Abstract
Importance
Focal cortical dysplasia (FCD), hemimegalencephaly (HMEG) and megalencephaly constitute a spectrum of malformations of cortical development with shared neuropathologic features. Collectively, these disorders are associated with significant childhood morbidity and mortality. FCD, in particular, represents the most frequent cause of intractable focal epilepsy in children.
Objective
To identify the underlying molecular etiology of FCD, HMEG, and diffuse megalencephaly.
Design, Setting and Participants
We performed whole exome sequencing (WES) on eight children with FCD or HMEG using standard depth (~50-60X) sequencing in peripheral samples (blood, saliva or skin) from the affected child and their parents, and deep (~150-180X) sequencing in affected brain tissue. We used both targeted sequencing and WES to screen a cohort of 93 children with molecularly unexplained diffuse or focal brain overgrowth (42 with FCD-HMEG, and 51 with diffuse megalencephaly). Histopathological and functional assays of PI3K-AKT-MTOR pathway activity in resected brain tissue and cultured neurons were performed to validate mutations.
Main Outcomes and Measures
Whole exome sequencing and targeted sequencing identified variants associated with this spectrum of developmental brain disorders.
Results
We identified low-level mosaic mutations of MTOR in brain tissue in four children with FCD type 2a with alternative allele fractions ranging from 0.012–0.086. We also identified intermediate level mosaic mutation of MTOR (p.Thr1977Ile) in three unrelated children with diffuse megalencephaly and pigmentary mosaicism in skin that resembles hypomelanosis of Ito. Finally, we identified a constitutional de novo mutation of MTOR (p.Glu1799Lys) in three unrelated children with diffuse megalencephaly and intellectual disability. Molecular and functional analysis in two children with FCD type 2a from whom multiple affected brain tissue samples were available revealed a gradient of alternate allele fractions with an epicenter in the most epileptogenic area. When expressed in cultured neurons, all MTOR mutations identified here drive constitutive activation of mTORC1 and enlarged neuronal size, establishing a link between the MTOR mutations and neuronal hypertrophy found in patients. The mTORC1 inhibitor RAD001 ameliorated these phenotypes.
Conclusions and Relevance
Our data show that mutations of MTOR are associated with a spectrum of brain overgrowth phenotypes extending from FCD type 2 to diffuse megalencephaly, distinguished by different mutations and levels of mosaicism. These mutations are sufficient to cause cellular hypertrophy in cultured neurons. Our data also provide a compelling demonstration of the pattern of mosaicism in brain, and substantiate the link between mosaic mutations of MTOR and pigmentary mosaicism in skin.
Keywords: megalencephaly, focal cortical dysplasia, MTOR, mosaicism, epilepsy
Introduction
Focal cortical dysplasia (FCD) is a malformation of cortical development found in almost half of children referred for epilepsy surgery, making it the most frequent known cause of intractable focal epilepsy.1 Histopathological changes defining FCD type 2 (FCD2) include cortical dyslamination and large dysmorphic neurons without (type 2a) or with (type 2b) balloon cells.2 Functional defects in the PI3K-AKT-MTOR pathway have been demonstrated by Western blot, immunohistochemistry, AKT kinase and RNA expression analyses,3–5 and mutations of DEPDC5, PTEN (germline) and PIK3CA (postzygotic or mosaic) have been found in FCD classified as FCD1 or FCD2.3,6–13 Further, three recent studies reported mosaic mutations in MTOR in patients with FCD2.14–16 Many studies have demonstrated links between diffuse megalencephaly (MEG), hemimegalencephaly (HMEG) and FCD2, which have historically been considered distinct disorders. A link is further supported by discovery of germline or mosaic mutations of the same genes in the PI3K-AKT-MTOR pathway in patients with MEG, HMEG or FCD2.3,6,17–19 These data suggest that further genetic studies of FCD as well as HMEG and MEG are warranted to define additional genetic causes, and explore links between the phenotypes and the levels and distribution of mosaic mutations.
We used massively parallel sequencing, histopathology and several functional assays of PI3K-AKT-MTOR activity in brain and peripheral tissues in FCD, HMEG and MEG, experiments that led us to discover low-level mutations of MTOR in FCD2a, compelling demonstration of the pattern of mosaicism in affected brain, a broad spectrum of brain overgrowth phenotypes distinguished by different mutations and levels of mosaicism, and a link between mosaic mutations of MTOR and pigmentary mosaicism.
Methods
Subjects
Patients with FCD, HMEG or MEG were recruited at Pediatric Hospital A. Meyer, the University of Hong Kong, and Seattle Children’s Research Institute with Institutional Review Board approval. Informed consent and research samples were obtained from all patients and selected family members. Brain tissue was obtained during clinically indicated epilepsy surgery.
Magnetic Resonance Imaging (MRI)
Patients in our epilepsy surgery cohort had brain magnetic resonance imaging (MRI) performed on Siemens 1.5T (Symphony or Avanto) or 3T (TrioTim) systems. Preoperative structural data included axial, coronal and sagittal T1 and T2, and axial fluid-attenuated inversion recovery (FLAIR) images. We performed skull stripping with the brain extraction tool in FSL then made 3-dimensional renderings with BioImage Suite. 20,21 Coordinates for brain tissue samples were created using the image guided surgery system (IGS). Intraoperative tissue coordinates were downloaded from IGS and transformed into MRI space using BioImage Suite.21 Tissue coordinates were overlaid onto 3D renderings of the brain to visualize sample locations. Patients in our megalencephaly cohort were scanned using standard brain MRI imaging including T1, T2 and FLAIR sequences in axial, sagittal and coronal planes at their respective centers.
