Abstract
Background
Increasing use of genetic testing raises questions about disclosing secondary findings, including pleiotropic information.
Objective
To determine the safety and behavioral impact of disclosing modest associations between APOE genotype and coronary artery disease (CAD) risk during APOE-based genetic risk assessments for Alzheimer’s disease (AD).
Design
Randomized, multicenter equivalence clinical trial
Setting
Four teaching hospitals
Participants
257 asymptomatic adults enrolled, 69% with one AD-affected first degree relative
Intervention
Disclosing AD and CAD genetic risk information (AD+CAD) versus disclosing only AD genetic risk (AD-only)
Measurements
Co-primary outcomes were Beck Anxiety Index (BAI) and Center for Epidemiologic Studies Depression Scale (CES-D) scores at 12 months. Secondary outcomes included test-related distress at 12 months, all measures at 6 weeks and 6 months, and health behavior changes at 12 months.
Results
12 months after disclosure, mean BAI scores were 3.5 and 3.5 in AD-only and AD+CAD arms (Δ=0.0, 95%CI: −1.0 to 1.0), and mean CES-D scores were 6.4 and 7.1 in AD-only and AD+CAD arms (Δ=0.7, 95%CI: −1.0 to 2.4). Both confidence bounds fell within the equivalence margin of +/−5 points. Among ε4-positive participants, distress was lower in AD+CAD arms than AD-only arms (Δ=−4.8, 95%CI: −8.6 to −1.0) (p=0.031 for disclosure arm x APOE genotype). AD+CAD participants also reported more health behavior changes, regardless of APOE genotype.
Limitations
Outcomes were self-reported from volunteers without severe anxiety, severe depression, or cognitive problems. Analyses omitted 33 randomized participants.
Conclusion
Disclosing pleiotropic information did not increase anxiety or depression, and may have decreased distress among those at increased risk for two conditions. Providing risk modification information regarding CAD improved health behaviors. Findings highlight potential benefits of secondary genetic findings disclosure when options exist for decreasing risk.
Keywords: genetics, genomics, pleiotropy, risk assessment, personalized medicine, secondary findings, Alzheimer, APOE
INTRODUCTION
Physicians of all specialties are increasingly using genomic tools, including whole genome and whole exome sequencing (1–3) and genotyping for risk variants and pharmacogenomics variants (4, 5). These tools often identify incidental or secondary findings that have important implications for disease, but are unrelated to the original purposes of testing. While recommendations exist for the management of secondary findings in genome sequencing (6, 7), this topic remains controversial (8–10). In particular, experts are concerned that disclosing such information to patients may increase psychological risks while providing minimal clinical benefits (11–15). Despite these concerns, few studies have empirically examined the benefits and harms of secondary genomic findings disclosure.
Pleiotropy, the association between genetic variants and multiple disease traits, provides a useful model for examining this issue. It is estimated that 17% of genes have pleiotropic effects (16). Pleiotropy poses challenges to communicating genetic test results, because disclosing a genetic variant associated with one disease may unexpectedly confer knowledge of a separate disease risk (17–19). The ε4 allele of the apolipoprotein E (APOE) gene, present in over 20% of most populations (20), is robustly associated with the risk of Alzheimer’s disease (AD) (21) and has a weaker, and less well known association with the risk of coronary artery disease (CAD) (22, 23). We previously conducted two randomized trials of APOE genotype disclosure during AD risk assessment, showing that such disclosure did not increase psychological risks to volunteer populations (24, 25) while motivating at-risk participants to change potential AD risk-reducing behaviors (26, 27). Neither trial addressed APOE-CAD associations.
Here, we describe an independent trial wherein we randomized participants seeking a genetic risk assessment for AD to receive a) only AD risk information or b) risk information for AD and CAD. We hypothesized that both groups would show equivalent levels of anxiety and depression one year after disclosure. We also conducted secondary analyses examining test-related distress and health behaviors.
SUBJECTS AND METHODS
Design Overview
The multidisciplinary REVEAL Study group designed the study and risk disclosure procedures (24, 25, 28, 29), including ethnicity-specific risk estimates (30, 31). An independent Ethics and Safety Board (ESB) and institutional review boards at each study site approved the protocol. Participants provided informed consent for initial steps during study enrollment, then again prior to the blood draw for genotyping. APOE was genotyped at a CLIA-certified facility.
