Abstract
In mammalian cells, chromatin poly(ADP-ribos)ylation (PARylation) at sites of DNA Double-Strand Breaks (DSBs) is mediated by two highly related enzymes, PARP1 and PARP2. However, enzyme-specific genetic interactions with other DSB repair factors remain largely undefined. In this context, it was previously shown that mice lacking PARP1 and H2AX, a histone variant that promotes DSB repair throughout the cell cycle, or the core nonhomologous end-joining (NHEJ) factor Ku80 are not viable, while mice lacking PARP1 and the noncore NHEJ factor DNA-PKcs are severely growth retarded and markedly lymphoma-prone. Here, we have examined the requirement for PARP2 in these backgrounds. We find that, like PARP1, PARP2 is essential for viability in mice lacking H2AX. Moreover, treatment of H2AX-deficient primary fibroblasts or B lymphocytes with PARP inhibitors leads to activation of the G2/M checkpoint and accumulation of chromatid-type breaks in a lineage- and gene-dose dependent manner. In marked contrast to PARP1, loss of PARP2 does not result in additional phenotypes in growth, development or tumorigenesis in mice lacking either Ku80 or DNA-PKcs. Altogether these findings highlight specific nonoverlapping functions of PARP1 and PARP2 at H2AX-deficient chromatin during replicative phases of the cell cycle and uncover a unique requirement for PARP1 in NHEJ-deficient cells.
Keywords: PARP1, PARP2, H2AX, DNA-PKcs, Ku80, double-strand breaks
Graphical abstract
1. Introduction
Poly(ADP-ribos)ylation (PARylation) is a ubiquitous posttranslational modification that regulates DNA damage signaling and repair, transcription and other cellular processes [1-3]. Among six bona fide cellular poly(ADP-ribose) polymerases (PARPs), PARP1 and PARP2 are nuclear proteins recruited to sites of DNA breaks to promote their repair [4]. While PARP2 provides the bulk of PARP activity in plants [5], PARP1 is thought to generate most PAR within mammalian cells in response to DNA damage [6-8]. PARP1 and PARP2 share a highly homologous carboxi-terminal PARP domain with bona fide polymerase activity [9] but display unique amino-terminal DNA binding domains. In this context, their substrates only partially overlap [10, 11] and the analysis of Parp2−/− mice has revealed unique functions for PARP2 in lymphoma suppression, spermatogenesis, metabolism and other processes ([12-14]; reviewed in [15]). Furthermore, depletion of both PARP1 and PARP2 leads to early embryonic lethality in the mouse [16], indicating that PARP2-dependent PARylation is not a mere redundant activity.
In the context of single-strand breaks (SSB) repair, both PARP1 and PARP2 PARylate polβ and XRCC1 to promote ligase III-dependent repair [7, 17]. In line with these findings, Parp1−/− or Parp2−/− mouse embryonic fibroblasts (MEFs) show delayed kinetics of SSB repair, radiosensitivity, hypersensitivity to alkylating agents and increased frequency of Sister Chromatid Exchanges [7, 16]. These functions of PARP1 and PARP2 in SSB repair have formed the basis for the use of PARP inhibitors (PARPi) as sensitizers to radiation or chemotherapy and for the treatment of cancers with deficiencies in HR factors [18-20].
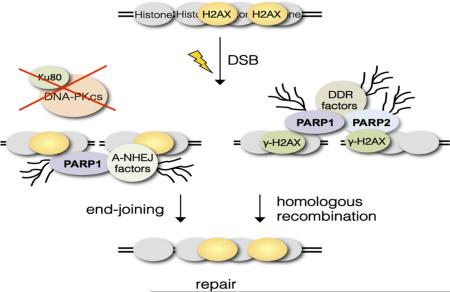
PARP1 and PARP2 are also recruited to chromatin at sites of DSBs [21], where they promote repair via PARylation of repair factors, histones and chromatin remodeling activities [10, 11, 21-24]. However, PARP1 and PARP2 are recruited to these lesions with different kinetics [25] and recognize differentially processed broken DNA ends [26], suggesting that they may have evolve specialized roles at DSBs. Specifically in the context of the DNA Damage Response (DDR), PARylation was previously shown to cooperate with γ-H2AX to promote BRCA1 recruitment to DSBs [27]. Consistent with this finding, we previously reported that mice lacking PARP1 and H2AX are not viable [28]. Although a role for PARP2 in this setting has not been reported, the observation that loss of PARP2 leads to embryonic lethality in ATM-deficient mice [29] suggests that both factors may become essential in DDR-deficient cells. In addition, PARP1 has been shown to play essential roles at DSBs arising in cells deficient in repair per se. Specifically, PARP1 disruption markedly aggravates the phenotypes of mice deficient for the NonHomologous End-Joining (NHEJ) factors Ku80 [30] or DNA-PKcs [31, 32]. Whether PARP2 may play a similar role in this setting has not been reported.
To better define common and unique functions for PARP1 and PARP2 at sites of DSBs, we have generated mice lacking histone H2AX or the NHEJ factors Ku80 and DNA-PKcs in a PARP2-deficient background. Their analyses demonstrate essential roles for both PARP1 and PARP2 at H2AX-deficient chromatin, while PARP1 is uniquely required at NHEJ-deficient chromatin.
