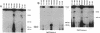Abstract
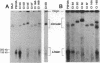
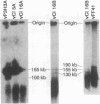
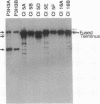
Epstein-Barr virus (EBV) genomes in the P3J-HR-1 (HR-1) Burkitt lymphoma cell line rearrange at a high rate. Previously described deletions and rearrangements in HR-1 cells have been found at sites of EBV replication in vivo, suggesting that DNA rearrangement may be an integral aspect of EBV biology and pathogenesis. We examined the structure of linear EBV genomes in subcultures of HR-1 cells using contour-clamped homogenous electric field gel electrophoresis. We developed a second pulsed electrophoretic technique to separate intracellular circular EBV plasmids. The standard, linear HR-1 EBV genome was approximately 155 kilobases in length. Linear molecules of less than unit length, presumably defective genomes, were seen in numerous subcultures. Linear intracellular genomes greater than 155 kilobases were also detected, but only linear genomes of 155 kilobases or less were packaged into virions. The size of circular EBV plasmids also varied greatly among HR-1 subcultures, some of which contained two plasmids of different size. The progeny of the unusual circular plasmids could be either standard or nonstandard linear genomes. No aberrant linear or circular form was detected in a subculture carrying the previously described het fragments. Pulsed-gel electrophoresis has provided two additional characteristics of mutant EBVs: abnormal linear and circular genome configurations.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A., Bjursell G., Kaschka-Dierich C., Lindahl T. Circular Epstein-Barr virus genomes of reduced size in a human lymphoid cell line of infectious mononucleosis origin. J Virol. 1977 May;22(2):373–380. doi: 10.1128/jvi.22.2.373-380.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A. Concentration of Epstein-Barr virus from cell culture fluids with polyethylene glycol. J Gen Virol. 1973 Sep;20(3):391–394. doi: 10.1099/0022-1317-20-3-391. [DOI] [PubMed] [Google Scholar]
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Bornkamm G. W., Delius H., Zimber U., Hudewentz J., Epstein M. A. Comparison of Epstein-Barr virus strains of different origin by analysis of the viral DNAs. J Virol. 1980 Sep;35(3):603–618. doi: 10.1128/jvi.35.3.603-618.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier-Greco A., Manet E., Chavrier P., Mosnier C., Daillie J., Sergeant A. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 1986 Dec 1;5(12):3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M. S., Bornkamm G. W., zur Hausen H. Structure of defective DNA molecules in Epstein-Barr virus preparations from P3HR-1 cells. J Virol. 1984 Jul;51(1):199–207. doi: 10.1128/jvi.51.1.199-207.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Cohen J. I., Wang F., Mannick J., Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Countryman J., Jenson H., Seibl R., Wolf H., Miller G. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J Virol. 1987 Dec;61(12):3672–3679. doi: 10.1128/jvi.61.12.3672-3679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Countryman J., Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D. K., Miller G., Gradoville L., Heston L., Westrate M. W., Maris W., Wright J., Brandsma J., Summers W. C. Genome of a mononucleosis Epstein-Barr virus contains DNA fragments previously regarded to be unique to Burkitt's lymphoma isolates. Cell. 1981 May;24(2):543–553. doi: 10.1016/0092-8674(81)90345-7. [DOI] [PubMed] [Google Scholar]
- Gardella T., Medveczky P., Sairenji T., Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J Virol. 1984 Apr;50(1):248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt W., Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989 Aug 3;340(6232):393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt W., Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988 Nov 4;55(3):427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- Heller M., Dambaugh T., Kieff E. Epstein-Barr virus DNA. IX. Variation among viral DNAs from producer and nonproducer infected cells. J Virol. 1981 May;38(2):632–648. doi: 10.1128/jvi.38.2.632-648.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heston L., Rabson M., Brown N., Miller G. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature. 1982 Jan 14;295(5845):160–163. doi: 10.1038/295160a0. [DOI] [PubMed] [Google Scholar]
