Abstract
Activation induced deaminase is the single B cell specific factor mediating class switch recombination and somatic hypermutation. Numerous studies have shown that AID preferentially targets Ig substrates and also attacks non-Ig substrates to create DNA damage that contributes to lymphomagenesis. AID targeting to Ig loci is linked to transcription but the mechanism governing this process has been obscure. Here we discuss research that illustrates the connection between AID targeting to DNA substrates and transcription processes to reveal rules governing the specificity of AID attack. These observations are woven together to provide a integrated view of AID function and a surprising linkage with global regulation of gene expression.
Keywords: B lymphocytes, Class switch recombination, Somatic hypermutation, Activation induced deaminase
Humoral immunity is mediated in B cells by antigen receptors (BCR) that are composed of immunoglobulin (Ig) heavy (H) and light (L) chains. The antigen receptor component of Ig is assembled from multiple gene segments via V(D)J recombination during early B cell development in the bone marrow and is mediated by the RAG1 and RAG2 recombinases [1]. In the peripheral lymphoid organs, B cells become activated by antigen and undergo somatic hypermutation (SHM) and class switch recombination (CSR) and upon terminal differentiation secrete Ig as antibody [2,3]. Antibody repertoires are diversified at high frequency during somatic hypermutation (SHM), by introducing mutations into Igh and Igl V(D)J exons. SHM occurs in germinal centers of the peripheral lymphoid organs where mutations are selected to produce BCR with increased affinity. Constant (CH) region genes encode the C-terminal domains of the Ig chains and determine IgH effector activities. IgH effector function is diversified through CSR, while retaining the original antigen binding specificity arising during V(D)J recombination. Activation induced deaminase (AID), a cytosine deaminase is essential for initiating both SHM and (CSR) in mature B cells [4]**.
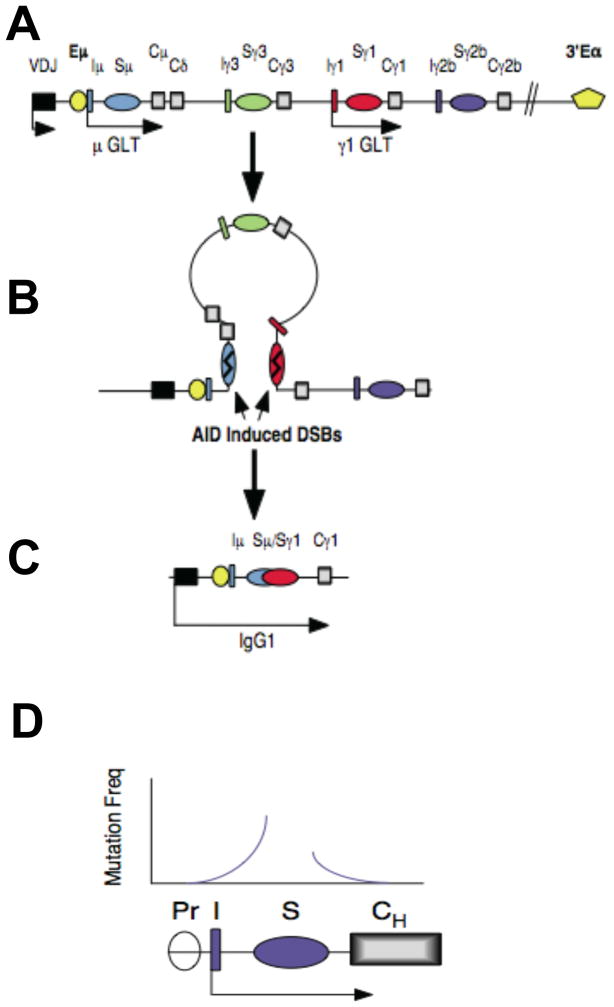
The mouse Igh locus includes eight CH genes, encoding μ, δ, γ3, γ1, γ2b, γ2a, ε, and α chains and each is paired with repetitive switch (S) DNA (with the exception of Cδ). The CH region genomic area spans 220 kb and is flanked by the intronic Eμ and 3′Eα enhancers [5]. CSR is focused on S regions and involves intra-chromosomal deletional rearrangements that replace the initial Cμ with a downstream CH region gene (fig. 1). Prior to and during CSR, a S-S synaptosome is formed to facilitate proximity between the donor Sμ and a downstream acceptor S region [6–9]. Formation of the S-S synaptosome is critically dependent on long range chromatin interactions that are tethered by key transcriptional elements (reviewed in [10]). AID initiates CSR by deaminating cytosines in S regions in donor Sμ and a downstream acceptor S region (Box 1) [11]. AID dependent DNA damage is focused to highly degenerate WRC hotspot motifs (W=A/T, R=A/G) that are found in V genes and at high density in S regions (reviewed in [12]). Conversion of AID induced lesions to DNA double strand breaks by general DNA repair factors has been extensively reviewed [2,3,11–17]. Degeneracy of the AID hotspot motif raises questions regarding the mechanism by which AID is targeted to its Ig substrates.