2. Materials and Methods
2.1. Mice
Mice deficient for H2AX [33], PARP1 [34], PARP2 [16], DNA-PKcs [35] or Ku80 [36] were previously described. For experiments with primary lymphocytes, mice were euthanized at 2-4 months of age. For experiments involving lymphomas, the health of mice cohorts was monitored clinically and mice were euthanized when moribund. All mouse experiments included similar numbers of males and females and were conducted in accordance with Institutional Animal Care and Use Committee (IACUC)-approved protocols. Mouse genotypes from tail biopsies were determined using real time PCR with specific probes designed for each gene and validated on positive and negative control tail DNA (Transnetyx, Cordova, TN).
2.2. Cells
Mouse embryonic fibroblasts (MEFs) were obtained from timed matings at E13.5 following standard procedures. B-lineage splenocytes were purified from the spleen by negative selection with CD43 beads (Miltenyi) and activated by incubation with the B cell mitogens α-CD40 antibody (1 μg/mL, BD Pharmingen) and IL-4 (20 ng/mL, R&D Systems), as described [37]. Fresh lymphoma tissue was disaggregated mechanically and cultured briefly in the presence of IL-2 (100 IU/mL) and IL-7 (5 ng/mL), as described [32].
2.3. Drug treatments
Olaparib/AZD2281 (Selleckchem, S1060) and veliparib/ABT-888 (Enzo Life Sciences, ALX-270-444-M001) were stored at −80° C in single use aliquots.
2.4. Telomere FISH
Activated B cells or primary MEFs were incubated in colcemid (KaryoMAX, Gibco), swollen in 30 mM sodium citrate, fixed in methanol/acetic acid (3/1), hybridized with a telomere PNA probe and imaged as described [37]. At least 30 metaphases per sample were scored for chromosomal aberrations.
2.5. Cell cycle analysis
Cells were fixed in cold 70% ethanol, permeabilized in Triton-X, digested with RNAse A and stained with propidium ioidide (PI), as described [38]. Data was acquired using a FACSCalibur and analyzed with FlowJo software.
2.6. Statistical analysis
To determine statistical significance, we performed Student's T test on 3-5 data points per experiment, from 3-5 independent experiments. P≥0.05, not significant (n.s.); P<0.05, *; P<0.01,**; P<0.001,***).
3. Results and Discussion
3.1. PARP2 is required for viability of H2AX-deficient mice
We previously showed that concomitant loss of PARP1 and H2AX results in embryonic lethality in the mouse [28]. To determine whether this genetic interaction is PARP1-specific, we interbred Parp2+/−/H2afx+/− mice and genotyped 107 pups from Parp2+/−/H2afx+/− intercrosses (n=18 litters; age at genotyping, 7-10 days). We failed to observe Parp2−/−/H2afx−/− pups, while all remaining genotypes were observed at near Mendelian ratios (Table 1). Additional breedings of Parp2−/−/H2afx+/− or Parp2+/−/H2afx−/− females to Parp2+/−/H2afx+/− males similarly failed to yield double mutants (not shown). In contrast, Parp2−/−/H2afx+/− and Parp2+/−/H2afx−/− mice were viable and showed no additional phenotypes on growth and development over their Parp2−/− or H2afx−/− control littermates (not shown). Moreover, cohorts of Parp2−/−/H2afx+/− (n=12 mice) and Parp2+/−/H2afx−/− mice (n=9 mice) followed for an average of 9 months (range, 6-13 months) appeared healthy and elective necropsies didn't reveal the appearance of tumors or other relevant macroscopic abnormalities (not shown). We conclude that both PARP1 and PARP2 are essential for embryonic development in an H2AX-deficient background, paralleling previous observations in the absence of ATM [29]. Together, these findings underscore unique functions for PARP2 at DSBs that cannot be compensated by PARP1.
Table 1.
