Abstract
Parasites are ubiquitous features of living systems and many parasites severely reduce the fecundity or longevity of their hosts. This parasite‐imposed selection on host populations should strongly favor the evolution of host resistance, but hosts typically face a trade‐off between investment in reproductive fitness and investment in defense against parasites. The magnitude of such a trade‐off is likely to be context‐dependent, and accordingly costs that are key in shaping evolution in nature may not be easily observable in an artificial environment. We set out to assess the costs of phage resistance for a plant pathogenic bacterium in its natural plant host versus in a nutrient‐rich, artificial medium. We demonstrate that mutants of Pseudomonas syringae that have evolved resistance via a single mutational step pay a substantial cost for this resistance when grown on their tomato plant hosts, but do not realize any measurable growth rate costs in nutrient‐rich media. This work demonstrates that resistance to phage can significantly alter bacterial growth within plant hosts, and therefore that phage‐mediated selection in nature is likely to be an important component of bacterial pathogenicity.
Keywords: Bacteriophage, coevolution, cost of resistance, phage resistance, plant pathogen
The presence of parasites that cause harm to their hosts, despite strong selection on host populations to evolve resistance, suggests the presence of trade‐offs between resistance and other traits such that host defenses are often costly to maintain in the absence of parasites (reviewed by Sheldon and Verhulst 1996). Costs of resistance are theoretically predicted to prevent the evolution of ever‐greater infectivity and resistance in parasites and hosts respectively when resources are finite and competition is an important selection pressure (Sasaki 2000). Associated costs of resistance to parasites have been quantified in a number of host systems, including plants (Parker 1991; Antonovics and Alexander 1995), birds (Nordling et al. 1998; Verhulst et al. 1999), mammals (Graham et al. 2010), and insects (Boots and Begon 1993; Kraaijeveld and Godfray 1997; Cotter et al. 2003). However, there have also been a number of published studies designed to measure costs of resistance that find little to no evidence for reduced fitness of resistant hosts (Purrington and Bergelson 1996; Labbé et al. 2010). In combination with the relative difficulty in publishing negative results, this may indicate that costs associated with resistance are often absent or relatively minor and therefore statistically difficult to detect (Lennon et al. 2007). Alternatively, it might be that costs associated with resistance are context‐dependent and therefore only detectable under certain abiotic or biotic conditions.
The context‐dependent nature of fitness costs can have important consequences for interactions among host organisms and other interacting species, such as competitors, predators, or other parasites. Such “ecological costs” can play critical roles in shaping host‐parasite coevolution across heterogeneous landscapes (Strauss et al. 2002), and can drive geographic selection mosaics, whereby the presence of one species in some patches can substantially alter the coevolution between two others (Thompson 2005). An example of this may be bacterial species that are either pathogenic or commensal on eukaryote hosts and are also themselves host to bacteriophage viruses (phages). In these cases any costs paid for resistance to phages may be exacerbated within the eukaryote “environment.”
Lytic phages (i.e. those phages that lyse their host cells to transmit to a new host) are highly abundant in all environments in which bacteria are found (Suttle 2005; Walsh et al. 2009). Given the estimated 10–20% of bacterial cells that are lysed through phage infection globally each day (Suttle 1994), the evolution of bacterial resistance mechanisms should be rapid and ongoing. Phage‐mediated selection has been shown, both empirically and theoretically, to play a central role in maintaining bacterial diversity (Buckling and Rainey 2002a; Banfield and Young 2009; Rodriguez‐Valera et al. 2009). At the community level, this is in part due to direct and indirect effects of phage populations on the competitive ability of their hosts (Bohannan and Lenski 2000). However, the prevalence of bacterial resistance remains unknown for many environments and appears to vary markedly by location (Fuhrman 1999; Jiang et al. 2003).
Costs of resistance to phage infection have previously been demonstrated during experimental evolution of bacteria in the presence of phage (Lenski and Levin 1985; Bohannan and Lenski 2000; Brockhurst et al. 2005; Koskella et al. 2012), but a number of studies have failed to find any costs associated with phage resistance (Lythgoe and Chao 2003; Mizoguchi et al. 2003). Furthermore, much of the work thus far relies on nutrient rich culture media and spatially homogenous environments, despite the evidence that microbial evolution is affected by varying spatial structure (Vos and Velicer 2008) and resource levels (Fierer et al. 2003). Moreover, the magnitude of costs associated with resistance is likely to vary depending on the mechanism of phage defense employed (Refardt and Kummerli 2013). Our primary goal in this study was to build on previous work demonstrating abiotic context‐dependent costs of resistance (Quance and Travisano 2009) to examine ecological costs of resistance. Specifically, we set out to test whether the costs paid by bacteria to resist phages differ between the laboratory environment and the natural plant host environment.