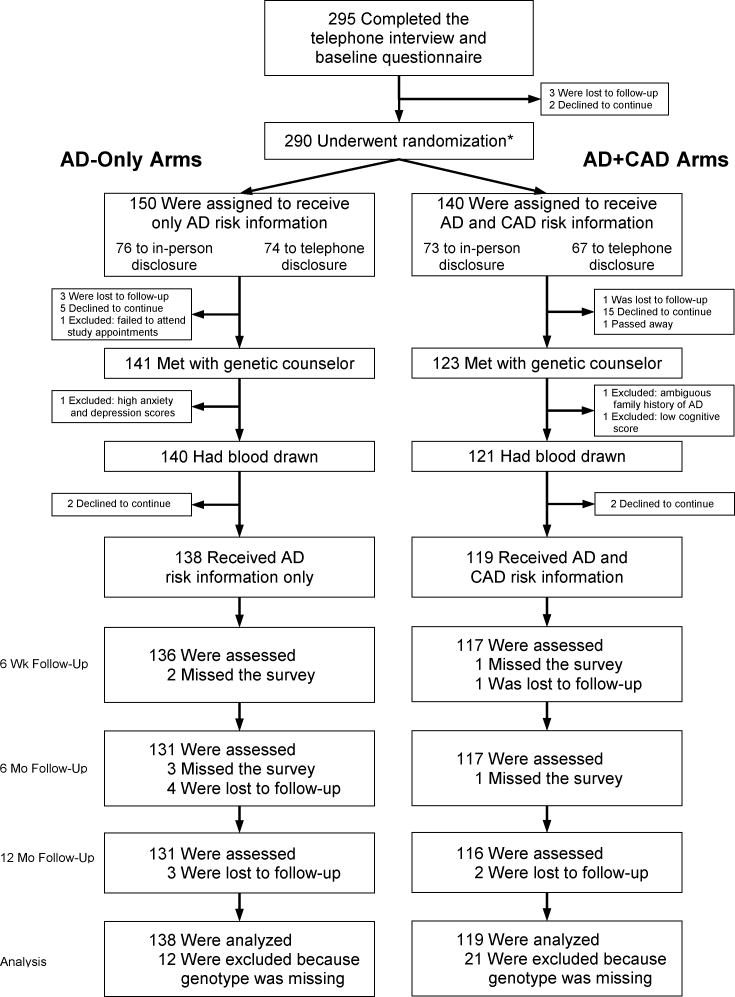
Figure 1 shows the study’s design and flow. After a phone interview and written questionnaire, participants received brochures that summarized known benefits, risks, and limitations of APOE testing, including potential difficulties coping with test results and the lack of “proven ways to prevent Alzheimer’s disease” (24, 29) (Appendix Figure 1). They then met with genetic counselors (GCs) who answered individual questions and had blood drawn for genotyping. Approximately one month after the blood draw, participants received scripted genetic risk information either in-person or by phone, depending on randomization, from one of seven GCs who also addressed any participant concerns. Participants were then followed for one year, with measurements at 6 weeks, 6 months and 12 months.
Figure 1.
Enrollment and outcomes
* A second randomization occurred here to determine whether subjects would receive in-person disclosure or telephone disclosure
Setting and Participants
We recruited cognitively normal adults from Boston, Cleveland, Washington, DC, and Ann Arbor using mailings to research registries, referrals from neurologists, and advertisements in local newspapers. To achieve greater sample diversity, we enrolled equal numbers of adults over and under age 60 years, and equal numbers of men and women. We also tried to enroll 75% of participants with a single AD-affected first degree relative (FDR), and 25% with no family history. We excluded individuals with two or more AD-affected FDRs; family members with average AD onset under age 60; scores below an education-adjusted 87 on the Modified Mini-Mental State Examination (32); or severe anxiety and depression, as defined in the Outcomes and Follow-Up section.
Randomization and Intervention
The primary goals of the trial focused upon the impact of pleiotropic disclosure, but the opportunity to address a key question regarding service delivery led to the addition of a second randomization to compare telephone and in-person disclosure of genotyping results. The telephone vs in-person disclosure results will be reported in a separate manuscript. Participants were randomized equally within strata, in blocks of size four, into “AD-only, in-person disclosure,” “AD-only, telephone disclosure,” “AD+CAD, in-person disclosure,” and “AD+CAD, telephone disclosure” arms. Randomization strata were defined by site, age (<60 vs ≥60), family history of AD, and gender. Serially-numbered envelopes concealed participants’ randomization status until needed. Prior to randomization, participants were only informed that they would receive “different types of genetic risk information.” Participants in AD-only arms were not informed about APOE-CAD associations. Participants in AD+CAD arms were told during a second consent step that they would receive information about CAD. Participants learned whether they would receive results in-person or via telephone during their blood draw appointment.
During genetic risk disclosure, all participants received scripted information about their APOE genotype, cumulative lifetime risk (range 6–73%), and remaining risk to age 85 for AD, along with AD risk curves (25, 30, 31). Participants randomized into AD+CAD arms were also provided with the following statement in oral and written form, regardless of genotype: “In addition to Alzheimer’s disease, APOE has been found to be connected to heart disease. Some studies have shown that people who carry e4 also have a higher risk of developing heart disease. Potential strategies to reduce the risk of coronary artery disease include smoking cessation, a healthy diet, weight loss, treatment of elevated cholesterol, and exercise (with your doctor’s permission).” This information was reiterated after each follow-up session. The statement was crafted to be appropriate for secondary findings disclosure during AD risk assessment by a study cardiologist (D.L.B.) after conferring with cardiologists unrelated to the REVEAL Study.
Outcomes and Follow-Up
Outcomes were assessed at 6 weeks, 6 months, and 12 months after disclosure, as summarized in Appendix Table 1. Co-primary outcomes were validated scales of anxiety and depression at one year, using the Beck Anxiety Inventory (BAI) (33) and the Center for Epidemiological Studies-Depression Scale (CES-D) (34). BAI scores range from 0–63 (>8: mild, >15: moderate, >25: severe). CES-D scores range from 0–60 (>10: mild, >16: moderate, >26: severe) (35). Anxiety and depression scores at 6 weeks and 6 months, as well as time-averaged, were secondary outcomes. Test-related distress specific to the genetic risk assessment at all time points was another secondary outcome, measured with the Impact of Event Scale (IES) (36, 37). IES scores range from 0–75 (≥20: significant distress). For safety purposes, an ESB-approved plan required immediate interview of participants whose BAI or CES-D scores exceeded 25 or 26, respectively, or increased by more than 15 points from baseline.
Secondary outcomes also included changes to health behaviors (diet, exercise, medications, dietary supplements, stress reduction, and mental activities) at 12 months after disclosure. At 6 weeks following disclosure, participants were asked, “Since you learned your APOE test results, have you made any health or wellness changes?” Participants responding affirmatively answered additional questions about the types of behavior change they had initiated. At 12 months, participants were asked a) if they had continued the changes reported at 6 weeks and b) about additional changes initiated since the 6-week survey. Participants were coded as having made a health behavior change at 12 months if they continued a behavior reported at 6 weeks or if they initiated a behavior between the 6-week and 12-month surveys. Physical activity was assessed using the Rapid Assessment of Physical Activity (RAPA) (38) which scored participants on a 1–7 scale for aerobic activity and a 0–3 scale for strength and flexibility training. Smoking status was assessed at baseline and 12 months by asking participants if they had smoked within the prior seven days. To assess recall of pleiotropic information, each follow-up survey asked participants in AD+CAD arms, “What other disease did we tell you is associated with the APOE gene?”
