correction methods that affect the CLTR. These could include population and demographic factors (eg, birth cohorts, founder mutations, mutation type, family history) and/or other methodological issues. For example, these include the events chosen for right censoring, the decision to censor at the age of ovarian cancer or age of risk-reducing salpingo-oophorectomy, and how many times these events occur in the study population.6,48 Differences in these choices are related to issues of competing risks and informative censoring, which will affect the occurrence of breast cancer. Some authors have therefore published risk estimates for developing breast or ovarian cancer, that is, with the cancer event defined as primary breast cancer or ovarian cancer instead of primary breast cancer only (ie, with or without censoring at ovarian cancer).6,14,21,30,44
Some researchers using the modified segregation analyses adopted a fixed population incidence24,25,30,41 as input for the model, whereas others used a birth cohort–specific incidence,54 which might have an effect on the estimated CLTR. However, in an additional sensitivity analysis on the modified segregation model, we found that the model was, in fact, quite robust for possible mis-specification of the population incidence input; a 10% increase in the input incidence resulted in a 1% to 3% increase in the CLTR.
The strength of our study was that it demonstrated the effects of a large number of bias-correction methods in one large, well-defined BRCA cohort and a simulated reference cohort. Some of the methods have been applied in several cohorts at the same time,20,37 but most authors present their CLTRs only with the inclusion and exclusion of the index patients.50–52 Women participating in a clinical cohort have already undergone genotype analysis, and data are gathered in the course of their standard care. This process makes this type of ascertainment the most straightforward and feasible manner for estimating breast cancer risk. However, this common design incorporates an ascertainment bias and a genetic testing bias, and these should both be avoided by properly correcting for them.
A limitation of our study is that the simulation was restricted to one scenario and tailored to the Dutch setting. Differences in genetic testing or ascertainment bias patterns (eg, as a result of different screening and referral guidelines) in other clinic-based cohorts, presumably from other countries, could affect performance of the methods. However, in simulations with a higher input value for the polygenic variance, ascertainment was more biased, but conclusions on the performance of the methods did not change (data not shown). Another limitation is that we applied only one approach for censoring events, whereas the estimation of the CLTRs could be affected by the chosen events.
In conclusion, when tested systematically in one retrospective clinical cohort of BRCA1/2 carriers, much of the variation in the CLTRs and CIs seems to be due to the method of bias correction used, whereas a smaller part is due to population differences. The modified segregation analysis is a complex method that concentrates on correcting all biases affecting the risk estimation.20 Our study shows that the modified segregation analysis, with ascertainment correction on the basis of the genotype of the indexes and all phenotypes in the family, yields estimates that most closely approach that of an unselected carrier in the general population. Most consistent estimates for carriers counseled in the clinic were estimated with the Kaplan-Meier method with bias corrected by including a proportion of untested FDRs. Compared with the other methods, this might be a simpler and more robust method to apply to clinical retrospective datasets. Future studies should focus on family-specific breast cancer risk estimates in BRCA carriers instead of population-averaged risks, and investigators should assess the effect of competing risks on the risk estimates and CIs.
Supplementary Material
Acknowledgment
We thank A. C. Antoniou, PhD, for advice on the modified segregation analysis and Jackie Senior for editing the manuscript.
Glossary Terms
- BRCA1:
a tumor suppressor gene known to play a role in repairing DNA breaks. Mutations in this gene are associated with increased risks of developing breast or ovarian cancer.
- BRCA2:
a tumor suppressor gene whose protein product is involved in repairing chromosomal damage. Although structurally different from BRCA1, BRCA2 has cellular functions similar to BRCA1. BRCA2 binds to RAD51 to fix DNA breaks caused by irradiation and other environmental agents. Also known as the breast cancer 2 early onset gene.
- competing risks:
events that prevent an event of interest from occurring. “Competing risks are said to be present when a patient is at risk of more than one mutually exclusive event, such as death from different causes, and the occurrence of one of these will prevent any other event from ever happening” (Hinchliffe SR: Presented at the University of Leicester, 2012).