STUDY SYSTEM
Bacterial resistance to phages is often gained through alteration or loss of phage‐binding sites located on the cell surface (reviewed in Labrie et al. 2010; Koskella and Brockhurst 2014), which can result in a number of pleiotropic effects. For example, in Escherichia coli, the loss of siderophore‐excreting channels or proteins involved in adhesion and biofilm formation are known to confer resistance to multiple phages but also to play key roles in shaping bacterial cooperation and stability (Langenscheid et al. 2004; Smith et al. 2007). The loss or reduction in expression of such metabolically important pathways therefore leads to the prediction that resistance to phages is intrinsically costly. Some mutations are expected to be more costly than others, and benefits may also be variable depending on the level of cross‐resistance to other phages conferred (Lenski 1988). Aside from receptor loss, a number of other mechanisms of phage resistance have recently been identified, including the CRISPR/Cas system for phage resistance (Barrangou et al. 2007), abortive infection (Labrie et al. 2012), and restriction modification systems within the host (Balado et al. 2013). These latter mechanisms may also carry costs, but the addition of new pathways may be less likely to generate trade‐offs than the de novo mutation of existing genes linked to phage receptors.
Many mechanisms conferring phage resistance are likely to influence bacterial fitness within their eukaryotic hosts (reviewed in Koskella and Taylor 2015). Understanding the magnitude of costs within hosts, such as plants, relative to those affecting growth in the laboratory is key to predicting the outcome of bacteria‐phage coevolution in natural host‐associated microbial communities. To investigate the contextual nature of costs of bacterial resistance to phages, we examined the growth patterns of resistant mutants of the plant pathogenic bacterium, Pseudomonas syringae pathovar tomato (Pst) both under standard laboratory conditions and during infection of its host plant. Pst is a natural pathogen of the tomato plant, Solanum lycopersicum, where infection typically comprises of an epiphytic existence until a threshold density is reached, whereby the plant's first line of defense (i.e., stomatal closure) is suppressed and bacteria invade the leaf (Melotto et al. 2006). The molecular basis of Pst infection and responsive plant defense is well understood (Chisholm et al. 2006), as are the ecological effects of humidity and bacterial density on infection (O'Brien and Lindow 1989; Roux et al. 2010). However, the role that phages might play in shaping plant infection by Pst remains relatively unexplored. This is perhaps surprising, as there are a number of reasons to predict that lytic phages will influence the ability of bacteria to grow in planta both directly, via infection and lysis of cells, and indirectly, via selection for costly resistance (Koskella et al. 2012).
Selection for resistance
We set out to compare the impact of lytic phages from the environment on Pst strain DC3000 growth in planta and in vitro. Pst DC3000 was first grown overnight from frozen stocks in King's Broth (KB) nutrient media on a rotary shaker at 28°C. A soft agar overlay seeded with 200 μl of culture was then spotted with 10 μl of each of six clonal lytic phage suspensions known to infect Pst DC3000 that had been previously isolated from various aquatic sources as potential agents for biopesticide control of the pathogen in an agricultural setting (collected by Omnilytics, Sandy, UT, USA; see Koskella et al. (2012) for further details). Using high phage titers, we were able to create zones where there was complete absence of bacterial growth. Within these zones, however, resistant colonies were occasionally observed, and we took advantage of this phenomenon to isolate resistant clones for each phage tested (i.e., one mutant per phage, creating six resistant lines). After isolation, we confirmed bacterial resistance with further plaque assays and subjected each line, as well as the sensitive, ancestral host, to multiple phenotypic assays to evaluate the costs of resistance, as detailed below. The breadth of resistance was also measured prior to each experiment, and we found that resistance was stable over all experiments, suggesting a genetic rather than plastic mechanism, and indicating that resistance to one phage conferred resistance to nearly all the other phages used, which are all likely to be highly related despite independent isolation from the environment.
Measuring costs of resistance in vitro
The first phenotypic assay was to measure bacterial density reached in a typical laboratory setting using conventional culture microcosms (30 ml glass universal with 6 ml King's B nutrient media on an orbital shaker at 28°C; Buckling and Rainey 2002b). Thirty microliters of overnights from each phage resistant line and six replicates of the ancestral line were standardized to ∼0.1 OD600 using a 96‐well plate reader (PowerWaveXS, Biotek, USA) prior to inoculation into independent microcosms. Samples were taken at 2, 24, and 72 hours postinoculation. A second experiment was run in parallel with the addition of 100 μl of the phage suspension from which resistance was selected (standardized at 1 × 104 PFU/ml), in order to confirm resistance of mutants relative to the phage‐sensitive genotype. Serial dilutions and plating were performed with sterile water using the drop plate method as described by Chen et al. (2003). In brief, four replicates per sample were diluted in 10‐fold increments to 105 and 10 μl from each was spotted onto KB agar. After 48‐hour incubation at room temperature (approximately 23°C), colonies were counted to determine bacterial density at each time point. In parallel with this assay we measured 48‐hour growth curves using a spectrophotometer (PowerWaveXS, Biotek, USA) using culture turbidity as a proxy for bacterial density. Twenty microliters of a 1:10 diluted overnight culture was added to 180 μl KB and optical density was recorded every 5 minutes for 48 hours. These data were used to compare the minimum bacterial doubling time in logarithmic phase (between 0.2 and 0.4 OD600) of sensitive versus resistant mutant strains.