Statistical Analysis
We estimated that 32 participants would need to receive genetic risk disclosure in each group to achieve 80% power to detect 5 point differences between AD+CAD and AD-only randomization arms (39). Enrollment targets of 70 at each study site were set to enroll 280 total participants and to achieve 256 total disclosures, assuming 10% dropout. The expanded sample size was set to allow for sub-analyses by APOE genotypes and demographic factors.
T-tests and chi-square tests compared demographic features and discontinuation rates of AD-only and AD+CAD groups, and participant variables associated with discontinuation. The protocol was initially designed under a superiority framework, but prior to seeing data and conducting data analyses, we concluded that the scientific aims were best served by use of equivalence comparisons. Data for telephone and in-person disclosure arms were pooled in analyses presented here because interactions between AD-only/AD+CAD randomization status and in-person/telephone disclosure randomization status were not observed (p-values for tests of interactions: BAI: p=0.18; CES-D: p=0.34; IES: p=0.68; p-values for tests of 3 way interactions between the two treatment arms and time were all ≥ 0.40). Two participants who did not receive genotype and AD risk disclosure and whose randomization status was mistakenly entered into the study database as AD-only were recoded as AD+CAD for analyses of study drop out.
We used longitudinal analyses for psychological outcomes, including all observed data and imputing data for the few missing observations from participants who received genetic risk disclosure. Since the distribution of these outcomes was skewed, we used generalized linear models fit with generalized estimating equations with log link and Gamma distribution to compare outcomes by AD-only/AD+CAD randomization status. We used an autoregressive working correlation structure with robust standard errors to account for the repeated measures within participant. A value of one was added to all measures to shift the distribution away from zero. Models included terms for AD-only vs AD+CAD randomization status, time as a categorical variable, interaction between time and randomization arm, corresponding baseline psychological measure where applicable and the GC conducting disclosure. Additional analyses further adjusted for age, gender, education, race, family history of AD, phone or in-person disclosure, and APOE genotype. We used contrasts to compare randomization arms at specific time points and overall for a time-averaged comparison. Equivalence was defined using a margin of 5 points per prior REVEAL Study trials (24, 25). We used 95% CIs based on recommendations to use CIs of (1–2α) x 100% for equivalence testing and using α=2.5% (0.05/2) to account for multiple testing across two primary outcomes (40, 41). To be conservative and consistent across psychological outcomes, we also used 95% CIs for all secondary analyses. We evaluated whether interaction effects existed between AD-only/AD+CAD randomization status and APOE genotype because pleiotropic information might concern participants only if they were at increased risk for both diseases. We used the same model as described above and added variables for APOE ε4, its interaction with time, its interaction with pleiotropy randomization arm and three-way interaction for APOE ε4, time and pleiotropy randomization. From these analyses we also obtained results for each APOE stratum for secondary analyses, comparing pleiotropy arms. Additionally, we used contrasts from these models to estimate the differences between APOE ε4-positive and APOE ε4-negative participants within randomization arms.
Secondary analyses tested for differences in health behavior change rates and physical activity levels between AD-only and AD+CAD arms and by APOE status. Health behavior change rates were compared between groups using logistic regression, allowing for interaction with APOE genotype and adjusting for disclosing GC. Changes in physical activity levels were compared between randomization groups using multiple linear regression, adjusting for APOE genotype, and genetic counselor providing disclosure. Changes to smoking status were assessed, but omitted from reporting due to small numbers of current smokers (13 enrolled).
Because APOE genotypes could not be reliably imputed, analyses included only participants receiving genetic risk information (genotype data for participants who provided blood but dropped out of the study before the disclosure session was destroyed per the IRB-approved protocol). Two study participants in the AD-only arms and 2 study participants of the AD+CAD arms were excluded from analysis for this reason. Twenty of the remaining 257 study participants were missing BAI, CES-D, and IES scores. We assumed data were missing at random and imputed missing values for these outcomes using multiple imputation (Markov Chain Monte Carlo procedures with 40 imputed data sets. See Appendix). All analyses were conducted using SAS, version 9.3 (SAS Institute, Cary, North Carolina).
Role of the Funding Source
This study was funded by the National Human Genome Research Institute of the National Institutes of Health, which had no role in study design; collection, analysis, or interpretation of data; writing, review, or approval of the manuscript; or the decision to submit the manuscript for publication.
RESULTS
Of 290 participants randomized, 257 (89%) received genetic risk disclosure (Figure 1). Four were screened out for the following reasons: cognitive score below eligibility criteria, high depression, ambiguous family history of AD, and failure to attend study appointments. Demographic characteristics did not vary by AD-only/AD+CAD randomization, other than by race (Table 1) and were similar to those of our previous trials (24, 29) except for the deliberate inclusion of participants without an affected FDR. GCs communicated results to between 9 and 82 participants apiece, and did not differ statistically in their likelihoods of being randomized to disclose AD-only or AD+CAD information (p=0.58). Drop out before disclosure occurred in 8% of AD-only and 15% of AD+CAD participants (p=0.06). Younger, female, unmarried and less educated participants were more likely to drop out (all p<0.05), regardless of AD-only/AD+CAD randomization (Appendix Table 2). At all time points, a high proportion of AD+CAD participants (81.3% at 6 weeks, 86.4% at 6 months, 84.4% at 12 months) correctly recalled receiving risk information about the association between APOE and CAD.
Table 1.