Appendix 1: Simulation Study
Structure of Families
Based on our clinical database, a general population was generated consisting of 100,000 families. All were three-generation families with a fixed pedigree structure similar to the current average Dutch families (Data Supplement; Jonker et al: J Med Genet 40:e25, 2003). Age at last contact in the first generation was generated following a normal distribution N (85,3). In the second and third generation, the age at contact was based on the mean age of the parents minus an age difference between the generations. This age difference was N (25,2) and N (26,2), respectively.
Genes and Polygenic Factors
BRCA1 and BRCA2 gene mutations were generated using gene dropping, with mutation frequencies of 0.003 and 0.001, respectively, and following a Mendelian transmission with an autosomal-dominant inheritance pattern (Jonker et al: J Med Genet 40:e25, 2003). Individuals could not have both mutations, mutation carriers were always heterozygote for the mutation, and in-laws were considered always negative for the mutation.
A polygenic risk factor, following a normal distribution, was generated to represent other familial risk factors affecting the individuals' cancer risk (Pankratz et al: Genet Epidemiol 28:97-109, 2005).38 In the first generation, the mean of the distribution was zero, in the second and third generations, mean of the distribution was equal to mean of the parents' polygenic component. In the first generation, the variance of the distribution was equal to input value 1.5, and in the second and third generation, the variation was equal to the half of the input value (Pankratz et al: Genet Epidemiol 28:97-109, 2005).38
Cancer and Censoring Events
The age at cancer (≥ 20 years) and death were generated following a Weibull distribution on the basis of Dutch population data, and for male carriers, a relative risk was used to generated the cancer incidence (Table 1; Thompson and Easton: Am J Hum Genet 68:410-419, 2001; Liede et al: J Clin Oncol 22:735-742, 2004; Netherlands Cancer Registry: http://www.cijfersoverkanker.nl/selecties/dataset_1/img54b79b31474b4; Statistics Netherlands: http://statline.cbs.nl/Statweb/publication/?DM=SLNL&PA=7052_95&D1=0&D2=1-2&D3=a&D4=56-61&VW=T).
An age-related probability for undergoing risk-reducing mastectomy or risk-reducing mastectomy salpingo-oophorectomy was applied using age-dependent probabilities on the basis of our clinical database. Female carriers ≥ 25 years old without breast cancer and ≥ 30 years old without ovarian cancer were eligible for risk-reducing mastectomy and risk-reducing mastectomy salpingo-oophorectomy, respectively.
Genetic Testing and the Index Person
When more individuals in the family fulfilled the Dutch referral criteria for genetic counseling and testing, the affected person with the youngest age at diagnosis was tested for the mutation (Mammacarcinoma, http://www.oncoline.nl/breastcancer). Then, when more individuals in the family fulfilled the referral criteria and only unaffected individuals were still alive at the time of referral, the individual most closely related to the cancer patient was tested.
When the index patient was positive for a BRCA1 or BRCA2 mutation, genetic testing was offered to the family. If the index patient tested negative for the gene mutation, the family was not offered further testing.
Affected family members were tested irrespective of any cascade protocol, and their probability of being tested was greater than their unaffected relatives. Unaffected family members were tested following a cascade protocol. The genetic testing probability was based on the clinical database, and was gender, phenotype, and age dependent.
Risk Analyses in a Clinic-Based Cohort
Dutch referral criteria for genetic counseling and testing were used to mimic the ascertainment bias seen in the family cancer clinic cohorts. Subsequently, the genetic testing bias was mimicked by making the probability for genetic testing in all relatives of the index carrier gender, age, and phenotype dependent (Oosterwijk et al: Maturitas 78:252-257, 2014).
In total, 50 datasets were generated with the same input values but with a different random seed.
