Abstract
Proteasome inhibitors are emerging as a new class of chemopreventive agents and have gained huge importance as potential pharmacological tools in breast cancer treatment. Improved understanding of the role played by proteases and their specific inhibitors in humans offers novel and challenging opportunities for preventive and therapeutic intervention. In this study, we demonstrated that the Bowman–Birk protease inhibitor from Vigna unguiculata seeds, named black-eyed pea trypsin/chymotrypsin Inhibitor (BTCI), potently suppresses human breast adenocarcinoma cell viability by inhibiting the activity of proteasome 20S. BTCI induced a negative growth effect against a panel of breast cancer cells, with a concomitant cytostatic effect at the G2/M phase of the cell cycle and an increase in apoptosis, as observed by an augmented number of cells at the sub-G1 phase and annexin V-fluorescin isothiocyanate (FITC)/propidium iodide (PI) staining. In contrast, BTCI exhibited no cytotoxic effect on normal mammary epithelial cells. Moreover, the increased levels of intracellular reactive oxygen species (ROS) and changes in the mitochondrial membrane potential in cells treated with BTCI indicated mitochondrial damage as a crucial cellular event responsible for the apoptotic process. The higher activity of caspase in tumoral cells treated with BTCI in comparison with untreated cells suggests that BTCI induces apoptosis in a caspase-dependent manner. BTCI affected NF-kB target gene expression in both non invasive and invasive breast cancer cell lines, with the effect highly pronounced in the invasive cells. An increased expression of interleukin-8 (IL-8) in both cell lines was also observed. Taken together, these results suggest that BTCI promotes apoptosis through ROS-induced mitochondrial damage following proteasome inhibition. These findings highlight the pharmacological potential and benefit of BTCI in breast cancer treatment.
Introduction
Breast cancer represents the most common cancer among women worldwide, with an estimated 1.67 million new cases diagnosed in 2012.1 A growing body of evidence suggests that Bowman–Birk inhibitors (BBIs), a major protease inhibitor family, can prevent or suppress carcinogenic processes that include colon,2–4 oral leukoplakia,5–9 esophageal tumors,10 leukemia,11 prostatic hyperplasia12 and breast cancer.13 However, despite acting on wide range of cancers, the underlying mechanism(s) of BBI activities as an anti-carcinogenic agent remain elusive. Nevertheless, the reliable explanation today is that BBIs are effective inhibitors of proteasomal chymotrypsin-like activities.14
The proteasome is a multi-subunit protease with three catalytic sites located in different subunits of the 20S core. These comprise the caspase-like, trypsin-like and chymotrypsin-like, β1, β2 and β5 subunits, respectively.15 The ubiquitin-proteasome proteolytic pathway is the principal mechanism in the cell for controlled protein degradation. As such, it controls the proteins that are crucial in cellular processes such as cell cycle progression, cell growth and cell development. It is therefore expected that dysfunction of these pathways will compromise cell survival. Indeed, it is well known that proteasome inhibition induces apoptosis.16–20 Moreover, high-proteasome activity has been reported in different malignancies.14 Because cancer cells are more sensitive than normal cells to the inhibition of proteasome activity,21,22 targeting the proteasome pathway has emerged as an ideal approach in anticancer therapy.
It has been shown that proteasome dysfunction causes damage to mitochondria, leading to increase in reactive oxygen species (ROS) production.23 Mitochondria have a major role in regulating cell death, which is mediated by outer membrane permeabilization in response to death triggers such as DNA damage.24 ROS are natural compounds which occur as chemical derivatives of metabolism. Performing key roles in apoptosis induction under both physiologic and pathologic conditions;25 elevated levels of ROS and downregulation of ROS scavengers lead to oxidative stress, which is associated with several human diseases, including cancers.26 It has been demonstrated that protease inhibitors promote cell cycle arrest and apoptosis.17,27–30 Moreover, proteasome impairment is associated with mitochondrial dysfunction, oxidative stress and death in neuronal cells.31,32 Therefore, it seems plausible to assume that proteasome inhibition induces apoptosis through mitochondrial dysfunction and oxidative stress. Recently, it has been shown that oxidative stress can convert estrogen-dependent non-aggressive breast cancer cells into an estrogen-independent aggressive form.33 Furthermore, mitochondrial ROS have been implicated in malignant cell transformation.34 Mitochondrial dysfunction was also shown to promote breast cancer cell migration and invasion through the accumulation of the transcription factor hypoxia-inducible factor 1α, via increased production of ROS.35 Nevertheless, the exact mechanisms controlling ROS involvement in carcinogenesis remain unclear.
The black-eyed pea trypsin/chymotrypsin inhibitor (BTCI) is a protease inhibitor isolated from Vigna unguiculata (Cowpea) seeds that belongs to the BBI family. BTCI is a stable double-headed Bowman–Birk protease inhibitor, inhibiting trypsin and chymotrypsin, simultaneously, with low-molecular mass (9071 Da) and seven disulfide bonds. The high-disulfide bond content of BTCI is responsible for its remarkable stability and also for the canonic conformation of loops containing the reactive sites of the inhibitor against proteases.36–38 The biochemical and biophysical properties of BTCI have been extensively characterized.36,39 Previously, we have reported that BTCI-induced significant cytostatic and cytotoxic effects on MCF-7 breast cancer cells; with these effects associated with changes in the morphology of the nucleus and mitochondria, increased number of cells with reduced mitochondrial membrane potential, DNA fragmentation and cells with altered plasma membrane integrity.13 We have also observed that during chemical induction of non-melanoma skin cancer in mice, topical application of BTCI significantly reduced the incidence and the volume of pre-malignant lesions (data to be published). In another study, we have shown that BTCI potentially inhibits the activity of trypsin-like, chymotrypsin-like and caspase-like sites of proteasome 20S, suggesting that BTCI is an effective proteasome inhibitor.40
To gain further insights into pleiotropic effects of BTCI, in the present study, we demonstrated that BTCI induced G2 phase/mitotic arrest and apoptotic cell breast cancer death. Furthermore, BTCI treatment caused mitochondrial membrane depolarization, triggering oxidative stress and increased caspase-3 activity. NF-kB target gene expression was also altered in both breast cancer cell lines and increased gene expression observed in interleukin-8 (IL-8) cancer cells. These finding suggest that BTCI induces apoptosis through mitochondrial impairment and oxidative damage following proteasome inhibition. Altogether, the results support the idea that the chemopreventive effect of BBIs may be attributable to their inhibitory effects toward proteasome function.
Results
Purification of BTCI
The purification of BTCI from V. unguiculata seeds and the analysis of its purity were performed before testing in cell assays. The DEAE-cellulose elution profile of V. unguiculata seeds crude extract (CE), as previously described,39,41 is shown in Figure 1a. BTCI was collected between fractions 67 and 87 and its molecular mass (9071.6 Da) and purity were confirmed by MALDI-TOFF spectrometry (Figure 1b).38
Figure 1.
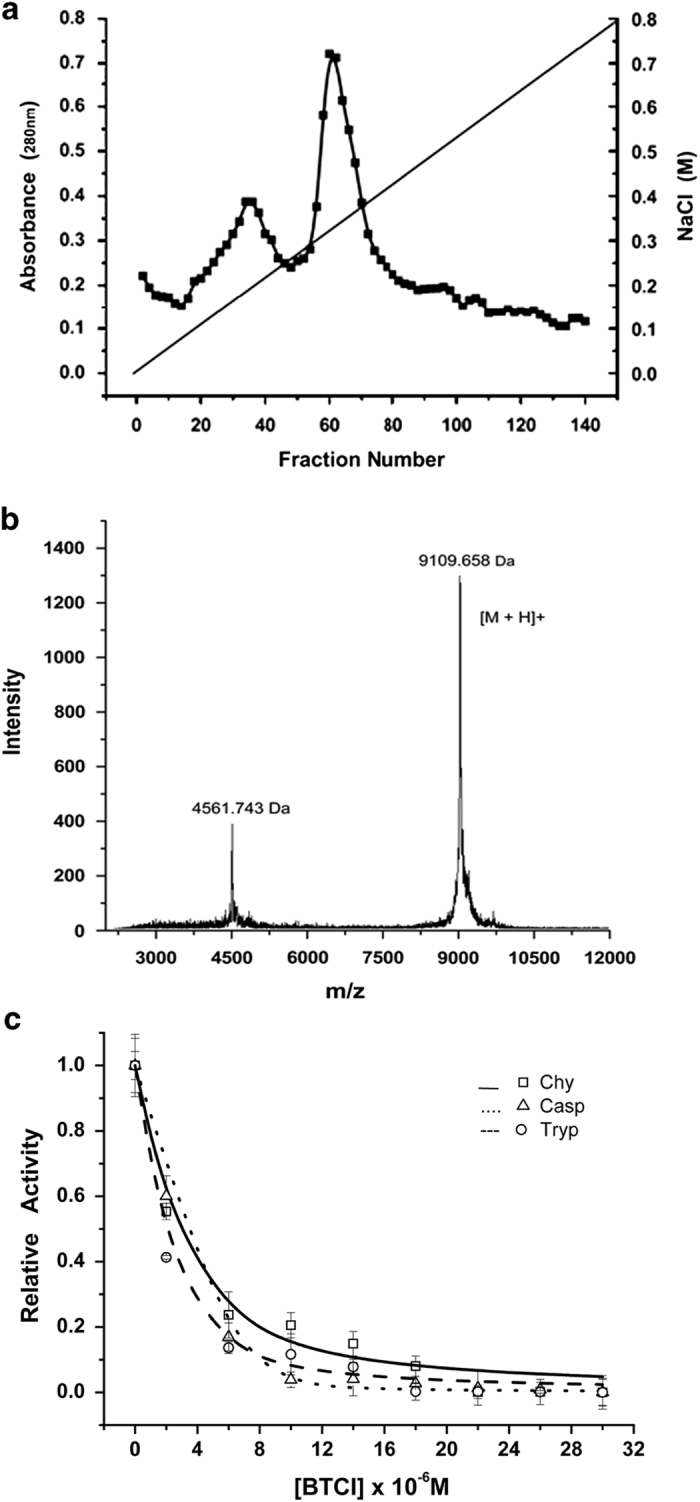
Purification and characterization of inhibitory activity of BTCI against proteasome 20S. (a) Elution protein profile from V. unguiculata seeds CE using DEAE-cellulose chromatography in accordance with Ventura and Xavier Filho (1996).39 BTCI was obtained from 67 to 87 eluted fractions. (b) MALDI-TOF MS spectrum showing BTCI as a pure protein of 9109.658 Da and its double charge [M + 2H] of 4561.743 Da.38 (c) Inhibition activity of trypsin-like, chymotrypsin-like and caspase-like sites by BTCI as previously described.40
Inhibition of proteasome 20S function by BTCI
BTCI was previously characterized as a potent inhibitor of the 20S proteasome through the inhibition of protease trypsin, chymotrypsin and caspase-like activities,40 as shown in Figure 1c. BTCI showed high affinity to the 20S proteasome, as indicated by inhibition constant values of 1.0×10−7 M, 7.0×10−7 M and 14.0×10-7 M, for trypsin-like, chymotrypsin-like and caspase-like, respectively. It is noteworthy that BTCI was a more potent inhibitor for trypsin than known proteasome inhibitor N-(benzyloxy—carbonyl) leucinyl-leucinyl-leucinal-Leu-Leu-Leu-al (MG132), with similar inhibition observed toward chymotrypsin and caspase.