Molecular methods
Genomic DNA was isolated with standard protocols. We hypothesized that FCD and HMEG are caused by low-level mosaic mutations, so designed a “tetrad-based” whole exome sequencing (WES) approach consisting of standard depth (~50-60X) sequencing in peripheral samples (blood, saliva or skin) from the affected child and both parents, plus deep (~150-180X) WES in DNA isolated from affected brain tissue. Data were analyzed using a standard trio approach with the peripheral sample, confirmation with the affected brain sample, and paired analysis between unaffected and affected brain sequence data (see eFigure 1). Mutations were confirmed using several targeted methods including single molecule molecular inversion probes (smMIPs), amplicon sequencing and Sanger sequencing. The same methods were used to screen our cohort of children with FCD, HMEG or MEG. Details regarding the sequencing methods are provided in the Supplementary Methods.
Neuropathology and functional analysis
Histologic sections of resected brain tissue were scored for features of FCD. Subjects with FCD2 or HMEG were selected for further study. Immunohistochemical staining was performed on formalin-fixed, paraffin-embedded sections by standard techniques using antibodies directed against mouse monoclonal MAP2 (Millipore), rabbit monoclonal Thr308-phospho-Akt and rabbit monoclonal phospho-S6 ribosomal protein (both from Cell Signaling Technology).3 Western blot analysis was performed on 100-200 mg frozen cortical tissue following standard protocols published previously.3
Neuronal culture models
Wild type or mutant hemagglutinin (HA)-tagged MTOR constructs were expressed by electroporation in embryonic day 18 cultured rat cortical neurons. Neurons aged 7‐14 days were treated with vehicle, amino acid/growth factor starvation conditions, or mTOR inhibitors, and then fixed and immunostained using standard techniques. Cultures were imaged with an automated confocal microscopy imaging system (General Electric InCell 6000) and high content automated analysis performed with InCell Analyzer software and Tibco Spotfire. Antibodies against phospho-S6 (240/244), HA tag, NeuN, or Map2 were used.
Results
Exome sequencing
We performed tetrad-based WES in 8 children with FCD (FCD2a x5, HMEG-FCD2a x1, FCD2b x2) to search for de novo germline and mosaic mutations. To identify de novo germline mutations, we filtered for variants observed in the patient but not the parents. To identify mosaic mutations, we looked for variants present in brain DNA that were not found in the matched peripheral sample. We successfully generated sequencing data for all eight samples from affected brain, seven of eight peripheral samples, and 15 of 16 parents (see eTable 1). We sequenced brain samples to a mean average read depth of 171X coverage across the targeted loci (range = 130X-204X) to facilitate identification of mosaic mutations at low allelic fractions, peripheral samples from seven of eight probands to a mean of 88X (range = 69X-103X), and parental samples to a mean of 82X (range = 59X-101X).
De novo deletion in DEPDC5
We performed variant identification and filtering for de novo single nucleotide variants (SNVs) and indels, and manually reviewed all filtered variants to remove artifacts not captured in our filtering scheme. In the seven patients with complete parental data, we identified eight putative de novo missense SNVs, one de novo in-frame insertion, and a de novo deletion in DEPDC5 leading to a frameshift (c.4187delC; see eTable 2 and eFigure 2). This girl with HMEG also had copy number neutral loss of heterozygosity on chromosome 22 suggesting possible loss of the other allele (see eFigure 3). Several other somatic indels were deemed not pathogenic based on multiple lines of evidence (see eTable 3).
Mosaic mutations in MTOR
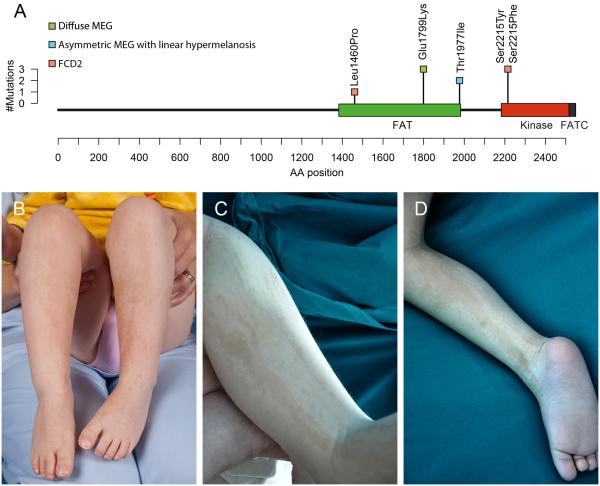
To identify mosaic SNVs, we used an algorithm sensitive to low allelic fractions. After filtering and manual review, we identified eight SNVs present at allelic fractions less than 0.1 (see eTable 3) including three missense mutations in MTOR in three unrelated children with FCD2a. The fraction of reads supporting the mutant allele was always less than 0.1 in brain and 0 in peripheral samples and parents. The p.Leu1460Pro mutation is located in the focal adhesion targeting (FAT) domain of MTOR, and the p.Ser2215Phe and p.Ser2215Tyr mutations in the kinase domain (see Table 1, Figure 1A). We next re-examined smMIP targeted sequencing data in 42 individuals with FCD or HMEG (including 21 with FCD2), and identified the p.Ser2215Phe mutation in a second unrelated patient with FCD2a (LR13-389).
Table.