Measuring costs of resistance in planta
The second assay measured bacterial density on tomato plants through the course of 3‐day infection trials. The experimental design is as described for the in vitro assay, with the substitution of glass microcosms for 24, 6‐week‐old tomato plants, S. lycopersicum cultivar Moneymaker, which were transferred to a controlled growth room for the infections (28°C, 60% humidity, 9:15 light/dark cycle). Overall, we ran three in planta experiments, each with slightly altered conditions. Excluding controls, experiments 1 and 2 used a total of 12 plants each, with six being inoculated with mutant lines (one per plant) and six being inoculated with the sensitive, ancestral line. Experiment 3 used a total of 24 plants, with each of the three mutation classes discovered after sequencing inoculated into four plants each, and 12 inoculated with the sensitive line. Bacterial inocula were prepared by overnight culturing of all lines in 25 ml of KB media at 28°C on an orbital shaker. These cultures were washed in 10 mM MgCl2 by centrifugation at 3000 g for 10 minutes then resuspending the pellet in MgCl2. This process was repeated to further remove any residual media, which can result in lesion formation on leaves. All suspensions were standardized by dilution to ∼0.05 OD600 using a 96‐well plate reader (PowerWaveXS, Biotek, USA), which corresponded to ∼7 × 105 colony forming units (CFU)/ml. Three leaves, excluding those on the highest and lowest branches, were selected haphazardly and infiltrated with bacteria using a blunt 5 ml syringe on the abaxial surface of the leaf (Wei et al. 2007). A MgCl2 suspension was inoculated into at least two plants to serve as negative controls. These control plants were interspersed amongst treatment plants in the growth chamber.
After 1 hour postinoculation the first batch of samples was collected from leaves (effectively 0 days postinoculation) using an ethanol sterilized hole punch with a diameter of 6 mm. One cutting was made per leaf per plant at each time point to ensure replicates were independent, and each cutting came from a separate leaf chosen haphazardly. Each leaf cutting was dipped in 0.1 M sterilization buffer (0.02% Tween 20, 1% Sodium hypochlorite) then sterile water to remove epiphytic microorganisms. Cuttings were placed in 1 ml of 0.1 M sodium phosphate buffer (pH 7) supplemented with peptone and glycerol, and frozen at –20°C until processing. Further samples were taken at 24 hours and 72 hours postinfection. CFU per ml were enumerated by homogenizing each frozen leaf sample using a Fast‐Prep 24 (MP Biomedicals) and two 0.25 inch ceramic spheres for 20 seconds. Serial dilutions were performed as described earlier, with the addition of 25 μg/ml of nystatin added to the agar to prevent fungal growth. Pst DC3000 colonies were identified by colony morphology and fluorescence on KB agar and confirmed as DC3000 through whole genome resequencing of four haphazardly selected colonies. Control plants yielded no colonies conforming to Pst DC3000 morphology. The second experiment was run within a glasshouse rather than CT room, with higher resolution of bacterial densities, and with an additional sampling at 48 hours postinoculation. Finally, in light of sequencing results, we designed the third experiment to include a more balanced design in which each mutation class was equally represented across replicates. Here, we used 24 plants in total, where 12 were inoculated with the phage sensitive ancestor and 12 plants inoculated with phage resistant lines (four plants per mutation type).
Sequencing and bioinformatics analysis
In order to identify the mutations underlying the observed phenotypic differences, whole genome resequencing was conducted for all six mutants and the ancestor to enable comparison between the ancestral, phage sensitive line, and the mutant, phage resistant lines. After Illumina paired‐end sequencing (MiSeq, 250bp, v2 chemistry), reads were quality filtered and mapped against a reference genome (Pst DC3000, NC‐004578). Unaligned reads were assembled with Velvet (Zerbino and Birney 2008) and SNP calling was performed using the SNPEff package (Cingolani et al. 2012). More details about the bioinformatics pipeline used can be found in the supplementary information. Based on the short‐read sequence results, custom primers were designed around each potential mutation site and PCR amplifications were Sanger sequenced by the Core Genomic Facility at the University of Sheffield and mapped to a reference genome in Geneious 6.0.6 (Biomatters) to verify mutations. Primers used for sequencing are available in the supplementary information.
