Abstract
Primary ciliary dyskinesia (PCD) is a recessively inherited disease that leads to chronic respiratory disorders owing to impaired mucociliary clearance. Conventional transmission electron microscopy (TEM) is a diagnostic standard to identify ultrastructural defects in respiratory cilia but is not useful in approximately 30% of PCD cases, which have normal ciliary ultrastructure. DNAH11 mutations are a common cause of PCD with normal ciliary ultrastructure and hyperkinetic ciliary beating, but its pathophysiology remains poorly understood. We therefore characterized DNAH11 in human respiratory cilia by immunofluorescence microscopy (IFM) in the context of PCD. We used whole-exome and targeted next-generation sequence analysis as well as Sanger sequencing to identify and confirm eight novel loss-of-function DNAH11 mutations. We designed and validated a monoclonal antibody specific to DNAH11 and performed high-resolution IFM of both control and PCD-affected human respiratory cells, as well as samples from green fluorescent protein (GFP)–left–right dynein mice, to determine the ciliary localization of DNAH11. IFM analysis demonstrated native DNAH11 localization in only the proximal region of wild-type human respiratory cilia and loss of DNAH11 in individuals with PCD with certain loss-of-function DNAH11 mutations. GFP–left–right dynein mice confirmed proximal DNAH11 localization in tracheal cilia. DNAH11 retained proximal localization in respiratory cilia of individuals with PCD with distinct ultrastructural defects, such as the absence of outer dynein arms (ODAs). TEM tomography detected a partial reduction of ODAs in DNAH11-deficient cilia. DNAH11 mutations result in a subtle ODA defect in only the proximal region of respiratory cilia, which is detectable by IFM and TEM tomography.
Keywords: left–right dynein, primary ciliary dyskinesia, normal ciliary ultrastructure, immunofluorescence microscopy, transmission electron microscopy
Clinical Relevance
Conventional transmission electron microscopy (TEM) is not diagnostic for approximately 30% of primary ciliary dyskinesia (PCD) cases because they have normal ciliary ultrastructure; DNAH11 mutations are a common cause of PCD with normal ciliary ultrastructure and hyperkinetic ciliary beating, but its molecular characterization in human respiratory cilia is completely lacking. We show that DNAH11 distinctly localizes to the proximal region of respiratory cilia, independently of all previously described factors governing dynein arm assembly. TEM tomography detects a partial reduction of outer dynein arms in only the proximal region of DNAH11-deficient cilia. This helps explain why DNAH11 mutations result in normal ciliary ultrastructure and hyperkinetic ciliary beating and suggests a novel mode of axonemal assembly in respiratory cilia.
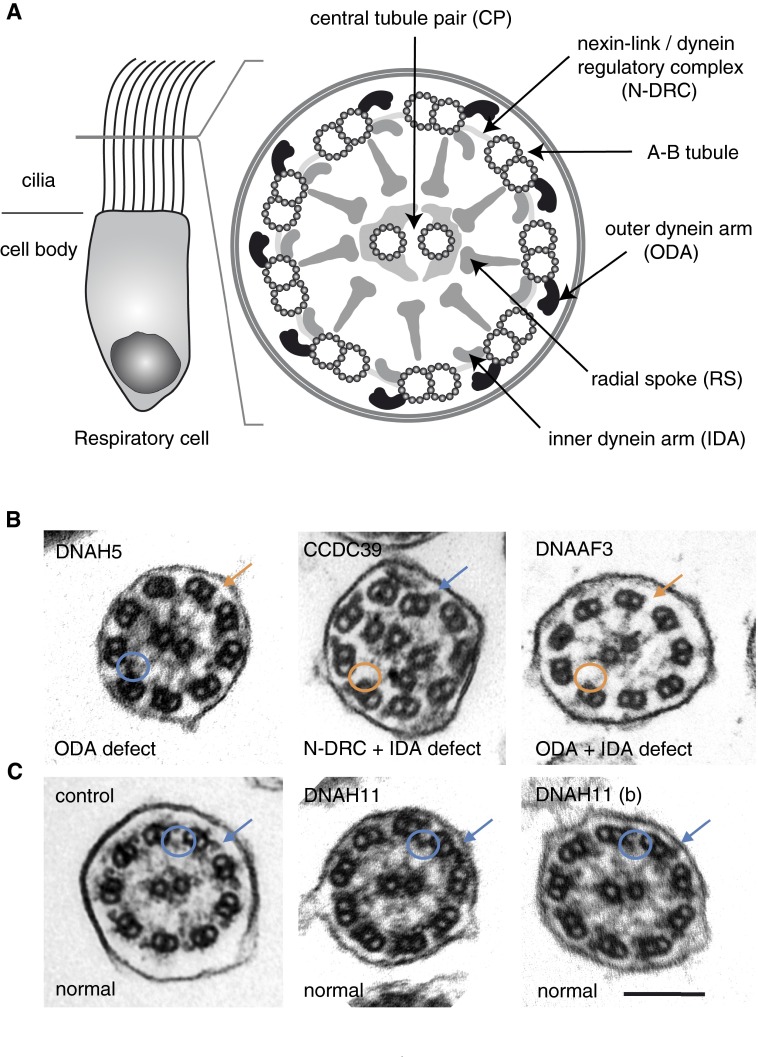
Primary ciliary dyskinesia (PCD; Mendelian Inheritance in Man or MIM no. 244400) is a recessively inherited disorder that affects at least 1 in 20,000 individuals (1). Individuals with PCD suffer from a chronic, destructive respiratory disorder characterized by bronchiectasis and progressive lung failure. This respiratory disorder results from defective mucociliary clearance, due to deficient ciliary beating of respiratory epithelial cells lining the airways (2). In almost half of individuals with PCD, left–right (LR) body patterning defects, such as situs inversus, are observed due to dysmotility of monocilia, which are present during early embryonic development at the LR organizer and produce a leftward fluid flow to establish body asymmetry (3). PCD is genetically heterogeneous, as mutations in over 30 distinct genes have been reported to cause this disease (see Table E1 in the online supplement). The majority of PCD-causing mutations result in gross ultrastructural defects of the respiratory ciliary axoneme that are detectable by transmission electron microscopy (TEM) analysis, such as the loss of outer dynein arms (ODAs) from mutations in DNAH5 (4) and DNAI2 (5) and microtubular disorganization with inner dynein arm (IDA) and nexin-link/dynein regulatory complex (N-DRC) defects from mutations in CCDC39 (6) and CCDC40 (7) (Figure 1). Indeed, TEM has been the gold standard to diagnose PCD (8) well before the first identification of mutations in a PCD-causing gene (9). In approximately 30% of PCD cases, however, there are no ultrastructural defects detectable by TEM (Figure 1), despite the clinical presentation of respiratory disease, including bronchiectasis and associated defects, such as situs inversus. The timely diagnosis of PCD and management of this respiratory disease may even be delayed in cases of normal ciliary ultrastructure without associated defects, such as situs inversus (10).
Figure 1.
Diagnostic transmission electron microscopy (TEM) is not useful in approximately 30% of primary ciliary dyskinesia (PCD) cases. (A) Schematic of respiratory cell cilia and its cross-section highlighting the “9 + 2” organization of the ciliary axoneme and associated structures, as visualized by TEM. (B) TEM is diagnostic for loss of outer dynein arms (ODAs; e.g., DNAH5 mutations), microtubular disorganization with nexin-link/dynein regulatory complex (N-DRC) and inner dynein arm (IDA) defects (e.g., CCDC39 mutations), and loss of both ODAs and IDAs (e.g., DNAAF3 mutations). (C) TEM is not diagnostic for DNAH11 mutations, as they are indistinguishable from healthy control axonemes. Blue arrows, ODAs; orange arrows, absent ODAs; blue circles, IDAs; orange circles, absent IDAs. Scale bar: 100 nm. Individuals with PCD OI-16 II2 (DNAH5; p.Gln1464* hom.) and OP-1482 (CCDC39, c.610-2A>G hom.) were identified by whole-exome sequencing; the DNAH5, p.Gln1464* variant is novel and not present in the 1,000 Genomes and ExAC databases (sequencing data not shown); the CCDC39, c.610–2A>G variant was previously described (40). Individuals with PCD OP-98 II1 (DNAH11 p.Trp2604*, p.Ile4445Asnfs*3 het.) and OP-235 II2 (DNAH11 [b] p.Gln4233*, p.Leu4327Ser het.) as well as OP-989 II2 (DNAAF3, p.Leu108Pro hom.) were reported previously (14, 26).