Characteristics of participants who received genetic risk disclosure.
| Characteristic | Randomization Arm | |
|---|---|---|
| AD Only (n=138) | AD+CAD (n=119) | |
| Age: years | ||
| Mean (SD) | 58.2 (12.4) | 58.2 (13.6) |
| Range | 27–82 | 21–83 |
| Female sex: n (%) | 76 (55) | 65 (55) |
| African American race: n (%)* | 29 (21)† | 9 (8)† |
| Education: years | ||
| Mean (SD) | 16.8 (2.2) | 16.8 (2.4) |
| Range | 12–20 | 10–20 |
| Currently married: n (%) | 81 (59) | 72 (61) |
| Mean BAI score (SD) | 3.8 (3.6) | 3.2 (3.3) |
| Mean CES-D score (SD) | 6.0 (5.3) | 5.3 (4.8) |
| Site: n (%) | ||
| Boston | 42 (30) | 36 (30) |
| Cleveland | 34 (25) | 30 (25) |
| Michigan | 33 (24) | 35 (29) |
| Washington, DC | 29 (21) | 18 (15) |
| Parent or sibling with AD: n (%) | 93 (67) | 85 (71) |
| ε4-positive | 51 (37) | 32 (27) |
| Has heart disease/ had heart attack: n (%) | 9 (7) | 13 (11) |
| Current smoker: n (%) | 8 (6) | 5 (4) |
Race was self-reported
p<0.01
Anxiety, Depression and Test-Related Distress
Mean anxiety, depression, and test-related distress scores were below cut-offs for mood disorders, regardless of disclosure protocol, at all time points and when time-averaged (Table 2). All 95% confidence bounds for between arm differences were within a margin of ±5 points. Equivalence was also supported in adjusted analyses (Appendix Table 3). Interactions between randomization status and time were not observed (BAI: p≥0.57; CES-D: p≥0.07; IES: p≥0.26).
Table 2.
Mean anxiety, depression and test-related distress scores by randomization arm and time after APOE genotype disclosure.*
| AD Only (n=138) | AD+CAD (n=119) | Difference† (95% CI) | |
|---|---|---|---|
| 12 month outcomes | |||
| BAI‡ | 3.5 | 3.5 | 0.0 (−1.0 to 1.0) |
| CES-D§ | 6.4 | 7.1 | 0.7 (−1.0 to 2.4) |
| IES|| | 4.0 | 2.6 | −1.4 (−3.3 to 0.5) |
| 6 month outcomes | |||
| BAI | 2.9 | 3.0 | 0.1 (−0.7 to 1.0) |
| CES-D | 6.0 | 5.2 | −0.8 (−2.3 to 0.7) |
| IES | 4.1 | 3.5 | −0.6 (−2.5 to 1.4) |
| 6 week outcomes | |||
| BAI | 3.0 | 3.0 | 0.0 (−0.8 to 0.7) |
| CES-D | 5.7 | 5.3 | −0.4 (−1.8 to 1.0) |
| IES | 4.4 | 3.8 | −0.6 (−2.6 to 1.4) |
| Time-averaged outcomes | |||
| BAI | 3.1 | 3.2 | 0.0 (−0.7 to 0.7) |
| CES-D | 6.0 | 5.8 | −0.2 (−1.4 to 1.0) |
| IES | 4.2 | 3.3 | −0.9 (−2.6 to 0.8) |
Scores were estimated using generalized estimating equations with log link and gamma distribution, adjusting for corresponding baseline values and the genetic counselor providing disclosure.
Difference refers to the difference between scores in the randomization arms.
Scores on the Beck Anxiety Inventory (BAI) range from 0 to 63, with higher scores indicating greater anxiety.
Scores on the Center for Epidemiological Studies Depression Scale (CES-D) range from 0 to 60, with higher scores indicating greater depression.
Scores on the Impact of Event Scale (IES) range from 0 to 75, with higher scores indicating greater distress.
Results on psychological scales by APOE group are presented in Table 3. Anxiety and depression scores remained well below cut-offs for concern regardless of ε4 status, and 95% confidence bounds for mean differences between AD-only and AD+CAD groups for each APOE genotype were within a margin of ±5 points at all time points. However, among APOE ε4-positive participants, mean IES scores were lower 12 months after AD+CAD disclosure, as compared to AD-only, whereas in APOE ε4-negative participants mean IES scores did not differ (mean Δ=−4.8, 95%CI −8.6 to −1.0, ε4-positive; 0.6, 95%CI −1.1 to 2.2, ε4-negative; p-interaction=0.031). Differences by APOE status were also observed at 6 months, when anxiety was modestly lower after AD+CAD disclosure in ε4-positive participants and modestly higher in ε4-negative participants (mean Δ=−1.8, 95%CI −3.2 to −0.4, ε4-positive; 1.1, 95%CI 0.1 to 2.0, ε4-negative; p-interaction=0.004). Findings from analyses stratified by APOE status were supported in adjusted analyses (Appendix Table 4).
Table 3.