Mendelian ratios from intercrosses between mice heterozygous for PARP2 (Parp2) and DNA repair factors H2AX (H2afx), DNA-PKcs (Prkdc) or Ku80 (Xrcc5)
| Parp2+/−/H2afx+/− intercrosses | Parp2+/−/Prkdc+/− intercrosses | Parp2+/−/Xrcc5+/− intercrosses | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
Parp2 genotype |
H2afx genotype |
Observed (%) |
Expected (%) |
Prkdc genotype |
Observed (%) |
Expected (%) |
Xrcc5 genotype |
Observed (%) |
Expected (%) |
| H2afx+/+ | 4 (3.7) | 6.7 (6.25) | Prkdc+/+ | 2 (4.7) | 2.7 (6.25) | Xrcc5+/+ | 14 (7.1) | 12.4 (6.5) | |
| Parp2+/+ | H2afx+/− | 16 (15.0) | 13.4 (12.5) | Prkdc+/− | 3 (7.0) | 5.4 (12.5) | Xrcc5+/− | 28 (14.1) | 24.8 (12.5) |
| H2afx−/− | 5 (4.7) | 6.7 (6.25) | Prkdc−/− | 0 (0.0) | 2.7 (6.25) | Xrcc5−/− | 10 (5.1) | 12.4 (6.5) | |
| H2afx+/+ | 17 (15.9) | 13.4 (12.5) | Prkdc+/+ | 5 (11.6) | 5.4 (12.5) | Xrcc5+/+ | 28 (14.1) | 24.8 (12.5) | |
| Parp2+/− | H2afx+/− | 39 (36.4) | 26.8 (25) | Prkdc+/− | 13 (30.2) | 10.8 (25) | Xrcc5+/− | 63 (31.8) | 49.5 (25) |
| H2afx−/− | 8 (7.5) | 13.4 (12.5) | Prkdc−/− | 4 (9.3) | 5.4 (12.5) | Xrcc5−/− | 16 (8.1) | 24.8 (12.5) | |
| H2afx+/+ | 6 (5.6) | 6.7 (6.25) | Prkdc+/+ | 1 (2.3) | 2.7 (6.25) | Xrcc5+/+ | 10 (5.1) | 12.4 (6.5) | |
| Parp2−/− | H2afx+/− | 12 (11.2) | 13.4 (12.5) | Prkdc+/− | 11 (25.6) | 5.4 (12.5) | Xrcc5+/− | 25 (12.6) | 24.8 (12.5) |
| H2afx−/− | 0 (0) | 6.7 (6.25) | Prkdc−/− | 4 (9.3) | 2.7 (6.25) | Xrcc5−/− | 4 (2.0) | 12.4 (6.5) | |
| Total | 107 | Total | 43 | Total | 198 | ||||
3.2. H2AX suppresses PARPi-induced genomic instability during replication
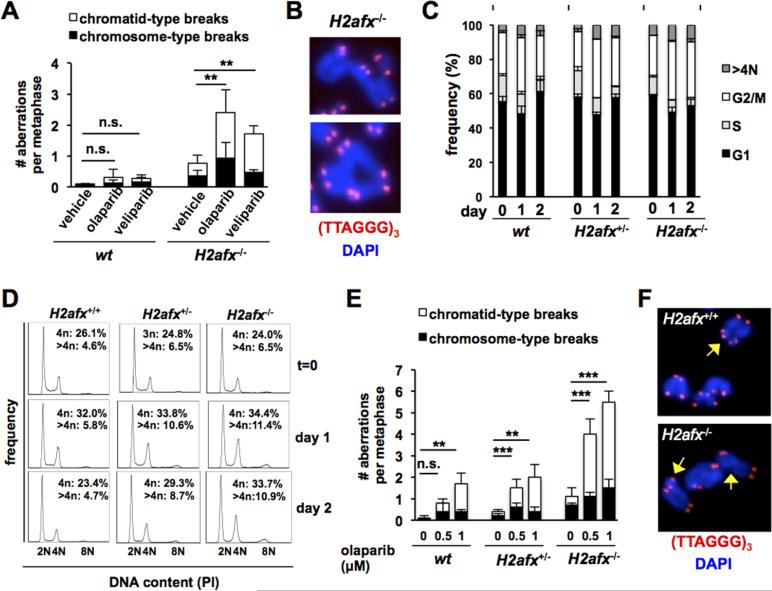
To gain insights into the mechanisms leading to embryonic lethality in mice lacking H2AX and either PARP1 or PARP2, we quantified genomic instability in metaphase spreads of wt and H2afx−/− activated B cells treated with PARPi olaparib [39] or veliparib [40], at concentrations known to inhibit both PARP1 and PARP2 (Table 2, Fig. 1). In HR-deficient cells, PARPi induce breaks during replication [18, 19], visualized as chromatid-type breaks in the first mitosis after treatment. We observed no statistically relevant increase in genomic instability H2afx+/+ B cells after PARPi exposure (Table 2; Fig. 1A). In contrast, both olaparib and veliparib induced a statistically significant increase in the frequency of metaphases containing aberrations and in the number of aberrations per cell in H2afx−/− B cells (Table 2 for quantification and p values; Fig. 1A). Importantly, most chromosomal aberrations in PARPi-treated H2afx−/− cells were chromatid-type (60% and 75% of all breaks after olaparib or veliparib, respectively; Table 2; Fig. 1B for examples). These observations were not specific to B cells, because concanavalin A-activated H2afx−/− T cells similarly harbored frequent chromatid-type chromosomal breaks after exposure to olaparib (Table S1).
Table 2.