We and others have shown that mutations in the gene encoding DNAH11 (MIM no. 611884) are a common cause of PCD with normal ciliary ultrastructure and a hyperkinetic beating pattern (11–14). We have established immunofluorescence microscopy (IFM) analysis of respiratory cilia as a complement to diagnostic measures, such as TEM and high-speed video microscopy analysis (HVMA), to identify axonemal defects of the ODAs and IDAs, as well as the N-DRC (5, 7, 15–17) and also tailored IFM to identify PCD variants with defective radial spoke (RS) head composure that do not exhibit gross ultrastructural abnormalities (18).
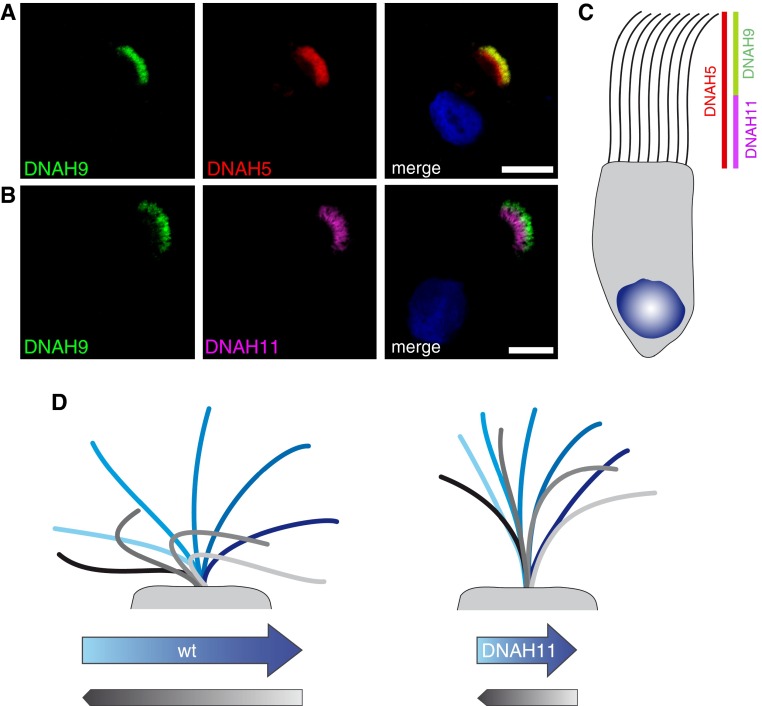
Studies addressing the molecular characterization of human DNAH11 in respiratory cilia, especially in the context of PCD, are completely lacking; only green fluorescent protein (GFP)-tagged mouse DNAH11 (GFP–left–right dynein [lrd]) localization has been shown previously (19). Therefore, we designed and validated a mouse monoclonal antibody specific to human DNAH11. Here, we show its specificity by matching its expression pattern with mouse GFP-DNAH11 (GFP-lrd) and confirming loss of DNAH11 in individuals with PCD with certain loss-of-function DNAH11 mutations. We demonstrate that DNAH11, the ODA protein orthologous to the Chlamydomonas β heavy chain (HC), is restricted spatially to only the proximal region of respiratory ciliary axonemes, whereas its β-HC paralogue, DNAH9, is restricted to only the distal region of respiratory ciliary axonemes, thus defining two compositionally distinct ciliary regions. DNAH11-mutant respiratory cilia demonstrate characteristic stiffness and reduced ciliary bending in the proximal axonemal region by HVMA, indicating that DNAH11 function is essential for proper ciliary bending in this newly defined ciliary region. TEM tomography detected a partial reduction of ODAs in the proximal, but not distal, regions of cilia lacking DNAH11. We demonstrate further that DNAH11 localizes to respiratory ciliary axonemes independently of gross ultrastructural defects caused by mutations in genes encoding the ODA assembly factors, DNAH5 and DNAI2, as well as the dynein axonemal assembly factors, DNAAF1 and DNAAF3, suggesting that a subtle ODA defect accompanies DNAH11 mutations, which is now detectable by IFM as well as TEM tomography.
Materials and Methods
Individuals with PCD
Signed and informed consent was received from patients fulfilling the diagnostic criteria of PCD and their family members, using protocols approved by the Institutional Ethics Review Board at the University of Muenster (Muenster, Germany) and collaborating institutions.
High-resolution immunofluorescence analyses of human respiratory cells were performed as described previously (5). Images were acquired using a Zeiss LSM 880 laser scanning microscope with Zen-2 software and Zeiss AxioObserver Z1 Apotome with AxioVision 4.8 software (Carl Zeiss, Jena, Germany) and processed using Adobe CS4 software (Adobe Systems Incorporated, San Jose, CA).
TEM analysis, Western blotting, homozygosity mapping, haplotype analysis, and high-speed videomicroscopy analysis were performed as described previously (5, 16, 20, 21).
Whole-exome and sanger sequencing were performed as described previously (18). Variant analysis of whole-exome data was performed using Varbank software (Cologne Center of Genomics, Cologne, Germany). The DNAH11 reference sequence is NM_001277115.1 (National Center for Biotechnology Information)/ENST00000409508 (Ensembl). Primer sequences used to confirm DNAH11 mutations by Sanger sequencing are listed in the online supplement.
Additional information describing mouse monoclonal anti-DNAH11 antibody design and production, antibodies, DNAH11 (GFP-lrd) localization in mouse, TEM tomography, and next-generation DNA sequencing and variant analysis are available in the online supplement.