All methods were applied to each dataset with these ascertainment and genetic testing biases (Table 2). Because the aim of the study was to assess the risk estimates for BRCA1/2 carriers seen in the family cancer clinic, we obtained the reference risk by Kaplan-Meier estimation using the same cohort with complete information on all genotypes. Thus, the reference estimate for the clinic would be a cohort affected by the same ascertainment bias but not by the genetic testing bias, whereas the general population cohort would not be affected by either.
Table A1.
Weibull Scale and Shape Input Parameters and Relative Risks
| Females |
Males |
|||
|---|---|---|---|---|
| Scale | Shape | Scale | Shape | |
| Breast cancer | ||||
| Noncarriers | 151.28 | 3.29 | 263.16 | 5.9 |
| BRCA1 carriers | 72.19 | 2.46 | RR | 3 |
| BRCA2 carriers | 70.59 | 3.12 | RR | 70 |
| Ovarian cancer | ||||
| Noncarriers | 242.72 | 4.06 | NA | |
| BRCA1 carriers | 84.22 | 3.58 | ||
| BRCA2 carriers | 91.86 | 4.52 | ||
| Death | 113.64 | 7.5 | 113.64 | 8.3 |
Abbreviations: NA, not applicable; RR, relative risk.
Table A2.
Estimated Cumulative Lifetime Risks and Relative Bias by Age 70 Years for 50 Simulated Datasets
| Bias Correction Method* |
BRCA1 Carriers |
BRCA2 Carriers |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CLTR |
Relative Bias (%) |
CLTR |
Relative Bias (%) |
|||||||
| Mean | SD of Means | Mean SD | Mean | SD | Mean | SD of Means | Mean SD | Mean | SD | |
| Reference estimate population | 48.1 | 1.8 | 1.6 | −15.6 | 2.1 | 54.4 | 2.9 | 3.6 | −18.2 | 4.0 |
| Reference estimate clinic | 57.0 | 2.3 | 2.9 | 0 | 0 | 66.6 | 3.9 | 7.6 | 0 | 0 |
| Kaplan-Meier analysis | ||||||||||
| Including index patients | 61.5 | 2.4 | 3.7 | 7.9 | 1.9 | 71.0 | 3.8 | 9.8 | 6.6 | 3.4 |
| Including index patients and including proportion of untested FDRs | 58.1 | 2.3 | 3.0 | 2.0 | 2.1 | 67.2 | 4.1 | 8.0 | 0.9 | 3.6 |
| Including index patients and including all untested FDRs | 49.5 | 2.0 | 1.8 | −13.1 | 1.8 | 58.6 | 3.4 | 4.7 | −11.9 | 3.5 |
| Excluding index patients | 46.0 | 3.8 | 3.2 | −19.4 | 4.6 | 55.4 | 6.7 | 9.7 | −17.0 | 8.5 |
| Excluding index patients and including proportion of untested FDRs | 44.2 | 3.5 | 2.5 | −22.4 | 4.5 | 51.2 | 7.3 | 6.9 | −23.2 | 9.5 |
| Excluding index patients and including all untested FDRs | 35.4 | 2.6 | 1.2 | −38.0 | 3.2 | 42.9 | 4.7 | 3.4 | −35.6 | 6.2 |
| Kaplan-Meier incident case analysis† | ||||||||||
| Excluding index patients | 51.9 | 4.5 | 4.7 | −9.0 | 6.5 | 57.9 | 8.3 | 12 | −13.2 | 11.1 |
| Excluding index patients and including proportion of untested FDRs | 51.4 | 4.2 | 4.3 | −9.9 | 6.1 | 55.9 | 7.5 | 10.4 | −16.1 | 10.2 |