Analysis of genomic instability via telomere-FISH in PARPi-treated H2afx−/− and control primary cells
| Genotype | Treatment | # mice | # metaphases | # metaphases with aberrations (%) | # aberrations per metaphase (# chromatid breaks per metaphase) | p valuesa |
|---|---|---|---|---|---|---|
| Activated B cells | ||||||
| H2afx+/+ | vehicle | 5 | 150 | 11 (7.3%) | 0.09 (0.01) | |
| olaparib (1 μM) | 5 | 150 | 28 (18.7%) | 0.31 (0.1) | p=0.18/0.17/0.20 | |
| veliparib (2.5 μM) | 3 | 90 | 13 (14.4%) | 0.3 (0.1) | p=0.21/0.36/0.26 | |
| H2afx−/− | vehicle | 5 | 150 | 55 (36.7%) | 0.76 (0.4) | |
| olaparib (1 μM) | 5 | 158 | 115 (74.1%) | 2.4 (1.4) | p=0.007/0.03/0.02 | |
| veliparib (2.5 μM) | 3 | 90 | 62 (68.9%) | 1.7 (1.3) | p=0.004/0.01/0.008 | |
| Primary MEFs | ||||||
| H2afx+/+ | vehicle | 3 | 90 | 8 (8.9%) | 0.1 (0.0) | |
| olaparib (0.5 μM) | 3 | 90 | 42 (46.7%) | 0.8 (0.4) | p=0.01/0.12/0.05 | |
| olaparib (1 μM) | 3 | 90 | 55 (61.1%) | 1.7 (1.3) | p=0.01/0.01/0.05 | |
| H2afx+/− | vehicle | 4 | 120 | 30 (25.0%) | 0.4 (0.2) | |
| olaparib (0.5 μM) | 4 | 120 | 65 (54.2%) | 1.4 (0.9) | p=0.01/0.004/0.02 | |
| olaparib (1 μM) | 4 | 120 | 75 (62.5%) | 1.9 (1.6) | p=0.002/0.02/0.02 | |
| H2afx−/− | vehicle | 3 | 67 | 42 (62.7%) | 1.3 (0.4) | |
| olaparib (0.5 μM) | 3 | 90 | 84 (93.3%) | 4.0 (2.9) | p=0.001/0.009/0.009 | |
| olaparib (1 μM) | 3 | 90 | 85 (94.5%) | 5.6 (4.0) | p=0.003/0.001/0.001 | |
the three indicated p values correspond to: % metaphases with aberrations / #chromosome breaks per metaphase / #chromatid breaks per metaphase, respectively.
Figure 1.
H2AX suppresses replication-dependent chromosomal breaks induced by PARP inhibitors in a lineage and gene-dose dependent manner. (A-B) H2afx−/− and control H2afx+/+ B cells were purified from the spleen, activated in vitro with cytokines and treated with olaparib (1 μM) or veliparib (3 μM) for 24 hours prior to metaphase analyses by telomere FISH. The number of chromosomal breaks per metaphase is shown and subdivided as “chromosome-type” or “chromatid-type”. Representative examples of complex “chromatid-type” rearrangements observed in H2AX-deficient cells after olaparib treatment are shown in B. (C-D) Primary P0-P1 MEFs of the indicated genotypes were treated with either 1 μM olaparib for 24 hours (day 1) or 48 hours (day 2) and harvested for cell cycle analysis after staining with propidium iodide (PI). The percentage of cells with 2N (G1), 2N>4N (S), 4N (G2) and >4N DNA content is shown in C. Each bar represents the average and standard deviation of three MEF cultures derived from 3 independent embryos. Representative examples for each genotype after either 1 day or 2 days of treatment are shown in D. (E-F) Primary P0-P1 MEFs of the indicated genotypes were treated with either 0.5 or 1 μM olaparib for 24 hours and metaphases were analyzed by telomere FISH. Bars represent the average and standard deviation of three independent cultures per genotype. Representative examples of chromosomal aberrations observed in wt and H2afx−/− MEFs exposed to olaparib for 24 hours are shown in F. Yellow arrows point to chromatid-type breaks.
To extend these analyses to a nonlymphoid lineage, we also examined the effect of PARPi on cell cycle distribution and genomic integrity in primary H2afx−/− and control wt MEFs (Table 2, Figure 1C-F). Because H2afx−/− MEFs senesce prematurely [41], these studies were done at early passage (P1), when the cell cycle distribution was similar across genotypes (Fig. 1C; “day 0”). Exposure to 1 μM olaparib led to a marked increase in the frequency of cells with 4N DNA content across all genotypes (Fig. 1C-D, “day 1”). However, while cell cycle distribution of wt MEFs had returned to baseline at day 2, persistent accumulation of cells with 4N DNA content was evident in H2afx−/− cultures (Fig. 1C-D; “day 2”). Because these experiments were done in primary cells with intact checkpoints, these observations strongly suggest persistent DNA damage in the absence of H2AX. In support of this notion, telomere FISH of metaphase spreads in the same cells revealed a marked, dose-dependent increase in the frequency of olaparib-induced chromosomal aberrations in H2afx−/− MEFs relative to wt controls (Table 2, Fig. 1E for quantification; Fig. 1F for examples). Similar to our observations in H2afx−/− B cells, olaparib-induced chromosomal aberrations were mostly chromatid-type (Table 2, Fig. 1E-F).
Because H2AX haploinsufficiency compromises DSB repair in some settings [33, 42], we also analyzed PARPi-induced genomic stability in H2afx+/− B cells and MEFs. We observed a small but statistically significant increase in the frequency of chromosomal aberrations in H2afx+/− MEFs, albeit only at the lower doses of olaparib (0.5 μM). In contrast, olaparib did not induce genomic instability in H2afx+/− B cells at the doses used here (data not shown). Altogether, these data suggest that decreased H2AX nucleosomal density may be sufficient to modestly sensitize some lineages to PARP inhibition.