Results
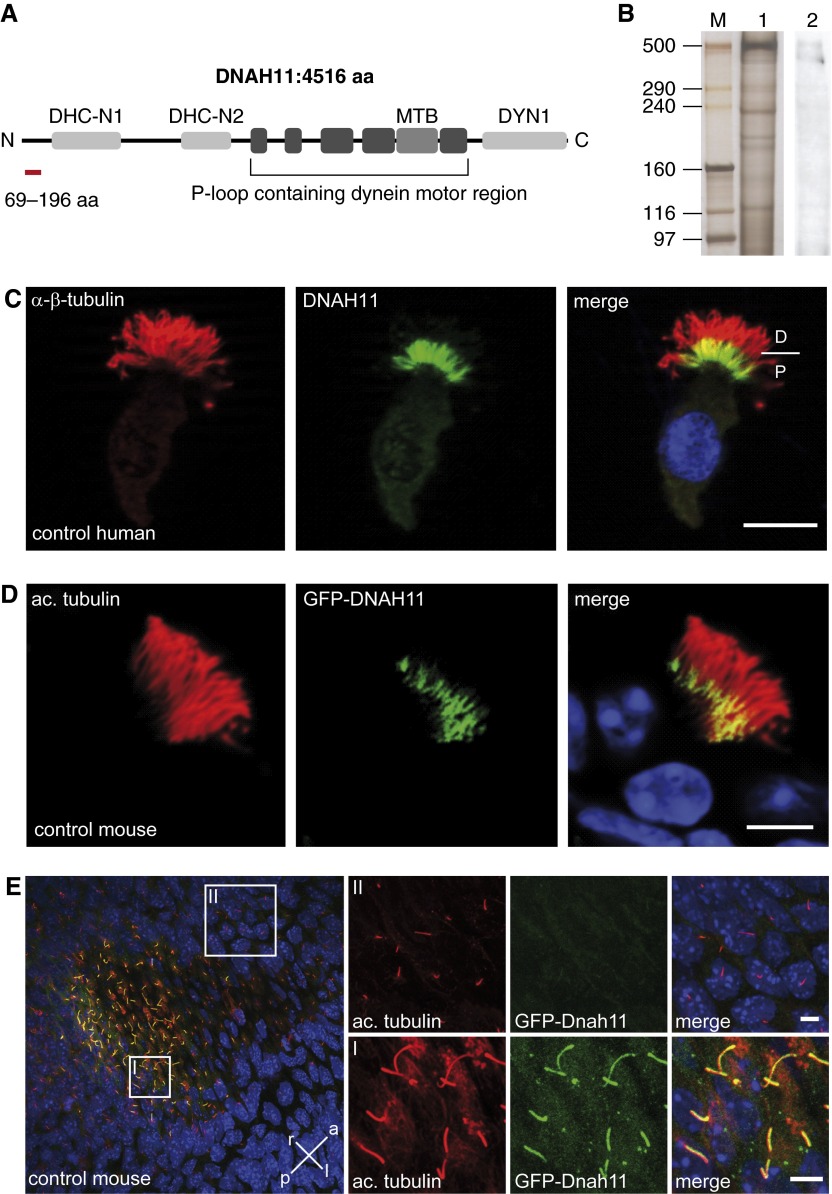
Axonemal dynein HC proteins (e.g., DNAH5, DNAH9, and DNAH11) show a high degree of sequence conservation, especially in the centrally located motor domain region. To generate a specific anti-human DNAH11 antibody, we selected an epitope with low sequence conservation to other axonemal dynein HC proteins, specifically in the aminoterminal region of DNAH11 (Figure 2A; see also Figure E1). Because DNAH11 has been identified in the proteome of human respiratory cilia (22), and mouse GFP-DNAH11 has been shown to be localized in motile cilia (19), we prescreened anti-DNAH11 hybridoma lines by IFM for ciliary localization in human respiratory cells. Western blotting of lysates from porcine respiratory cells with anti-human DNAH11 (Figure 2B) demonstrated a cross-reactive, albeit weak, band of approximately 500 kD, consistent with the expected molecular weight of 520 kD. An immunoreactive band slightly below 500 kD may represent an isoform of DNAH11. Respiratory cells from healthy control individuals were costained with anti-DNAH11 and anti–α/β-tubulin. As shown in Figure 2C, DNAH11 localized only to the proximal region of ciliary axonemes, as compared with panaxonemal localization of α/β-tubulin. We determined that a commercially available, polyclonal anti-DNAH11 antibody also identified DNAH11 in the proximal region of human respiratory cilia by IFM, and provided a robust cross-reactive signal by Western blotting of porcine axonemal extracts (see Figure E13). The mouse ortholog to DNAH11, also known as the lrd, was shown to be expressed in tissues containing motile cilia, including the LR organizer (19). Consistent with findings in human respiratory cells, mouse GFP-DNAH11 localized to the proximal region of respiratory cilia (Figure 2D). In contrast, mouse GFP-DNAH11 localization in monocilia of the LR organizer of GFP-lrd mice was panaxonemal rather than proximal (Figure 2E). These data demonstrate that human DNAH11 and mouse GFP-DNAH11 have a restricted, proximal localization in respiratory cilia, and that mouse GFP-DNAH11 has an unrestricted, panaxonemal localization pattern in the embryonic LR organizer; this distinction was not reported previously (19).
Figure 2.
DNAH11 localizes exclusively to the proximal region of respiratory cilia. (A) Schematic of DNAH11 protein domains that are conserved among other DNAH proteins per the Conserved Domain Database. The P-loop region contains several ATPase domains as well as the microtubule binding (MTB) domain. DHC-N, dynein heavy chain, N-terminal region; DYN, dynein heavy chain/region D6 of dynein motor. Red highlight, the epitope of DNAH11 spanning amino acids 69–196. (B) Anti-DNAH11Western blotting of porcine axonemal extracts indicates immunoreactive bands of approximately 500 kD. M, marker lane; 1, silver stain of lysate; 2, anti-DNAH11. (C) Immunofluorescence microscopy (IFM) of human control respiratory cells shows that native DNAH11 (green) localizes to the proximal region of ciliary axonemes; α/β-tubulin (red) was used as counterstain to detect ciliary axonemes. D, distal; P, proximal; scale bar: 10 µm. (D) IFM of mouse respiratory cells expressing green fluorescent protein (GFP)-DNAH11 (GFP–left–right dynein [lrd]) demonstrates that GFP-DNAH11 localizes to the proximal region of ciliary axonemes, similar to the pattern of human DNAH11 localization. Acetylated α-tubulin was used as counterstain to detect ciliary axonemes (red). Scale bar: 10 μm. (E) In contrast to mouse respiratory cilia, GFP-DNAH11 localizes throughout the entire axoneme in motile monocilia of the left–right organizer (I) in early mouse embryos but is not detectable in nonmotile cilia of the node or primary cilia of endodermal cells (II). Body axis is specified as right (r), left (l), anterior (a), and posterior (p). Scale bars in (I) and (II): 10 μm. ac., acetylated.
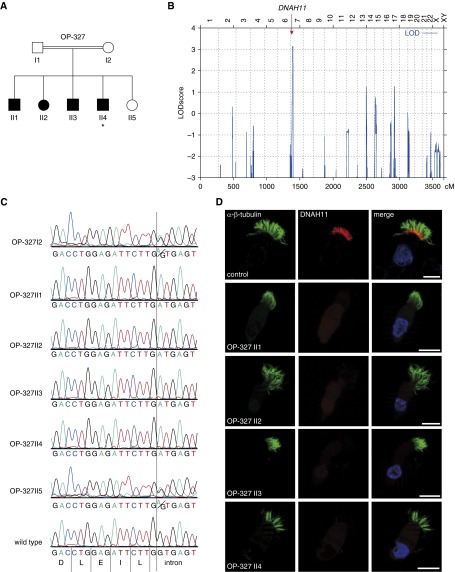
To validate the specificity of this anti-DNAH11 antibody, we examined individuals with PCD harboring novel loss-of-function DNAH11 mutations. We identified a consanguineous Iranian family (OP-327) with five siblings, of whom four presented with recurrent upper and lower respiratory tract infections, and one also displayed situs inversus totalis (Figure 3A). Genome-wide linkage analysis of OP-327 identified chromosome 7 as the only genomic region with a significant logarithm of odds (LOD) score (above 3.0). Haplotype analysis demonstrated an 11.9-Mb region of homozygosity among the PCD siblings within the chromosomal interval 7p15.2–7p21.3 (physical positions 13,201,036–25,126,009; National Center for Biotechnology Information Map Viewer), consistent with homozygosity by descent. This genomic interval contains 136 genes, and the only known PCD-causing gene within this locus is DNAH11 (Figure 3B). We subsequently analyzed DNA from OP-327 II4 by whole-exome sequencing and filtered pathogenic variants (Varbank software) located within this interval on chromosome 7. We identified a homozygous splice-site mutation in DNAH11 (c.11,739 + 1 G>A, intron 73); this variant affects a canonical donor splice-site, predicts aberrant splicing, and is absent from both the 1,000 Genomes and ExAC databases. We confirmed by Sanger sequencing that this homozygous DNAH11 mutation was present in all affected siblings, but was heterozygous in the mother and unaffected sibling (DNA from the father was not available). Thus, segregation was consistent with autosomal recessive inheritance (Figure 3C). Next, we performed IFM on respiratory cells from all affected siblings using this anti-DNAH11 antibody and determined that DNAH11 was absent in respiratory cilia from these siblings (Figure 3D). These data demonstrate that the DNAH11 c.11,739 + 1 G>A donor splice-site mutation results in the loss of DNAH11 and validated the specificity of this anti-DNAH11 antibody. Consistent with the reported DNAH11-mutant phenotype, the affected siblings had normal ciliary composition for the ODA HC protein DNAH5, the IDA light chain (LC) protein DNALI1, and the RS head protein (RSPH) 9, as determined by IFM (see Figure E2). Respiratory cells from the mother and unaffected sibling were not available for IFM; TEM and HVMA were not available for the affected siblings.