BTCI-induced cytotoxicity in breast adenocarcinoma cells
The effect of BTCI on viability of MDA.MB.231 (highly invasive human breast cells), MCF-7 (human breast adenocarcinoma cells) and MCF-10A (normal mammary epithelial cells) were determined using the MTT assay as described in the Methods section. Cells were incubated with BTCI at concentrations ranging from 0 to 300 μM for 24 h. Results showed that BTCI exerts its cytotoxic effect in a dose-dependent manner. BTCI at the concentration of 150 μM markedly induced MCF-7 death (P=0.014) (Figure 2a), meanwhile, the same effect on MDA.MB.231 was observed at a concentration of 100 μM (P<0.001; Figure 2b). In contrast, BTCI did not induce a cytotoxic effect on MCF-10A cells. The results of the MTT assay demonstrated a cytotoxic effect of BTCI on breast cancer cell lines in a dose-dependent manner, absent in normal mammary epithelial cells (Figure 2c).
Figure 2.
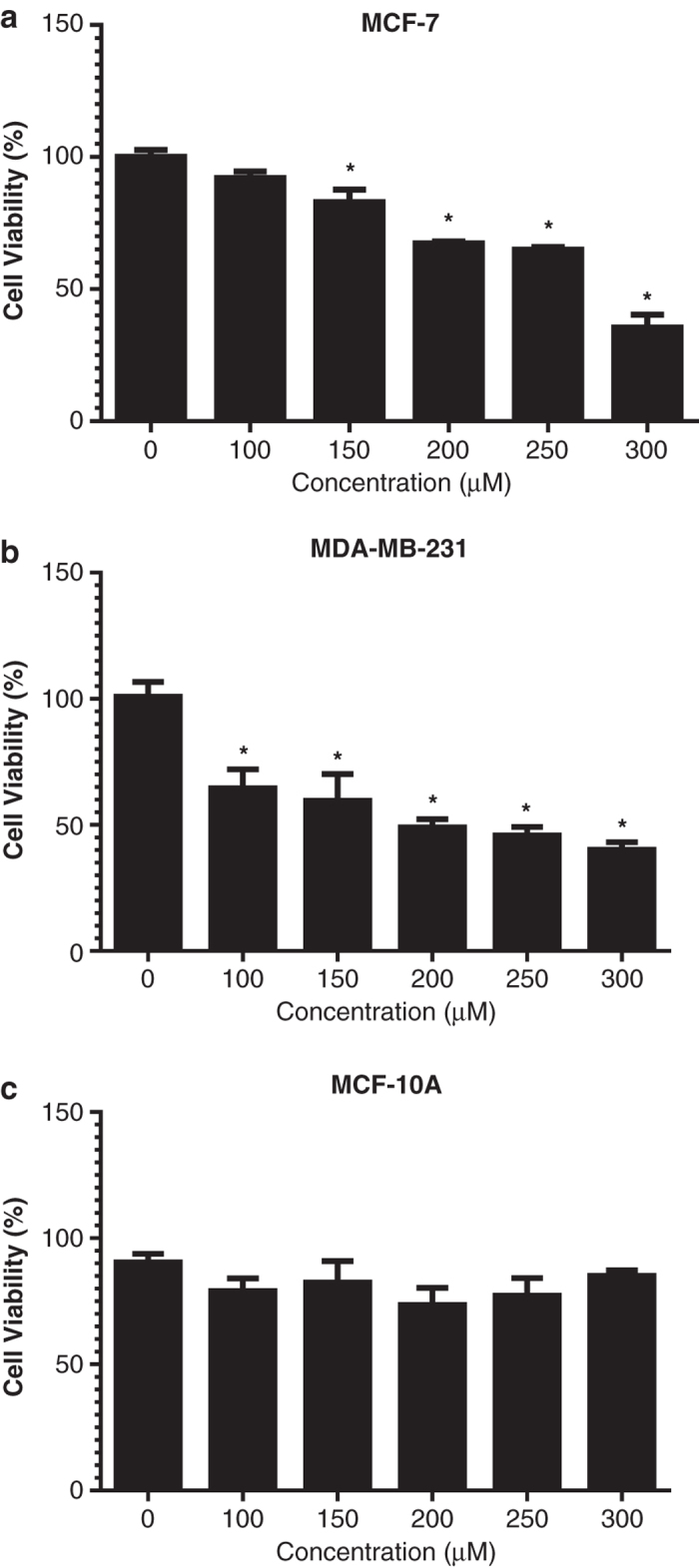
Cytotoxic effects induced by BTCI. (a) MCF-7, (b) MDA-MB-231 and (c) MCF-10 cells were incubated with BTCI (0-300 μM) for 24 h. Cell viability was determined by MTT assay. Results are presented as mean±S.D. of two separate experiments conducted in triplicate, *P<0.05 versus untreated cells.
BTCI-induced cell cycle arrest at G2/M phase and apoptotic cell death
We previously showed that BTCI significantly induces cell cycle arrest at G2/M phase, DNA fragmentation and enhanced numbers of cells marked by annexin-V in breast cancer MCF-7, suggesting apoptosis as a possible mechanism for the cell death process.13 To explore whether apoptosis is involved in BTCI-induced MDA.MB.231, the amount of sub-G1 DNA in cancer cells treated with BTCI was investigated. As shown in Figure 3, the treatment of MDA.MB.231 cells with BTCI resulted in a marked accumulation of cells in the sub-G1 phase, suggesting a massive apoptotic, but not necrotic, cell death. In addition, BTCI arrested cell cycle at G2/M phase after 24 h. The apoptotic effect of BTCI was confirmed by an annexin V-fluorescin isothiocyanate (FITC)/propidium iodide (PI) staining assay. BTCI at the concentration of 100 μM for 24 h induced apoptosis in MDA-MB-231 cells (Figure 4). Taken together, these results confirm that BTCI-induced apoptotic cell death in breast adenocarcinoma cells.
Figure 3.