Clinical, neuroimaging, neuropathologic and molecular findings of MTOR (N=10) and DEPDC5 (N =1) mutation-positive patients
| Patient DB# | Diagnosis | Seizures | Mutation | Alternative allele fractiona | ||||
|---|---|---|---|---|---|---|---|---|
| Clinical | Imaging | Neuro- pathologic |
cDNA change |
Amino acid change |
Non-brain tissue |
Brain tissue |
||
| DEPDC5 | ||||||||
| LR12-250 | HMEG | PQD | FCD2a | ++ | c.4187delC | p.Ala1396Valfs*7 8 |
0.28s | 0.35 |
| MTOR Group 1: FCD | ||||||||
| LR12-243 | FCD | FCD | FCD2a | ++ | c.4379T>C | p.Leu1460Pro | 0.00s | 0.06 |
| LR13-354 | FCD | FCD | FCD2a | ++ | c.6644C>T | p.Ser2215Phe | 0.00s | 0.055 |
| LR13-389 | FCD | FCD | FCD2a | ++ | c.6644C>T | p.Ser2215Phe | 0.00s | 0.012-0.086 |
| LR12-245 | FCD | FCD | FCD2a | ++ | c.6644C>A | p.Ser2215Tyr | 0.00s | 0.035 |
| MTOR Group 2: Asymmetric MEG with polymicrogyria and cutaneous pigmentary mosaicismb | ||||||||
| LR13-310 | MEG-PIGMb | MEG-PMG | − | + | c.5930C>T | p.Thr1977Ile | 0.23fb
0.07wbc |
NA |
| LR14-326 | MEG-PIGMb | MEG-PMG | − | + | c.5930C>T | p.Thr1977Ile | 0.55fb+
0.42fb− 0.20wbc 0.10s |
NA |
| LR15-044 | MEG-PIGMb | MEG-PMG | − | − | c.5930C>T | p.Thr1977Ile | 0.20wbc | NA |
| MTOR Group 3: Diffuse MEG | ||||||||
| LR12-379a1/a2 | MEG-AUTc | MEG | − | + | c.5395G>A | p.Glu1799Lys | 0.50wbc | NA |
| LR14-012 | MEGd | MEG | − | − | c.5395G>A | p.Glu1799Lys | 0.47s | NA |
| LR15-065 | MEGe | MEG | − | − | c.5395G>A | p.Glu1799Lys | 0.50wbc
0.50s |
NA |
Transcript IDs: DEPDC5, NM_001242897.1; MTOR, NM_004958.3.
Abbreviations: AUT, autism; DB, database number; FCD, focal cortical dysplasia; MEG, megalencephaly; PIGM, pigmentary mosaicism of skin; PMG, polymicrogyria; PQD, posterior quadrantic dysplasia. +, seizures; ++, intractable seizures leading to epilepsy surgery. Tissue samples: fb, skin fibroblasts; fb+, fibroblasts grown from hyperpigmented streaks; fb−, fibroblasts grown from hypopigmented streaks; s, saliva; wbc, white blood cells.
Additional clinical details:
Further details regarding the specific alternative allele levels in various tissues are provided in eTables 3 and 4.
Skin color pattern consistent with “cutis tricolor of the Blaschko-linear type”, which consists of diffuse alternating hypo- and hyper-pigmented streaks.
a1 and a2 denote two identical twin brothers. Both had autism at 17 years of age with severe impairment of social reciprocity, no eye contact and absent speech. They were found to meet the criteria for Autism Spectrum Disorders (ASD) on formal longitudinal neurodevelopmental assessments.
This girl has early autistic features at 3 years of age, a capillary malformation on the scalp that has faded with age, and a small hemangioma on her chest wall that completely involuted in infancy.
This boy also has a small liver hemangioma measuring 2×1 cm in size.
Figure 1. MTOR mutations (A) and clinical photographs of patients with MTOR mutations, megalencephaly and cutis tricolor of the Blaschko-linear type (B–D).
Panel A shows the positions of MTOR coding mutations identified in this report relative to their locations within the MTOR protein domain structure. The height of the bar indicates the number of patients with mutations at that amino acid position, and the color of the square indicates the phenotype of the patient(s). Green=diffuse MEG; blue=asymmetric MEG with linear hyper‐ and hypomelanosis; pink=FCD2. Panels B (patient LR13-310) and C‐D (LR14-326) show alternating, predominatly linear streaks of hyper‐ and hypopigmented skin, which correpond to cutis tricolor of the Blaschko-linear type.
MTOR mutations in megalencephaly
Prompted by these findings, we re-examined sequencing data in 51 individuals with unexplained MEG including two with trio-based WES, and identified six additional patients with MTOR mutations who segregated into two groups. The first consists of three children with asymmetric MEG, cortical dysplasia including polymicrogyria, and linear streaks of hypo‐ and hyperpigmented skin following the lines of Blaschko (Figure 1B-D). All three had the same MTOR mutation, p.Thr1977Ile, with levels of mosaicism varying from 0.23 to 0.55 in affected skin and 0.07 to 0.20 in blood or saliva (see Table 1). Their skin markings correspond to cutis tricolor of the Blaschko-linear type, a form of pigmentary mosaicism (personal communication, Prof. Rudolf Happle).22 This is a more specific dermatologic diagnosis than “hypomelanosis of Ito”, which has been used for almost any form of pigmentary mosaicism. The second group includes four children (including two identical twins) with symmetric MEG, intellectual disability, autism (in the two oldest), and limited or no polymicrogyria. All four had the same de novo constitutional MTOR mutation, p.Glu1799Lys (Table 1). The alternative allele fractions of all mutations identified by various methods in these three groups are further shown in eTable 4. The neuroimaging and neuropathologic features for all three groups are summarized in eTables 5 and 6, and represented in eFigures 4, 5 and 6.