Figure 3.
DNAH11 is absent in respiratory cilia of individuals with PCD with a homozygous DNAH11 splice-site mutation. (A) Pedigree of the consanguineous PCD family OP-327. PCD siblings are shaded black, and the unaffected sibling is shaded white. *Situs inversus totalis. (B) Genome-wide linkage analysis of family OP-327 indicates chromosome 7 (x-axis represents genomic position in centimorgans or cM) as the only significant logarithm of odds (LOD) score (y-axis); this genomic interval contains DNAH11 (red arrow). (C) Sanger sequencing confirmed a homozygous splice donor variant in DNAH11 (intron 73, c.11739 + 1G>A) in four PCD siblings (OP-327 II1–II4). The unaffected parent (OP-327 I2) and sibling (OP-327 II5) are heterozygous (A/G) for this variant, whereas wild-type DNA shows the G nucleotide at this position. The triplet nucleotide sequence (color) is shown above the translated amino acid sequence (single letters). (D) IFM shows that, in contrast to healthy human respiratory cells, DNAH11 (red) is absent in PCD siblings OP-327 II1–II4. α/β-tubulin (green) is used as counterstain to detect ciliary axonemes. Scale bar: 10 µm.
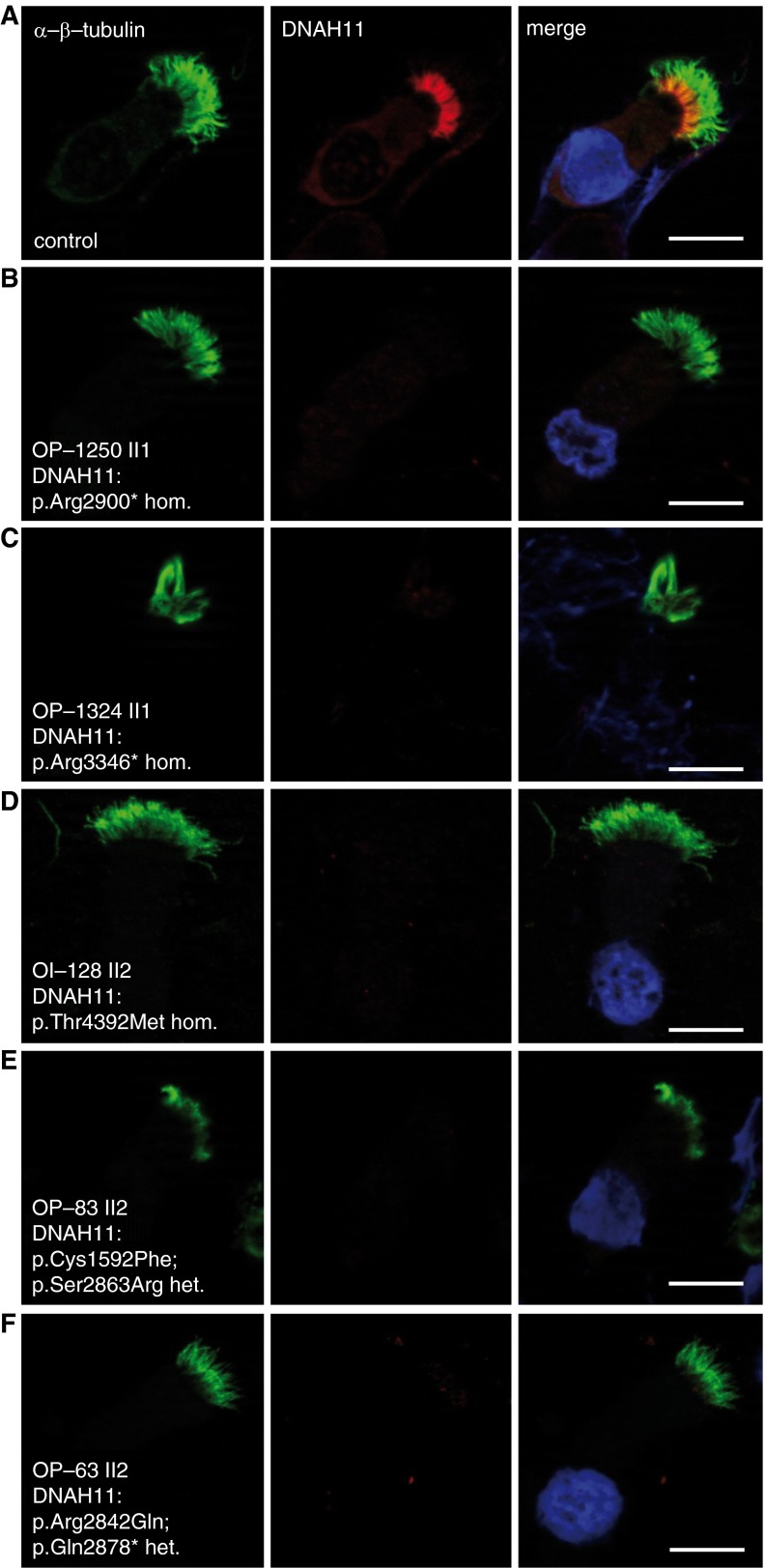
We examined DNAH11 localization in additional individuals with PCD with biallelic DNAH11 mutations identified by either whole-exome or targeted next-generation sequencing (NGS) and verified by Sanger sequencing (see Table E2 and Figures E3–E7). All mutations were either absent or showed a frequency below 0.01 in the 1,000 Genomes and ExAC databases, and were scored as pathogenic (see Table E3). Consistent with classical loss-of-function DNAH11 mutations in individuals with PCD OP-1250 II1 (exon 53, c.8698C>T, p.Arg2900* hom.) and OP-1324 II1 (exon 62, c.10036C>T, p.Arg3346* hom.), we found that DNAH11 was absent in respiratory cells from these individuals (Figures 4A–4C). TEM of both OP-1250 II1 and OP-1324 II1 showed normal ciliary ultrastructure (see Figures E3 and E4). Thus, anti-DNAH11 IFM can detect certain loss-of-function DNAH11 mutations in cases with normal TEM. We next examined whether respiratory cells harboring either homozygous or compound heterozygous DNAH11 missense mutations can exhibit aberrant DNAH11 localization, including respiratory cells from individuals with PCD OI-128 II2 (exon 81, c.13175C>T, p.Thr4392Met, hom.), OP-83 II2 (exon 27, c.4775G>T, p.Cys1592Phe; exon 52, c.8589C>G, p.Ser2863Arg), and OP-63 II2 (exon 52, c.8525G>A, p.Arg2842Gln; exon 52, c.8632C>T, p.Gln2878*) (Figures 4D–4F). Interestingly, we found that DNAH11 was also undetectable in these respiratory cilia, indicating that these DNAH11-mutant proteins are either not expressed or unable to be assembled in the ciliary axonemes. OI-128 II1 showed normal ciliary ultrastructure by TEM (see Figure E5). In contrast, IFM of respiratory cells from individuals with PCD OP-98 II1 (exon 48, c.7914G>C, p.Trp2604*; exon 82, c.13333_34insACCA, p.Ile4445Asnfs*3) and OP-235 II1 (exon 77, c.12697C>T, p.Gln4233*; exon 79, c.12980T>C, p.Leu4327Ser), which were reported previously (14), have normal ciliary ultrastructure by TEM (Figure 1C), and demonstrated normal proximal DNAH11 localization (see Figure E8). We conclude that anti-DNAH11 IFM can detect DNAH11-mutant cilia and therefore be helpful to establish the correct diagnosis (Figures 3D and 4A–4E; see also Figure E13). However, IFM is not always abnormal in individuals with PCD with DNAH11 mutations (see Figure E8), demonstrating that certain DNAH11 variants do not significantly alter DNAH11 protein expression and/or its assembly to the proximal ciliary region.
Figure 4.