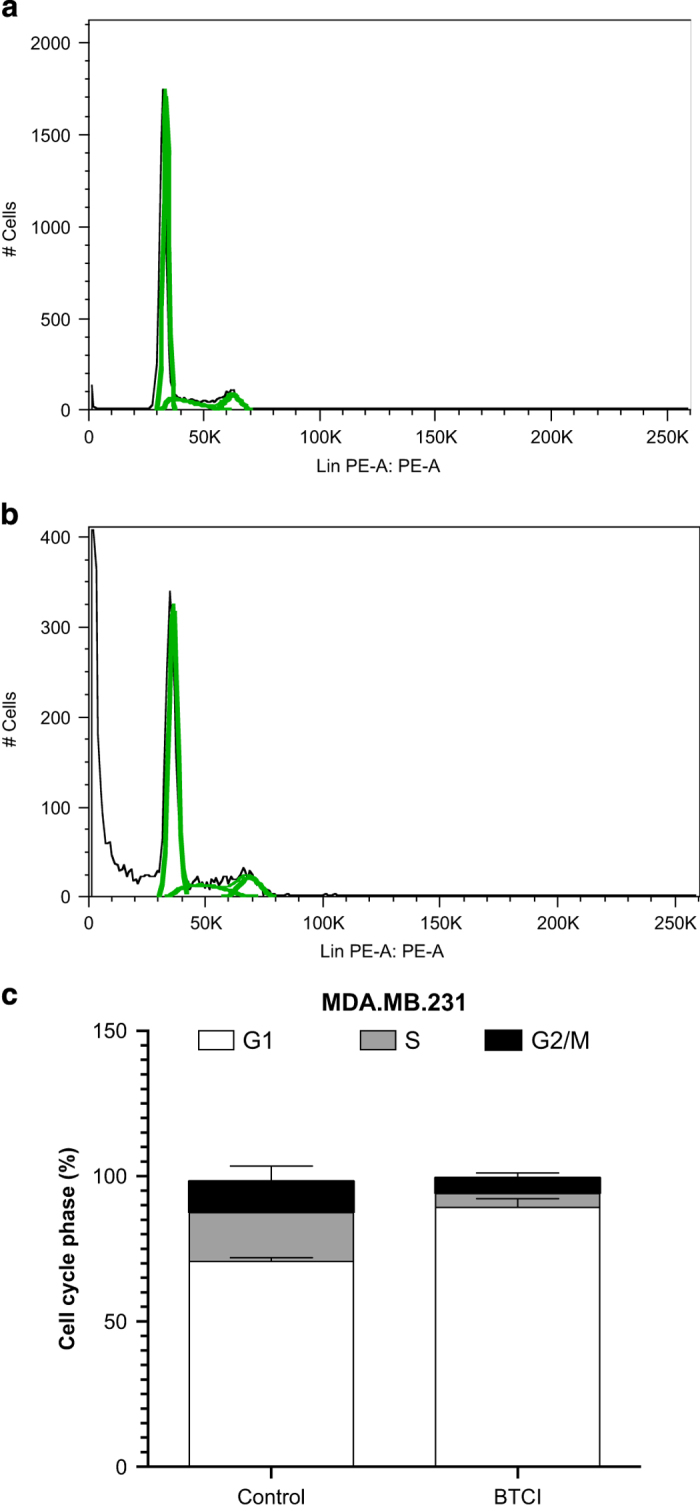
Effects of BTCI on DNA content. MDA-MB-231 cells were incubated with 100 μM of BTCI for 24 h. DNA contents in Sub-G1 phase were analyzed using flow cytometry. Representative flow cytometry images were shown (a and b). (c) The DNA profile representing cells in sub-G1/G1, S and G2/M phases. Results are represented as means±S.D. of two separate experiments conducted in triplicate, P<0.05 versus untreated cells.
Figure 4.
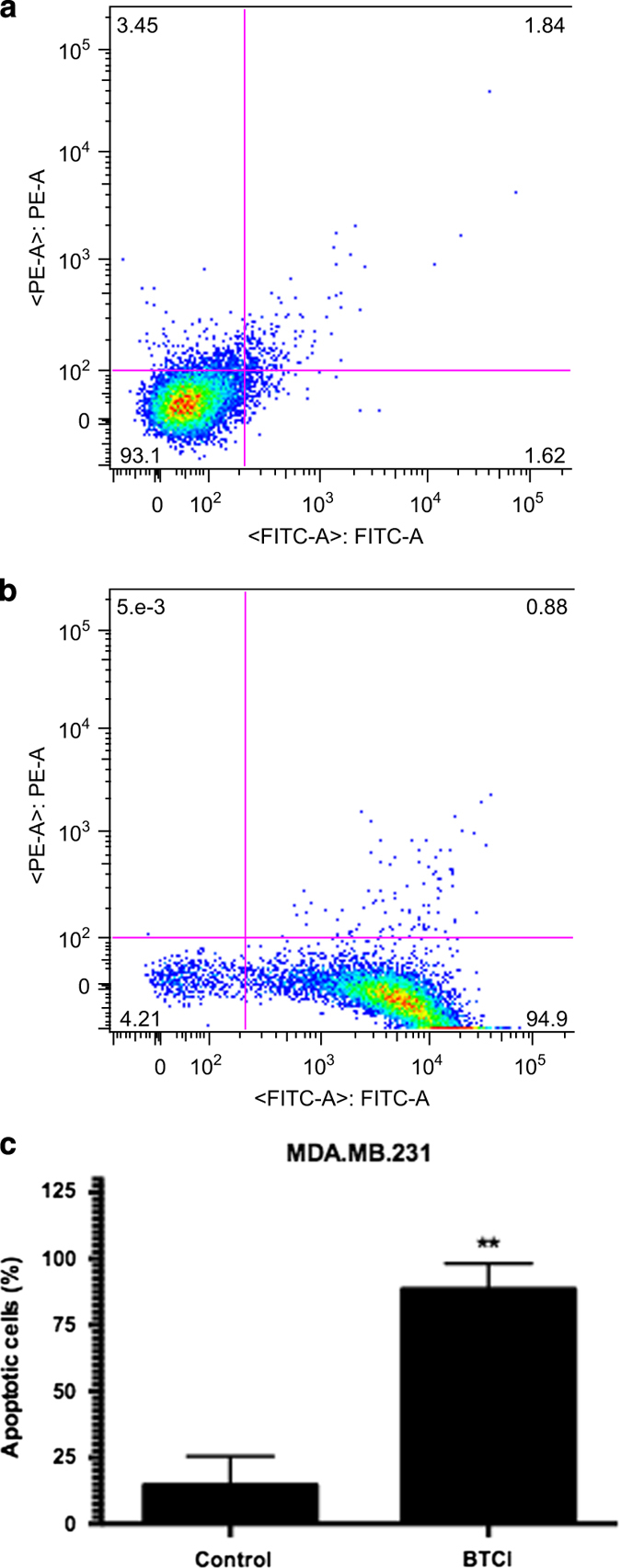
Apoptosis induced by BTCI MDA-MB-231. Cells were incubated with BTCI (100 μM) for 24 h, and then stained with annexin V-FITC/PI and flow cytometry analysis. (a and b) representative images of flow cytometry analysis were displayed. (c) The percentage of apoptotic cells was statistically analyzed. Results are presented as mean±S.D. of two separate experiments conducted in triplicate, *P<0.001 versus untreated cells.
Mitochondrial membrane depolarization is involved in BTCI-induced cytotoxicity
Proteasome inhibition is known to induce apoptosis through mitochondrial cytochrome c release. As BTCI inhibits the proteasome function, its involvement in mitochondrial dysfunction in cancer cells was investigated. A sensitive cationic and lipophilic JC-10 fluorescent probe was used to monitor the mitochondrial membrane potential change in cells based on the presence of the JC-10 dye. JC-10 concentrates in the mitochondrial matrix where it forms red fluorescent aggregates. However, in apoptotic and necrotic cells, JC-10 diffuses out of mitochondria and changes from the aggregate to a monomeric form, causing a shift in fluorescence emission from ~590 to 515 nm, staining cells in green fluorescence. As shown in Figure 5, the mitochondrial membrane potential in both MCF-7 and MDA.MB.231 cells dropped, as indicated by increasing in monomer/aggregate ratio (P=0.016 and P=0.009, respectively). These data indicate that mitochondrial membrane depolarization and mitochondrial swelling precede an increase in oxidative stress and loss of cell viability in cells exposed to BTCI.
Figure 5.
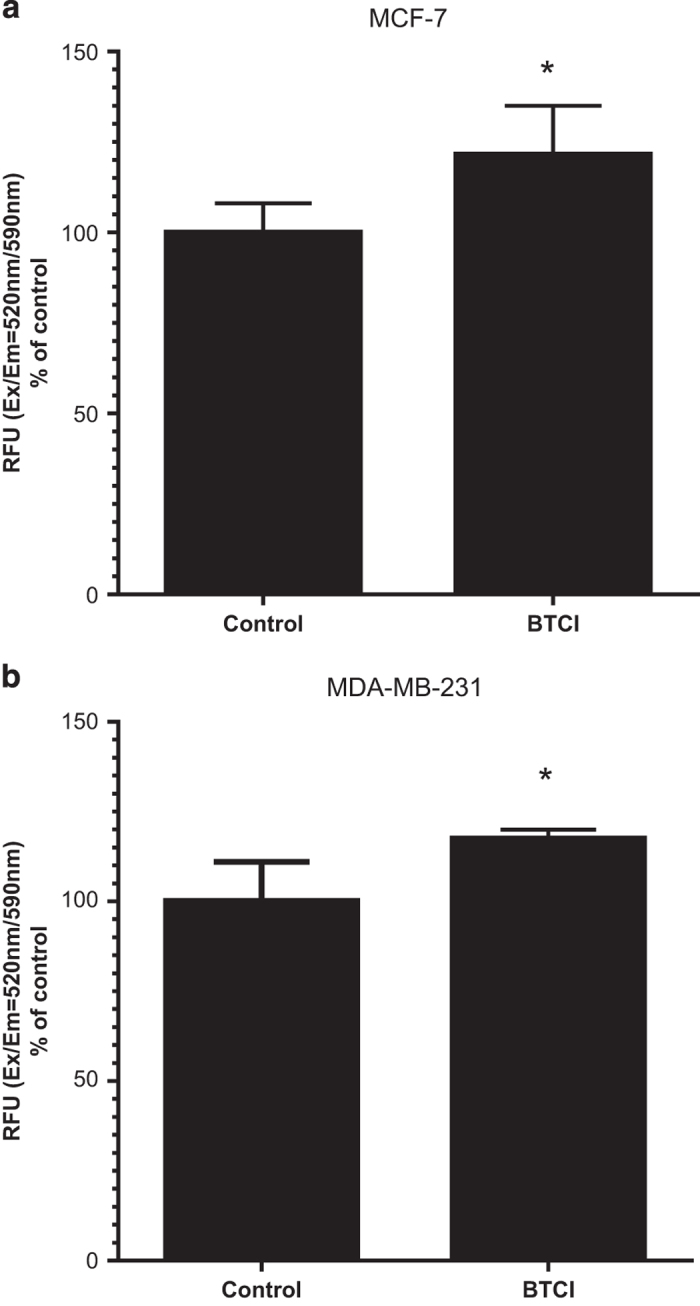
BTCI-induced mitochondrial membrane depolarization. (a) MCF-7 and (b) MDA-MB-231 cells were incubated with BTCI at the concentration of 150 μM and 100 μM, respectively, for 24 h. Dye-loading solutions were added to cells and incubated for 30 min. The change of mitochondrial membrane potential was measured by using Mitochondrial Membrane Potential Kit (Sigma). Results are presented as mean±S.D. of two separate experiments conducted in triplicate, *P<0.01 versus untreated cells.