Mutation gradient and epileptogenesis
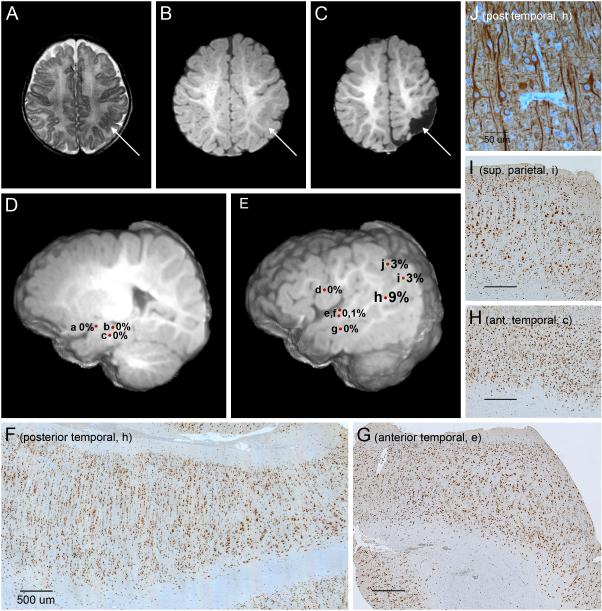
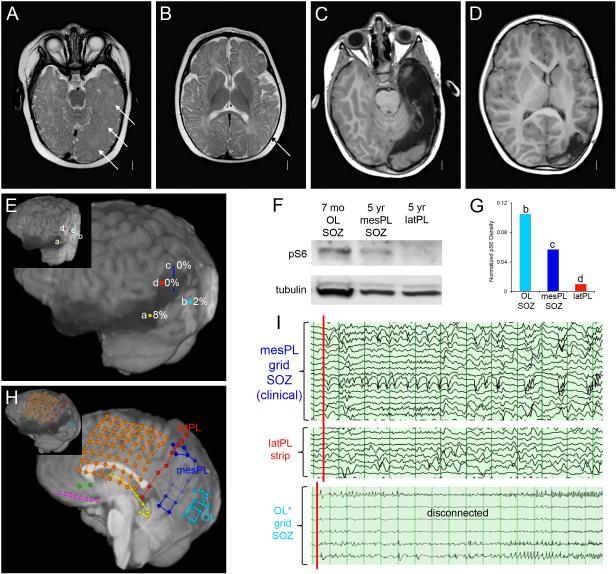
For one patient (LR13-389; p.Ser2215Phe), we had DNA from 10 different brain regions following parieto-temporal lobectomy with intra-operative electrocorticography (see Figure 2). The mutation level (alternate allele fraction) varied from 0 to 0.086 and demonstrated a striking gradient with an epicenter in the posterior temporal lobe and lower levels at the periphery. We were unable to detect the mutation in six of seven samples from the anterior edge of the resected area with average read depth of 269X (range 178-351X) even though all tissue samples showed clear FCD2a pathology. The patient underwent a functional hemispherectomy six months later due to recurrent epilepsy. Another patient (LR12-245, p.Ser2215Tyr) had several surgical resections. The first consisted of temporal and occipital lobectomy with intra-operative electrocorticography performed at 7 months, followed by a staged redo craniotomy for left parietal resection with electrocorticography at 5 years of age, with resolution of epilepsy following the second procedure (Figure 3, A-E). Western blots from highly epileptogenic posterior temporal/occipital cortex resected during the first surgery had higher phospho-S6 expression than parietal cortex resected during the second surgery (see Figure 3, F-G; eFigure 7). Intracranial EEG by depth electrodes and grids prior to the second surgery showed seizure onset from the mesial parietal region, which exhibited elevated phospho-S6 levels, but not from the lateral parietal region, which exhibited low phospho-S6 expression (Figure 3, H-I). Strikingly, the mutation was not detected in either sample (see eTable 4).
Figure 2. Brain imaging and histopathology in patient LR13-389 with MTOR p.Ser2215Phe mutation.
Panels A‐C show an area of cortical infolding and thickening in the left posterior temporal and parietal lobes (arrows) on preoperative T2 (A) and T1-weighted (B) images, that has been excised on a postsurgical T2-weighted image (C). Panels D and E are preoperative deep (D) and superficial (E) 3-dimensional images that show the locations and levels of mosaicism (alternate allele fractions) of specimens collected during surgery. These include the amygdala (a), hippocampus (b), deep anterior temporal lobe (c), frontal operculum (d), anterior temporal lobe – superior temporal gyrus (e and f), anterior temporal lobe – middle temporal gyrus (g), posterior temporal lobe (h), inferior parietal lobe (i), and superior parietal lobe (j). The levels of mosaicism, while all low, are highest in the center of the dysplasia (h at 9%), intermediate along the posterior border (i and j at 3%), and too low to detect consistently along the anterior border of the lesion (a-g at 0-1%) even though all sections show changes of focal cortical dysplasia type 2a. Panels F through I are brain sections stained with NeuN, which all show loss of cortical lamination, excessive tall vertical columns of neurons (especially prominent in F), numerous maloriented large neurons, and blurring of the cortical-white matter boundary. The proportion of large dysplastic neurons appears higher in sections with higher mutation levels (F and I) compared to regions with undetected mutations (G and H). In the section from the center of the dysplasia with the highest level of mutation, a transition can be seen with less severe dysplasia on the left of the image and more severe dysplasia with more numerous large dysplastic neurons on the right (F). Panel J is the same section as panel F stained with MAP2 at higher power, and shows several large dysplastic neurons with disorganized processes, excessive cytoskeletal elements within cell bodies and abnormally oriented dendrites that often crowd together.
Figure 3. Differential MTOR mutation burden and phosphorylated ribosomal protein S6 expression in a girl (LR12-245) with both early- and later-onset seizures due to FCD2a.