Overall, these data is in line with previous findings in chicken DT40 cells [20] and mouse ES cells [43] that deficiency for H2AX sensitizes cells to PARP inhibitors and demonstrates for the first time that hypersensitivity relates to increased genomic instability. Moreover, cell cycle and cytogenetic analyses demonstrate that all or most PARPi-dependent chromosomal instability in H2AX-deficient cells occurs in the context of replication, leading to frequent chromatid-type aberrations. These findings suggest that PARP1 and PARP2-dependent PARylation and γ-H2AX provide compensatory mechanisms specifically in S and/or G2/M phases of the cell cycle. These findings were unanticipated, because H2AX-deficient cells show primarily a defect in NHEJ [37]. Similarly, treatment of ATM-deficient cells with PARP inhibitors leads to replication-dependent DSBs [44]. Altogether, these findings provide a rationale for the development of PARP1- or PARP2-specific inhibitors for the treatment of cancers lacking ATM [45-50] and/or haploinsufficient for H2AX [51].
3.3. Unlike PARP1, PARP2 does not compromise growth and development of DNA-PKcs-deficient mice
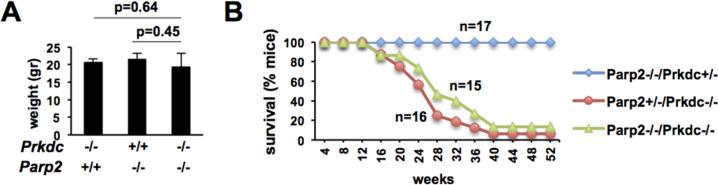
Loss of PARP1 results in embryonic lethality or severe growth defects in NHEJ-deficient backgrounds [30, 31, 38], but a role for PARP2 in this context has not been investigated. To address this issue, we first generated mice doubly deficient for PARP2 and DNA-PKcs, a noncore NHEJ factor. In marked contrast to the severe growth retardation observed in Parp1−/−/Prkdc−/− mice [31, 32], Parp2−/−/Prkdc−/− mice were born at mendelian ratios and showed no additional phenotypes in growth and development over single mutants (Table 1, Fig. 2A).
Figure 2.
Phenotypic analysis of mice deficient for DNA-PKcs and PARP2. (A) Weight of mice deficient for DNA-PKcs and/or PARP2. Bars represent the average and standard deviation of 3-5 8-12 week-old females. Similar observations were made for males (not shown). (B) Kaplan Meier analyses of cohorts of Parp2−/−/Prkdc+/− (n=17), Parp2+/−/Prkdc−/− (n=16) and Parp2−/−/Prkdc−/− (n=15) mice followed for one year.
PARP2 was previously shown to promote survival of thymocytes by suppressing apoptosis in response to programmed DSBs [52]. Moreover, loss of PARP1 markedly increases the penetrance and decreases the latency of thymic lymphomas in DNA-PKcs-deficient mice [31, 32], but a similar role for PARP2 has not been examined. To address this question, we followed cohorts of Parp2−/−/Prkdc−/− and control Parp2+/−/Prkdc−/− and Parp2−/−/Prkdc+/− littermates until moribund. In contrast to Parp1−/−/Prkdc−/− mice, we found that the survival of Parp2+/−/Prkdc−/− and Parp2−/−/Prkdc+/− mice was comparable (Fig. 2B). Specifically, 14/15 Parp2−/−/Prkdc−/− mice and 15/16 Parp2+/−/Prkdc−/− mice died within the first year of life. The average age at death was 26.9 weeks (range, 13-40 weeks) and 24.5 weeks (range, 13-42 weeks) for Parp2−/−/Prkdc−/− and Parp2+/−/Prkdc−/− mice, respectively. Necropsies of moribund animals revealed thymic lymphoma as cause of death in approximately 7/14 and 7/15 Parp2−/−/Prkdc−/− and Parp2+/−/Prkdc−/− mice, respectively. In this subset of lymphoma-bearing mice, the average age at death was 29.2 weeks (range, 13-40 weeks; n=7) and 23.1 weeks (range, 14-27 weeks; n=7) for Parp2−/−/Prkdc−/− and Parp2+/−/Prkdc−/− mice, respectively. Consistent with previous observations in Parp2−/− mice [16], 17/17 Parp2−/−/Prkdc+/− littermate mice appeared healthy at the end of the one-year observation period (Fig. 2B) and elective necropsies failed to reveal tumors or other macroscopic alterations (not shown). We conclude that, unlike PARP1, PARP2 does not mediate tumor suppression in thymocytes deficient for DNA-PKcs.
3.4. PARP2 is dispensable for the fusion of DNA-PKcs-deficient telomeres
DNA-PKcs functions to suppress telomere fusions in mouse and human cells [53, 54] and, consistent with these previous observations, control Parp2+/−/Prkdc−/− thymic lymphomas accumulate telomere fusions (Fig. S1A for quantification; Fig. S1B for representative metaphases). Because PARP2 binds the shelterin component TRF2 [55] and suppresses chromosomal translocations of programmed DSBs in B cells undergoing Class Switch Recombination (CSR) [56], we investigated whether PARP2 functions to suppress the fusion of DNA-PKcs-deficient telomeres. To this end, we quantified telomere fusions in five Parp2−/−/Prkdc−/− thymic lymphomas (Figs. S1A-B). Three out of five double mutant tumors contained telomere fusions in all or most cells, while the remaining two tumors contained fusions in a smaller fraction of cells. Moreover, the frequency and patterns of telomere fusions in Parp2−/−/Prkdc−/− thymic lymphomas were similar to those observed in Parp2−/−/Prkdc−/− lymphomas.