BTCI increases intracellular ROS generation in breast adenocarcinoma
There is increasing evidence that ROS induced by apoptotic stimuli leads to mitochondrial dysfunction. To investigate the involvement of oxidative stress in BTCI-induced mitochondrial impairment as well as by proteasome inhibition, intracellular ROS levels were measured. Cells were treated with BTCI for 30, 60, 90, 120 and 180 min. As shown in Figure 5, treatment of breast adenocarcinoma cells with BTCI significantly increased intracellular ROS generation in a cell type and time point dependent manner. It is noteworthy that these results are strongly related with proteasome dysfunction-induced ROS and mitochondrial membrane depolarization in human breast adenocarcinoma.6
Figure 6.
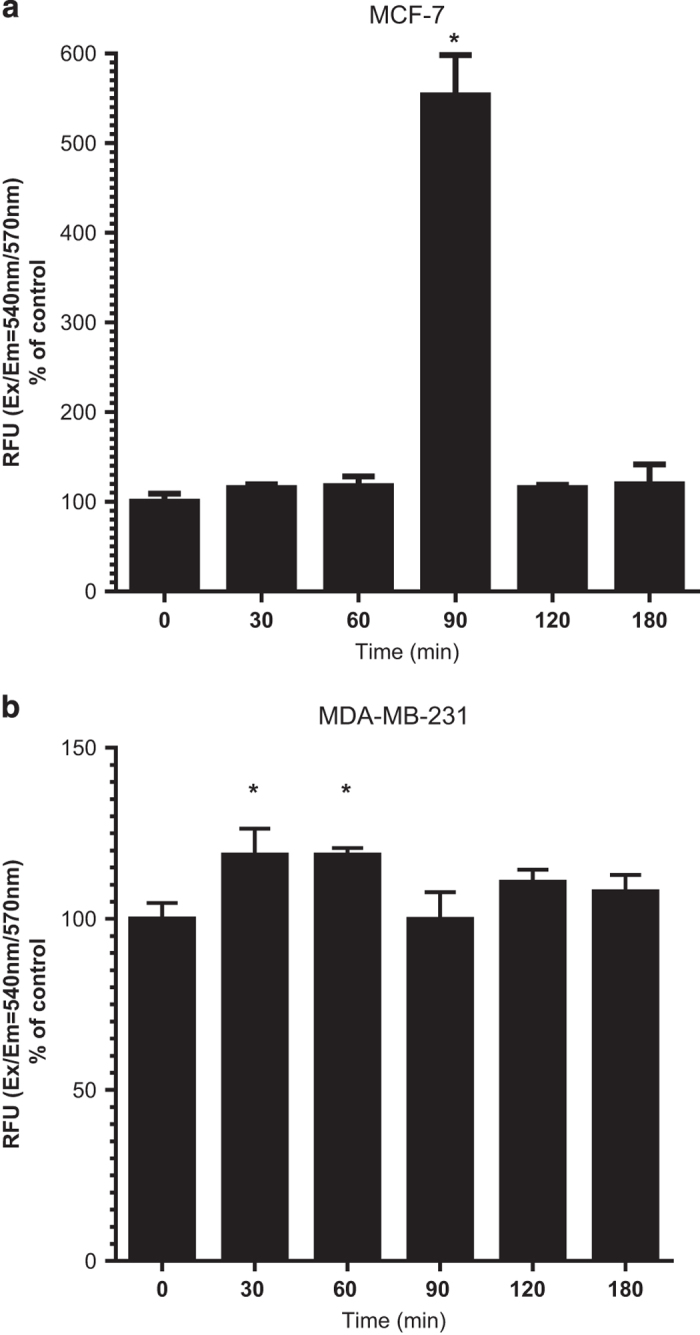
BTCI enhanced the generation of ROS in human breast adenocarcinoma cells. (a) MCF-7 and (b) MDA-MB-231 cells were incubated with BTCI at the concentration of 150 μM and 100 μM, respectively, for 30, 60, 90 and 180 min. The production of intracellular ROS was estimated fluorimetrically using Fluorometric Intracellular Ros Kit. Results are presented as mean±S.D. of two separate experiments conducted in triplicate, *P<0.001 versus untreated cells.
BTCI treatment led to increased caspase-3 activity
To analyze whether BTCI triggers apoptosis in a caspase-dependent manner, the activity of caspase-3 was measured. As shown in Figure 7, the activity of caspase-3 in MCF-7 and MDA.MB.231 cells was increased after treatment with BTCI. Although MCF-7 does not express caspase-3, the increased activity observed in MCF-7 may be due to the assessment method via DEVD-dependent caspase activity. The assay is based on detection of the cleaved substrate DEVD-AFC (AFC: 7-amino-4-trifluoromethyl coumarin) that also can detect caspase-7 activity, which is expressed in MCF-7 cells and exhibits a certain degree of functional redundancy with caspase-3.
Figure 7.
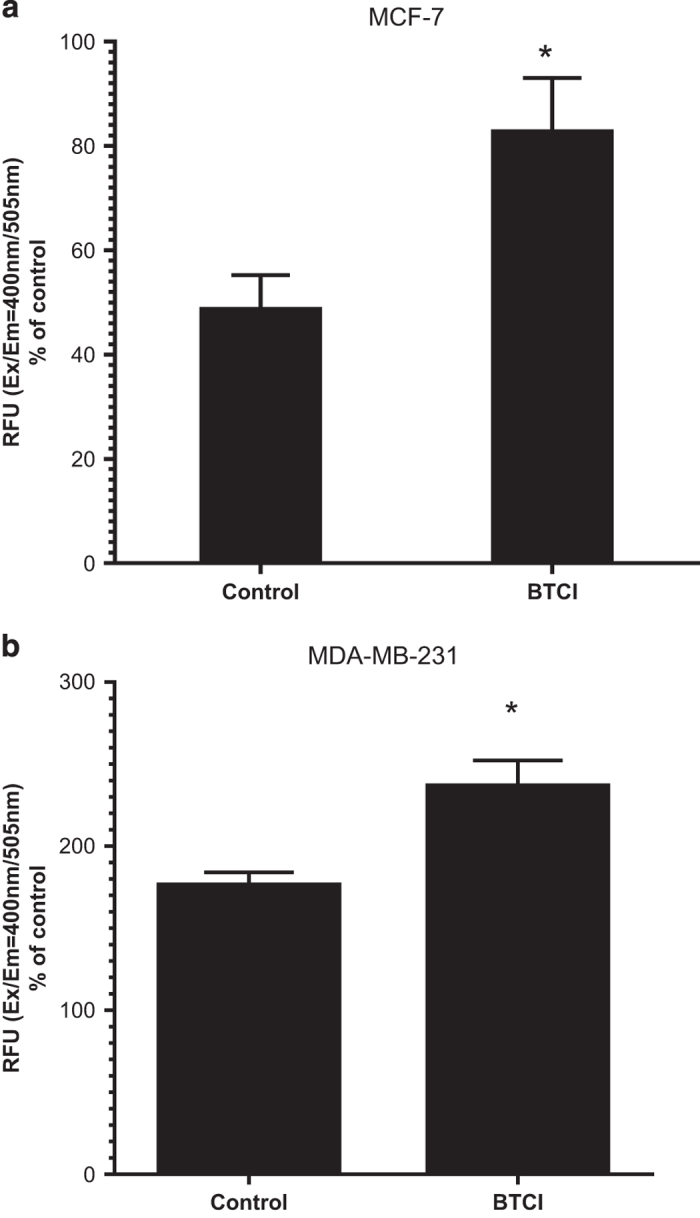
Effect of BTCI on caspase-3 activity. (a) MCF-7 and (b) MDA-MB-231 cells were incubated with BTCI at the concentration of 150 μM and 100 μM, respectively for 24 h; the activity of caspase-3 was evaluated by using caspase-3 activity kit. Results are presented as mean±S.D. of two separate experiments conducted in triplicate, *P<0.001 versus untreated cells.