DNAH11 is absent in respiratory cilia of individuals with PCD with both homozygous nonsense and homozygous or compound heterozygous missense DNAH11 mutations. We confirmed additional novel DNAH11 loss-of-function mutations (see Figures E3–E7 and Tables E2 and E3) and determined that, in contrast to healthy control (A), these cilia lack DNAH11 (B–F). α/β-tubulin (green) is used as counterstain to detect ciliary axonemes. Scale bars: 10 µm.
The ODAs are large, multisubunit molecular motors formed by the combined assembly of polypeptides of different sizes: HC (∼400–500 kD), intermediate chain (∼45–110 kD) and LC (∼8–55 kD) (23, 24). In the unicellular alga, Chlamydomonas, the ODAs comprise three distinct HCs (α-, β-, and γ-HC). We reported previously that at least two distinct types of ODA complexes are present in human respiratory cilia (15): the ODA HC DNAH5 (orthologous to the Chlamydomonas γ-HC) is present throughout the entire length of the ciliary axoneme, whereas the ODA HC DNAH9 (orthologous to the Chlamydomonas β-HC) localizes exclusively to the distal ciliary compartment. Thus in respiratory cilia, at least two ODA types are present: type 1 (DNAH9 negative and DNAH5 positive; proximal ciliary axoneme) and type 2 (DNAH9 and DNAH5 positive; distal ciliary axoneme). To characterize further these ODA subtypes in human respiratory cilia, we costained control respiratory cells with anti-DNAH11 and anti-DNAH9. As shown in Figure 5, both DNAH11 and DNAH9 have distinct localization patterns in only the proximal or distal axonemal regions of respiratory cilia, respectively. This supports and clarifies our previous finding that distinct ODA subtypes are present in human respiratory cilia: a proximal axonemal region positive for DNAH11 (and negative for DNAH9, type 1), and a distal axonemal region positive for DNAH9 (and negative for DNAH11, type 2); DNAH5, which is localized along the entire respiratory axoneme, possibly functions as a heterodimeric dynein complex with the Chlamydomonas β-HC orthologs DNAH11 in the proximal region or with DNAH9 in the distal region. This proximal ciliary region containing DNAH11 is also distinct from the ciliary transition zone (see Figure E9). We next analyzed ciliary beating of DNAH11-mutant respiratory cells from individuals with PCD OP-1250 II1, OP-1324 II1, and OP-1324 II2 by HVMA. These respiratory cells characteristically exhibited reduced ciliary bending in the proximal (but not distal) region of the ciliary axonemes, subsequently reducing the amplitude of the waveform and manifesting as hyperkinetic beating (Figure 5D; see also Figure E10 and Videos E1–E8). We propose that loss or dysfunction of DNAH11 in the proximal region of respiratory cilia reduces dynein-mediated force generation in this area, resulting in reduced ciliary bending and increased stiffness in the proximal axonemal region.
Figure 5.
DNAH11 and DNAH9 distinctly localize to proximal and distal regions of respiratory cilia, respectively. (A) IFM shows that DNAH9 (green) localizes to the distal region of respiratory cilia; DNAH5 (red) localizes throughout the entire cilium. Scale bar: 10 µm. (B) IFM shows that DNAH11 (purple) localizes distinctly in the proximal region of respiratory cilia in contrast to DNAH9 (green), which localizes distinctly in the distal region of respiratory cilia. Scale bar: 10 µm. (C) A schematic shows that DNAH11 and DNAH9 localize to distinct regions in respiratory cilia, thus defining distinct proximal and distal ODA subtypes. (D) Model of hyperkinetic ciliary beating. Whereas healthy respiratory cilia display an average beat frequency of approximately 6 Hz, cilia from individuals with DNAH11 mutations have an average beat frequency above 9 Hz, owing to reduced bending in the proximal ciliary region (see Figure E10 and Videos E1–E8). Blue shades indicate forward effective stroke; gray shades indicate recovery stroke. wt, wild type.
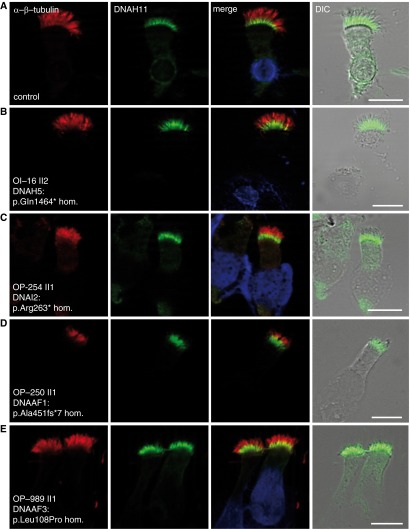
To characterize further DNAH11 in diverse PCD subtypes, we examined respiratory cilia from individuals with PCD with defined genetic/phenotypic defects by IFM. Previous studies have demonstrated that loss-of-function mutations in the genes DNAH5 and DNAI2 (encoding structural components of type 1 and type 2 ODA complexes) result in lack of axonemal ODA assembly (4, 5, 15); these DNAH5- and DNAI2-mutant cilia are deficient for the ODA proteins, DNAH5, DNAH9, and DNAI2 (5, 15), and lack the ODA structure, as determined by TEM (Figure 1B). Interestingly, we found normal proximal DNAH11 localization in DNAH5- and DNAI2-mutant respiratory cilia, indicating that the integrity of the ODA macromolecular complex is not essential for proximal DNAH11 assembly in respiratory axonemes (Figures 6A–6C). Previous studies have also demonstrated that loss-of-function mutations in DNAAF1 (25) and DNAAF3 (26) cause loss of both ODA and IDA assembly; DNAAF1- and DNAAF3-mutant cilia are deficient for the same ODA proteins as DNAH5- and DNAI2-mutant cilia, as well as the inner arm LC protein DNALI1, and they lack both the ODA and IDA structures, as determined by TEM (Figure 1B). We also observed normal proximal DNAH11 localization in these DNAAF1- and DNAAF3-mutant cilia (Figures 6D and 6E), demonstrating that the integrity of neither the ODA nor IDA macromolecular complex is essential for DNAH11 assembly in respiratory axonemes. Further supporting this observation, we also determined that DNAH11 retains proximal ciliary localization in CCDC151-mutant respiratory cilia, which also have ODA defects, due to functional loss of the ODA axonemal docking complex (27) (see Figure E11).
Figure 6.
DNAH11 localizes to the proximal region of respiratory cilia in individuals with PCD with mutations affecting ODA and IDA assembly. Similar to healthy control respiratory cells (A), DNAH11 localizes to the proximal region of respiratory cilia in individuals with PCD harboring homozygous nonsense mutations in DNAH5 (B) and DNAI2 (C) that lack ODA structures, as well as individuals with PCD harboring homozygous loss-of-function mutations in DNAAF1 (D) and DNAAF3 (E) that lack both ODA and IDA structures. Anti-DNAH11 (green) is costained with α/β-tubulin (red) to detect ciliary axonemes. Scale bar: 10 µm. Individuals with PCD OP-254 II1, OP-250 II1 and OP-989 II1 were reported previously (5, 25, 26). TEM of OI-16 II2 and OP-989 II1 are shown in Figure 1B. DIC, differential interference contrast microscopy.
We next analyzed whether DNAH11 localization depends on the integrity of defined axonemal multiprotein complexes that intercalate with the A-B tubules, such as the N-DRC and RS structures (Figure 1A). We observed proximal DNAH11 localization in healthy control respiratory cilia as well as those harboring N-DRC (CCDC164-mutant cilia) (28), N-DRC and IDA (CCDC39-mutant cilia) (6), and RS (RSPH4A-mutant cilia lacking the entire RS head structure) (18) defects stemming from loss-of-function mutations in these respective PCD-causing genes (see Figures E11 and E12). We conclude that axonemal assembly of the β-HC ortholog, DNAH11, does not depend on the integrity of the N-DRC and RS ciliary complexes or the ODAs and IDAs in respiratory cilia.