Mean anxiety, depression and test-related distress scores by randomization arm, APOE status, and time after APOE genotype disclosure.*
| ε4-negative | ε4-positive | p interact | |||||
|---|---|---|---|---|---|---|---|
| AD Only (n=87) | AD+ CAD (n=87) | Difference† (95% CI) | AD Only (n=51) | AD+ CAD (n=32) | Difference† (95% CI) | ||
| 12 month outcomes | |||||||
| BAI‡ | 2.8 | 3.6 | 0.9 (−0.2 to 2.0) | 4.8 | 3.1 | −1.7 (−3.5 to 0.1) | 0.048 |
| CES-D§ | 5.5 | 6.8 | 1.3 (−0.5 to 3.1) | 7.8 | 7.7 | −0.1 (−3.6 to 3.4) | 0.14 |
| IES|| | 2.1 | 2.7 | 0.6 (−1.1 to 2.2) | 7.1 | 2.3 | −4.8 (−8.6 to −1.0) | 0.031 |
| 6 month outcomes | |||||||
| BAI | 2.3 | 3.4 | 1.1 (0.1 to 2.0) | 3.8 | 2.0 | −1.8 (−3.2 to −0.4) | 0.004 |
| CES-D | 5.5 | 5.5 | 0.0 (−1.7 to 1.7) | 6.9 | 4.6 | −2.3 (−4.8 to 0.2) | 0.29 |
| IES | 2.4 | 3.0 | 0.7 (−1.3 to 2.6) | 7.0 | 4.7 | −2.3 (−5.9 to 1.4) | <0.001 |
| 6 week outcomes | |||||||
| BAI | 2.8 | 2.9 | 0.2 (−0.8 to 1.1) | 3.4 | 3.1 | −0.3 (−1.6 to 1.0) | 0.67 |
| CES-D | 5.9 | 5.6 | −0.3 (−2.1 to 1.5) | 5.3 | 4.6 | −0.7 (−2.7 to 1.3) | 0.60 |
| IES | 2.5 | 3.6 | 1.1 (−0.7 to 2.9) | 7.7 | 4.3 | −3.4 (−7.2 to 0.5) | 0.002 |
| Time-averaged outcomes | |||||||
| BAI | 2.6 | 3.3 | 0.7 (−0.1 to 1.5) | 4.0 | 2.7 | −1.3 (−2.3 to −0.2) | 0.006 |
| CES-D | 5.6 | 5.9 | 0.3 (−1.0 to 1.7) | 6.6 | 5.5 | −1.1 (−3.1 to 0.9) | 0.23 |
| IES | 2.3 | 3.1 | 0.8 (−0.7 to 2.3) | 7.2 | 3.6 | −3.6 (−7.0 to −0.2) | 0.005 |
Scores were estimated using generalized estimating equations with log link, gamma distribution and robust standard errors, adjusting for baseline values and the genetic counselor providing disclosure.
Difference refers to the mean scores among participants in the AD+CAD arms minus mean scores among participants in the AD-only arms.
Scores on the Beck Anxiety Inventory (BAI) range from 0 to 63, with higher scores indicating greater anxiety.
Scores on the Center for Epidemiological Studies Depression Scale (CES-D) range from 0 to 60, with higher scores indicating greater depression.
Scores on the Impact of Event Scale (IES) range from 0 to 75, with higher scores indicating greater distress.
Overall, 24% of study participants reported moderate anxiety, depression, or test-related distress at one or more follow-up time points, with no differences by AD-only/AD+CAD randomization over time (p=0.53). As in prior REVEAL Study trials (24, 25), mean IES scores were greater among APOE ε4-positive participants than APOE ε4-negative participants when only AD risk information was disclosed (12 month mean Δ=3.8, 95%CI 0.7 to 6.9) while differences in mean depression scores by ε4 status within AD-only arms were not observed (12 month mean Δ=1.6, 95%CI −0.9 to 4.0). Anxiety scores were higher among APOE ε4-positive than A-POE ε4-negative participants when only AD risk information was disclosed (12 month mean Δ=1.9, 95%CI 0.1 to 3.7). No differences were noted by ε4 status in AD+CAD arms.
Health Behavior Responses
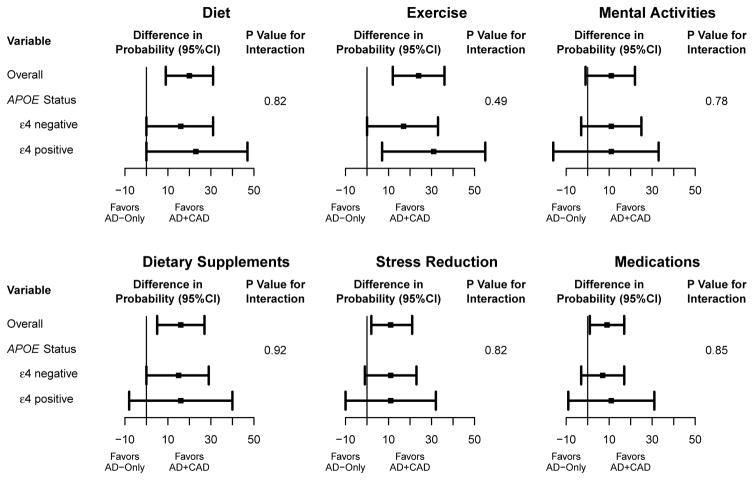
Among all participants, 57% reported changing at least one health behavior at 12 months in response to genetic risk disclosure. Percentages reporting specific health behavior changes are reported in Appendix Table 5. Participants in AD+CAD arms were more likely than participants in AD-only arms to report changes to most queried health behaviors with differences being independent of APOE genotype (Figure 2). Among the 36 participants who reported a medication change, 9 (25%) made a change related to CAD (e.g., blood pressure or cholesterol medications, fiber supplements). Secondary analyses also showed that participants who were APOE ε4-positive were more likely to report changes than participants who were APOE ε4-negative on all health behaviors outcomes (Appendix Table 5). Differences were not observed between AD+CAD and AD-only arms on 12 month aerobic activity scores (mean Δ=0.34, 95%CI −0.09 to 0.76) or strength and flexibility scores (mean Δ=−0.01, 95%CI −0.35 to 0.33). Of additional note, 33% of participants in the AD+CAD arms reported sharing results with a health professional, compared to 22% of participants in the AD-only arms (p=0.063).
Figure 2.