Treatment with BTCI results in alteration on NF-kB dependent gene expression
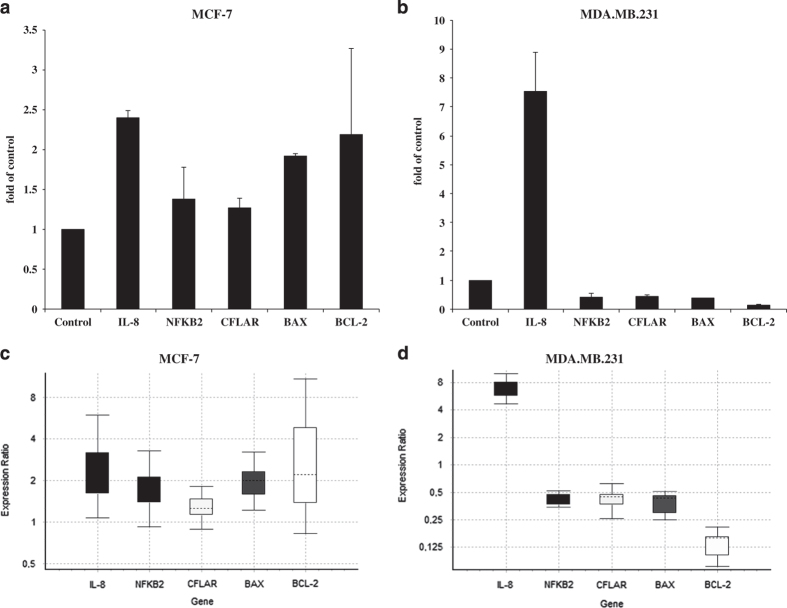
Because of the ability of proteasome inhibitors to block NF-kB activity, we investigated expression of some established NF-kB target genes, including BAX (BCL2-Associated X Protein), BCL-2 (B-Cell CLL/Lymphoma 2), NFKB2 (Nuclear Factor Of Kappa Light Polypeptide Gene Enhancer In B-Cells 2 (P49/P100)), c-FLIP (CASP8 And FADD-Like Apoptosis Regulator) and IL-8. As shown in Figure 8, BTCI treatment affected gene expression in both breast cancer cell lines, although the effect of BTCI was more pronounced in MDA.MB.231. Interestingly, BTCI-induced IL-8 upregulation in both cell lines, albeit with expression of other NF-kB target genes downregulated and/or unchanged. Although the expression of BAX increased slightly in MCF-7 treated with BTCI in comparison with untreated; however, BTCI treatment did not cause any change in expression of BCL-2, NFKB2 and c-FLIP in MCF-7 cells; while in MDA.MB.231 cells treated with BTCI expression of those genes was markedly downregulated.
Figure 8.
mRNA expression of IL-8, NFKB2, CFLAR, BAX and BCL-2 quantified by RT-PCR in human breast adenocarcinoma cells. Cells were treated with BTCI for 24 h, and RNA was extracted. This RNA was used to prepare cDNA that was subject to real-time PCR to quantify the mRNA level of each gene. Each sample was run in triplicate, and the relative amount of mRNA was normalized to the β-actin content in each sample. Normalized expression values (specific gene expression levels/β-actin expression levels) are presented. Data points are represented by the expression ratio and mean±S.D. fold of control in MCF-7 (a and b) and MDA.MB.231 (c and d). Each RNA sample represents a pool of RNA samples from two independent biological experiments conducted in triplicate, *P<0.05 versus untreated cells.
Discussion
Evidence based on animal and epidemiological studies indicates that BBIs, the most extensively studied group of proteasome inhibitors, could act as anti-carcinogenic agents.13,14,42–45 The idea of chemopreventive effects of BBIs came from epidemiological studies demonstrating that consumption of soybean products, which are particularly rich in protease inhibitors, is associated with decreased cancer incidence rates.7,14,42,43 Although the suppressive effect of BBIs on carcinogenesis has been suggested in a human phase IIa clinical trial,7 the underlying mechanisms by which they exert their chemopreventive feature remains unclear.
BTCI is a natural plant protease inhibitor from V. unguiculata seeds which belongs to the BBI family. As shown previously40 and confirmed in the present study, BTCI exerts inhibitory activity toward the 20S proteasome. The proteasome is phylogenetically ancient and has remained highly conserved throughout eukaryotic evolution, as well as ubiquitously distributed and conserved among distantly related species such as archaea, bacteria and eukaryotes.46–50 Given the high-evolutionary conservation of the 20S proteasome, data of its proteolytic activity inhibition can be applicable to those expected in the human 20S proteasome, considering any related eukaryotic source. BTCI was characterized as a potent 20S proteasome inhibitor and may be related to ubiquitin-proteasome pathway inhibition, which was investigated in the present work.
Previously, we showed that BTCI-induced cytotoxic and cytostatic effects on MCF-7 cells associated with DNA fragmentation, lysosome membrane permeabilization and apoptosis.13 In the present study, we confirm that BTCI has potent in vitro anti-tumor activity due to its ability to induce apoptosis against a panel of breast cancer cells. The ubiquitin-proteasome pathway controls the proteins that are crucial in cellular processes such as cell cycle progression. Recently, it has been reported that under proteasome stress, cell cycle progression was impaired at G2/M phases.51 Also, the activation of apoptosis at mitotic arrest depends on the phosphorylation of p38 MAPK.52,53 Thus, we suggest that p38/MAPK signaling might be involved in BTCI-induced apoptosis and G2/M arrest in breast cancer cells under proteasome inhibition.
Our findings also demonstrated that BTCI induced a rapid increase in ROS and loss of mitochondrial membrane potential, probably through the proteasome dysfunction; however, the order of these events remain unclear. These observations are consistent with earlier reports showing that inhibition of proteasome activity causes ROS generation.31,54,55 In the same line of evidence, it has been reported that proteasome inhibition by MG132 leads to mitochondrial damage and increased ROS production, providing new insights into the mechanism of cell death caused by proteasome dysfunction.23
Several lines of evidence suggest a crucial role for ROS as a common and critical denominator in stimulatory and inhibitory cellular signaling events involving NF-κB, which is directly involved in gene regulation associated with various cellular processes such as cell growth, differentiation and apoptosis.56,57 Previous studies demonstrated that proteasome inhibition may interact with NF-kB, resulting in cell death in malignant cells.58–60 On the other hand, it has been reported that bortezomib, the first proteasome inhibitor used as anticancer drug,61 could not inhibit NF-kB p65 nuclear translocation leading to NF-kB activation.62–64 We then hypothesized about the possibility that BTCI-induced proteasome impairment may knockdown the transcriptional activity of NF-κB as a possible pathway of apoptotic process. To this end, we investigated expression of some NF-kB-dependent genes. Our findings showed that BTCI treatment inhibited and/or unaltered expression of NF-kB target genes in a cell-type manner; whilst increasing expression of IL-8 in both cell lines.64,65 As a consequence of proteasome inhibition, a drop in cellular FLICE-inhibitory protein levels, which is a truncated and inactive form of caspase-8, has been reported in prostate cancer, renal cancer and leukemia.66,67 Given this, c-FLIP as a target for cancer therapy seems reasonable. Taken together, our results are in good agreement with previous reports and support the idea that NF-kB may not be a key mechanism of the proteasome inhibitor’s anticancer activity.
Proteasome dysfunction has been shown to induce either the intrinsic or extrinsic pathway for apoptosis, consisting of early release of mitochondrial cytochrome c into the cytosol, caspase-9 activation and consequently the activation of caspase-3 and 7; or followed by independent activation of the caspase-8 pathway.68 Here, we have observed BTCI-induced apoptosis through the mitochondrial pathway involving caspase-3 activation in MDA-MB-231 cells. Although MCF-7 does not express caspase 3, the increased activity observed in MCF-7 may be due to the method of assessment via DEVD-dependent caspase activity. The assay is based on detection of cleavage of substrate DEVD-AFC that also can detect caspase-7 activity, which is expressed in MCF-7 cells and exhibits a certain degree of functional redundancy with caspase-3. Even though we did not investigate caspase-7 activity, the increase caspase activity observed in MCF-7 might be attributable to the caspase-7 activity. Indeed, mitochondrial membrane permeabilization is a point of no return in many pathways to apoptosis. On the other hand, the caspase-independent cell death resulting from mitochondrial membrane permeabilization does not exhibit the typical hallmarks of apoptosis.69 Therefore, it seems plausible to assume that BTCI induces apoptosis in a caspase-dependent manner following mitochondrial damage.
In summary, the present findings showed that BTCI might inhibit the cancer cell function directly by blocking the 20S proteasome core cavity, and indirectly by the generation of molecular intermediates responsible for oxidative stress. As cancer cells are generally more sensitive than normal cells to the inhibition of proteasome activity, proteasome inhibitors are being employed as chemopreventive agents. BTCI is safe, of low cost and can be taken as a part of a daily diet. Thus, the potential of BTCI for combinational therapies for breast cancer is promising.