We previously showed that most Parp1−/−/Prkdc−/− lymphomas harbor oncogenic mutations in p53, leading to aneuploidy and amplification of telomere fusions [32]. In contrast, cytogenetic analysis of Parp2−/−/Prkdc−/− lymphomas revealed that all tumors were euploid or near-euploid, similar to control Parp2-+-/Prkdc−/− tumors (Fig. S2A for quantification; Fig. S2B for examples of representative metaphases). These data strongly suggest that PARP2 does not play a significant role in the suppression of oncogenic p53 mutations in DNA-PKcs-deficient thymocytes.
3.5. Mice lacking PARP2 and Ku80 are viable and show no significant additional phenotypes in growth and development
Because DNA-PKcs is required for a subset of NHEJ reactions, we next extended our analysis to a core NHEJ factor, Ku80 (gene symbol, Xrcc5) [57]. To this end, we bred Parp2+/−/Xrcc5+/− males and females to generate double mutants (expected ratio, 1/16). Genotyping of n=198 pups at day 7 of life revealed similar numbers of Ku80-deficient mice regardless of Parp2 copy number (Table 1). Like control Parp2+/+/Xrcc5−/− mice [36], Parp2+/−/Xrcc5−/− and Parp2−/−/Xrcc5−/− littermate mice were markedly growth retarded and about half of the mice died prior to weaning. However, weaned double mutant mice appeared robust and a few mice survived when followed for up to six months (not shown). Together with our analysis of Parp2−/−/Prkdc−/− mice above and previous observations of early embryonic lethality in Parp1−/−/Xrcc5−/− mice [30], these findings indicate that PARP1 plays unique, nonoverlapping functions in the signaling and/or repair of DSBs that arise in the absence of NHEJ. In this regard, PARP1 has been implicated in end-joining via an alternative, ligase III-dependent pathway (A-NHEJ) [58-60] and cooperates with DNA-PKcs in the restart of a subset of stalled replication forks [61]. In the future, it will be important to determine PARP1 unique substrates and functions in these contexts.
4. Conclusions
PARP2 is closely related to PARP1, yet their unique amino-terminal domains confer them with unique substrates and functions. To define these in the context of DNA double-strand break (DSB) repair, we have generated here three novel mouse models of PARP2 deficiency, Parp2−/−/H2afx−/−, Parp2−/−/Prkdc−/− and Parp2−/−/Xrcc5−/− mice. The analyses of these compound models have uncovered a novel essential function for PARP2 in concert with H2AX during replicative phases of the cell cycle. In contrast, we find that the genetic interaction between PARP1 and NHEJ, a DSB repair pathway that operates primarily in pre-replicative phases of the cell cycle, is not shared with PARP2. Overall, these findings have broad implications for understanding how PARylation cooperates with other cellular pathways to maintain genomic integrity, thereby suppressing organismal phenotypes of aging and cancer. In the future, the analyses of novel genetic models for conditional inactivation of PARP1 and/or PARP2 in somatic cells deficient for H2AX or NHEJ factors will further define the mechanisms underlying their common and unique functions.
Supplementary Material
Acknowledgements
We thank Drs. Fred Alt, Ted Dawson and Mary Armanios for mouse strains; Ada Tam at the Flow Cytometry Core for expert assistance and the staff at the Cancer Research Building I vivarium for excellent animal care.
Funding Information
This research was funded by grants R21 CA181417-01A1 from the National Cancer Institute (NCI) and a Basser Innovation Award from the Basser Center for BRCA Research at the Abramson Cancer Center, University of Pennsylvania to Sonia Franco. The funders had no role in study design, data collection and interpretation or the decision to submit the work for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there is no conflict of interest
Appendix A. Supplementary Data
Supplementary Data associated to this article includes one table (Table S1) and two Figures (Figs. S1-S2).