Panels A and B show MRI images performed at 6 months just before the first surgical procedure. The findings include increased volume of the left mid-temporal lobe, mild thickening and irregularity of the cortex (arrows in A) and similar but more focal changes in the superior temporal lobe (arrow in B). Panels C and D show MRI images performed just before the second surgery at 5 years. They demonstrate the surgical defect and subtle changes in the left parietal lobe (C‐D). Panel E shows the locations of the four surgical specimens used for both genetic and tissue analysis as indicated by colored circles: temporal lobe from the first surgery (a, yellow), occipital lobe seizure onset zone (SOZ) disconnected during the first surgery but not removed till the second surgery (b, light blue), medial parietal SOZ from the second surgery (c, dark blue) and lateral parietal cortex also from second surgery (d, red) that was not involved in seizure onset. The inset shows the locations in 3 dimensions. Panels F and G show Western blots for phospho-S6 from the occipital lobe (OL, location b in panel E), mesial parietal lobe (mesPL, c) and lateral parietal lobe (latPL, d). This analysis demonstrated a higher level of phospho-S6 in the occipital lobe compared to the mesial and lateral parietal lobes. Panel H is a 3D brain rendering trimmed to midline to display medial electrodes, and shows grid and strip placement for intracranial EEG monitoring at 5 years. Dark blue = medial parietal grid, red = lateral parietal strip, light blue = occipital grid. The inset provides a better 3D view of the locations. Panel I shows EEG tracings from intraoperative grids placed over the temporal, parietal and occipital regions just prior to the second resection. The tracings show several seizure onset zones (SOZ). The most active interictal discharges came from the most inferior leads on the mesial parietal grid (dark blue grid in H, leads 1-3 and 9-11), marking a SOZ. These events spread quickly to other mesial parietal leads as well as the lateral parietal strip (red strip in E, leads 1-8). Several independent SOZ were seen in the occipital lobe, which had been disconnected several years before. The voltages are lower as the grid was not as closely apposed to the brain surface. Overall, the highest levels of phospho-S6 expression were seen in the occipital lobe (OL) SOZ responsible for early-onset seizures, with lower expression in the medial parietal cortex SOZ responsible for later-onset seizures, and nearly undetectable levels in the lateral parietal cortex that was not involved in seizure generation. These findings correlate with MTOR mutation burden determined from these same samples (panel E and Supplementary Table 4).
Functional effects
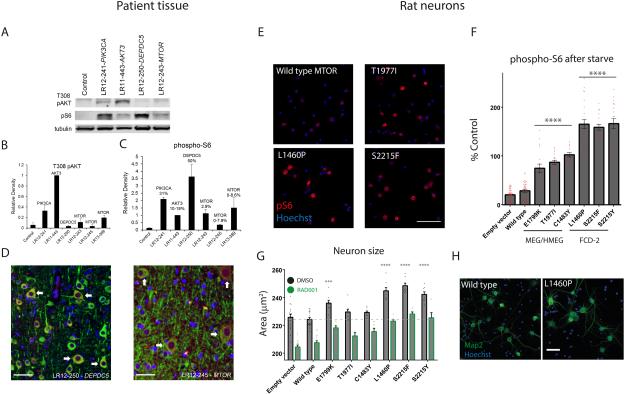
To evaluate the functional impact of identified mutations, we queried the Catalogue of Somatic Mutations In Cancer (COSMIC) database and found that four of five MTOR mutations (plus Thr1977Arg but not Thr1977Ile) have been observed in tumors.23 All activate mTORC1 signaling in vitro.24 We assessed PI3K-AKT-MTOR activation by Western blot for mutations in MTOR, DEPDC5, PIK3CA, AKT and control brain tissue (Fig. 4, A‐C). All samples with mutations showed elevated phospho-S6 expression, while T308 phospho-AKT was elevated only in samples with PIK3CA and AKT3 mutations. Immunohistochemistry in MTOR p.Ser2215Tyr and DEPDC5 samples revealed increased expression of phospho-S6 in large dysmorphic neurons (Fig. 4, D).
Figure 4. Functional consequences of PI3K-AKT-mTOR pathway mutations in patient tissue and rodent neurons.
Panel A shows representative Western blots comparing levels of T308 AKT phosphorylation (PI3K-PDK1-dependent) to ribosomal protein S6 phosphorylation (mTORC1-dependent) in control and dysplastic brain specimens containing the indicated mutations. Specimens with “upstream” pathway mutations (PIK3CA, AKT3) have the highest levels of AKT phosphorylation, while specimens with “downstream” mutations (DEPDC5, MTOR) exhibit elevation of phospho-S6 with lesser elevation of T308 phospho-AKT. Panels B and C show averaged results from five blots for T308 phospho-AKT (B) and phospho-S6 (C). Panels in D show phosphorylated ribosomal protein S6 expression at high power, indicating activation of the PI3K-AKT-mTOR pathway, in a subset of neurons in dysplastic human cortex with DEPDC5 or MTOR mutations. Green = MAP2 neuronal marker, Red = phospho-S6, Blue = DAPI nuclear stain. Arrows indicate dysmorphic neurons co-expressing MAP2 and phospho-S6, which appear orange in color. Scale bars = 50 μm.
Panel E shows representative images of phospho-S6 indirect immunofluorescence of rat neurons electroporated with indicated wild-type or mutant MTOR constructs. Scale bar = 100 μm. Panel F quantifies average phospho-S6 immunofluorescence intensity for DIV12 neurons transfected (NeuN positive, HA-tag/ZsGreen positive) with MTOR or empty vector constructs as indicated, and treated with amino acid and growth factor starvation for 2 hours. Data are baseline subtracted (phospho-S6 values in empty vector transfected neurons treated with 200 nM RAD001) and normalized to phospho-S6 immunofluorescence intensities in wild-type MTOR neurons in normal media. Data points represent averages per individual wells; columns and error bars are means across wells and standard error of means. Asterisks and lines represent significant differences between genotypes (ANOVA, Tukey’s test for multiple comparisons, comparing each condition to every other condition). Within each group (E1799K, T1977I, C1483Y; or S2215F, S2215Y, L1460P), data for individual genotypes are significantly different from genotypes in each other group or controls (empty vector and wild type MTOR electroporated neurons) at a significance level of p < 10-4. Panel G shows average neuronal size (cell body area encompassed by NeuN stain) for transfected neurons treated from DIV7-DIV14 with DMSO or 200 nM RAD001. 1 nM RAD001 intermediately reduced size (not shown). Asterisks denote significant difference of neuronal size for a given genotype compared to wild-type MTOR (ANOVA, Dunnett’s test for multiple comparisons). For every genotype, RAD001 significantly reduced neuronal size (p < 10-3 or less). See also figure 8 in supplemental appendix for size analysis of neurons at DIV12, distributions of phospho-S6 and cell sizes across cells, and representative NeuN immunostaining. Panel H shows representative fields of immunostaining for Map2 and Hoechst. Scale bar = 50 μm.