| Excluding index patients and including all untested FDRs | 46.7 | 3.9 | 2.9 | −18.2 | 5.7 | 53.1 | 5.4 | 7.4 | −20.3 | 7.0 |
| Kaplan-Meier analysis with bootstrapping at family-level | ||||||||||
| Including index patients and including proportion of untested FDRs | 66.0 | 2.7 | 2.4 | 15.8 | 2.2 | 74.1 | 4.0 | 3.8 | 11.2 | 3.6 |
| Excluding index patients and including proportion of untested FDRs | 57.0 | 4.4 | 3.9 | 0.0 | 5.7 | 65.4 | 6.1 | 6.6 | −1.8 | 8.7 |
| Frailty model analysis | ||||||||||
| Including index patients | 61.5 | 2.2 | 2.2 | 7.9 | 2 | 70.6 | 3.9 | 3.8 | 6.0 | 3.4 |
| Including index patients and including all untested FDRs | 50.0 | 1.8 | 1.8 | −12.3 | 1.9 | 58.6 | 3.5 | 3.2 | −12.0 | 3.6 |
| Excluding index patients | 46.1 | 4 | 3.6 | −19.3 | 5.3 | 55.5 | 6.8 | 7.4 | −16.7 | 8.9 |
| Excluding index patients and including all untested FDRs | 36.5 | 2.5 | 2.3 | −36.0 | 3.1 | 43.8 | 4.9 | 4.6 | −34.3 | 6.3 |
| Modified segregation analysis‡ | ||||||||||
| Joint likelihood conditioned on genotype of index carriers and all phenotypes | 43.3 | 4.7 | 7.7 | −23.9 | 8.5 | 52.4 | 10.9 | 14.2 | −21.1 | 17.0 |
| Joint likelihood conditioned on genotype of index patients and all phenotypes | 43.3 | 4.7 | 7.7 | −23.9 | 8.5 | 52.4 | 10.9 | 14.2 | −21.2 | 16.9 |
| Joint likelihood conditioned on genotype of index patients and phenotypes at time of index patients' DNA test | 51.5 | 3.5 | 3.7 | −9.7 | 5.1 | 59.0 | 5.7 | 7.2 | −11.5 | 6.8 |
| Retrospective likelihood conditioned only on all phenotypes | 58.5 | 2.3 | 3.4 | 2.7 | 2.2 | 68.3 | 4.1 | 4.3 | 2.5 | 3.4 |
Abbreviations: CLTR, cumulative lifetime risk; FDRs, first-degree relatives; NA, not applicable.
Right censoring at date of first event (which might be diagnosis of breast cancer, ovarian cancer, risk-reducing mastectomy, risk-reducing salpingo-oophorectomy, or last contact or death).
Incident case analysis includes only years at risk and events after the date of the first positive DNA test in the family.
Modeling the probability of breast cancer conditioned on the genotype and phenotype of the index patients or index carriers, and/or the phenotype of relatives.
Footnotes
Supported in part by a grant from the Dutch Cancer Society (to J.R.V.).
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Presented in part at the 5th International Symposium on Hereditary Breast and Ovarian Cancer: Twenty Years of Advances, Montreal, Quebec, Canada, April 23-25, 2014.
Authors' disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Disclosures provided by the authors are available with this article at www.jco.org.