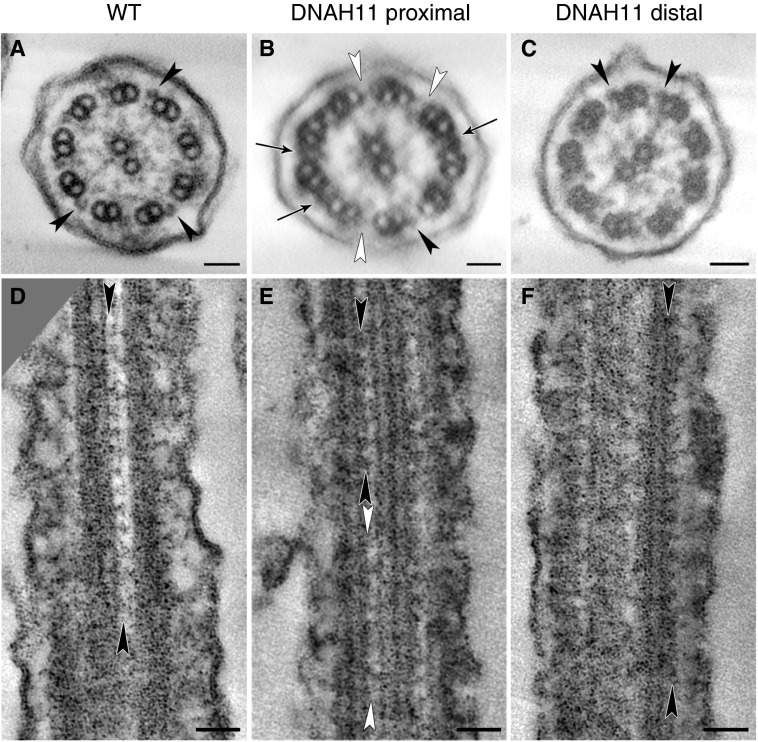
Studies in the Chlamydomonas β-HC mutant, oda4-s7 (a truncated β-HC protein lacking its carboxyl terminus), have demonstrated a variable reduction in electron density in the midportion of ODAs and occasional reduction in ODA number by averaged TEM cross-section analysis (29–31). Therefore, we carefully re-evaluated the proximal region of DNAH11-deficient cilia (OP-1250 II1) by TEM tomography and detected a partial reduction in ODAs in the proximal, but not distal, ciliary region of these cilia (Figure 7). This indicates that DNAH11-mutant cilia harbor subtle ODA defects in only the proximal ciliary region, and clarifies the apparently normal ciliary ultrastructure of these cilia: this subtle ODA defect in DNAH11-mutant cilia may be easily overlooked by conventional TEM analysis (visualization of a single, structural projection in two dimensions), but is detectable by careful analysis of three-dimensional models of the proximal ciliary region obtained by TEM tomography (detailed visualization of a structure inside a reconstructed three-dimensional volume).
Figure 7.
TEM tomography detects a partial reduction of ODAs in the proximal, but not distal, region of DNAH11-deficient cilia. Tomographic reconstruction of ciliary cross-sections from (A) wild-type (WT) and (B and C) DNAH11-deficient cilia (OP-1250 II1; see also Figure 4B and Figures E3 and E14, and Videos E1–E4) identify a partial reduction of ODAs (white arrowheads) in the proximal (B), but not distal (C), region of DNAH11-deficient cilia. Black arrowheads, structured ODAs; black arrows in B indicate the electron-dense material sometimes visible between microtubule doublets in the proximal ciliary region, approximately 400 nm distal to the transition zone and 100 nm distal to the base of the central pair microtubules. Tomographic reconstructions of ciliary longitudinal sections from (D) WT and (E and F) DNAH11-deficient cilia (OP-1250 II1) also identify a partial reduction of ODAs (white arrowheads) in the proximal (E), but not distal (F), region of DNAH11-deficient cilia. Scale bars: 50 nm.
Discussion
The diagnosis of PCD in cases of normal ciliary ultrastructure and normal visceral arrangement (situs solitus) remains challenging and requires additional supportive assays (10). Despite several reports describing DNAH11 mutations as a cause of PCD with normal ciliary ultrastructure, its molecular characterization and mechanistic contribution to this PCD subtype remains poorly understood. Here, we have validated a monoclonal antibody specific to DNAH11; DNAH11 is localized to the proximal region of respiratory cilia, and this is supported by the observation that GFP-DNAH11 is also localized proximally in mouse respiratory cilia. The differential localization pattern of GFP-DNAH11 between mouse respiratory cilia (restricted, proximal) and motile monocilia of the embryonic LR organizer (panaxonemal) underscores the observation that DNAH11/Dnah11 mutations result in LR body patterning defects, and supports a critical function for DNAH11 in nodal ciliary beating (12, 14, 32–34).
Because DNAH11 mutations are a common cause of PCD with normal ciliary ultrastructure, and conventional TEM is not useful in these cases, anti-DNAH11 IFM of respiratory cilia should facilitate diagnosis of this PCD subtype (Figures 3–4). However, we also show that some DNAH11 mutations are compatible with axonemal assembly of a mutant DNAH11 protein (see Figure E8). This antibody likely would not detect carboxyl-terminal missense and/or truncating mutations (OP-98 II1, p.Ile4445Asnfs*3; OP-235 II2, p.Gln4233*, p.Leu4327Ser) unless they affect overall protein stability. Thus, a normal DNAH11 staining pattern using this antibody cannot rule out the presence of certain DNAH11 mutations, and a negative DNAH11 staining pattern is not diagnostic per se. However, due to current limitations in routine genetic testing (e.g., low coverage, detection of indels), anti-DNAH11 IFM analysis could support a PCD diagnosis in unresolved genetic cases, for example, when only a single heterozygous DNAH11 mutation is detected (14). We showed recently that IFM can help to establish a PCD diagnosis in cases with RS defects, using anti-RSPH1, RSPH4A, and RSPH9 antibodies (18). Furthermore, we have previously established IFM-based diagnostic screening to identify ODA defects using anti-DNAH5, anti-DNAH9, and anti-DNAI2 antibodies (5, 15, 16). We also showed that IFM using anti-CCDC39, anti-GAS8, and anti-DNALI1 antibodies can detect N-DRC and IDA defects (6, 7, 17, 25). Thus, the majority of known PCD types are now detectable by IFM, which is helpful to establish the correct diagnosis.
The restricted localization of human DNAH11 in the proximal region of respiratory cilia can help us to understand why DNAH11 mutations result in PCD with hyperkinetic ciliary beating and reduced beating amplitude. A previous report that systematically analyzed ciliary beating of respiratory cilia (at 25°C) with diverse genetic PCD variants (20) found that DNAH11 mutations resulted in a higher ciliary beat frequency with a mean value of 8.7 Hz, in contrast to reported mean values for healthy controls (6.36 Hz) and respiratory disease controls (5.32 Hz). In contrast to other PCD variants, including those harboring ODA defects (e.g., DNAH5 mutations), DNAH11-mutant respiratory cilia characteristically showed increased stiffness and reduced bending in the proximal axonemal region (see Videos E1–E8). The distinct proximal ciliary localization of DNAH11 and the characteristic beating deficiency observed in DNAH11 mutations suggest that DNAH11 is critical for bending of the proximal ciliary region during the effective and recovery strokes of the ciliary beat cycle (Figure 5D). To our knowledge, this is the first study to describe a well defined compartment in the proximal region of respiratory cilia. Previous studies using Chlamydomonas have described proximal axonemal localization of IDA proteins, including dynein HC DHC11, the ortholog to human ciliary dynein HC DNAH7 (35, 36) and Fa2p, an ortholog to human NIMA–related kinase (NEK) family proteins (37). These studies also proposed a specialized role for the proximal axonemal region in bending and waveform initiation (35, 36), as well as mechanosensation and regulation of intraflagellar transport at the site of flagellar assembly/autotomy (37).