Initial sequencing uncovered what appeared to be parallel mutations conferring resistance to multiple phages and therefore being isolated across multiple phage backgrounds (Table 1). To determine whether this result was driven by convergent evolution or by selection acting on standing variation within the single Pst DC3000 colony used to seed the experiment, we conducted a further experiment. An overnight culture of the stock used for the first experiment was grown then diluted 100‐fold to reduce any standing variation. We then exposed this diluted culture to the three phages that led to the same mutation in the rfbA gene previously (FRS, NOI, and SNK) on a soft agar overlay. For each phage we isolated three colonies from within the clearance zone and one from an area of the plate not exposed to these phages to serve as a control line. We then sequenced each with the primers previously used to determine whether the same mutation arose again in the rfbA gene.
Table 1.
Phage resistance mutations in Pst DC3000
| Bacterial line | Mutation | Position | Gene | Selection experiment | Mutation effect |
|---|---|---|---|---|---|
| FRS | C → T | 1184566 | rfbA | 1 | Methionine → Isoleucine |
| NOI | C → T | 1184566 | rfbA | 1 | Methionine → Isoleucine |
| SNK | C → T | 1184566 | rfbA | 1 | Methionine → Isoleucine |
| SHL | C → T | 1185429 | rfbD | 1 | Tryptophan → Stop codon |
| WIL | C → T | 1185429 | rfbD | 1 | Tryptophan → Stop codon |
| REC | CAGC insertion | 47222 | No predicted function | 1 | Frameshift |
| NOI | T → G | 1184510 | rfbA | 2 | Histidine → Proline |
Each three‐letter code refers to a phage strain. Selection Experiment 1 was the generation of the phage resistant lines used in the assays, while Experiment 2 was to test for convergent molecular evolution.
Statistical analysis
All statistical analyses were conducted in R v2.15 (http://www.r‐project.org/). General linear models and generalized linear‐mixed models were used where appropriate with a quassipoisson error structure to account for overdispersion. Nonsignificant terms were removed and the resultant minimum adequate model was used for F tests for significant interactions (Venables and Ripley 1994).
Results
As expected given the resistance observed within soft agar overlays, we found a clear benefit of phage resistance when measured in vitro and when bacteria were grown in the presence of phage, with resistant bacterial lines reaching much higher densities at 24 hours than the sensitive ancestral genotype (Fig. 1A; General Linear Model, F1, 9 = 116.86, P < 0.001). Conversely, and more surprisingly, in the absence of phages we found no difference in density between phage resistant and sensitive ancestor (Fig. 1B; GLM, F1,10 = 0.323, P = 0.58). When bacterial densities within the plant host were measured, however, we found a clear cost of resistance in the absence of phages such that phage resistant bacteria attained lower densities after 24 hours than did the sensitive populations (Experiment 1, Fig. 2A; GLM, F1,10 = 25.88, P < 0.001). When the in planta experiment was repeated in the glasshouse, this time sampling after 1 and 2 days, we found a difference in densities after 24 hours, but not 48 hours (Experiment 2, Fig. 2B; Kruskal–Wallis rank sum test, Chi = 4.33, df = 1, P = 0.037, and Chi = 1.64, df = 1, P = 0.2, respectively).
Figure 1.
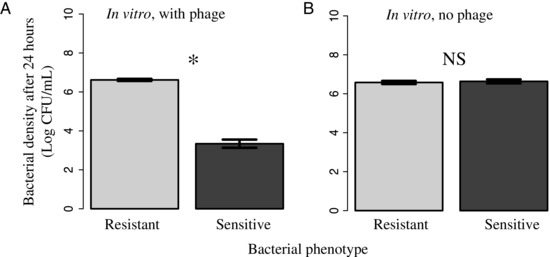
In vitro bacterial densities of resistant mutants (light grey) versus their sensitive ancestors (dark grey) after 24 hours in the presence (A) or absence (B) of phages. Densities are Log 10 transformed and error bars equal 1 standard error of the mean.
Figure 2.
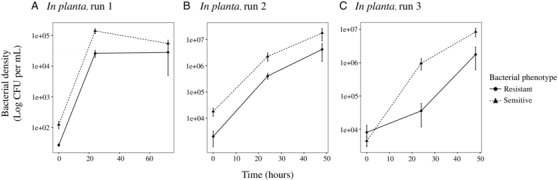
In planta bacterial densities when grown in the absence of phages. (A) Experiment 1, where the mutant lines exhibit reduced densities after 24 hours but not 72 (B) Experiment 2, where mutant densities are consistently lower than the phage sensitive ancestor, and (C) Experiment 3, where the sensitive ancestor also exhibit lower densities. Densities are Log 10 transformed and error bars equal 1 standard error of the mean. Experiments differed in the time points at which density was measured and Experiment 3 (C) was conducted after sequencing using only the three mutation classes.