References
- 1.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nature reviews. Molecular cell biology. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 2.Hottiger MO. Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annual review of biochemistry. 2015 doi: 10.1146/annurev-biochem-060614-034506. [DOI] [PubMed] [Google Scholar]
- 3.Gagne JP, Ethier C, Defoy D, Bourassa S, Langelier MF, Riccio AA, Pascal JM, Moon KM, Foster LJ, Ning Z, Figeys D, Droit A, Poirier GG. Quantitative site-specific ADP-ribosylation profiling of DNA-dependent PARPs. DNA repair. 2015;30:68–79. doi: 10.1016/j.dnarep.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: Focus on PARP1, PARP2 and PARP3. Experimental cell research. 2014 doi: 10.1016/j.yexcr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Song J, Keppler BD, Wise RR, Bent AF. PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses. PLoS genetics. 2015;11:e1005200. doi: 10.1371/journal.pgen.1005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. The Journal of biological chemistry. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 7.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. The Journal of biological chemistry. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 8.Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, Jacobson EL. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. The Journal of biological chemistry. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 9.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Molecular cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Isabelle M, Moreel X, Gagne JP, Rouleau M, Ethier C, Gagne P, Hendzel MJ, Poirier GG. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci. 2010;8:22. doi: 10.1186/1477-5956-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter-O'Connell I, Jin H, Morgan RK, David LL, Cohen MS. Engineering the substrate specificity of ADP-ribosyltransferases for identifying direct protein targets. J Am Chem Soc. 2014;136:5201–5204. doi: 10.1021/ja412897a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leger K, Bar D, Savic N, Santoro R, Hottiger MO. ARTD2 activity is stimulated by RNA. Nucleic acids research. 2014 doi: 10.1093/nar/gku131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 [corrected] controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma [corrected] heterodimer. The Journal of biological chemistry. 2007;282:37738–37746. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- 14.Nicolas L, Martinez C, Baro C, Rodriguez M, Baroja-Mazo A, Sole F, Flores JM, Ampurdanes C, Dantzer F, Martin-Caballero J, Aparicio P, Yelamos J. Loss of poly(ADP-ribose) polymerase-2 leads to rapid development of spontaneous T-cell lymphomas in p53-deficient mice. Oncogene. 2010;29:2877–2883. doi: 10.1038/onc.2010.11. [DOI] [PubMed] [Google Scholar]
- 15.Yelamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol Med. 2008;14:169–178. doi: 10.1016/j.molmed.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. The EMBO journal. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dantzer F, Schreiber V, Niedergang C, Trucco C, Flatter E, De La Rubia G, Oliver J, Rolli V, Menissier-de Murcia J, de Murcia G. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie. 1999;81:69–75. doi: 10.1016/s0300-9084(99)80040-6. [DOI] [PubMed] [Google Scholar]
- 18.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 19.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 20.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer research. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. The Journal of biological chemistry. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 22.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. The Journal of biological chemistry. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 23.Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Lu LY, Yang CY, Wang S, Yu X. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes & development. 2013;27:1752–1768. doi: 10.1101/gad.226357.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic acids research. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steffen JD, Brody JR, Armen RS, Pascal JM. Structural Implications for Selective Targeting of PARPs. Front Oncol. 2013;3:301. doi: 10.3389/fonc.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23:693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orsburn B, Escudero B, Prakash M, Gesheva S, Liu G, Huso DL, Franco S. Differential requirement for H2AX and 53BP1 in organismal development and genome maintenance in the absence of poly(ADP)ribosyl polymerase 1. Molecular and cellular biology. 2010;30:2341–2352. doi: 10.1128/MCB.00091-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber A, Bai P, de Murcia JM, de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA repair. 2004;3:1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Henrie MS, Kurimasa A, Burma S, Menissier-de Murcia J, de Murcia G, Li GC, Chen DJ. Lethality in PARP-1/Ku80 double mutant mice reveals physiological synergy during early embryogenesis. DNA repair. 2003;2:151–158. doi: 10.1016/s1568-7864(02)00199-4. [DOI] [PubMed] [Google Scholar]
- 31.Morrison C, Smith GC, Stingl L, Jackson SP, Wagner EF, Wang ZQ. Genetic interaction between PARP and DNA-PK in V(D)J recombination and tumorigenesis. Nature genetics. 1997;17:479–482. doi: 10.1038/ng1297-479. [DOI] [PubMed] [Google Scholar]