AUTHOR CONTRIBUTIONS
Conception and design: Janet R. Vos, Marian J.E. Mourits, Jan C. Oosterwijk, Geertruida H. de Bock
Collection and assembly of data: Janet R. Vos, Jan C. Oosterwijk
Data analysis and interpretation: Janet R. Vos, Li Hsu, Richard M. Brohet, Jakob de Vries, Kathleen E. Malone, Geertruida H. de Bock
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Bias Correction Methods Explain Much of the Variation Seen in Breast Cancer Risks of BRCA1/2 Mutation Carriers
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Janet R. Vos
No relationship to disclose
Li Hsu
No relationship to disclose
Richard M. Brohet
No relationship to disclose
Marian J.E. Mourits
No relationship to disclose
Jakob de Vries
No relationship to disclose
Kathleen E. Malone
No relationship to disclose
Jan C. Oosterwijk
No relationship to disclose
Geertruida H. de Bock
No relationship to disclose
REFERENCES
- 1.Easton DF, Ford D, Bishop DT. Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am J Hum Genet. 1995;56:265–271. [PMC free article] [PubMed] [Google Scholar]
- 2.Easton DF, Steele L, Fields P, et al. Cancer risks in two large breast cancer families linked to BRCA2 on chromosome 13q12-13. Am J Hum Genet. 1997;61:120–128. doi: 10.1086/513891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Struewing JP, Hartge P, Wacholder S, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. New Engl J Med. 1997;336:1401–1408. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- 4.Fodor FH, Weston A, Bleiweiss IJ, et al. Frequency and carrier risk associated with common BRCA1 and BRCA2 mutations in Ashkenazi Jewish breast cancer patients. Am J Hum Genet. 1998;63:45–51. doi: 10.1086/301903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorlacius S, Struewing JP, Hartge P, et al. Population-based study of risk of breast cancer in carriers of BRCA2 mutation. Lancet. 1998;352:1337–1339. doi: 10.1016/s0140-6736(98)03300-5. [DOI] [PubMed] [Google Scholar]
- 6.Dorum A, Heimdal K, Hovig E, et al. Penetrances of BRCA1 1675delA and 1135insA with respect to breast cancer and ovarian cancer. Am J Hum Genet. 1999;65:671–679. doi: 10.1086/302530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong G, Whittemore AS. Estimating genetic influence on disease from population-based casecontrol data: Application to cancers of the breast and ovary. Stat Med. 1999;18:3321–3336. doi: 10.1002/(sici)1097-0258(19991215)18:23<3321::aid-sim319>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 8.Hopper JL, Southey MC, Dite GS, et al. Population-based estimate of the average agespecific cumulative risk of breast cancer for a defined set of protein-truncating mutations in BRCA1 and BRCA2: Australian Breast Cancer Family Study. Cancer Epidemiol Biomarkers Prev. 1999;8:741–747. [PubMed] [Google Scholar]
- 9.Warner E, Foulkes W, Goodwin P, et al. Prevalence and penetrance of BRCA1 and BRCA2 gene mutations in unselected Ashkenazi Jewish women with breast cancer. J Natl Cancer Inst. 1999;91:1241–1247. doi: 10.1093/jnci/91.14.1241. [DOI] [PubMed] [Google Scholar]
- 10.Antoniou AC, Gayther SA, Stratton JF, et al. Risk models for familial ovarian and breast cancer. Genet Epidemiol. 2000;18:173–190. doi: 10.1002/(SICI)1098-2272(200002)18:2<173::AID-GEPI6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 11.Baffoe-Bonnie AB, Beaty TH, Bailey-Wilson JE, et al. Genetic epidemiology of breast cancer: segregation analysis of 389 Icelandic pedigrees. Genet Epidemiol. 2000;18:81–94. doi: 10.1002/(SICI)1098-2272(200001)18:1<81::AID-GEPI6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 12.Anglian Breast Cancer Study Group. Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases: Anglian Breast Cancer Study Group. Br J Cancer. 2000;83:1301–1308. doi: 10.1054/bjoc.2000.