Materials and methods
Materials
MDA.MB.231 (highly invasive human breast cells), MCF-7 (human breast adenocarcinoma cells) and MCF-10A (normal mammary epithelial cells) were purchased from Rio de Janeiro Cell Bank (RJCB). Caspase 3 Assay Kit and annexin V-FITC Apoptosis Detection Kit were obtained from Abcam Inc. (Cambridge, MA, USA). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), PI, Fluorometric Intracellular ROS Kit, Mitochondrial Membrane Potential Kit and all cell culture reagents were purchased from Sigma (St Louis, MO, USA).
Cell culture
Human breast cancer cell lines MCF-7 and MDA.MB.231 were maintained in Eagle’s minimum essential medium and Leibovitz’s L-15 medium, respectively. The media were supplemented with 10–20% (v/v) of fetal bovine serum and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin). MCF-10A cells were maintained in Mammary Epithelial Cell Growth Medium supplemented with 5% horse serum, 20 ng/ml epidermal growth factor, 10 lg/ml insulin, 0.5 lg/ml hydrocortisone, 100 ng/ml cholera toxin and antibiotics (100 IU/ml penicillin and 100 lg/ml streptomycin). Cells were maintained at 37 °C under humidified air containing 5% CO2 for MCF-7 and MCF-10A and without CO2 in the case of MDA.MB.231.
BTCI and proteasome 20S purification
BTCI was purified as previously described.39,41 In brief, V. unguiculata seeds were triturated, homogenized in water and submitted to 2.5% (v/v) trichloroacetic acid and 70% (w/v) ammonium sulfate precipitation. The obtained CE was dialyzed, lyophilized and stored at −20 oC. BTCI was purified from CE by ion exchange chromatography using DEAE-cellulose column (Sigma). The purity and molecular mass of BTCI were evaluated by MALDI-TOF/MS using an UltraFlex III (Bruker Daltonics, Bremen, Germany). BTCI concentration was determined using its specific absorption coefficient and molecular mass of 9.1 kDa. Mammal 20S proteasome was purified from horse erythrocytes and used to analyze cytotoxic effects of BTCI on human breast adenocarcinoma cells, given its phylogenetic conservation in the biogenesis,70,71 as well as in the tridimensional structure in all eukaryotes organisms.47 Moreover, these cells were chosen based on available facilities and the well-established purification method for the 20S proteasome in large amounts.72 In brief, horse erythrocytes lysates were obtained in 1 mM dithiothreitol (DTT) after removal of membranes. Cell lysates were applied onto sequential ion exchange DEAE-Sepharose and mono-Q columns, using an FPLC facility (GE Healthcare, Pharmacia, Bio-Rad Laboratories, Inc., Foster City, CA, USA) and sucrose density gradient ultracentrifugation.72
Effect of BTCI on enzymatic activity of the 20S proteasome
Fluorogenic substrates t-butyloxycarbonyl-L-leucyl-L-arginyl-L-arginine-4-methylcoumaryl-7-amide (Boc-Leu-ArgArg-AMC), benzyloxycarbonyl-L-leucyl-L-leucyl-L-glutamyl- naphthylamide (Z-Leu-Leu-Glu-βNA) and N-succinyl- Leu-Leu-Val-Tyr-AMC (Suc-Leu-Leu-Val-Tyr) were used for trypsin, caspase and chymotrypsin-like activities, respectively as previously described.40 In brief, purified 20S proteasome (2.0 μg/ml) was incubated with BTCI (2.0 to 30.0 μg/ml) and 62.5 μg/ml of the fluorogenic substrates in 20.0 mM Tris-HCl pH 7.5, 1.0 mM EDTA, 1.0 mM NaN3, 1.0 mM DTT. The assays were carried out in triplicate, at room temperature for 60 min. The hydrolysis of fluorogenic substrates was monitored at 480 nm for chymotrypsin-like and trypsin-like activities, and 410 nm for caspase-like activities.40 The inhibition constant, KI was calculated by nonlinear regression using the GRAFIT program version 3 (Erithacus Software Ltd., Middlesex, UK). The control for this experiment was established using the proteasome inhibitor MG132 (carbobenzoxylleucyl-leucyl-leucinaI-H) at a similar concentration to that used when BTCI displayed 80–95% of protease inhibition.
MTT assay
Cells were digested with trypsin, harvested, adjusted to a density of 2×104 cells/ml and seeded to 96-well plates at volume 200μl per well. After 24 h when cells formed a monolayer, BTCI was added to the medium at different concentrations. After incubation for 24 h, MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium Bromide; 5 mg/ml) was added (10 μl per well). Cells were then incubated at 37 °C for a further 3 h, culture supernatant removed and dimethyl sulfoxide added (100 μl per well). Cells were incubated on a shaker at 37 °C for 10 min until crystals were completely dissolved. The absorbance at 570 nm was determined using a SpectraMax M3 microplate reader (Molecular Devices, Sunnyvale, CA, USA), then correlated with cell viability. Each experiment was conducted in quintuplicate at least twice.
Annexin V-FITC and PI staining
Subsequent to treatment, cells were harvested and washed twice with phosphate-buffered saline (PBS). A total of 5 μl annexin V-FITC and 5 μl PI were then added according to the manufacturer’s instructions. Cells were then incubated at room temperature, in the dark, for 15 min and analyzed using a flow cytometer system (FACScan; BD Biosciences, San Jose, CA, USA).
DNA flow cytometric analysis
After treatment with BTCI, cells were digested with trypsin, harvested, washed twice with ice-cold PBS, fixed with 75% ethanol at −20 °C overnight, washed again and then incubated with RNase A (25 μg/ml, Bio Basic Inc., Markham, Ontario, Canada) at 37 °C for 30 min. Cells were washed once with PBS and incubated with PI (50 μg/ml) for 30 min at room temperature in the dark. The cells were re-suspended in 500 μl PBS and subjected to flow cytometry on a Cytomics FC500 flow cytometer (FACScan; BD Biosciences). A sub G0/G1 population seen to the left of the G0/G1 peak represents DNA fragmentation caused by apoptosis.
JC-10 mitochondrial membrane potential assay
Mitochondrial membrane potential was measured using JC-10 Molecular Probes staining from Sigma (MAK159) as per manufacturer´s instructions. In brief, cells were cultured on 96-well white-walled clear-bottom plates and treated with BTCI for 24 h. Then, 50 μl of JC-10 dye-loading solution was added to each well and incubated for 30 min. The samples were read on a SpectraMax M3 microplate reader (Molecular Devices), measuring fluorescence intensities (Ex/Em= 485/515 nm and Ex/Em= 540/590 nm). The change of mitochondrial membrane potential was measured as the ratio between aggregate (Em=515 nm) and monomeric forms (Em= 590 nm) of JC-10. Increasing ratios indicate mitochondrial membrane depolarization.
Caspase-3 activity assay
Caspase-3 activity was measured using the Caspase-3 assay kit from Abcam Inc. (ab39383) as per manufacturer´s instructions. In brief, samples were trypsinized and re-suspended in cell lysis buffer and allowed to lyse for 10 min. Reaction buffer containing 5 μl (DTT) and the 10 μl (DEVD-AFC) substrate was added and the mixture was incubated at 37 °C for 2 h. The samples were read on a SpectraMax M3 microplate reader (Molecular Devices), using a 400 nm excitation and 505 nm emission.
Determination of intracellular generation of ROS
The production of intracellular ROS was estimated fluorimetrically using the oxidation sensitive fluorescent kit from Sigma (MAK144) that provides a sensitive, one-step fluorometric assay to detect intracellular ROS (especially superoxide and hydroxyl radicals) in live cells. In brief, cells were treated with BTCI for 30, 60, 90, 120 and 180 min. Then, 100 μl of Master Reaction Mix was added and samples were incubated at 37 °C for 60 min. The samples were read on a SpectraMax M3 microplate reader (Molecular Devices), using a 540 nm excitation and 570 nm emission.
Real-time polymerase chain reaction
The amount of BAX (BCL2-Associated X Protein), BCL-2(B-Cell CLL/Lymphoma 2), NFKB2 (Nuclear Factor of Kappa Light Polypeptide Gene Enhancer in B-Cells 2 (P49/P100)), CFLAR (CASP8 and FADD-Like Apoptosis Regulator) and IL-8 mRNA was assessed by real-time reverse transcription-PCR (RT-PCR) using a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer´s protocol. RNA (1 μg) was used for cDNA synthesis using QuantiTect Reverse Transcription Kit (QIAGEN, Valencia, CA, USA). Real-time PCR was carried out using the Fast SYBR as a detection method. Primer sequences, as presented in Table 1, were purchased from Exxtend (Rio de Janeiro, Brazil). In addition, the amount of β-actin mRNA was quantified in all samples as an internal constitutive control. The concentration of gene-specific mRNA in treated samples relative to untreated samples was calculated using the 2 –ΔΔCT formula as follows: 2–ΔΔCT=[(CT gene of interest—CT internal control) treatment group]—[(CT gene of interest—CT internal control) control group]. Results are shown as fold changes compared with the control.73,74 All experiments were conducted in triplicate.