Figure 1. The looping-out and deletion model of Ig switch recombination.
A) A partial schematic map of Igh locus before CSR is shown not to scale. A productive V(D)J rearrangement has occurred allowing expression of the γ and ™ IgH chains. Intact Sμ and Sγ1 are separated by approximately 70 kb. Stimulation of B cells with antigen or mitogen induces germline transcription through the Iγ1-Sγ1-Cγ1 region prior to recombination. B) The Sμ and Sγ1 regions are aligned causing the intervening genomic DNA to form a loop. C) A reciprocal crossover between Sμ and Sγ1 results in the formation of a new hybrid transcriptional unit containing the original VDJ exons contiguous with Cγ1 and the formation of hybrid Sμ/Sγ1 molecules. D) A schematic diagram depicts a generic I-S-CH region downstream of the GLT promoter (Pr). Above the diagram a summary of mutation frequencies 5′ and 3′ of the S region is shown.
Box 1.
AID introduces DNA damage by converting deoxycytidine (dC) to deoxyuracil (dU) [14]. The AID initiated DNA lesions are processed by engagement with base excision repair (BER) and mismatch repair (MMR) pathways to create mutations required for SHM and DNA double strand break (DSB) intermediates that consumed in CSR (reviewed in [2,3,11,14]). The observations that AID dependent dU residues are detected in the 5′Sμ region [99] and that uracil DNA glycosylase (UNG) is required for formation of double strand breaks (DSB) and mutations demonstrate that AID deamination initiates SHM and CSR [100,101]. Conversion of AID induced lesions to DNA double strand breaks is mediated by general DNA repair factors [17].
Transcription is a hallmark of SHM and CSR and transcriptional elements are candidates for providing specificity to AID targeting [5,18]. V region transcription initiates from a promoter 5′ proximal to the rearranged V(D)J exon and terminates 3′ of the CH region exons. CSR is focused to specific S regions by differential activation of germline transcription [10,19]. CH gene transcription units are comprised of the noncoding intervening (I) exon, an S region and a CH coding region. Germline transcription initiates at a transcription start site (TSS) 5′ of each I exon, proceeds through the S region and terminates downstream of the corresponding CH gene (fig. 1A). The V(D)J mutation profile has a sharp 5′ boundary ~120 bp downstream of the TSS and a less defined 3′ border ~1 kb downstream of the promoter. Alteration of the V region promoter position displaces transcription initiation and perturbs the mutation distribution [20*,21] thereby linking induction of AID dependent mutations with transcription. Similarly, AID induced DNA lesions in S regions begin ~150 bp downstream of the I exon TSS [22]. A recent study shows that sequence intrinsic features target AID dependent DNA lesions to Ig templates [23]. Although AID targeting to Ig substrates requires transcription, the unique transcriptional features that determine preferential AID attack at Ig templates have been difficult to discern.
AID induced double strand breaks (DSBs) in normal B cells occur at hundreds of non-Ig sites many of which are syntenic with sites of translocations, deletions, and amplifications found in human B cell lymphomas [24]. Physiological levels of AID in GC B cells have been linked to deamination of a large cohort of non-Ig genes [25*–28] where the mutation rate is 20–100 fold lower than at Ig loci [26]**. The findings that SHM and CSR are tightly linked to transcription [20*,29**,30] and AID interacts with RNA Pol II (RNAP II) [27*,31] have led to the notion that transcription and RNAP II-associated proteins might facilitate the binding of AID to target DNA sequences. Based on provocative new studies, we address three important interrelated questions regarding AID targeting. What is the mechanism by which AID preferentially targets Ig substrates and its corollary, how is AID attack at non-Ig substrates directed? Finally, is there an adaptive advantage for AID attack at non-Ig templates? We focus primarily on CSR as this area has been the most intensively investigated. Recent work implies that AID attack on non-Ig substrates may mediate functional outcomes that contribute to a biological “jackpot”.