AD+CAD vs. AD-Only differences in the percent reporting health behavior changes 12 months after genetic risk disclosure.*
* Plots display between arm (AD+CAD – AD-Only) differences in the percentage of participants reporting a health behavior change. Estimates are from an analysis using logistic regression, accounting for APOE status, its interaction with pleiotropy randomization arm and the genetic counselor providing disclosure (except for stress reduction, where genetic counselor was omitted because some combinations of randomization status, APOE status, and genetic counselor had no events). Adjusted percentages are conditional probabilities estimated from the logistic model with all covariates set to their mean values (SAS lsmeans). P-values for interaction correspond to the p-values from the randomization arm x APOE status interaction terms. Unadjusted number of participants reporting changes to each health behavior: diet, 86; exercise, 91; mental activities, 76; dietary supplements, 71; stress reduction, 57; medications, 37.
DISCUSSION
We report a randomized trial of disclosing pleiotropic risk information during a genetic risk assessment for AD. Participants receiving AD risk plus secondary information about CAD risk responded equivalently to participants receiving only AD risk on primary outcomes of anxiety and depression, with no differences in mean scores at any time point and confidence intervals within conservative margins for clinical significance. However, participants at increased risk for disease (APOE ε4-positive) appeared to have experienced less test-related distress at 12 months if they also received CAD information. The vast majority of other studies in disclosure of genetic risk have shown no impact of genetic risk disclosure on general measures of mood, but occasional short-term increases in test-related distress among individuals at increased risk for disease (42). Our results build on those findings by suggesting that “positive pleiotropic disclosure,” i.e., return of unsolicited risk information about a modifiable condition like CAD, may reduce distress experienced when receiving risk information about a less readily modifiable condition like AD. These findings prompt the interesting speculation that worry about medically non-actionable genetic risk results may be mitigated by simultaneously providing actionable genetic risk results. Our study did not address “negative pleiotropic disclosure,” such as choosing to learn APOE genotype for CAD risk and incidentally discovering its implications for AD risk, which could have yielded different results.
Nearly every health behavior assessed was reportedly improved in response to AD+CAD information, regardless of APOE status. The statement about strategies to reduce coronary artery disease risk, given to all participants in the AD+CAD arms, may explain differences in reported changes in health behavior. However, participants receiving AD+CAD information tended to be more likely to report sharing results with a health professional, who may have in turn encouraged health behavior changes. Pleiotropic disclosure may also have prompted individuals who had been focused on AD to attend to a more modifiable and prevalent condition in CAD. APOE ε4-positive participants in both randomization arms were more likely to report changes to all health behaviors than APOE ε4-negative participants despite receiving education that highlighted a lack of proven AD risk-reducing options. It is possible that learning about an increased risk for AD and addressing pleiotropic outcomes are motivating individuals to be healthier in general rather than motivating steps to reduce risk for a specific disease.
Our participants were generally well-educated individuals who volunteered for genetic risk assessment for AD, were not representative of the general population and were more likely to have known about APOE-CAD associations independent of our study. Our study focused on pleiotropic information disclosure during single-gene testing for AD, and may not generalize to other conditions or to contexts like genomic sequencing that can explore broader sets of genetic variants and diseases. Our study excluded one individual with low cognitive testing score as well as one participant with severe depression, raising the possibility that results could be different among more vulnerable populations. We additionally omitted 33 randomized participants who dropped out of the study before being genotyped. Self-reported outcomes, particularly those measuring health behaviors, are subject to bias where participants respond in ways they expect investigators want them to respond (43). Some of our health behavior measures have not been validated and do not provide insight about whether changes were clinically meaningful, although our physical activity measure has demonstrated validity for older adults (38). Finally, clinical outcomes associated with health behavior measures (e.g., weight loss) were not assessed.
Our study examined only one strategy for communicating APOE-CAD associations. Because of questions about the strength of the APOE-CAD relationship at the time of our study, our disclosure statement deliberately omitted quantified risk estimates for CAD that may have made pleiotropic disclosure more impactful. Indeed, meta-analyses published during our study suggest that ε4 carriers’ increased CAD risk may be modest (23). Further complicating the issue, APOE may be associated with other neurological and ocular disorders (44). As the field transitions to technologies that identify a wider array of secondary and incidental genomic findings, laboratories and clinicians will need to make difficult decisions about what kinds of findings merit disclosure, as well as how to do so.
Nevertheless, our data support the safety of disclosing secondary, pleiotropic information about a modifiable condition such as CAD during genetic risk assessment for AD, and suggest the counter-intuitive inference that such disclosure may mitigate test-related distress among those who learn that they are at increased risk for not one, but two life-threatening conditions.
Supplementary Material
Acknowledgments
Primary Funding Source: National Human Genome Research Institute
This work was supported by NIH grants HG002213, HG006500, HD077671, HG006993, AG013846, RR000533, RR010284, and TR001102.
Footnotes
Trial Registration: ClinicalTrials.gov number NCT00462917 (http://clinicaltrials.gov/ct2/show/NCT00462917)
Contributor Information
Dr. Kurt D. Christensen, Brigham and Women’s Hospital, EC Alumnae Building, Suite 301, 41 Avenue Louis Pasteur, Boston, MA 02115.
Dr. J. Scott Roberts, University of Michigan School of Public Health, 3854 SPH I, 1415 Washington Heights, Ann Arbor, MI 48109.
Dr. Peter J. Whitehouse, Case Western Reserve University, University Foley Elderhealth Center, 12200 Fairhill Rd, Cleveland, OH 44120.
Dr. Charmaine D. M. Royal, Duke University, Office of Undergraduate Scholars and Fellows, Smith Warehouse, Room B209, 114 S. Buchanan Street, Box 90756, Durham, NC 27701.
Dr. Thomas O. Obisesan, Howard University Hospital, 2041 Georgia Ave NW, Towers Building 5000, Washington, DC 20060.
Dr. L. Adrienne Cupples, Boston University School of Public Health, 801 Massachusetts Ave, Boston, MA 02118.