We next electroporated constructs containing wild-type or mutant MTOR into cultured rodent neurons, and measured mTORC1 activation using high content imaging of phospho-S6 (240/244) immunofluorescence. Neurons expressing mutant MTOR constructs showed hyperactivation of mTORC1 compared to wild type (see eFigure 8). To determine if these mutations impact regulation of mTORC1 signaling by nutrients and growth factors, we subjected mutant and wild type neurons to starvation with PBS/glucose solution. While starvation completely suppressed mTORC1 activity in wild type neurons, neurons expressing the MTOR mutations displayed constitutive activation (see Fig. 4, E-F), consistent with the effect of these mutations in human cell lines (see eFigure 9).24 Interestingly, mutants found in megalencephaly and hemimegalencephaly patients induced an intermediate level of mTORC1 hyperactivation between wild-type and FCD mutations. To assess whether the FCD or MEG MTOR mutations induced cellular hypertrophy, a hallmark of FCD histopathology, we measured neuronal cell size using automated analysis of a combination of nuclei staining (Hoechst) and the NeuN neuronal marker that outlines the cell soma. Consistent with mTORC1 hyper-activation, we observed significant increases in cell size in mutant neurons (see Fig. 4, G-H; eFigure 8) that could be reversed by 7 days of mTOR inhibition with the rapamycin analogue RAD001 (see Fig. 4G).
Discussion
In our cohort, we have identified five de novo MTOR mutations at four amino acid residues in ten unrelated patients with FCD, HMEG and MEG. Our data confirm the occurrence of low level mosaic MTOR mutations in a subset of patients with FCD2 and provide evidence for (1) association of MTOR with a spectrum of brain malformations ranging from FCD2a to diffuse MEG; (2) a causal link between MTOR mutations and induction of elevated mTORC1 activity and neuronal hypertrophy; (3) a gradient of mutation in brain in which levels of mosaicism below the detection level of NextGen sequencing are still sufficient to cause FCD2 pathology; (4) a strong link between functional expression of the mutation and epilepsy; (5) a new association between MTOR mutations and autism; (6) a new association between mosaic MTOR mutations and pigmentary mosaicism; and (7) variation in phospho-AKT and phospho-S6 expression in brain tissue that correlates with mutations of different core components of the PI3K-AKT-MTOR pathway.
MTOR and DEPDC5
MTOR is a serine/threonine protein kinase that integrates many intracellular and extracellular signals and serves as a central regulator of cell proliferation, growth, metabolism, and survival.25,26 Upregulation of MTOR mediates the cellular and molecular changes leading to both the cortical malformation and epilepsy in FCD and Tuberous Sclerosis Complex.3,5 DEPDC5 is a repressor of mTORC1 signaling, so that loss-of-function mutations of DEPDC5 also upregulate MTOR activity. 27,28 Adding to our report, mutations of DEPDC5 have been observed with HMEG, FCD2a, FCD2b and probably FCD type 1.6,7 Loss of heterozygosity of chromosome 22 was observed in the patient with the DEPDC5 alteration, which suggests inactivation of both copies of the gene in affected (brain) tissue.
Three MTOR-related phenotypes
Our 10 patients with MTOR mutations segregate into 3 distinct groups with emerging genotype-phenotype correlations (Table 1). The first consists of FCD2a. All four children with FCD2a had low-level mosaic mutations detectable only in brain at levels ranging from 0.012-0.086 of reads. The second consists of asymmetric MEG, cortical dysplasia, and pigmentary mosaicism. The skin phenotype could be designated as hypomelanosis of Ito, but the more specific diagnosis of cutis tricolor of the Blaschko-linear type reflects the association with both hypo- and hyperpigmented streaks.22 Indeed, review of photographs of children with HMEG or FCD and hypomelanosis of Ito usually show both hypo- and hyperpigmented streaks.29–33 All three children in this group had the p.Thr1977Ile MTOR mutation at intermediate mutation levels. The third group includes children with diffuse megalencephaly, intellectual disability and (when old enough to be evaluated) autism. None had dysmorphic features or altered skin pigmentation, but 3 were noted to have patchy areas of cortical dysplasia. All three had the recurrent de novo constitutional MTOR mutation, p.Glu1799Lys.
Other groups recently reported low level mosaic MTOR mutations in brain in 12 of 77 (16%) subjects with FCD214, or 6 of 13 subjects with FCD2b 15, similar to our data. Additionally, MTOR mutations have been reported in brain but not blood-derived DNA in two patients with HMEG.6,19
Patient phenotypes and the PI3K-AKT-MTOR pathway
Our results combined with published data demonstrate that FCD2 and HMEG are disorders of the PI3K-AKT-MTOR pathway. Mutations of pathway genes – PIK3CA, PTEN, AKT3, MTOR, DEPDC5 and TSC2 – have been identified in 19 of 53 (36%) patients with HMEG,3,6,19 and 23 out of 116 (20%) with FCD2 (including this report).14–16 The most frequently mutated genes so far have been PIK3CA in HMEG (9/53 or 17%) and MTOR in FCD2 (22/116 or 19%).3,6,14,16,18,19
Pathologic studies have separated FCD into 9 subgroups including FCD2a and 2b based on the absence or presence of balloon cells.2 Adding our data to other reports, mutations of PTEN, MTOR and DEPDC5 have been associated with FCD2a or FCD2b (and possibly FCD type 1) in different patients, observations that weaken the rationale for separating them. We propose instead that separation using functional assays and mutation analysis will be more useful. We found that PIK3CA, PTEN and AKT3 mutation-positive samples demonstrated elevations of T308 phospho-AKT expression, while MTOR and DEPDC5 samples did not. This is consistent with upstream AKT activation in patients with pathway mutations upstream of mTORC1. Expression of phospho-S6 was increased in all mutation-positive specimens, possibly correlating with the level of mosaicism.