Transcribed S regions are specialized targets of AID
S regions are repetitive, nonidentical, 1–12 kb long and guanine rich on the nontemplate strand [32]. Deletion, inversion or replacement of S regions reduces CSR frequency indicating that S regions are specialized targets of CSR [33–36]. However, CSR S/S junctions are notable for their lack of consensus sequence or homology originating from site specific or homologous recombination. The degeneracy of the S region repeats and the absence of discernable recombination signal motifs have led to models in which higher order structures provide recognition motifs for the CSR machinery. In vitro transcription studies indicated that the looped out ssDNA nontemplate strand can assume specialized structures including stem loops [37], four-stranded G quartets [38] and R-loops (Box 2) [39,40]. In vivo studies confirm that transcription through S regions in vivo creates G quartets [41,42] and generates long stretches of R-loops that form on the nontemplate strand [33,43*,44] and that enhance CSR efficiency.
Box 2.
R-loops are composed of RNA:DNA hybrids in which the nontemplate ssDNA strand is looped out while the template strand is stably annealed to nascent S region RNA transcripts [43,102]. The presence of R-loops within a transcription unit can cause RNAP II stalling [103]. Frequent R-loop mediated RNAP II stalling may account, at least in part, for enrichment of initiating RNAP II occupancy in S regions that in turn mediates the introduction of activating histone modifications and increased chromatin accessibility [45,104]. Accordingly, targeted inversion of an S region leads to loss of R-loop formation and reduction of CSR frequency [33]. Furthermore, S region deletion leads to reduced RNAP II occupancy at flanking sites [45,104] and loss of activating histone marks [45]. The enrichment of initiating RNAP II at promoter distal sites provides a plausible explanation for increased levels of histone activating marks and chromatin accessibility throughout the S region. Biochemical studies indicated that transcription generated R-loops within dsDNA permit AID directed deamination on the ssDNA nontemplate strand while the template strand remained blocked by the nascent RNA transcript (reviewed in [10,11]).
Epigenetic studies indicate that the initiating form of RNAP II phosphortylated on serine 5 (p-ser5) and activating histone marks decorate the length of transcribed S regions [45*,46]. In contrast, genome wide studies show that transcriptionally active genes are associated with promoter proximal enrichment of RNAP II p-ser5 coupled with activating histone modifications [47–51]. R-loops within a transcription unit can act as a structural impediment for RNAP II elongation and cause stalling (reviewed in [52]). S region inversion led to loss of R-loops and reduced CSR in vivo thereby linking R-loops and CSR [33]. However, the contribution of RNAP II stalling in S regions to the CSR reaction has remained unclear.
AID specifically binds to components of the transcription machinery
Several AID binding proteins that facilitate association of AID with transcribed S DNA and modulate CSR have been identified in genetic screens, including the adaptor 14-3-3 protein [53] and the RNA processing and/or splicing factors, polypyrimidine-tract binding protein (PTBP2) [54], RNAP II stalling cofactor, Spt5 [27] and the 11-subunit cellular noncoding RNA 3′–5′ exonucleolytic processing complex, RNA exosome [55]**. SPT5 collaborates with the DSIF complex and negative elongation factor (NEF) to stall RNAP II p-ser5 at promoter proximal sites [56] and links RNAP II to splicing factors [57], capping enzyme [57,58], and the RNA exosome (reviewed in [52]). The RNA exosome contains 3′–5′ exoribonucleases that process structural RNA, degrade improperly processed pre-mRNAs and some long noncoding (lnc) RNAs of which GLT RNA hybridized to the S region in an R-loop is an example [52,59]. The RNA exosome interacts indirectly with RNAP II via SPT5 and SPT6 [55]**. Deletion of PTBP2, SPT5 and RNA exosome components impair CSR and link transcription processes with AID recruitment and function at S DNA (reviewed in [11]). These observations directly demonstrate that the transcriptional machinery associated with RNAP II stalling, the presence of RNA:DNA hybrid structures as impediments to RNAP II elongation and the release of those obstacles by means of the RNA exosome are integral to targeting AID to S region DNA.