Dr. Jacqueline A. Vernarelli, Fairfield University, 1073 North Benson Rd, Fairfield, CT 06824.
Dr. Deepak L. Bhatt, Brigham and Women’s Hospital, 75 Francis St, Boston, MA 02115.
Dr. Erin Linnenbringer, Washington University School of Medicine, Division of Public Health Sciences, Department of Surgery, 660 S Euclid Ave, Campus Box 8100, St. Louis, MO 63110.
Dr. Melissa B. Butson, Case Western Reserve University, University Foley Elderhealth Center, 12200 Fairhill Rd, Cleveland, OH 44120.
Ms. Grace-Ann Fasaye, Inova Fairfax Hospital Cancer Center, 3600 Joseph Siewick Dr, Fairfax, VA 22033.
Ms. Wendy R. Uhlmann, University of Michigan Medical School, Department of Human Genetics, 300 North Ingalls Bldg, NI3 A03, SPC 5419, Ann Arbor, MI 48109.
Ms. Susan Hiraki, GeneDx, 207 Perry Parkway, Gaithersburg, MD 20877.
Ms. Na Wang, Boston University School of Public Health, 801 Massachusetts Ave, CT340C, Boston, MA 02118.
Dr. Robert Cook-Deegan, Sanford School of Public Policy, Duke Box 90239, Durham, NC 27708.
Dr. Robert C. Green, Brigham and Women’s Hospital, EC Alumnae Building, Suite 301, 41 Avenue Louis Pasteur, Boston, MA 02115.
References
- 1.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–9. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green RC, Rehm HL, Kohane IS. Clinical genome sequencing. In: Ginsberg GS, Willard HF, editors. Genomic and Personalized Medicine. 2. San Diego: Academic Press; 2013. pp. 102–22. [Google Scholar]
- 3.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370:2418–25. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 4.Lee JW, Aminkeng F, Bhavsar AP, Shaw K, Carleton BC, Hayden MR, et al. The emerging era of pharmacogenomics: current successes, future potential, and challenges. Clin Genet. 2014;86:21–8. doi: 10.1111/cge.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korf BR. Integration of genomics into medical practice. Discov Med. 2013;16:241–8. [PubMed] [Google Scholar]
- 6.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med. 2015;17:68–9. doi: 10.1038/gim.2014.151. [DOI] [PubMed] [Google Scholar]
- 8.Burke W, Matheny Antommaria AH, Bennett R, Botkin J, Clayton EW, Henderson GE, et al. Recommendations for returning genomic incidental findings? We need to talk! Genet Med. 2013;15:854–9. doi: 10.1038/gim.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green RC, Lupski JR, Biesecker LG. Reporting genomic sequencing results to ordering clinicians: incidental, but not exceptional. JAMA. 2013;310:365–6. doi: 10.1001/jama.2013.41703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGuire AL, Joffe S, Koenig BA, Biesecker BB, McCullough LB, Blumenthal-Barby JS, et al. Ethics and genomic incidental findings. Science. 2013;340:1047–8. doi: 10.1126/science.1240156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Presidential Commision for the Study of Bioethical Issues. Anticipate and communicate ethical management of incidental and secondary findings in the clinical, research and direct-to-consumer contexts. 2013 doi: 10.1093/aje/kwu217. [DOI] [PubMed] [Google Scholar]
- 12.Janssens ACJW. The hidden harm behind the return of results from personal genome services: a need for rigorous and responsible evaluation. Genet Med. 2014 doi: 10.1038/gim.2014.169. [DOI] [PubMed] [Google Scholar]
- 13.Yu J-H, Harrell TM, Jamal SM, Tabor HK, Bamshad MJ. Attitudes of genetics professionals toward the return of incidental results from exome and whole-genome sequencing. Am J Hum Genet. 2014;95:77–84. doi: 10.1016/j.ajhg.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf SM, Annas GJ, Elias S. Patient autonomy and incidental findings in clinical genomics. Science. 2013 doi: 10.1126/science.1239119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson L, Goldsmith L, O’Connor A, Skirton H. Incidental findings in genetic research and clinical diagnostic tests: a systematic review. Am J Med Genet A. 2012;158A:3159–67. doi: 10.1002/ajmg.a.35615. [DOI] [PubMed] [Google Scholar]
- 16.Sivakumaran S, Agakov F, Theodoratou E, Prendergast JG, Zgaga L, Manolio T, et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet. 2011;89:607–18. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrikson NB, Burke W, Veenstra DL. Ancillary risk information and pharmacogenetic tests: social and policy implications. Pharmacogenomics J. 2008;8:85–9. doi: 10.1038/sj.tpj.6500457. [DOI] [PubMed] [Google Scholar]
- 18.Kocarnik JM, Fullerton SM. Returning pleiotropic results from genetic testing to patients and research participants. JAMA. 2014;311:795–6. doi: 10.1001/jama.2014.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wachbroit R. The question not asked: the challenge of pleiotropic genetic tests. Kennedy Inst Ethics J. 1998;8:131–44. doi: 10.1353/ken.1998.0013. [DOI] [PubMed] [Google Scholar]
- 20.Corbo RM, Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a ‘thrifty’ allele? Ann Hum Genet. 1999;63:301–10. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- 21.Morris JC. Dementia update 2005. Alzheimer Dis Assoc Disord. 2005;19:100–17. doi: 10.1097/01.wad.0000167923.56275.d8. [DOI] [PubMed] [Google Scholar]
- 22.Song Y, Stampfer MJ, Liu S. Meta-analysis: apolipoprotein E genotypes and risk for coronary heart disease. Ann Intern Med. 2004;141:137–47. doi: 10.7326/0003-4819-141-2-200407200-00013. [DOI] [PubMed] [Google Scholar]