Sequencing of isolated mutants revealed a single mutation in each line (Table 1). In some lines these mutations were found to be identical, suggesting standing genetic variation in the ancestral generation. To test this idea further we selected another nine phage resistant mutants and sequenced the same rfbA gene. This revealed a mutation in the same gene, although at a different position, in only one of the eight lines (one line failed to grow following freezing, preventing sequencing). This result supports the idea that mutations in the same gene may occur, but mutations at identical positions likely reflect some standing variation in the initial population despite bottlenecking at the single colony stage. In terms of gene function, the mutations found in rfbA and rfbD are likely linked to LPS synthesis (Dasgupta and Lam 1995; Nian et al. 2007). In one case this represent a loss of gene function due to mutation into a stop codon while in the other an amino acid substitution (Table 1). The third mutation, however, was found to be an insertion in a region with no predicted function.
To account for the uneven experimental design of the previous experiments as revealed by the sequencing (i.e., some mutations were represented in more replicates than others, as they were initially thought to be independent mutations), we repeated the assays using just one line representing each class of mutations. We again found a clear reduction in phage‐resistant bacterial densities in planta relative to the ancestral phage‐sensitive phenotype when plant and time were controlled for, with an average reduction in density of around 24% (Experiment 3, Fig. 2C; Generalized mixed effect model, Chi = 6.55, df = 1,5, P = 0.0105). We also repeated the in vitro element of the experiment using the same representative bacterial lines, this time recording optical densities over the course of 24 hours. We again found no differences in the maximum growth rate achieved relative to the sensitive line (Fig. 3; GLM, F = 1.52, df = 1, 29, P = 0.23), confirming an absence of costs when grown in vitro. In addition, post‐hoc testing did not reveal any difference in growth rates among the three mutations.
Figure 3.
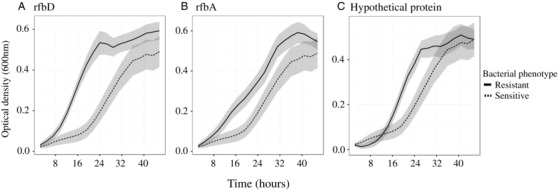
Forty‐eight hour growth curves of each mutant line relative to the ancestral, phage sensitive line recorded with optical density readings at 600 nm. Shaded areas represent 95% confidence intervals.
Discussion
Our key finding is that the evolution of resistance to phages can carry context‐dependent costs with direct relevance to plant‐bacterial interactions. Under standard laboratory conditions, we found no differences in density or maximum growth rates of sensitive versus resistant mutants when grown in the absence of phages. In contrast, when grown in planta the resistant mutants exhibited reduced densities relative to the sensitive population in each of three experiments. The observed cost of resistance in the absence of phages is in keeping with a body of theoretical work that predicts such costs can prevent resistance mutations from reaching fixation within a population (Flor 1971; Sasaki 2000; Agrawal and Lively 2002). Although this makes intuitive sense, such costs can be nuanced by the presence of different local fitness peaks, the evolution of compensatory mutations, and the background evolutionary contexts, all of which have been studied more extensively in the context of antibiotic resistance (Comas et al. 2012; Melnyk et al. 2014; Tazzyman and Hall 2015). While there are parallels to be drawn between phage resistance and antibiotic resistance, there are few studies empirically demonstrating the effects of phage‐resistance evolution in eukaryotic hosts. Phage resistance has been shown to attenuate virulence in Yersinia pestis infected mice (Filippov et al. 2011) and Erwinia carotovora infected potato tubers (Evans et al. 2010). However in these studies phage resistance was achieved through transposon mutagenesis, rather than exposure to a population of phages, which could bias towards the selection of loss of function mutations rather than subtler receptor modifications. Moreover, as our test of fitness focused on bacterial growth rather than competitive ability (i.e., head‐to‐head competition), our measure of fitness cost is likely to be conservative.
Mechanistically, the costs we have observed seem likely to have arisen through the alteration or loss of a phage‐binding site in the bacterial lipopolysaccharide (LPS) layer as a result of mutations in the rfbD and rfbA genes and an insertion into a gene of unknown function. Both the rfbD and rfbA genes identified through our genomic work are involved in the LPS biosynthesis pathway based on the KEGG orthology database (Kanehisa et al. 2014). Such LPS mutations are frequently associated with both phage resistance and a reduction in fitness (Lenski and Levin 1985; Bohannan et al. 2002; Brockhurst et al. 2005). Our results therefore have implications for the use of phages as a biocontrol agent, or phage therapy. Broadly, however these results emphasize that even when phage application “fails” in the sense that bacteria are able to rapidly evolve resistance, the evolved population is likely to have lower fitness and therefore more likely to either be out‐competed by commensals or more readily dealt with by the host immune system.