A recent provocative study has identified another regulatory feature contributing to AID targeting specificity that is directly dependent on expression of the long noncoding GLT RNA from transcribed S regions. Evidence indicates that AID functions as an RNA binding protein with specificity for S region GLTs through G-quadruplexes structures and is required for CSR [42]. Indeed, AID is one of 12 members of the APOBEC family of DNA/RNA cytidine deaminases and has recently been shown to mutate small RNA genes when expressed in yeast [60] suggesting a role for RNA in the recruitment of AID to S regions. Furthermore, short RNA segments have been shown to function as specific guides for nuclease modification of the genome [61] and to regulate DNA rearrangements in ciliates [62]. The requirement for G rich S region guide-RNAs for AID targeting implies that AID off target genes might also express non-coding RNA that functions in a similar capacity. Thus, AID targeting to Ig loci is dependent on transcription machine components.
AID targeting and mistargeting to genome wide substrates
Mistargeting of AID to non-Ig substrates has been implicated in the pathogenesis of B cell lymphomas and chromosomal translocations [63,64]. In the Western world, the vast majority of lymphomas arise in B cells that actively engage in SHM and CSR [65] that in turn generate recurrent chromosomal translocations [66]. Normal mature B cells are particularly prone to dynamic AID dependent chromosomal translocations that juxtapose Ig genes and proto-oncogenes. The high frequency of these events allows detection in the absence of selection (reviewed in [17]). The preferential focus of AID to Ig loci and the nonrandom aspect of AID attack on non-Ig genes implies a determinant set of rules governing AID targeting. Recent GRO-seq studies indicate that most AID induced off-target translocations occur at defined regions of target genes in which sense and anti-sense transcription converge [67]**. Strikingly, convergent transcription is due to antisense transcription originating from super-enhancers within sense transcribed gene bodies [67]. Super-enhancers are clusters of enhancers that have prominent roles in cell type specific processes [68,69] and express relatively high levels of enhancer RNAs (eRNA) [70]. Super-enhancers have the propensity to associate in three dimensional nuclear space via long range chromatin interactions [71] that are mediated by lineage specific transcription factors (TFs) [70]. Hence, lineage specific TFs indirectly facilitate AID initiated chromosomal translocations [67]**.
The RNA exosome is an RNA surveillance complex that degrades a variety of non-coding RNAs (reviewed in [52]. RNA species arising in response to exosome subunit deficiencies have revealed the identities of non-Ig genes and intergenic regions that are the focus of AID mediated translocations [52,72]. Several types of non-coding exosome substrate RNAs accumulate in the transcriptomes of exosome-deficient B cells including, transcription start site (TSS)-associated antisense transcripts (xTSS-RNAs). The xTSS-RNAs are divergently transcribed from cognate coding gene transcripts that accumulate R-loops, AID-mediated mutations, and/or are frequent translocation partners of Igh DNA DSBs in B cells [59]. A subset of xTSS-RNAs originate as overlapping sense and antisense x-eRNAs at super-enhancers and are located at recurrent translocation hotspots [72]. It is not clear whether the characterization of these transcripts as divergent represents a real or semantic difference with the “convergent” transcripts defined by Meng et al. described above. Collectively, these findings begin to define a set of predictive transcriptional features that characterize Ig– and non-Ig substrates of AID.
Noncanonical AID Expression
The prevalence of AID off-target sites implies that this phenomenon may have adaptive advantages. Low levels of AID have been detected in a number of tissues and cell types including early stage B cells [73,74], and in pluripotent cells such as embryonic stem cells [75]* and spermatocytes [76] where it may have an unanticipated function in cellular reprogramming [77–80]. AID expression can be upregulated in some instances of infection [81] and under conditions of chronic inflammation that promote tumorigenesis [81–85]. Intriguingly, inflammatory stimuli (LPS) robustly induce AID in primary pro-B cells and Abelson transformed B cell lines leading to CSR prior to V(D)J joining [86] and mutations associated with acute lymphoblastic leukemia [87,88]. Strikingly, AID expression in transitional B cells appears to mediate tolerance [89,90] by a B cell intrinsic mechanism [91]. It is intriguing to speculate that by analogy to new gene expression linked to RAG recombinase induced DNA DSBs [92], AID induced DSBs direct new gene expression leading to functional outcomes including immune tolerance.