- Kinchington D., Griffin B. E. Size heterogeneity of EBV and mitochondrial DNAs in Burkitt's lymphoma lines. Nucleic Acids Res. 1987 Dec 23;15(24):10345–10354. doi: 10.1093/nar/15.24.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe D. M. The role of viral and cellular nuclear proteins in herpes simplex virus replication. Adv Virus Res. 1989;37:85–123. doi: 10.1016/s0065-3527(08)60833-7. [DOI] [PubMed] [Google Scholar]
- Kolman C. J., Snyder M., Söll D. Genomic organization of tRNA and aminoacyl-tRNA synthetase genes for two amino acids in Saccharomyces cerevisiae. Genomics. 1988 Oct;3(3):201–206. doi: 10.1016/0888-7543(88)90080-8. [DOI] [PubMed] [Google Scholar]
- Manet E., Gruffat H., Trescol-Biemont M. C., Moreno N., Chambard P., Giot J. F., Sergeant A. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 1989 Jun;8(6):1819–1826. doi: 10.1002/j.1460-2075.1989.tb03576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Heller M., Petti L., O'Shiro E., Kieff E. Persistence of the entire Epstein-Barr virus genome integrated into human lymphocyte DNA. Science. 1984 Dec 14;226(4680):1322–1325. doi: 10.1126/science.6095452. [DOI] [PubMed] [Google Scholar]
- Miller G., Heston L., Countryman J. P3HR-1 Epstein-Barr virus with heterogeneous DNA is an independent replicon maintained by cell-to-cell spread. J Virol. 1985 Apr;54(1):45–52. doi: 10.1128/jvi.54.1.45-52.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Rabson M., Heston L. Epstein-Barr virus with heterogeneous DNA disrupts latency. J Virol. 1984 Apr;50(1):174–182. doi: 10.1128/jvi.50.1.174-182.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Robinson J., Heston L., Lipman M. Differences between laboratory strains of Epstein-Barr virus based on immortalization, abortive infection, and interference. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4006–4010. doi: 10.1073/pnas.71.10.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonoyama M., Pagano J. S. Separation of Epstein-Barr virus DNA from large chromosomal DNA in non-virus-producing cells. Nat New Biol. 1972 Aug 9;238(84):169–171. doi: 10.1038/newbio238169a0. [DOI] [PubMed] [Google Scholar]
- Patton D. F., Shirley P., Raab-Traub N., Resnick L., Sixbey J. W. Defective viral DNA in Epstein-Barr virus-associated oral hairy leukoplakia. J Virol. 1990 Jan;64(1):397–400. doi: 10.1128/jvi.64.1.397-400.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Traub N., Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986 Dec 26;47(6):883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- Rabson M., Gradoville L., Heston L., Miller G. Non-immortalizing P3J-HR-1 Epstein-Barr virus: a deletion mutant of its transforming parent, Jijoye. J Virol. 1982 Dec;44(3):834–844. doi: 10.1128/jvi.44.3.834-844.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabson M., Heston L., Miller G. Identification of a rare Epstein-Barr virus variant that enhances early antigen expression in Raji cells. Proc Natl Acad Sci U S A. 1983 May;80(9):2762–2766. doi: 10.1073/pnas.80.9.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney C., Taylor N., Countryman J., Jenson H., Kolman J., Miller G. Genome rearrangements activate the Epstein-Barr virus gene whose product disrupts latency. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9801–9805. doi: 10.1073/pnas.85.24.9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixbey J. W., Shirley P., Sloas M., Raab-Traub N., Israele V. A transformation-incompetent, nuclear antigen 2-deleted Epstein-Barr virus associated with replicative infection. J Infect Dis. 1991 May;163(5):1008–1015. doi: 10.1093/infdis/163.5.1008. [DOI] [PubMed] [Google Scholar]
- Taylor N., Countryman J., Rooney C., Katz D., Miller G. Expression of the BZLF1 latency-disrupting gene differs in standard and defective Epstein-Barr viruses. J Virol. 1989 Apr;63(4):1721–1728. doi: 10.1128/jvi.63.4.1721-1728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M. The importance of circular DNA in mammalian gene amplification. Cancer Res. 1989 Mar 15;49(6):1333–1340. [PubMed] [Google Scholar]