Conclusion
In the natural environment phages can outnumber bacteria 20:1 (Wommack and Colwell 2000) and therefore selection for bacterial resistance to phages should be strong. Despite this, infective phages are routinely isolated from bacterial communities (Lu et al. 2003; Suttle 2005; Koskella et al. 2011), suggesting that the evolution of resistance may reduce competitive ability and therefore not be maintained under relaxed phage‐mediated selection. Our results from bacterial populations grown within the plant leaf support this hypothesis, and show that resistance to phage infection does indeed appear to be costly. The fact that we only observed these costs within the plant host, and not in vitro implies they may be more frequently observed in natural systems than has been reported in laboratory systems. As such, laboratory estimates of fitness may miss crucial mutational effects not just quantitatively but qualitatively.
Supporting information
Supplementary Information
ACKNOWLEDGMENTS
We thank Ben Longdon for valuable discussion of the results. We would also like to thank the editors and three anonymous reviewers for helpful comments and suggestions on previous versions of this manuscript. This work was funded in part by a NERC research fellowship to B.K. (NE/K00879X/1) and in part by a studentship to S.M. funded by the University of Exeter.
DATA ARCHIVING
http://dx.doi.org/10.5061/dryad.2qg86.
LITERATURE CITED
Associate Editor: D. Rozen
Handling Editor: J. Conner
- Agrawal, A. , and Lively C. M. 2002. Infection genetics: gene‐for‐gene versus matching‐alleles models and all points in between. Evol. Ecol. Res. 4:79–90. [Google Scholar]
- Antonovics, J. , and Alexander H. M.. 1995. Spread of anther‐smut disease (Ustilago violacea) and character correlations in a genetically variable experimental population of Silene alba . J. Ecol. 83:783–794. [Google Scholar]
- Balado, M. , Lemos M. L., and Osorio C. R.. 2013. Integrating conjugative elements of the SXT/R391 family from fish‐isolated Vibrios encode restriction‐modification systems that confer resistance to bacteriophages. FEMS Microbiol. Ecol. 83:457–467. [DOI] [PubMed] [Google Scholar]
- Banfield, J. F. , and Young M.. 2009. Microbiology. Variety—the splice of life—in microbial communities. Science 326:1198–1199. [DOI] [PubMed] [Google Scholar]
- Barrangou, R. , Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., and Horvath P.. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- Bohannan, B. J. M. , Kerr B., Jessup C. M., Hughes J. B., and Sandvik G.. 2002. Trade‐offs and coexistence in microbial microcosms. Antonie Van Leeuwenhoek 81:107–115. [DOI] [PubMed] [Google Scholar]
- Bohannan, B. J. M. , and Lenski R. E.. 2000. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3:362–377. [Google Scholar]
- Boots, M. , and Begon M.. 1993. Trade‐offs with resistance to a granulosis virus in the Indian meal moth, examined by a laboratory evolution experiment. Funct. Ecol. 7:528–534. [Google Scholar]
- Brockhurst, M. A. , Buckling A., and Rainey P. B.. 2005. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa . Proc. Biol. Sci. 272:1385–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling, A. , and Rainey P. B.. 2002a. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. Biol. Sci. 269:931–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling, A. . 2002b. The role of parasites in sympatric and allopatric host diversification. Nature 420:496–499. [DOI] [PubMed] [Google Scholar]
- Chen, C.‐Y. , Nace G. W., and Irwin P. L.. 2003. A 6 × 6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli . J. Microbiol. Methods 55:475–479 [DOI] [PubMed] [Google Scholar]
- Chisholm, S. T. , Coaker G., Day B., and Staskawicz B. J.. 2006. Host‐microbe interactions: shaping the evolution of the plant immune response. Cell 124:803–814. [DOI] [PubMed] [Google Scholar]
- Cingolani, P. , Platts A., Wang L. L., Coon M., Nguyen T., Wang L., Land S. J., Lu X., and Ruden D. M.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso‐2; iso‐3. Fly (Austin) 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas, I. , Borrell, S. , Roetzer, A. , Rose, G. , Malla, B. , Kato‐Maeda, M. Galaganm J, Niemann, S. and Gagneux, S. 2012. Whole‐genome sequening of rifampicin‐resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat. Genet. 44:106–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter, S. C. , Kruuk L. E. B., and Wilson K.. 2003. Costs of resistance: genetic correlations and potential trade‐offs in an insect immune. System. J. Evol. Biol. 17:421–429. [DOI] [PubMed] [Google Scholar]