Role for AID in DNA Demethylation
One provocative explanation for AID expression during development and in early B cells is related to DNA demethylation. DNA methylation is an epigenetic modification that is considered central to the establishment and maintenance of stable cellular identities (reviewed in [93]). The processes by which methylation is removed from cytosine were unclear until recent studies indicated active modes of DNA demethylation that involve modification of the meC base coupled to DNA repair. One pathway proceeds through oxidation catalyzed by the TET (ten eleven translocation) enzymes [93,94]. A second pathway uses AID, which promotes DNA demethylation through direct deamination of meC to thymidine [75]*, and subsequent repair of the resultant T:G mismatch by classical repair pathways [77,79,80,95]. Evidence suggests that AID’s demethylation activity is required for reprogramming in zebrafish embryos [80] and in mice [79,96]. AID interacts with and demethylates the promoters of the OCT4 and NANOG genes during reprogramming of human fibroblasts fused to mouse ES cells [78]. In mice, DNA demethylation is mediated by base excision repair (BER) through AID/Apobec deamination of 5meC to thymidine followed by G:T mismatch repair by TDG [75*,79,95] (Box 1). Notably, differentially methylated cytosines between naïve and GC B cells are enriched in non-Ig genes that are targeted by AID for SHM, and these genes form networks required for B cell development and proliferation [97]. Thus, emerging evidence implies that genome wide DNA demethylation by AID may be an important mechanism toward global regulation of gene expression programs [98].
Concluding Thoughts
Recent discoveries indicate that AID targeting to Ig and non-Ig loci is highly integrated with the transcription machinery and non-coding RNA biogenesis and degradation implying that off-target events are regulated and might be adaptive. Parallel studies link AID to active genome wide DNA demethylation. The final link in this chain is a new study that shows that gene loci hotspots for AID dependent mutagenesis are also DNA demethylated and their expression is networked. This raises the intriguing possibility that what we had previously considered AID off-targeting is actually integral to functional gene regulation. More work is necessary to define the molecular mechanism mediating apparently non-canonical AID effects at non-Ig loci.
Highlights.
AID is guided by non-coding germline transcipts to switch region targets
Integration of transcription and AID targeting with class switch recombination
AID deamination function intersects with global DNA demethylation
AID regulation of genomic DNA methylation may regulate gene expression
Acknowledgments
This work was supported in part by the National Institutes of Health (R01AI121286, R21AI117687, R21AI117687) to A.L.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nature reviews Immunology. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 2.Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 3.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Perlot T, Alt FW. Cis-regulatory elements and epigenetic changes control genomic rearrangements of the IgH locus. Adv Immunol. 2008;99:1–32. doi: 10.1016/S0065-2776(08)00601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW, Cogne M, Pinaud E, Kenter AL. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldman S, Achour I, Wuerffel R, Kumar S, Gerasimova T, Sen R, Kenter AL. Constraints Contributed by Chromatin Looping Limit Recombination Targeting during Immunoglobulin Class Switch Recombination. J Immunol. 2015 doi: 10.4049/jimmunol.1401170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sellars M, Reina-San-Martin B, Kastner P, Chan S. Ikaros controls isotype selection during immunoglobulin class switch recombination. J Exp Med. 2009;206:1073–1087. doi: 10.1084/jem.20082311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong J, Panchakshari RA, Zhang T, Zhang Y, Hu J, Volpi SA, Meyers RM, Ho YJ, Du Z, Robbiani DF, et al. Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature. 2015;525:134–139. doi: 10.1038/nature14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenter AL. AID targeting is dependent on RNA polymerase II pausing. Semin Immunol. 2012;24:281–286. doi: 10.1016/j.smim.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthews AJ, Zheng S, DiMenna LJ, Chaudhuri J. Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair. Adv Immunol. 2014;122:1–57. doi: 10.1016/B978-0-12-800267-4.