Table 1. Human gene-targeting primers used in this study.
| Primer | Sequence (5'—3') |
|---|---|
| IL-8 forward | ACTGAGAGTGATTGAGAGTGGAC |
| IL-8 reverse | AACCCTCTGCACCCAGTTTTC |
| NFKB2 forward | ATGGAGAGTTGCTACAACCCA |
| NFKB2 reverse | CTGTTCCACGATCACCAGGTA |
| CFLAR forward | AGAGTGAGGCGATTTGACCTG |
| CFLAR reverse | GTCCGAAACAAGGTGAGGGTT |
| BAX forward | CCCGAGAGGTCTTTTTCCGAG |
| BAX reverse | CCAGCCCATGATGGTTCTGAT |
| BCL-2 forward | TACAGGCTGGCTCAGGACTAT |
| BCL-2 reverse | CGCAACATTTTGTAGCACTCTG |
| ACTB forward | GGATGCAGAAGGAGATCACTG |
| ACTB reverse | CGATCCACACGGAGTACTTG |
Statistical analysis
All data are presented as the mean±S.D. of at least two independent experiments conducted in triplicate. Statistical analysis was performed using one-way ANOVA followed by Dunnett’s multiple comparison test using SPSS 19.0 software (SPSS Inc., Chicago, IL, USA). Differences were considered to be significant at a value of P<0.05.
Acknowledgments
The authors wish to express their gratitude to Professor Manuel Mateus Ventura. The authors dedicate this work to him. This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação de Apoio a Pesquisa no Distrito Federal (FAPDF), Brazil. The authors would like to thank Dr. Luciano P. Silva for assistance and scientific support in MALDI-TOF spectrometry and Marco Oliveira for his help in RT-PCR analysis.
Footnotes
The authors declare no conflict of interest.
References
- Globocan. Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012; 2012.
- St Clair WH, Billings PC, Carew JA, Keller-McGandy C, Newberne P, Kennedy AR. Suppression of dimethylhydrazine-induced carcinogenesis in mice by dietary addition of the Bowman-Birk protease inhibitor. Cancer Res 1990; 50: 580–586. [PubMed] [Google Scholar]
- Billings PC, Newberne PM, Kennedy AR. Protease inhibitor suppression of colon and anal gland carcinogenesis induced by dimethylhydrazine. Carcinogenesis 1990; 11: 1083–1086. [DOI] [PubMed] [Google Scholar]
- Clemente A, Moreno FJ, Marin-Manzano Mdel C, Jimenez E, Domoney C. The cytotoxic effect of Bowman-Birk isoinhibitors, IBB1 and IBBD2, from soybean (Glycine max) on HT29 human colorectal cancer cells is related to their intrinsic ability to inhibit serine proteases. Mol Nutr Food Res 2010; 54: 396–405. [DOI] [PubMed] [Google Scholar]
- Kennedy AR, Billings PC, Maki PA, Newberne P. Effects of various preparations of dietary protease inhibitors on oral carcinogenesis in hamsters induced by DMBA. Nutr Cancer 1993; 19: 191–200. [DOI] [PubMed] [Google Scholar]
- Messadi DV, Billings P, Shklar G, Kennedy AR. Inhibition of oral carcinogenesis by a protease inhibitor. J Natl Cancer Inst 1986; 76: 447–452. [PubMed] [Google Scholar]
- Armstrong WB, Kennedy AR, Wan XS, Atiba J, McLaren CE, Meyskens FL Jr. Single-dose administration of Bowman-Birk inhibitor concentrate in patients with oral leukoplakia. Cancer Epidemiol Biomarkers Prev 2000; 9: 43–47. [PubMed] [Google Scholar]
- Armstrong WB, Kennedy AR, Wan XS, Taylor TH, Nguyen QA, Jensen J et al. Clinical modulation of oral leukoplakia and protease activity by Bowman-Birk inhibitor concentrate in a phase IIa chemoprevention trial. Clin Cancer Res 2000; 6: 4684–4691. [PubMed] [Google Scholar]
- Armstrong WB, Taylor TH, Kennedy AR, Melrose RJ, Messadi DV, Gu M et al. Bowman birk inhibitor concentrate and oral leukoplakia: a randomized phase IIb trial. Cancer Prev Res 2013; 6: 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hofe E, Newberne PM, Kennedy AR. Inhibition of N-nitrosomethylbenzylamine-induced esophageal neoplasms by the Bowman-Birk protease inhibitor. Carcinogenesis 1991; 12: 2147–2150. [DOI] [PubMed] [Google Scholar]
- An WG, Hwang SG, Trepel JB, Blagosklonny MV. Protease inhibitor-induced apoptosis: accumulation of wt p53, p21WAF1/CIP1, and induction of apoptosis are independent markers of proteasome inhibition. Leukemia 2000; 14: 1276–1283. [DOI] [PubMed] [Google Scholar]
- Malkowicz SB, McKenna WG, Vaughn DJ, Wan XS, Propert KJ, Rockwell K et al. Effects of Bowman-Birk inhibitor concentrate (BBIC) in patients with benign prostatic hyperplasia. Prostate 2001; 48: 16–28. [DOI] [PubMed] [Google Scholar]
- Joanitti GA, Azevedo RB, Freitas SM. Apoptosis and lysosome membrane permeabilization induction on breast cancer cells by an anticarcinogenic Bowman-Birk protease inhibitor from Vigna unguiculata seeds. Cancer Lett 2010; 293: 73–81. [DOI] [PubMed] [Google Scholar]
- Chen YW, Huang SC, Lin-Shiau SY, Lin JK. Bowman-Birk inhibitor abates proteasome function and suppresses the proliferation of MCF7 breast cancer cells through accumulation of MAP kinase phosphatase-1. Carcinogenesis 2005; 26: 1296–1306. [DOI] [PubMed] [Google Scholar]
- Zhang L, Littlejohn JE, Cui Y, Cao X, Peddaboina C, Smythe WR. Characterization of bortezomib-adapted I-45 mesothelioma cells. Mol Cancer 2010; 9: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoul R, Fernandez PA, Quiquerez AL, Martinou I, Maki M, Schroter M et al. Involvement of the proteasome in the programmed cell death of NGF-deprived sympathetic neurons. EMBO J 1996; 15: 3845–3852. [PMC free article] [PubMed] [Google Scholar]
- Drexler HC. Activation of the cell death program by inhibition of proteasome function. Proc Natl Acad Sci USA 1997; 94: 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes UG, Erhardt P, Yao R, Cooper GM. p53-dependent induction of apoptosis by proteasome inhibitors. J Biol Chem 1997; 272: 12893–12896. [DOI] [PubMed] [Google Scholar]
- Jana NR, Zemskov EA, Wang G, Nukina N. Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum Mol Genet 2001; 10: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Jana NR, Dikshit P, Goswami A, Nukina N. Inhibition of proteasomal function by curcumin induces apoptosis through mitochondrial pathway. J Biol Chem 2004; 279: 11680–11685. [DOI] [PubMed] [Google Scholar]
- Adams J. Proteasome inhibitors as new anticancer drugs. Curr Opin Oncol 2002; 14: 628–634. [DOI] [PubMed] [Google Scholar]
- Adams J. The proteasome: a suitable antineoplastic target. Nat Rev Cancer 2004; 4: 349–360. [DOI] [PubMed] [Google Scholar]
- Maharjan S, Oku M, Tsuda M, Hoseki J, Sakai Y. Mitochondrial impairment triggers cytosolic oxidative stress and cell death following proteasome inhibition. Sci Rep 2014; 4: 5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R, Emi M, Tanabe K. Role of mitochondria as the gardens of cell death. Cancer Chemother Pharmacol 2006; 57: 545–553. [DOI] [PubMed] [Google Scholar]
- Simon HU, Haj-Yehia A, Levi-Schaffer F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis 2000; 5: 415–418. [DOI] [PubMed] [Google Scholar]
- Loft S, Poulsen HE. Cancer risk and oxidative DNA damage in man. J Mol Med (Berl) 1996; 74: 297–312. [DOI] [PubMed] [Google Scholar]
- An B, Goldfarb RH, Siman R, Dou QP. Novel dipeptidyl proteasome inhibitors overcome Bcl-2 protective function and selectively accumulate the cyclin-dependent kinase inhibitor p27 and induce apoptosis in transformed, but not normal, human fibroblasts. Cell Death Differ 1998; 5: 1062–1075. [DOI] [PubMed] [Google Scholar]
- Fan XM, Wong BC, Wang WP, Zhou XM, Cho CH, Yuen ST et al. Inhibition of proteasome function induced apoptosis in gastric cancer. Int J Cancer 2001; 93: 481–488. [DOI] [PubMed] [Google Scholar]
- Xiang T, Du L, Pham P, Zhu B, Jiang S. Nelfinavir, an HIV protease inhibitor, induces apoptosis and cell cycle arrest in human cervical cancer cells via the ROS-dependent mitochondrial pathway. Cancer Lett 2015; 364: 79–88. [DOI] [PubMed] [Google Scholar]
- Oliveira CS, Pereira H, Alves S, Castro L, Baltazar F, Chaves SR et al. Cathepsin D protects colorectal cancer cells from acetate-induced apoptosis through autophagy-independent degradation of damaged mitochondria. Cell Death Dis 2015; 6: e1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa L, Gomes E, Rockwell P. Reactive oxygen species induced by proteasome inhibition in neuronal cells mediate mitochondrial dysfunction and a caspase-independent cell death. Apoptosis 2007; 12: 1389–1405. [DOI] [PubMed] [Google Scholar]
- Papa L, Rockwell P. Persistent mitochondrial dysfunction and oxidative stress hinder neuronal cell recovery from reversible proteasome inhibition. Apoptosis 2008; 13: 588–599. [DOI] [PubMed] [Google Scholar]
- Mahalingaiah PK, Ponnusamy L, Singh KP. Chronic oxidative stress causes estrogen-independent aggressive phenotype, and epigenetic inactivation of estrogen receptor alpha in MCF-7 breast cancer cells. Breast Cancer Res Treat 2015. [DOI] [PubMed]