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moslehi R, Chu W, Karlan B, et al. BRCA1 and BRCA2 mutation analysis of 208 Ashkenazi Jewish women with ovarian cancer. Am J Hum Genet. 2000;66:1259–1272. doi: 10.1086/302853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Einbeigi Z, Bergman A, Kindblom L, et al. A founder mutation of the BRCA1 gene in Western Sweden associated with a high incidence of breast and ovarian cancer. Eur J Cancer. 2001;37:1904–1909. doi: 10.1016/s0959-8049(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 15.Eerola H, Pukkala E, Pyrhnen S, et al. Risk of cancer in BRCA1 and BRCA2 mutation-positive and -negative breast cancer families (Finland) Cancer Causes Control. 2001;12:739–746. doi: 10.1023/a:1011272919982. [DOI] [PubMed] [Google Scholar]
- 16.Risch HA, McLaughlin JR, Cole DEC, et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet. 2001;68:700–710. doi: 10.1086/318787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satagopan JM, Offit K, Foulkes W, et al. The lifetime risks of breast cancer in Ashkenazi Jewish carriers of BRCA1 and BRCA2 mutations. Cancer Epidemiol Biomarkers Prev. 2001;10:467–473. [PubMed] [Google Scholar]
- 18.Antoniou AC, Pharoah PD, McMullan G, et al. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer. 2002;86:76–83. doi: 10.1038/sj.bjc.6600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brose MS, Rebbeck TR, Calzone KA, et al. Cancer risk estimates for BCRA1 mutation carriers identified in a risk evaluation program. J Natl Cancer Inst. 2002;94:1365–1372. doi: 10.1093/jnci/94.18.1365. [DOI] [PubMed] [Google Scholar]
- 20.Antoniou A, Pharoah PD, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: A combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heimdal K, Maehle L, Apold J, et al. The Norwegian founder mutations in BRCA1: High penetrance confirmed in an incident cancer series and differences observed in the risk of ovarian cancer. Eur J Cancer. 2003;39:2205–2213. doi: 10.1016/s0959-8049(03)00548-3. [DOI] [PubMed] [Google Scholar]
- 22.Kaufman DJ, Beaty TH, Struewing JP. Segregation analysis of 231 Ashkenazi Jewish families for evidence of additional breast cancer susceptibility genes. Cancer Epidemiol Biomarkers Prev. 2003;12:1045–1052. [PubMed] [Google Scholar]
- 23.King MC, Marks JH, Mandell JB, et al. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 24.Scott CL, Jenkins MA, Southey MC, et al. Average age-specific cumulative risk of breast cancer according to type and site of germline mutations in BRCA1 and BRCA2 estimated from multiple-case breast cancer families attending Australian family cancer clinics. Hum Genet. 2003;112:542–551. doi: 10.1007/s00439-003-0908-6. [DOI] [PubMed] [Google Scholar]
- 25.Southey MC, Tesoriero A, Young MA, et al. A specific GFP expression assay, penetrance estimate, and histological assessment for a putative splice site mutation in BRCA1. Hum Mutat. 2003;22:86–91. doi: 10.1002/humu.10224. [DOI] [PubMed] [Google Scholar]
- 26.Marroni F, Aretini P, D'Andrea E, et al. Penetrances of breast and ovarian cancer in a large series of families tested for BRCA1/2 mutations. Eur J Hum Genet. 2004;12:899–906. doi: 10.1038/sj.ejhg.5201256. [DOI] [PubMed] [Google Scholar]
- 27.Antoniou AC, Pharoah PD, Narod S, et al. Breast and ovarian cancer risks to carriers of the BRCA1 5382insC and 185delAG and BRCA2 6174delT mutations: A combined analysis of 22 population based studies. J Med Genet. 2005;42:602–603. doi: 10.1136/jmg.2004.024133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroiss R, Winkler V, Bikas D, et al. Younger birth cohort correlates with higher breast and ovarian cancer risk in European BRCA1 mutation carriers. Hum Mutat. 2005;26:583–589. doi: 10.1002/humu.20261. [DOI] [PubMed] [Google Scholar]