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maul RW, Gearhart PJ. AID and somatic hypermutation. Adv Immunol. 2010;105:159–191. doi: 10.1016/S0065-2776(10)05006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chahwan R, Edelmann W, Scharff MD, Roa S. AIDing antibody diversity by error-prone mismatch repair. Seminars in immunology. 2012;24:293–300. doi: 10.1016/j.smim.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 15.Saribasak H, Rajagopal D, Maul RW, Gearhart PJ. Hijacked DNA repair proteins and unchained DNA polymerases. Philos Trans R Soc Lond B Biol Sci. 2009;364:605–611. doi: 10.1098/rstb.2008.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 17.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storb U. Why does somatic hypermutation by AID require transcription of its target genes? Adv Immunol. 2014;122:253–277. doi: 10.1016/B978-0-12-800267-4.00007-9. [DOI] [PubMed] [Google Scholar]
- 19.Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 20.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 21.Bachl J, Carlson C, Gray-Schopfer V, Dessing M, Olsson C. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J Immunol. 2001;166:5051–5057. doi: 10.4049/jimmunol.166.8.5051. [DOI] [PubMed] [Google Scholar]
- 22.Xue K, Rada C, Neuberger MS. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J Exp Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeap LS, Hwang JK, Du Z, Meyers RM, Meng FL, Jakubauskaite A, Liu M, Mani V, Neuberg D, Kepler TB, et al. Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell. 2015;163:1124–1137. doi: 10.1016/j.cell.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staszewski O, Baker RE, Ucher AJ, Martier R, Stavnezer J, Guikema JE. Activation-induced cytidine deaminase induces reproducible DNA breaks at many non-Ig Loci in activated B cells. Mol Cell. 2011;41:232–242. doi: 10.1016/j.molcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 26.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 27.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, Reina San-Martin B, Barreto V, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stavnezer-Nordgren J, Sirlin S. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. EMBO J. 1986;5:95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yancopoulos GD, DePinho RA, Zimmerman KA, Lutzker SG, Rosenberg N, Alt FW. Secondary genomic rearrangement events in pre-B cells: VHDJH replacement by a LINE-1 sequence and directed class switching. EMBO J. 1986;5:3259–3266. doi: 10.1002/j.1460-2075.1986.tb04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 32.Gritzmacher CA. Molecular aspects of heavy-chain class switching. Crit Rev Immunol. 1989;9:173–200. [PubMed] [Google Scholar]
- 33.Shinkura R, Tian M, Smith M, Chua K, Fujiwara Y, Alt FW. The influence of transcriptional orientation on endogenous switch region function. Nat Immunol. 2003;4:435–441. doi: 10.1038/ni918. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharya P, Wuerffel R, Kenter AL. Switch region identity plays an important role in Ig class switch recombination. J Immunol. 2010;184:6242–6248. doi: 10.4049/jimmunol.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zarrin AA, Tian M, Wang J, Borjeson T, Alt FW. Influence of switch region length on immunoglobulin class switch recombination. Proc Natl Acad Sci U S A. 2005;102:2466–2470. doi: 10.1073/pnas.0409847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khamlichi AA, Glaudet F, Oruc Z, Denis V, Le Bert M, Cogne M. Immunoglobulin class-switch recombination in mice devoid of any S mu tandem repeat. Blood. 2004;103:3828–3836. doi: 10.1182/blood-2003-10-3470. [DOI] [PubMed] [Google Scholar]
- 37.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 38.Dempsey LA, Sun H, Hanakahi LA, Maizels N. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, A role for G-G pairing in immunoglobulin switch recombination. J Biol Chem. 1999;274:1066–1071. doi: 10.1074/jbc.274.2.1066. [DOI] [PubMed] [Google Scholar]
- 39.Reaban ME, Griffin JA. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature. 1990;348:342–344. doi: 10.1038/348342a0. [DOI] [PubMed] [Google Scholar]
- 40.Reaban ME, Lebowitz J, Griffin JA. Transcription induces the formation of a stable RNA. DNA hybrid in the immunoglobulin alpha switch region. J Biol Chem. 1994;269:21850–21857. [PubMed] [Google Scholar]