- Haigis MC, Deng CX, Finley LW, Kim HS, Gius D. SIRT3 is a mitochondrial tumor suppressor: a scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res 2012; 72: 2468–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Zhang Q, Chen S, Fang B, Yang Q, Chen C et al. Mitochondrial dysfunction promotes breast cancer cell migration and invasion through HIF1alpha accumulation via increased production of reactive oxygen species. PloS One 2013; 8: e69485. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Barbosa JA, Silva LP, Teles RC, Esteves GF, Azevedo RB, Ventura MM et al. Crystal structure of the Bowman-Birk Inhibitor from Vigna unguiculata seeds in complex with beta-trypsin at 1.55 A resolution and its structural properties in association with proteinases. Biophys J 2007; 92: 1638–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Freitas SM, de Mello LV, da Silva MC, Vriend G, Neshich G, Ventura MM. Analysis of the black-eyed pea trypsin and chymotrypsin inhibitor-alpha-chymotrypsin complex. FEBS Lett 1997; 409: 121–127. [DOI] [PubMed] [Google Scholar]
- Silva LP, Liete JR SA, Bloch C Jr, Freitas SM. Thermal stability of a black eyed pea trypsin/chymotrypsin inhibitor (BTCI). Protein Pept Lett 2001; 7: 397–401. [Google Scholar]
- Ventura MM X-FJ. Trypsin and chymotrypsin inhibitor from black-eyed pea (V. Sinensis L.) I. Purification and partial characterization. An Acad Bras Cienc 1966; 38: 553–566. [Google Scholar]
- Souza Lda C, Camargo R, Demasi M, Santana JM, de Sa CM, de Freitas SM. Effects of an anticarcinogenic Bowman-Birk protease inhibitor on purified 20S proteasome and MCF-7 breast cancer cells. PloS One 2014; 9: e86600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura MM, Xavier Filho J, Moreira RA, Aquino Ade M, Pinheiro PA. A trypsin and chymotrypsin inhibitor from black-eyed pea (Vigna sinensis L.). II. Further studies on its characterization and a reevaluation of earlier results. An Acad Bras Cienc 1971; 43: 233–242. [PubMed] [Google Scholar]
- Kennedy AR. The evidence for soybean products as cancer preventive agents. J Nutr 1995; 125(3 Suppl): 733S–743S. [DOI] [PubMed] [Google Scholar]
- Kennedy AR. The Bowman-Birk inhibitor from soybeans as an anticarcinogenic agent. Am J Clin Nutr 1998; 68(6 Suppl): 1406S–1412S. [DOI] [PubMed] [Google Scholar]
- Lee HP, Gourley L, Duffy SW, Esteve J, Lee J, Day NE. Dietary effects on breast-cancer risk in Singapore. Lancet 1991; 337: 1197–1200. [DOI] [PubMed] [Google Scholar]
- Barnes S, Grubbs C, Setchell KD, Carlson J. Soybeans inhibit mammary tumors in models of breast cancer. Prog Clin Biol Res 1990; 347: 239–253. [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 1997; 386: 463–471. [DOI] [PubMed] [Google Scholar]
- Kunjappu MJ, Hochstrasser M. Assembly of the 20S proteasome. Biochim Biophys Acta 2014; 1843: 2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 1995; 268: 533–539. [DOI] [PubMed] [Google Scholar]
- Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N et al. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure 2002; 10: 609–618. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 1999; 68: 1015–1068. [DOI] [PubMed] [Google Scholar]
- Gavilan E, Giraldez S, Sanchez-Aguayo I, Romero F, Ruano D, Daza P. Breast cancer cell line MCF7 escapes from G1/S arrest induced by proteasome inhibition through a GSK-3beta dependent mechanism. Sci Rep 2015; 5: 10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust D, Schmitt C, Oesch F, Oesch-Bartlomowicz B, Schreck I, Weiss C et al. Differential p38-dependent signalling in response to cellular stress and mitogenic stimulation in fibroblasts. Cell Commun Signal 2012; 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pundir S, Vu HY, Solomon VR, McClure R, Lee H. VR23: a quinoline-sulfonyl hybrid proteasome inhibitor that selectively kills cancer via cyclin E-mediated centrosome amplification. Cancer Res 2015. [DOI] [PubMed]
- Keller JN, Hanni KB, Markesbery WR. Possible involvement of proteasome inhibition in aging: implications for oxidative stress. Mech Ageing Dev 2000; 113: 61–70. [DOI] [PubMed] [Google Scholar]
- Ling YH, Liebes L, Zou Y, Perez-Soler R. Reactive oxygen species generation and mitochondrial dysfunction in the apoptotic response to Bortezomib, a novel proteasome inhibitor, in human H460 non-small cell lung cancer cells. J Biol Chem 2003; 278: 33714–33723. [DOI] [PubMed] [Google Scholar]
- Ding Q, Keller JN. Proteasome inhibition in oxidative stress neurotoxicity: implications for heat shock proteins. J Neurochem 2001; 77: 1010–1017. [DOI] [PubMed] [Google Scholar]
- Morgan MJ, Liu ZG. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res 2011; 21: 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack JC Jr, Liu R, Houston M, Abendroth K, Elliott PJ, Adams J et al. Enhanced chemosensitivity to CPT-11 with proteasome inhibitor PS-341: implications for systemic nuclear factor-kappaB inhibition. Cancer Res 2001; 61: 3535–3540. [PubMed] [Google Scholar]
- Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J et al. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res 2001; 61: 3071–3076. [PubMed] [Google Scholar]
- Teicher BA, Ara G, Herbst R, Palombella VJ, Adams J. The proteasome inhibitor PS-341 in cancer therapy. Clin Cancer Res 1999; 5: 2638–2645. [PubMed] [Google Scholar]
- Richardson PG, Hideshima T, Anderson KC. Bortezomib (PS-341): a novel, first-in-class proteasome inhibitor for the treatment of multiple myeloma and other cancers. Cancer Control 2003; 10: 361–369. [DOI] [PubMed] [Google Scholar]
- Dolcet X, Llobet D, Encinas M, Pallares J, Cabero A, Schoenenberger JA et al. Proteasome inhibitors induce death but activate NF-kappaB on endometrial carcinoma cell lines and primary culture explants. J Biol Chem 2006; 281: 22118–22130. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Ikeda H, Chauhan D, Okawa Y, Raje N, Podar K et al. Bortezomib induces canonical nuclear factor-kappaB activation in multiple myeloma cells. Blood 2009; 114: 1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna S, Singha B, Phyo SA, Gatla HR, Chang TP, Sanacora S et al. Proteasome inhibition by bortezomib increases IL-8 expression in androgen-independent prostate cancer cells: the role of IKKalpha. J Immunol 2013; 191: 2837–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singha B, Gatla HR, Manna S, Chang TP, Sanacora S, Poltoratsky V et al. Proteasome inhibition increases recruitment of IkappaB kinase beta (IKKbeta), S536P-p65, and transcription factor EGR1 to interleukin-8 (IL-8) promoter, resulting in increased IL-8 production in ovarian cancer cells. J Biol Chem 2014; 289: 2687–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang X, Olumi AF. MG-132 sensitizes TRAIL-resistant prostate cancer cells by activating c-Fos/c-Jun heterodimers and repressing c-FLIP(L). Cancer Res 2007; 67: 2247–2255. [DOI] [PubMed] [Google Scholar]
- Sayers TJ, Brooks AD, Koh CY, Ma W, Seki N, Raziuddin A et al. The proteasome inhibitor PS-341 sensitizes neoplastic cells to TRAIL-mediated apoptosis by reducing levels of c-FLIP. Blood 2003; 102: 303–310. [DOI] [PubMed] [Google Scholar]
- Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D, Fanourakis G, Gu X et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc Natl Acad Sci USA 2002; 99: 14374–14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Adrain C, Martin SJ. Proteases, proteasomes and apoptosis: breaking Ub is hard to do. Cell Death Differ 2005; 12: 1213–1217. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Trondle N, Albrecht G, Wolf DH. PRE5 and PRE6, the last missing genes encoding 20S proteasome subunits from yeast? Indication for a set of 14 different subunits in the eukaryotic proteasome core. Biochemistry 1994; 33: 12229–12237. [DOI] [PubMed] [Google Scholar]
- Schmidtke G, Kraft R, Kostka S, Henklein P, Frommel C, Lowe J et al. Analysis of mammalian 20S proteasome biogenesis: the maturation of beta-subunits is an ordered two-step mechanism involving autocatalysis. EMBO J 1996; 15: 6887–6898. [PMC free article] [PubMed] [Google Scholar]
- Demasi M, Shringarpure R, Davies KJ. Glutathiolation of the proteasome is enhanced by proteolytic inhibitors. Arch Biochem Biophys 2001; 389: 254–263. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 2002; 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3: 1101–1108. [DOI] [PubMed] [Google Scholar]