- 23.Bennet AM, Di Angelantonio E, Ye Z, Wensley F, Dahlin A, Ahlbom A, et al. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA. 2007;298:1300–11. doi: 10.1001/jama.298.11.1300. [DOI] [PubMed] [Google Scholar]
- 24.Green RC, Christensen KD, Cupples LA, Relkin NR, Whitehouse PJ, Royal CDM, et al. A randomized non-inferiority trial of condensed protocols for genetic risk disclosure of Alzheimer’s disease. Alzheimers Dement. doi: 10.1016/j.jalz.2014.10.014. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green RC, Roberts JS, Cupples LA, Relkin NR, Whitehouse PJ, Brown T, et al. Disclosure of APOE genotype for risk of Alzheimer’s disease. N Engl J Med. 2009;361:245–54. doi: 10.1056/NEJMoa0809578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chao S, Roberts JS, Marteau TM, Silliman R, Cupples LA, Green RC. Health behavior changes after genetic risk assessment for Alzheimer disease: the REVEAL Study. Alzheimer Dis Assoc Disord. 2008;22:94–7. doi: 10.1097/WAD.0b013e31815a9dcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vernarelli JA, Roberts JS, Hiraki S, Chen CA, Cupples LA, Green RC. Effect of Alzheimer disease genetic risk disclosure on dietary supplement use. Am J Clin Nutr. 2010;91:1402–7. doi: 10.3945/ajcn.2009.28981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts JS, Cupples LA, Relkin NR, Whitehouse PJ, Green RC. Genetic risk assessment for adult children of people with Alzheimer’s disease: the Risk Evaluation and Education for Alzheimer’s Disease (REVEAL) Study. J Geriatr Psychiatry Neurol. 2005;18:250–5. doi: 10.1177/0891988705281883. [DOI] [PubMed] [Google Scholar]
- 29.Roberts JS, Chen CA, Uhlmann WR, Green RC. Effectiveness of a condensed protocol for disclosing APOE genotype and providing risk education for Alzheimer disease. Genet Med. 2012;14:742–8. doi: 10.1038/gim.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cupples LA, Farrer LA, Sadovnick AD, Relkin N, Whitehouse P, Green RC. Estimating risk curves for first-degree relatives of patients with Alzheimer’s disease: the REVEAL Study. Genet Med. 2004;6:192–6. doi: 10.1097/01.gim.0000132679.92238.58. [DOI] [PubMed] [Google Scholar]
- 31.Christensen KD, Roberts JS, Royal CDM, Fasaye G-A, Obisesan T, Cupples LA, et al. Incorporating ethnicity into genetic risk assessment for Alzheimer disease: the REVEAL Study experience. Genet Med. 2008;10:207–14. doi: 10.1097/GIM.0b013e318164e4cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teng EL, Chui HC. The Modified Mini-Mental State (3MS) Examination. J Clin Psychiatry. 1987;48:314–8. [PubMed] [Google Scholar]
- 33.Beck AT, Epstein N, Brown G, Steer RA. An inventory for measuring clinical anxiety: psychometric properties. J Consult Clin Psychol. 1988;56:893–7. doi: 10.1037//0022-006x.56.6.893. [DOI] [PubMed] [Google Scholar]
- 34.Radloff LS. The CES-D Scale: A self report depression scale for research in the general population. Appl Psychol Meas. 1977;1:385–401. [Google Scholar]
- 35.Santor DA, Zuroff DC, Ramsay JO, Cervantes P, Palacios J. Examining scale discriminability in the BDI and CES-D as a function of depressive severity. Psychol Assess. 1995;7:131–9. [Google Scholar]
- 36.Horowitz M, Wilner N, Alvarez W. Impact of Event Scale: a measure of subjective stress. Psychosom Med. 1979;41:209–18. doi: 10.1097/00006842-197905000-00004. [DOI] [PubMed] [Google Scholar]
- 37.Payne K, Nicholls S, McAllister M, MacLeod R, Donnai D, Davies LM. Outcome measurement in clinical genetics services: a systematic review of validated measures. Value Health. 2008;11:497–508. doi: 10.1111/j.1524-4733.2007.00259.x. [DOI] [PubMed] [Google Scholar]
- 38.Topolski TD, LoGerfo J, Patrick DL, Williams B, Walwick J, Patrick MB. The Rapid Assessment of Physical Activity (RAPA) among older adults. Prev Chronic Dis. 2006;3:A118. [PMC free article] [PubMed] [Google Scholar]
- 39.Julious SA. Sample sizes for clinical trials with normal data. Stat Med. 2004;23:1921–86. doi: 10.1002/sim.1783. [DOI] [PubMed] [Google Scholar]
- 40.Jones B, Jarvis P, Lewis JA, Ebbutt AF. Trials to assess equivalence: the importance of rigorous methods. BMJ. 1996;313:36–9. doi: 10.1136/bmj.313.7048.36. Erratum, BMJ 1996, 313, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker E, Nowacki AS. Understanding equivalence and noninferiority testing. J Gen Intern Med. 2011;26:192–6. doi: 10.1007/s11606-010-1513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heshka JT, Palleschi C, Howley H, Wilson B, Wells PS. A systematic review of perceived risks, psychological and behavioral impacts of genetic testing. Genet Med. 2008;10:19–32. doi: 10.1097/GIM.0b013e31815f524f. [DOI] [PubMed] [Google Scholar]
- 43.Orne MT. On the social psychology of the psychological experiment: with particular reference to demand characteristics and their implications. Am Psychol. 1962;17:776–83. [Google Scholar]
- 44.Online Mendelian Inheritance in Man OMIM®. MIM Number: 107741. APOLIPOPROTEIN E; APOE [Internet] Baltimore, MD: Johns Hopkins University; 2015. cited 2015 Apr 7 Available from: http://www.omim.org/entry/107741. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.