With our rodent functional expression system, we evaluated MTOR mutants and found hyperactivation patterns that segregated by patient groups: mutations found in megalencephaly or HMEG patients with broad distributions of MTOR mutant neurons (Glu1799Lys, Thr1977Ile, and Cys1483Tyr) exhibited a level of elevated mTORC1 signaling that was intermediate between wild-type levels and those observed in mutants found in FCD2a patients (Ser2215Phe, Ser2215Tyr, Leu1460Pro). These data suggest that small lesions of FCD2a can be epileptogenic because of strong elevation of mTORC1 activity. Only weaker mutations may be observed in patients with broad distribution of mutant cells in brain and peripheral tissues because more severe mutations may be lethal. Common to all mutations, however, was the finding that MTOR inhibitors decreased pathological neuronal enlargement.
In summary, our discovery of MTOR activating mutations in the MEG-HMEG-FCD2 spectrum of malformations has important clinical implications. Everolimus, an inhibitor of mTORC1 activity is an approved treatment for Tuberous Sclerosis Complex-associated subependymal giant cell astrocytomas, and is currently in a phase 3 clinical trial as an adjunctive therapy in patients with TSC and refractory partial-onset epilepsy (EXIST-3; ClinicalTrials.gov ID NCT01713946). Several other molecules that inhibit PI3K-AKT-MTOR components are undergoing clinical development for cancer, and alone or in combination offer a new approach to the treatment of intractable epilepsy that involves direct correction of the underlying molecular defect rather than simply reducing membrane excitability. Multiple studies have shown that 25-40% of intractable epilepsy requiring resective surgery is caused by FCD. While the contribution of MTOR and other PI3K-AKT pathway genes to intractable epilepsy not amenable to surgical treatment is unknown, our data suggest that it may be high. Thus, mutations of the PI3K-AKT-MTOR pathway including MTOR itself may prove to be a common and treatable cause of intractable focal epilepsy.
Supplementary Material
Acknowledgements
We wish to thank the patients and their families for their important contribution to our research, and Prof. Dr. Rudolf Happle for review of the cutaneous phenotype. This study was funded by the US National Institutes of Health under NINDS grants K08NS092898 (to G.M. M.), NS058721, NS092772 (to W.B.D.), NS072162 (to L.A.J.), and HG006542 (to the Baylor-Hopkins Center for Mendelian Genomics research program), by Citizens United for Research in Epilepsy (CURE; to L.A.J.), by EU Seventh Framework Program under the project DESIRE grant agreement N602531, and E-RareJTC2011 (both to R.G.), and by the SK Yee Medical Research Fund (to K.S.Y. and B.C.). C.D.C., N.S., S.M., J.D.B., R.L., S.M., S.L., W.R.S., M.M., L.O.M., and W.W. are employees of Novartis, Inc. Novartis Institutes for BioMedical Research funded exome sequencing and neuronal culture experiments. J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, Inc. is a member of the Scientific Advisory Board of Baylor Miraca Genetics Laboratories, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis (CMA) and clinical exome sequencing offered in the Baylor Miraca Genetics Laboratory (BMGL: http://www.bmgl.com/BMGL/Default.aspx). J.S. and E.A.B. have a patent and copyright for systems, algorithms, and software for molecular inversion probe (MIP) design with royalties paid by Roche. All other authors declare no competing interests. G.M. and W.B.D. had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. The funding sources have no role in the design and conduct of the study, collection, management, analysis and interpretation of the data, preparation, review or approval of the manuscript, or decision to submit the manuscript for publication.