The Chlamydomonas β-HC gene show ambiguous BLAST homologies to three human genes: DNAH9, DNAH11, and DNAH17. This suggests that β-HC has diverged for specialized functions in higher eukaryotes during evolution. Our data demonstrate that certain differences between proximal type 1 ODAs and distal type 2 ODAs have evolved because β-HC DNAH9 assembly depends on the integrity of the ODA complex (5, 15, 25), whereas β-HC DNAH11 assembly does not (Figure 6; see also Figure E11). Recently, in vitro studies using Chlamydomonas demonstrated that β-HC is the main force generator coupled to ATP hydrolysis, in contrast to α-HC (for which there is no clear human ortholog) and γ-HC (human orthologs DNAH5 and DNAH8) (38). Although we cannot exclude a functional role for DNAH17 in respiratory cilia, it has not been identified in several transcriptomic and proteomic analyses of eukaryotic cilia (39), in contrast to both DNAH11 and DNAH9. Interestingly, anti-DNAH9 IFM analysis of both DNAH11-deficient (OP-1250 II1) and DNAH11-mutant (OP-235 II2) cilia demonstrated that the β-HC DNAH9 shifts from a distinctly distal to a panaxonemal localization (see Figure E14), in an identical distribution pattern to the γ-HC ortholog, DNAH5. Presumably, DNAH9 can heterodimerize with DNAH5 in the proximal ciliary region when DNAH11 is either defective or absent. However, DNAH9 cannot functionally replace the partial ODA defect in the proximal ciliary region (Figure 7) and characteristic hyperkinetic beating (Figure 5D; see also Figure E10 and Videos E1–E8) in DNAH11-mutant cilia. Based on our observation that DNAH11 and DNAH9 occupy distinct proximal and distal regions of respiratory cilia, we conclude that DNAH11 and DNAH9 are paralogs: DNAH11 has diverged from an ancestral β-HC as the principal effector of proximal ciliary bending in respiratory cilia, whereas DNAH9 functions in the distal axonemal region.
We and others have demonstrated previously that DNAH11 mutations do not affect assembly of ODAs (12, 14). We now show by TEM tomography that a partial reduction of ODAs is detectable in only the proximal region of DNAH11-deficient cilia. In addition, we show by anti-DNAH11 IFM that the axonemal assembly of DNAH11 does not depend on the integrity of the macromolecular ODA and IDA complexes as well as the axonemal N-DRC and RS complexes. This demonstrates that the ODA defect underlying DNAH11 mutations is subtle, and helps explain why these PCD cases demonstrate normal ciliary ultrastructure by conventional TEM. Furthermore, this predicts that a novel pathway of axonemal assembly and as-yet unidentified genetic factors may be responsible for this relatively common (over 30%) PCD subtype.
In summary, these studies use a validated anti-DNAH11 antibody to define a distinct proximal region in respiratory cilia and provide insight into the pathophysiology of a common cause of PCD with normal ciliary ultrastructure and hyperkinetic ciliary beating.
Supplementary Material
Acknowledgments
Acknowledgments
The authors thank the individuals with primary ciliary dyskinesia and their families for participating in this study, and acknowledge the German patient support group “Kartagener Syndrom und Primaere Ciliaere Dyskinesie e.V.” The authors also thank M. Herting, B. Lechtape, S. Fleige-Menzen, D. Ernst, L. Overkamp, K. Wohlgemuth, and F. J. Seesing for excellent technical assistance, G. Nürnberg and P. Nürnberg for homozygosity mapping data, and the Exome Aggregation Consortium for creating their exome variant database.
Footnotes
This work was supported by funding from the Deutsche Forschungsgemeinschaft grants OL/450-1 (H. Olbrich) and OM 6/4, OM 6/7, and OM6/8 (H. Omran), Interdisziplinären Zentrum für Klinische Forschung Muenster grant Om2/009/12 (H. Omran), European Commission grant FP7/2007–2013 grant agreement (GA) 262,055 (Y.M. and H. Omran) as a Transnational Access project of the European Sequencing and Genotyping Infrastructure, EU-FP7 programs SYSCILIA GA 241,955 (H. Omran and N.K.), BESTCILIA GA 305,404 (H. Omran), Wellcome Trust grant WT098051 (R.D. and A.K.-K.), Chief Scientist Office of the Ministry of Health, Israel grant 3-6176 (I.A.), European Respiratory Society Long-Term Research Fellowship 2014-3574 (V.M.), and by U.S. National Institutes of Health grants DK072301 (N.K. and E.E.D.) and R01HL093280 (M.B.). N.K. is a distinguished George W. Brumley Professor.
Author Contributions: G.W.D. wrote the manuscript and performed immunofluorescence analysis with N.T.L., J.A.K., V.M., and R.H. and performed Sanger sequencing analysis with J.A.K. and H. Olbrich; G.W.D. and N.T.L. performed Western blotting and with H. Olbrich designed and characterized the anti–axonemal dynein heavy chain 11 antibody; P.P. and T.M. analyzed green fluorescent protein (GFP)–left–right dynein (lrd) mice; M.B. provided GFP-lrd mice and performed data analysis; C. Westermann performed transmission electron microscopy (TEM); J.R., J.W., C. Werner, and K.H. performed clinical analysis of patients with primary ciliary dyskinesia, including high-speed video microscopy; M.A.K., S.K., F.E., P.V., G.B., M.A., L.B., and I.A. provided clinical data; Y.M., R.D., and A.K.-K. performed whole-exome sequencing; C.R., K.P., E.E.D., N.K., and B.D. performed targeted next-generation sequencing and variant analysis; G.P. and A.S. performed TEM tomography; H. Omran designed and co-wrote the manuscript and analyzed all data.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2015-0353OC on February 24, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Werner C, Onnebrink JG, Omran H. Diagnosis and management of primary ciliary dyskinesia. Cilia. 2015;4:2. doi: 10.1186/s13630-014-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 3.Pennekamp P, Menchen T, Dworniczak B, Hamada H. Situs inversus and ciliary abnormalities: 20 years later, what is the connection? Cilia. 2015;4:1. doi: 10.1186/s13630-014-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olbrich H, Häffner K, Kispert A, Völkel A, Volz A, Sasmaz G, Reinhardt R, Hennig S, Lehrach H, Konietzko N, et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left–right asymmetry. Nat Genet. 2002;30:143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 5.Loges NT, Olbrich H, Fenske L, Mussaffi H, Horvath J, Fliegauf M, Kuhl H, Baktai G, Peterffy E, Chodhari R, et al. DNAI2 mutations cause primary ciliary dyskinesia with defects in the outer dynein arm. Am J Hum Genet. 2008;83:547–558. doi: 10.1016/j.ajhg.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merveille AC, Davis EE, Becker-Heck A, Legendre M, Amirav I, Bataille G, Belmont J, Beydon N, Billen F, Clément A, et al. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat Genet. 2011;43:72–78. doi: 10.1038/ng.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker-Heck A, Zohn IE, Okabe N, Pollock A, Lenhart KB, Sullivan-Brown J, McSheene J, Loges NT, Olbrich H, Haeffner K, et al. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left–right axis formation. Nat Genet. 2011;43:79–84. doi: 10.1038/ng.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Afzelius BA. A human syndrome caused by immotile cilia. Science. 1976;193:317–319. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- 9.Pennarun G, Escudier E, Chapelin C, Bridoux AM, Cacheux V, Roger G, Clément A, Goossens M, Amselem S, Duriez B. Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 result in primary ciliary dyskinesia. Am J Hum Genet. 1999;65:1508–1519. doi: 10.1086/302683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuehni CE, Frischer T, Strippoli MP, Maurer E, Bush A, Nielsen KG, Escribano A, Lucas JS, Yiallouros P, Omran H, et al. ERS Task Force on Primary Ciliary Dyskinesia in Children. Factors influencing age at diagnosis of primary ciliary dyskinesia in European children. Eur Respir J. 2010;36:1248–1258. doi: 10.1183/09031936.00001010. [DOI] [PubMed] [Google Scholar]