- 41.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng S, Vuong BQ, Vaidyanathan B, Lin JY, Huang FT, Chaudhuri J. Non-coding RNA Generated following Lariat Debranching Mediates Targeting of AID to DNA. Cell. 2015;161:762–773. doi: 10.1016/j.cell.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 44.Zhang ZZ, Pannunzio NR, Han L, Hsieh CL, Yu K, Lieber MR. The strength of an Ig switch region is determined by its ability to drive R loop formation and its number of WGCW sites. Cell Rep. 2014;8:557–569. doi: 10.1016/j.celrep.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Whang N, Wuerffel R, Kenter AL. AID-dependent histone acetylation is detected in immunoglobulin S regions. J Exp Med. 2006;203:215–226. doi: 10.1084/jem.20051774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 49.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pefanis E, Basu U. RNA Exosome Regulates AID DNA Mutator Activity in the B Cell Genome. Adv Immunol. 2015;127:257–308. doi: 10.1016/bs.ai.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Z, Fulop Z, Wu G, Pone EJ, Zhang J, Mai T, Thomas LM, Al-Qahtani A, White CA, Park SR, et al. 14-3-3 adaptor proteins recruit AID to 5′-AGCT-3′-rich switch regions for class switch recombination. Nat Struct Mol Biol. 2010;17:1124–1135. doi: 10.1038/nsmb.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nowak U, Matthews AJ, Zheng S, Chaudhuri J. The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nat Immunol. 2011;12:160–166. doi: 10.1038/ni.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pei Y, Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J Biol Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 58.Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pefanis E, Wang J, Rothschild G, Lim J, Chao J, Rabadan R, Economides AN, Basu U. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014;514:389–393. doi: 10.1038/nature13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor BJ, Wu YL, Rada C. Active RNAP pre-initiation sites are highly mutated by cytidine deaminases in yeast, with AID targeting small RNA genes. Elife. 2014;3:e03553. doi: 10.7554/eLife.03553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, 3rd, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 65.Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nature reviews Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 66.Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- 67.Meng FL, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon KR, Meyers RM, Amor C, Wasserman CR, Neuberg D, et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell. 2015;58:362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ing-Simmons E, Seitan VC, Faure AJ, Flicek P, Carroll T, Dekker J, Fisher AG, Lenhard B, Merkenschlager M. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res. 2015;25:504–513. doi: 10.1101/gr.184986.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–789. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han JH, Akira S, Calame K, Beutler B, Selsing E, Imanishi-Kari T. Class switch recombination and somatic hypermutation in early mouse B cells are mediated by B cell and Toll-like receptors. Immunity. 2007;27:64–75. doi: 10.1016/j.immuni.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mao C, Jiang L, Melo-Jorge M, Puthenveetil M, Zhang X, Carroll MC, Imanishi-Kari T. T cell-independent somatic hypermutation in murine B cells with an immature phenotype. Immunity. 2004;20:133–144. doi: 10.1016/s1074-7613(04)00019-6. [DOI] [PubMed] [Google Scholar]
- 75.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 76.Schreck S, Buettner M, Kremmer E, Bogdan M, Herbst H, Niedobitek G. Activation-induced cytidine deaminase (AID) is expressed in normal spermatogenesis but only infrequently in testicular germ cell tumours. J Pathol. 2006;210:26–31. doi: 10.1002/path.2014. [DOI] [PubMed] [Google Scholar]
- 77.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhutani N, Decker MN, Brady JJ, Bussat RT, Burns DM, Corbel SY, Blau HM. A critical role for AID in the initiation of reprogramming to induced pluripotent stem cells. FASEB J. 2013;27:1107–1113. doi: 10.1096/fj.12-222125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matsumoto Y, Marusawa H, Kinoshita K, Endo Y, Kou T, Morisawa T, Azuma T, Okazaki IM, Honjo T, Chiba T. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat Med. 2007;13:470–476. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- 82.Endo Y, Marusawa H, Kinoshita K, Morisawa T, Sakurai T, Okazaki IM, Watashi K, Shimotohno K, Honjo T, Chiba T. Expression of activation-induced cytidine deaminase in human hepatocytes via NF-kappaB signaling. Oncogene. 2007;26:5587–5595. doi: 10.1038/sj.onc.1210344. [DOI] [PubMed] [Google Scholar]