- 29.Tesoriero AA, Wong EM, Jenkins MA, et al. Molecular characterization and cancer risk associated with BRCA1 and BRCA2 splice site variants identified in multiple-case breast cancer families. Hum Mutat. 2005;26:495. doi: 10.1002/humu.9379. [DOI] [PubMed] [Google Scholar]
- 30.Antoniou AC, Durocher F, Smith P, et al. BRCA1 and BRCA2 mutation predictions using the BOADICEA and BRCAPRO models and penetrance estimation in high-risk French-Canadian families. Breast Cancer Res. 2006;8:R3. doi: 10.1186/bcr1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S, Iversen ES, Friebel T, et al. Characterization of BRCA1 and BRCA2 mutations in a large United States sample. J Clin Oncol. 2006;24:863–871. doi: 10.1200/JCO.2005.03.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gronwald J, Huzarski T, Byrski B, et al. Cancer risks in first degree relatives of BRCA1 mutation carriers: Effects of mutation and proband disease status. J Med Genet. 2006;43:424–428. doi: 10.1136/jmg.2005.036921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Risch HA, McLaughlin JR, Cole DEC, et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: A kin-cohort study in Ontario, Canada. J Natl Cancer Inst. 2006;98:1694–1706. doi: 10.1093/jnci/djj465. [DOI] [PubMed] [Google Scholar]
- 34.Tryggvadottir L, Sigvaldason H, Olafsdottir GH, et al. Population-based study of changing breast cancer risk in Icelandic BRCA2 mutation carriers, 1920-2000. J Natl Cancer Inst. 2006;98:116–122. doi: 10.1093/jnci/djj012. [DOI] [PubMed] [Google Scholar]
- 35.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25:1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu R, Harris EL, Helfand M, et al. Estimating risk of breast cancer in carriers of BRCA1 and BRCA2 mutations: a meta-analytic approach. Stat Med. 2007;26:1775–1787. doi: 10.1002/sim.2811. [DOI] [PubMed] [Google Scholar]
- 37.Vogl FD, Badzioch MD, Steele L, et al. Risks of cancer due to a single BRCA1 mutation in an extended Utah kindred. Fam Cancer. 2007;6:63–71. doi: 10.1007/s10689-006-9106-8. [DOI] [PubMed] [Google Scholar]
- 38.Antoniou AC, Cunningham AP, Peto J, et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer. 2008;98:1457–1466. doi: 10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Begg CB, Haile RW, Borg A, et al. Variation of breast cancer risk among BRCA1/2 carriers. JAMA. 2008;299:194–201. doi: 10.1001/jama.2007.55-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans DG, Shenton A, Woodward E, et al. Penetrance estimates for BRCA1 and BRCA2 based on genetic testing in a clinical cancer genetics service setting: risks of breast/ovarian cancer quoted should reflect the cancer burden in the family. BMC Cancer. 2008;8:155. doi: 10.1186/1471-2407-8-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milne RL, Osorio A, Cajal TRY, et al. The average cumulative risks of breast and ovarian cancer for carriers of mutations in BRCA1 and BRCA2 attending genetic counseling units in Spain. Clin Cancer Res. 2008;14:2861–2869. doi: 10.1158/1078-0432.CCR-07-4436. [DOI] [PubMed] [Google Scholar]
- 42.Chen L, Hsu L, Malone K. A frailty-model-based approach to estimating the age-dependent penetrance function of candidate genes using population-based case-control study designs: an application to data on the BRCA1 gene. Biometrics. 2009;65:1105–1114. doi: 10.1111/j.1541-0420.2008.01184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hashemian AH, Hajizadeh E, Kazemnejad A, et al. Penetrance of BRCA1/BRCA2 specific gene mutations in Iranian women with breast cancer. Saudi Med J. 2009;30:41–44. [PubMed] [Google Scholar]
- 44.Cardenosa EE, Bolufer GP, de Juan Jimenez I, et al. Relationship of BRCA1 and BRCA2 mutations with cancer burden in the family and tumor incidence. Fam Cancer. 2010;9:291–295. doi: 10.1007/s10689-010-9327-8. [DOI] [PubMed] [Google Scholar]