- 11.Bartoloni L, Blouin JL, Pan Y, Gehrig C, Maiti AK, Scamuffa N, Rossier C, Jorissen M, Armengot M, Meeks M, et al. Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia. Proc Natl Acad Sci USA. 2002;99:10282–10286. doi: 10.1073/pnas.152337699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwabe GC, Hoffmann K, Loges NT, Birker D, Rossier C, de Santi MM, Olbrich H, Fliegauf M, Failly M, Liebers U, et al. Primary ciliary dyskinesia associated with normal axoneme ultrastructure is caused by DNAH11 mutations. Hum Mutat. 2008;29:289–298. doi: 10.1002/humu.20656. [DOI] [PubMed] [Google Scholar]
- 13.Pifferi M, Michelucci A, Conidi ME, Cangiotti AM, Simi P, Macchia P, Boner AL. New DNAH11 mutations in primary ciliary dyskinesia with normal axonemal ultrastructure. Eur Respir J. 2010;35:1413–1416. doi: 10.1183/09031936.00186209. [DOI] [PubMed] [Google Scholar]
- 14.Knowles MR, Leigh MW, Carson JL, Davis SD, Dell SD, Ferkol TW, Olivier KN, Sagel SD, Rosenfeld M, Burns KA, et al. Genetic Disorders of Mucociliary Clearance Consortium. Mutations of DNAH11 in patients with primary ciliary dyskinesia with normal ciliary ultrastructure. Thorax. 2012;67:433–441. doi: 10.1136/thoraxjnl-2011-200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fliegauf M, Olbrich H, Horvath J, Wildhaber JH, Zariwala MA, Kennedy M, Knowles MR, Omran H. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am J Respir Crit Care Med. 2005;171:1343–1349. doi: 10.1164/rccm.200411-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hornef N, Olbrich H, Horvath J, Zariwala MA, Fliegauf M, Loges NT, Wildhaber J, Noone PG, Kennedy M, Antonarakis SE, et al. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am J Respir Crit Care Med. 2006;174:120–126. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Omran H, Kobayashi D, Olbrich H, Tsukahara T, Loges NT, Hagiwara H, Zhang Q, Leblond G, O’Toole E, Hara C, et al. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456:611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frommer A, Hjeij R, Loges NT, Edelbusch C, Jahnke C, Raidt J, Werner C, Wallmeier J, Große-Onnebrink J, Olbrich H, et al. Immunofluorescence analysis and diagnosis of primary ciliary dyskinesia with radial spoke defects. Am J Respir Cell Mol Biol. 2015;53:563–573. doi: 10.1165/rcmb.2014-0483OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGrath J, Somlo S, Makova S, Tian X, Brueckner M. Two populations of node monocilia initiate left–right asymmetry in the mouse. Cell. 2003;114:61–73. doi: 10.1016/s0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- 20.Raidt J, Wallmeier J, Hjeij R, Onnebrink JG, Pennekamp P, Loges NT, Olbrich H, Häffner K, Dougherty GW, Omran H, et al. Ciliary beat pattern and frequency in genetic variants of primary ciliary dyskinesia. Eur Respir J. 2014;44:1579–1588. doi: 10.1183/09031936.00052014. [DOI] [PubMed] [Google Scholar]
- 21.Olbrich H, Schmidts M, Werner C, Onoufriadis A, Loges NT, Raidt J, Banki NF, Shoemark A, Burgoyne T, Al Turki S, et al. UK10K Consortium. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left–right body asymmetry. Am J Hum Genet. 2012;91:672–684. doi: 10.1016/j.ajhg.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, Boucher RC. A proteomic analysis of human cilia: identification of novel components. Mol Cell Proteomics. 2002;1:451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- 23.Ibañez-Tallon I, Heintz N, Omran H. To beat or not to beat: roles of cilia in development and disease. Hum Mol Genet. 2003;12:R27–R35. doi: 10.1093/hmg/ddg061. [DOI] [PubMed] [Google Scholar]
- 24.King SM. Integrated control of axonemal dynein AAA+ motors. J Struct Biol. 2012;179:222–228. doi: 10.1016/j.jsb.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loges NT, Olbrich H, Becker-Heck A, Häffner K, Heer A, Reinhard C, Schmidts M, Kispert A, Zariwala MA, Leigh MW, et al. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am J Hum Genet. 2009;85:883–889. doi: 10.1016/j.ajhg.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchison HM, Schmidts M, Loges NT, Freshour J, Dritsoula A, Hirst RA, O’Callaghan C, Blau H, Al Dabbagh M, Olbrich H, et al. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia Nat Genet 201244381–389.S1–S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hjeij R, Onoufriadis A, Watson CM, Slagle CE, Klena NT, Dougherty GW, Kurkowiak M, Loges NT, Diggle CP, Morante NF, et al. UK10K Consortium. CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am J Hum Genet. 2014;95:257–274. doi: 10.1016/j.ajhg.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wirschell M, Olbrich H, Werner C, Tritschler D, Bower R, Sale WS, Loges NT, Pennekamp P, Lindberg S, Stenram U, et al. The nexin–dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat Genet. 2013;45:262–268. doi: 10.1038/ng.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakakibara H, Takada S, King SM, Witman GB, Kamiya R. A Chlamydomonas outer arm dynein mutant with a truncated beta heavy chain. J Cell Biol. 1993;122:653–661. doi: 10.1083/jcb.122.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z, Takazaki H, Nakazawa Y, Sakato M, Yagi T, Yasunaga T, King SM, Kamiya R. Partially functional outer-arm dynein in a novel Chlamydomonas mutant expressing a truncated gamma heavy chain. Eukaryot Cell. 2008;7:1136–1145. doi: 10.1128/EC.00102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takazaki H, Liu Z, Jin M, Kamiya R, Yasunaga T. Three outer arm dynein heavy chains of Chlamydomonas reinhardtii operate in a coordinated fashion both in vitro and in vivo. Cytoskeleton (Hoboken) 2010;67:466–476. doi: 10.1002/cm.20459. [DOI] [PubMed] [Google Scholar]
- 32.Hummel KP, Chapman DB. Visceral inversion and associated anomalies in the mouse. J Hered. 1959;50:9–13. [Google Scholar]
- 33.Supp DM, Witte DP, Potter SS, Brueckner M. Mutation of an axonemal dynein affects left–right asymmetry in inversus viscerum mice. Nature. 1997;389:963–966. doi: 10.1038/40140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Supp DM, Brueckner M, Kuehn MR, Witte DP, Lowe LA, McGrath J, Corrales J, Potter SS. Targeted deletion of the ATP binding domain of left–right dynein confirms its role in specifying development of left–right asymmetries. Development. 1999;126:5495–5504. doi: 10.1242/dev.126.23.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yagi T, Uematsu K, Liu Z, Kamiya R. Identification of dyneins that localize exclusively to the proximal portion of Chlamydomonas flagella. J Cell Sci. 2009;122:1306–1314. doi: 10.1242/jcs.045096. [DOI] [PubMed] [Google Scholar]
- 36.Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol. 2012;198:913–925. doi: 10.1083/jcb.201201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahjoub MR, Qasim Rasi M, Quarmby LM. A NIMA-related kinase, Fa2p, localizes to a novel site in the proximal cilia of Chlamydomonas and mouse kidney cells. Mol Biol Cell. 2004;15:5172–5186. doi: 10.1091/mbc.E04-07-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furuta A, Yagi T, Yanagisawa HA, Higuchi H, Kamiya R. Systematic comparison of in vitro motile properties between Chlamydomonas wild-type and mutant outer arm dyneins each lacking one of the three heavy chains. J Biol Chem. 2009;284:5927–5935. doi: 10.1074/jbc.M807830200. [DOI] [PubMed] [Google Scholar]
- 39.Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- 40.Antony D, Becker-Heck A, Zariwala MA, Schmidts M, Onoufriadis A, Forouhan M, Wilson R, Taylor-Cox T, Dewar A, Jackson C, et al. Uk10k. Mutations in CCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Hum Mutat. 2013;34:462–472. doi: 10.1002/humu.22261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.