Footnotes
Web Links:
COSMIC, http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/
dbSNP, http://www.ncbi.nlm.nih.gov.offcampus.lib.washington.edu/SNP/
ExAC, http://exac.broadinstitute.org/
Freebayes, https://github.com/ekg/freebayes/
NHLBI Exome Variant Server, http://evs.gs.washington.edu/EVS/
References
- 1.Harvey AS, Cross JH, Shinnar S, Mathern BW. Defining the spectrum of international practice in pediatric epilepsy surgery patients. Epilepsia. 2008;49:146–55. doi: 10.1111/j.1528-1167.2007.01421.x. [DOI] [PubMed] [Google Scholar]
- 2.Blümcke I, Thom M, Aronica E, et al. The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia. 2011;52(1):158–174. doi: 10.1111/j.1528-1167.2010.02777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jansen LA, Mirzaa GM, Ishak GE, et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain J. Neurol. 2015 doi: 10.1093/brain/awv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ljungberg MC, Bhattacharjee MB, Lu Y, et al. Activation of mammalian target of rapamycin in cytomegalic neurons of human cortical dysplasia. Ann. Neurol. 2006;60(4):420–429. doi: 10.1002/ana.20949. [DOI] [PubMed] [Google Scholar]
- 5.Wong M. Mammalian target of rapamycin (mTOR) activation in focal cortical dysplasia and related focal cortical malformations. Exp. Neurol. 2013;244:22–26. doi: 10.1016/j.expneurol.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Gama AM, Geng Y, Couto JA, et al. mTOR pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann. Neurol. 2015 doi: 10.1002/ana.24357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baulac S, Ishida S, Marsan E, et al. Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Ann. Neurol. 2015 doi: 10.1002/ana.24368. [DOI] [PubMed] [Google Scholar]
- 8.Cheung KM, Lam CW, Chan YK, Siu WK, Yong L. Atypical focal cortical dysplasia in a patient with Cowden syndrome. Hong Kong Med. J. Xianggang Yi Xue Za Zhi Hong Kong Acad. Med. 2014;20(2):165–167. doi: 10.12809/hkmj133863. [DOI] [PubMed] [Google Scholar]
- 9.Child ND, Cascino GD. Mystery case: Cowden syndrome presenting with partial epilepsy related to focal cortical dysplasia. Neurology. 2013;81(13):e98–99. doi: 10.1212/WNL.0b013e3182a55ef0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elia M, Amato C, Bottitta M, et al. An atypical patient with Cowden syndrome and PTEN gene mutation presenting with cortical malformation and focal epilepsy. Brain Dev. 2012;34(10):873–876. doi: 10.1016/j.braindev.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Merks JHM, de Vries LS, Zhou X-P, et al. PTEN hamartoma tumour syndrome: variability of an entity. J. Med. Genet. 2003;40(10):e111. doi: 10.1136/jmg.40.10.e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Rourke DJ, Twomey E, Lynch S-A, King MD. Cortical dysplasia associated with the PTEN mutation in Bannayan Riley Ruvalcaba syndrome: a rare finding. Clin. Dysmorphol. 2012;21(2):91–92. doi: 10.1097/MCD.0b013e328351639d. [DOI] [PubMed] [Google Scholar]
- 13.Schick V, Majores M, Engels G, et al. Activation of Akt independent of PTEN and CTMP tumor-suppressor gene mutations in epilepsy-associated Taylor-type focal cortical dysplasias. Acta Neuropathol. (Berl.) 2006;112(6):715–725. doi: 10.1007/s00401-006-0128-y. [DOI] [PubMed] [Google Scholar]
- 14.Lim JS, Kim W-I, Kang H-C, et al. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat. Med. 2015 doi: 10.1038/nm.3824. [DOI] [PubMed] [Google Scholar]
- 15.Nakashima M, Saitsu H, Takei N, et al. Somatic Mutations in the MTOR gene cause focal cortical dysplasia type IIb. Ann. Neurol. 2015;78(3):375–386. doi: 10.1002/ana.24444. [DOI] [PubMed] [Google Scholar]
- 16.Leventer RJ, Scerri T, Marsh APL, et al. Hemispheric cortical dysplasia secondary to a mosaic somatic mutation in MTOR. Neurology. 2015 doi: 10.1212/WNL.0000000000001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivière J-B, Mirzaa GM, O’Roak BJ, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet. 2012;44(8):934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirzaa GM, Parry DA, Fry AE, et al. De novo CCND2 mutations leading to stabilization of cyclin D2 cause megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome. Nat. Genet. 2014 doi: 10.1038/ng.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Huynh M, Silhavy JL, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat. Genet. 2012;44(8):941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith SM. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papademetris X, Jackowski MP, Rajeevan N, et al. BioImage Suite: An integrated medical image analysis suite: An update. Insight J. 2006;2006:209. [PMC free article] [PubMed] [Google Scholar]
- 22.Happle R. Mosaicism in Human Skin: Understanding Nevi, Nevoid Skin Disorders, and Cutaneous Neoplasia. 2014. [publisher]
- 23.Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer. 2004;91(2):355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grabiner BC, Nardi V, Birsoy K, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4(5):554–563. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laplante M, Sabatini DM. mTOR signaling at a glance. J. Cell Sci. 2009;122(20):3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bar-Peled L, Chantranupong L, Cherniack AD, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340(6136):1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Kranenburg M, Hoogeveen-Westerveld M, Nellist M. Preliminary functional assessment and classification of DEPDC5 variants associated with focal epilepsy. Hum. Mutat. 2015;36(2):200–209. doi: 10.1002/humu.22723. [DOI] [PubMed] [Google Scholar]
- 29.Assogba K, Ferlazzo E, Striano P, et al. Heterogeneous seizure manifestations in Hypomelanosis of Ito: report of four new cases and review of the literature. Neurol. Sci. Off. J. Ital. Neurol. Soc. Ital. Soc. Clin. Neurophysiol. 2010;31(1):9–16. doi: 10.1007/s10072-009-0160-5. [DOI] [PubMed] [Google Scholar]
- 30.Auriemma A, Agostinis C, Bianchi P, et al. Hemimegalencephaly in hypomelanosis of Ito: early sonographic pattern and peculiar MR findings in a newborn. Eur. J. Ultrasound Off. J. Eur. Fed. Soc. Ultrasound Med. Biol. 2000;12(1):61–67. doi: 10.1016/s0929-8266(00)00099-9. [DOI] [PubMed] [Google Scholar]
- 31.Chapman K, Cardenas JF. Hemimegalencephaly in a patient with a neurocutaneous syndrome. Semin. Pediatr. Neurol. 2008;15(4):190–193. doi: 10.1016/j.spen.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 32.Manjila S, Miller BR, Goodman A, et al. Pharmacoresistant epilepsy in hypomelanosis of Ito: palliative surgical treatment with modified anatomic posterior quadrantic resection. Clin. Neurol. Neurosurg. 2014;123:15–17. doi: 10.1016/j.clineuro.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 33.Sharma S, Sankhyan N, Kabra M, Kumar A. Hypomelanosis of Ito with hemimegalencephaly. Dermatol. Online J. 2009;15(11):12. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.