- 83.Endo Y, Marusawa H, Kou T, Nakase H, Fujii S, Fujimori T, Kinoshita K, Honjo T, Chiba T. Activation-induced cytidine deaminase links between inflammation and the development of colitis-associated colorectal cancers. Gastroenterology. 2008;135:889–898. 898 e881–883. doi: 10.1053/j.gastro.2008.06.091. [DOI] [PubMed] [Google Scholar]
- 84.Komori J, Marusawa H, Machimoto T, Endo Y, Kinoshita K, Kou T, Haga H, Ikai I, Uemoto S, Chiba T. Activation-induced cytidine deaminase links bile duct inflammation to human cholangiocarcinoma. Hepatology. 2008;47:888–896. doi: 10.1002/hep.22125. [DOI] [PubMed] [Google Scholar]
- 85.Kou T, Marusawa H, Kinoshita K, Endo Y, Okazaki IM, Ueda Y, Kodama Y, Haga H, Ikai I, Chiba T. Expression of activation-induced cytidine deaminase in human hepatocytes during hepatocarcinogenesis. Int J Cancer. 2007;120:469–476. doi: 10.1002/ijc.22292. [DOI] [PubMed] [Google Scholar]
- 86.Kumar S, Wuerffel R, Achour I, Lajoie B, Sen R, Dekker J, Feeney AJ, Kenter AL. Flexible ordering of antibody class switch and V(D)J joining during B-cell ontogeny. Genes Dev. 2013;27:2439–2444. doi: 10.1101/gad.227165.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Swaminathan S, Klemm L, Park E, Papaemmanuil E, Ford A, Kweon SM, Trageser D, Hasselfeld B, Henke N, Mooster J, et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16:766–774. doi: 10.1038/ni.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meyers G, Ng YS, Bannock JM, Lavoie A, Walter JE, Notarangelo LD, Kilic SS, Aksu G, Debre M, Rieux-Laucat F, et al. Activation-induced cytidine deaminase (AID) is required for B-cell tolerance in humans. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11554–11559. doi: 10.1073/pnas.1102600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuraoka M, Holl TM, Liao D, Womble M, Cain DW, Reynolds AE, Kelsoe G. Activation-induced cytidine deaminase mediates central tolerance in B cells. Proc Natl Acad Sci U S A. 2011;108:11560–11565. doi: 10.1073/pnas.1102571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cantaert T, Schickel JN, Bannock JM, Ng YS, Massad C, Oe T, Wu R, Lavoie A, Walter JE, Notarangelo LD, et al. Activation-Induced Cytidine Deaminase Expression in Human B Cell Precursors Is Essential for Central B Cell Tolerance. Immunity. 2015 doi: 10.1016/j.immuni.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bredemeyer AL, Helmink BA, Innes CL, Calderon B, McGinnis LM, Mahowald GK, Gapud EJ, Walker LM, Collins JB, Weaver BK, et al. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 2008;456:819–823. doi: 10.1038/nature07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Franchini DM, Schmitz KM, Petersen-Mahrt SK. 5-Methylcytosine DNA demethylation: more than losing a methyl group. Annu Rev Genet. 2012;46:419–441. doi: 10.1146/annurev-genet-110711-155451. [DOI] [PubMed] [Google Scholar]
- 95.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Palermo A, Doyonnas R, Bhutani N, Pomerantz J, Alkan O, Blau HM. Nuclear reprogramming in heterokaryons is rapid, extensive, and bidirectional. FASEB J. 2009;23:1431–1440. doi: 10.1096/fj.08-122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dominguez PM, Teater M, Chambwe N, Kormaksson M, Redmond D, Ishii J, Vuong B, Chaudhuri J, Melnick A, Vasanthakumar A, et al. DNA Methylation Dynamics of Germinal Center B Cells Are Mediated by AID. Cell Rep. 2015;12:2086–2098. doi: 10.1016/j.celrep.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Illingworth RS, Bird AP. CpG islands--‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 99.Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, Gramlich HS, Schatz DG, Woodgate R, Wilson DM, 3rd, et al. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat Immunol. 2010;12:70–76. doi: 10.1038/ni.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schrader CE, Linehan EK, Mochegova SN, Woodland RT, Stavnezer J. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J Exp Med. 2005;202:561–568. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 102.Huang FT, Yu K, Balter BB, Selsing E, Oruc Z, Khamlichi AA, Hsieh CL, Lieber MR. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol Cell Biol. 2007;27:5921–5932. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tous C, Aguilera A. Impairment of transcription elongation by R-loops in vitro. Biochem Biophys Res Commun. 2007;360:428–432. doi: 10.1016/j.bbrc.2007.06.098. [DOI] [PubMed] [Google Scholar]
- 104.Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, Gearhart PJ. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med. 2009;206:1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]