- Dasgupta, T. and Lam J. S.. 1995. Identification of rfbA, involved in B‐band lipopolysaccharide biosynthesis in Pseudomonas aeruginosa serotype O5. Infect Immun. 63:1674–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, T. J. , Ind A., Komitopoulou E., and Salmond G. P.. 2010. Phage‐selected lipopolysaccharide mutants of Pectobacterium atrosepticum exhibit different impacts on virulence. J. Appl. Microbiol. 109:505–514 [DOI] [PubMed] [Google Scholar]
- Fierer, N. , Schimel J. P., and Holden P. A.. 2003. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 35:167–176. [Google Scholar]
- Filippov, A. A. , Sergueev K. V, He Y., Huang X.‐Z., Gnade B. T., Mueller A. J., Fernandez‐Prada C. M., and Nikolich M. P.. 2011. Bacteriophage‐resistant mutants in Yersinia pestis: identification of phage receptors and attenuation for mice. PLoS One 6:e25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H. H. 1971. Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9:275–296. [Google Scholar]
- Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541–548. [DOI] [PubMed] [Google Scholar]
- Graham, A. L. , Hayward A. D., Watt K. A., Pilkington J. G., Pemberton J. M., and Nussey D. H.. 2010. Fitness correlates of heritable variation in antibody responsiveness in a wild mammal. Science 330:662–625. [DOI] [PubMed] [Google Scholar]
- Jiang, S. , Fu W., Chu W., and Fuhrman J. A.. 2003. The vertical distribution and diversity of marine bacteriophage at a station off Southern California. Microb. Ecol. 45:399–410. [DOI] [PubMed] [Google Scholar]
- Kanehisa, M. , Goto S., Sato Y., Kawashima M., Furumichi M., and Tanabe M.. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42:199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella, B. and Brockhurst M. A.. 2014. Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 38:916–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella, B. , Lin D. M., Buckling A., and Thompson J. N.. 2012. The costs of evolving resistance in heterogeneous parasite environments. Proc. Biol. Sci. 279:1896–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella, B. , Thompson J. N., Preston G. M., and Buckling A.. 2011. Local biotic environment shapes the spatial scale of bacteriophage adaptation to bacteria. Am. Nat. 177:440–451. [DOI] [PubMed] [Google Scholar]
- Koskella, B. , and Taylor, T. B. 2015. The potential role of bacteriophages in shaping plant‐bacterial interactions. In Murillo J., Jackson R. W., Arnold D., and Vinatzer B., eds. Bacteria‐plant interactions: advanced research and future trends. Caister Academic Press, Poole, UK. [Google Scholar]
- Kraaijeveld, A. R. , and Godfray H. C.. 1997. Trade‐off between parasitoid resistance and larval competitive ability in Drosophila melanogaster . Nature 389:278–280. [DOI] [PubMed] [Google Scholar]
- Labbé, P. , Vale P. F., and Little T. J.. 2010. Successfully resisting a pathogen is rarely costly in Daphnia magna . BMC Evol. Biol. 10:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie, S. J. , Samson J. E., and Moineau S.. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327. [DOI] [PubMed] [Google Scholar]
- Labrie, S. J. , Tremblay D. M., Moisan M., Villion M., Magadán A. H., Campanacci V., Cambillau C., and Moineau S.. 2012. Involvement of the major capsid protein and two early‐expressed phage genes in the activity of the lactococcal abortive infection mechanism AbiT. Appl. Environ. Microbiol. 78:6890–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenscheid, J. , Killmann H., and Braun V.. 2004. A FhuA mutant of Escherichia coli is infected by phage T1‐independent of TonB. FEMS Microbiol. Lett. 234:133–137. [DOI] [PubMed] [Google Scholar]
- Lennon, J. T. , Khatana S. A. M., Marston M. F., and Martiny J. B. H.. 2007. Is there a cost of virus resistance in marine cyanobacteria?ISME J. 1:300–312. [DOI] [PubMed] [Google Scholar]
- Lenski, R. E. Experimental studies of pleiotropy and epistasis in Escherichia coli . 1988. I. Variation in competitive fitness among mutants resistant to virus T4. Evolution 42:425–432. [DOI] [PubMed] [Google Scholar]
- Lenski, R. E. , and Levin B. R.. 1985. Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am. Nat. 125:585–602. [Google Scholar]
- Lu, Z. , Breidt F., Plengvidhya V., and Fleming H. P.. 2003. Bacteriophage ecology in commercial sauerkraut fermentations. Appl. Environ. Microbiol. 69:3192–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lythgoe, K. A. , and Chao L.. 2003. Mechanisms of coexistence of a bacteria and a bacteriophage in a spatially homogeneous environment. Ecol. Lett. 6:326–334. [Google Scholar]