- 32.Rybanska I, Ishaq O, Chou J, Prakash M, Bakhsheshian J, Huso DL, Franco S. PARP1 and DNA-PKcs synergize to suppress p53 mutation and telomere fusions during T-lineage lymphomagenesis. Oncogene. 2013;32:1761–1771. doi: 10.1038/onc.2012.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 34.Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes & development. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 35.Taccioli GE, Amatucci AG, Beamish HJ, Gell D, Xiang XH, Torres Arzayus MI, Priestley A, Jackson SP, Marshak Rothstein A, Jeggo PA, Herrera VL. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 1998;9:355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- 36.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig MC, Li GC. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 37.Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, Yan C, Tepsuporn S, Morales JC, Adams MM, Lou Z, Bassing CH, Manis JP, Chen J, Carpenter PB, Alt FW. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Molecular cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Rybanska-Spaeder I, Reynolds TL, Chou J, Prakash M, Jefferson T, Huso DL, Desiderio S, Franco S. 53BP1 is limiting for NHEJ repair in ATM-deficient model systems that are subjected to oncogenic stress or radiation. Molecular cancer research : MCR. 2013;11:1223–1234. doi: 10.1158/1541-7786.MCR-13-0252-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. The New England journal of medicine. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 40.Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, Ferguson DC, Ghoreishi-Haack NS, Grimm DR, Guan R, Han EK, Holley-Shanks RR, Hristov B, Idler KB, Jarvis K, Johnson EF, Kleinberg LR, Klinghofer V, Lasko LM, Liu X, Marsh KC, McGonigal TP, Meulbroek JA, Olson AM, Palma JP, Rodriguez LE, Shi Y, Stavropoulos JA, Tsurutani AC, Zhu GD, Rosenberg SH, Giranda VL, Frost DJ. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 41.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rass E, Chandramouly G, Zha S, Alt FW, Xie A. ATM is dispensable for I-SceI-induced homologous recombination in mouse embryonic stem cells. The Journal of biological chemistry. 2013 doi: 10.1074/jbc.M112.445825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weston VJ, Oldreive CE, Skowronska A, Oscier DG, Pratt G, Dyer MJ, Smith G, Powell JE, Rudzki Z, Kearns P, Moss PA, Taylor AM, Stankovic T. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116:4578–4587. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- 45.Stilgenbauer S, Winkler D, Ott G, Schaffner C, Leupolt E, Bentz M, Moller P, Muller-Hermelink HK, James MR, Lichter P, Dohner H. Molecular characterization of 11q deletions points to a pathogenic role of the ATM gene in mantle cell lymphoma. Blood. 1999;94:3262–3264. [PubMed] [Google Scholar]
- 46.Au WY, Gascoyne RD, Viswanatha DS, Connors JM, Klasa RJ, Horsman DE. Cytogenetic analysis in mantle cell lymphoma: a review of 214 cases. Leuk Lymphoma. 2002;43:783–791. doi: 10.1080/10428190290016890. [DOI] [PubMed] [Google Scholar]
- 47.Stilgenbauer S, Liebisch P, James MR, Schroder M, Schlegelberger B, Fischer K, Bentz M, Lichter P, Dohner H. Molecular cytogenetic delineation of a novel critical genomic region in chromosome bands 11q22.3-923.1 in lymphoproliferative disorders. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:11837–11841. doi: 10.1073/pnas.93.21.11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lens D, Matutes E, Catovsky D, Coignet LJ. Frequent deletions at 11q23 and 13q14 in B cell prolymphocytic leukemia (B-PLL) Leukemia. 2000;14:427–430. doi: 10.1038/sj.leu.2401644. [DOI] [PubMed] [Google Scholar]
- 49.Cuneo A, Bigoni R, Rigolin GM, Roberti MG, Milani R, Bardi A, Minotto C, Agostini P, De Angeli C, Narducci MG, Sabbioni S, Russo G, Negrini M, Castoldi G. Acquired chromosome 11q deletion involving the ataxia teleangiectasia locus in B-cell non-Hodgkin's lymphoma: correlation with clinicobiologic features. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2000;18:2607–2614. doi: 10.1200/JCO.2000.18.13.2607. [DOI] [PubMed] [Google Scholar]
- 50.Austen B, Barone G, Reiman A, Byrd PJ, Baker C, Starczynski J, Nobbs MC, Murphy RP, Enright H, Chaila E, Quinn J, Stankovic T, Pratt G, Taylor AM. Pathogenic ATM mutations occur rarely in a subset of multiple myeloma patients. Br J Haematol. 2008;142:925–933. doi: 10.1111/j.1365-2141.2008.07281.x. [DOI] [PubMed] [Google Scholar]
- 51.Caren H, Kryh H, Nethander M, Sjoberg RM, Trager C, Nilsson S, Abrahamsson J, Kogner P, Martinsson T. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4323–4328. doi: 10.1073/pnas.0910684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yelamos J, Monreal Y, Saenz L, Aguado E, Schreiber V, Mota R, Fuente T, Minguela A, Parrilla P, de Murcia G, Almarza E, Aparicio P, Menissier-de Murcia J. PARP-2 deficiency affects the survival of CD4+CD8+ double-positive thymocytes. The EMBO journal. 2006;25:4350–4360. doi: 10.1038/sj.emboj.7601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goytisolo FA, Samper E, Edmonson S, Taccioli GE, Blasco MA. The absence of the dna-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Molecular and cellular biology. 2001;21:3642–3651. doi: 10.1128/MCB.21.11.3642-3651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bailey SM, Meyne J, Chen DJ, Kurimasa A, Li GC, Lehnert BE, Goodwin EH. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14899–14904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dantzer F, Giraud-Panis MJ, Jaco I, Ame JC, Schultz I, Blasco M, Koering CE, Gilson E, Menissier-de Murcia J, de Murcia G, Schreiber V. Functional interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Molecular and cellular biology. 2004;24:1595–1607. doi: 10.1128/MCB.24.4.1595-1607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robert I, Dantzer F, Reina-San-Martin B. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. The Journal of experimental medicine. 2009;206:1047–1056. doi: 10.1084/jem.20082468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 58.Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic acids research. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. The Journal of biological chemistry. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 60.Wray J, Williamson EA, Singh SB, Wu Y, Cogle CR, Weinstock DM, Zhang Y, Lee SH, Zhou D, Shao L, Hauer-Jensen M, Pathak R, Klimek V, Nickoloff JA, Hromas R. PARP1 is required for chromosomal translocations. Blood. 2013;121:4359–4365. doi: 10.1182/blood-2012-10-460527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ying S, Chen Z, Medhurst AL, Neal JA, Bao Z, Mortusewicz O, McGouran J, Song X, Shen H, Hamdy FC, Kessler BM, Meek K, Helleday T. DNA-PKcs and PARP1 Bind to Unresected Stalled DNA Replication Forks Where They Recruit XRCC1 to Mediate Repair. Cancer research. 2016;76:1078–1088. doi: 10.1158/0008-5472.CAN-15-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.