- 45.Mavaddat N, Peock S, Frost D, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst. 2013;105:812–822. doi: 10.1093/jnci/djt095. [DOI] [PubMed] [Google Scholar]
- 46.Evans DG, Harkness E, Lalloo F, et al. Long-term prospective clinical follow-up after BRCA1/2 presymptomatic testing: BRCA2 risks higher than in adjusted retrospective studies. J Med Genet. 2014;51:573–580. doi: 10.1136/jmedgenet-2014-102336. [DOI] [PubMed] [Google Scholar]
- 47.De Bock GH, Mourits MJ, Oosterwijk JC. One risk fits all? J Clin Oncol. 2007;25:3383–3384. doi: 10.1200/JCO.2007.12.3489. [DOI] [PubMed] [Google Scholar]
- 48.Kramer JL, Velazquez IA, Chen BE, et al. Prophylactic oophorectomy reduces breast cancer penetrance during prospective, long-term follow-up of BRCA1 mutation carriers. J Clin Oncol. 2005;23:8629–8635. doi: 10.1200/JCO.2005.02.9199. [DOI] [PubMed] [Google Scholar]
- 49.Gadzicki D, Evans DG, Harris H, et al. Genetic testing for familial/hereditary breast cancer-comparison of guidelines and recommendations from the UK, France, the Netherlands and Germany. J Community Genet. 2011;2:53–69. doi: 10.1007/s12687-011-0042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beristain E, Ibanez B, Vergara I, et al. Breast and ovarian cancer risk evaluation in families with a disease-causing mutation in BRCA1/2. J Community Genet. 2010;1:91–99. doi: 10.1007/s12687-010-0014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van der Kolk DM, de Bock GH, Leegte BK, et al. Penetrance of breast cancer, ovarian cancer and contralateral breast cancer in BRCA1 and BRCA2 families: high cancer incidence at older age. Breast Cancer Res Treat. 2010;124:643–651. doi: 10.1007/s10549-010-0805-3. [DOI] [PubMed] [Google Scholar]
- 52.Vos JR, Teixeira N, Van der Kolk DM, et al. Variation in mutation spectrum partly explains regional differences in the breast cancer risk of female BRCA mutation carriers in the Netherlands. Cancer Epidemiol Biomarkers Prev. 2014;23:2482–2491. doi: 10.1158/1055-9965.EPI-13-1279. [DOI] [PubMed] [Google Scholar]
- 53.Tea MK, Kroiss R, Muhr D, et al. Central European BRCA2 mutation carriers: Birth cohort status correlates with onset of breast cancer. Maturitas. 2014;77:68–72. doi: 10.1016/j.maturitas.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 54.Brohet RM, Velthuizen ME, Hogervorst FB, et al. Breast and ovarian cancer risks in a large series of clinically ascertained families with a high proportion of BRCA1 and BRCA2 Dutch founder mutations. J Med Genet. 2014;51:98–107. doi: 10.1136/jmedgenet-2013-101974. [DOI] [PubMed] [Google Scholar]
- 55.Chatterjee N, Wacholder S. A marginal likelihood approach for estimating penetrance from kin-cohort designs. Biometrics. 2001;57:245–252. doi: 10.1111/j.0006-341x.2001.00245.x. [DOI] [PubMed] [Google Scholar]
- 56.Hsu L, Chen L, Gorfine M, et al. Semiparametric estimation of marginal hazard function from case-control family studies. Biometrics. 2004;60:936–944. doi: 10.1111/j.0006-341X.2004.00249.x. [DOI] [PubMed] [Google Scholar]
- 57.Rondeau V, Mazroui Y, Gonzalez JR. Frailtypack: An R package for the analysis of correlated survival data with frailty models using penalized likelihood estimation or parametric estimation. J Stat Softw. 2012;47:1–28. [Google Scholar]
- 58.R Foundation for Statistical Computing. R: A language and environment for statistical computing. 2013. http://www.R-project.org.
- 59.Lange K, Papp JC, Sinsheimer JS, et al. Mendel: The Swiss Army knife of genetic analysis programs. Bioinformatics. 2013;29:1568–1570. doi: 10.1093/bioinformatics/btt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lange K, Weeks D, Boehnke M. Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol. 1988;5:471–472. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.