- Melnyk, A.H. , Wong, A. and Kassen, R. 2014. The fitness costs of antibiotic resistance mutations. Evol. Appl. 8:273‐283. DOI: 10.1111/eva.12196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto, M. , Underwood W., Koczan J., Nomura K., and He S. Y.. 2006. Plant stomata function in innate immunity against bacterial invasion. Cell 126:969–980. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, K. , Morita M., Fischer C. R., Yoichi M., Tanji Y., and Unno H.. 2003. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Appl. Environ. Microbiol. 69:170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nian, H. , Zhang J., Song F., Fan L., and Huang D.. 2007. Isolation of transposon mutants and characterization of genes involved in biofilm formation by Pseudomonas fluorescens TC222. Arch Microbiol. 188:205–213 [DOI] [PubMed] [Google Scholar]
- Nordling, D. , Andersson M., Zohari S., and Lars G.. 1998. Reproductive effort reduces specific immune response and parasite resistance. Proc. R. Soc. B Biol. Sci. 265:1291–1298. [Google Scholar]
- O'Brien, R. D. , and Lindow S. E.. 1989. Effect of plant species and environmental conditions on epiphytic population sizes of Pseudomonas syringae and other bacteria. Phytopathology 79:619–627. [Google Scholar]
- Parker, M. A. 1991. Local genetic differentiation for disease resistance in a selfing annual. Biol. J. Linn. Soc. 42:337–349. [Google Scholar]
- Purrington, C. B. and Bergelson J.. 1996. Surveying patterns in the cost of resistance in plants. Am. Nat. 148:536–558. [Google Scholar]
- Quance, M. A. , and Travisano M.. 2009. Effects of temperature on the fitness cost of resistance to bacteriophage T4 in Escherichia coli . Evolution 63:1406–1416. [DOI] [PubMed] [Google Scholar]
- Refardt, D. , and Kummerli, R. 2013. Defying bacteriophages: contrasting altruistic with individual‐based resistance mechanisms in Escherichia coli. Commun. Integr. Biol. 6:e25159. DOI:10.4161/cib.25159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Valera, F. , Martin‐Cuadrado A.‐B., Rodriguez‐Brito B., Pasić L., Thingstad T. F., Rohwer F., and Mira A.. 2009. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7:828–836. [DOI] [PubMed] [Google Scholar]
- Roux, F. , Gao L., and Bergelson J.. 2010. Impact of initial pathogen density on resistance and tolerance in a polymorphic disease resistance gene system in Arabidopsis thaliana . Genetics 185:283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A. 2000. Host‐parasite coevolution in a multilocus gene‐for‐gene system. Proc. Biol. Sci. 267:2183–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, B. C. , and Verhulst S.. 1996. Ecological immunology: costly parasite defences and trade‐offs in evolutionary ecology. Trends Ecol. Evol. 11:317–321. [DOI] [PubMed] [Google Scholar]
- Smith, S. G. , Mahon V., Lambert M.A., and Fagan R.P.. 2007. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 273:1–11. [DOI] [PubMed] [Google Scholar]
- Strauss, S. Y. , Rudgers J. A., Lau J. A., and Irwin R. E.. 2002. Direct and ecological costs of resistance to herbivory. Trends Ecol. Evol. 17:278–285. [Google Scholar]
- Suttle, C. A. 1994. The significance of viruses to mortality in aquatic microbial communities. Microb. Ecol. 28:237–243. [DOI] [PubMed] [Google Scholar]
- Suttle, C. A. 2005. Viruses in the sea. Nature 437:356–361. [DOI] [PubMed] [Google Scholar]
- Tazzyman, S. J. and Hall, A. R. 2015. Lytic phages obscure the cost of antibiotic resistance in Escherichia coli . Isme J. 9:809–820. DOI: 10.1038/ismej.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. N. 2005. The geographic mosaic of coevolution. Chicago Univ. Press, Chicago, IL. [Google Scholar]
- Venables, W. D. , and Ripley B. D.. 1994. Modern applied statistics with S‐PLUS, 2nd edition. Springer Verlag, New York. [Google Scholar]
- Verhulst, S. , Dieleman S. J., and Parmentier H. K.. 1999. A tradeoff between immunocompetence and sexual ornamentation in domestic fowl. Proc. Natl. Acad. Sci. 96:4478–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, M. , and Velicer G. J.. 2008. Isolation by distance in the spore‐forming soil Bacterium myxococcus xanthus. Curr. Biol. 18:386–391. [DOI] [PubMed] [Google Scholar]
- Walsh, C. T. , Wright G. D., Buchrieser C., Cole S. T., and Thurber R. V.. 2009. Current insights into phage biodiversity and biogeography. Curr. Opin. Microbiol. 12:582–587. [DOI] [PubMed] [Google Scholar]
- Wei, C.‐F. , Kvitko B. H., Shimizu R., Crabill E., Alfano J. R., Lin N.‐C., Martin G. B., Huang H.‐C., and Collmer A.. 2007. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1–1 is able to cause disease in the model plant Nicotiana benthamiana . Plant J. 51:32–46. [DOI] [PubMed] [Google Scholar]
- Wommack, K. E. , and Colwell R. R.. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino, D. R. , and Birney E.. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


