Summary
The single‐stranded, positive‐sense RNA genome of viruses in the genus Potyvirus encodes a large polyprotein that is cleaved to yield 10 mature proteins. The first three cleavage products are P1, HCpro and P3. An additional short open reading frame (ORF), called pipo, overlaps the P3 region of the polyprotein ORF. Four related potyviruses infecting sweet potato (Ipomoea batatas) are predicted to contain a third ORF, called pispo, which overlaps the 3′ third of the P1 region. Recently, pipo has been shown to be expressed via polymerase slippage at a conserved GA6 sequence. Here, we show that pispo is also expressed via polymerase slippage at a GA6 sequence, with higher slippage efficiency (∼5%) than at the pipo site (∼1%). Transient expression of recombinant P1 or the ‘transframe’ product, P1N‐PISPO, in Nicotiana benthamiana suppressed local RNA silencing (RNAi), but only P1N‐PISPO inhibited short‐distance movement of the silencing signal. These results reveal that polymerase slippage in potyviruses is not limited to pipo expression, but can be co‐opted for the evolution and expression of further novel gene products.
Keywords: frameshift, gene expression, overlapping gene, RNA silencing suppression, RNA virus, transcriptional slippage
Introduction
Several viruses of the genus Potyvirus (family Potyviridae) infect sweet potato (Ipomoea batatas). Currently, most fall within the monophyletic ‘Sweet potato feathery mottle virus (SPFMV) group’ (Untiveros et al., 2008). These include SPFMV, Sweet potato virus C (SPVC), Sweet potato virus G (SPVG) and Sweet potato virus 2 (SPV2), but not Sweet potato latent virus. SPFMV is the most common virus infecting sweet potatoes worldwide. In mixed infections with Sweet potato chlorotic stunt virus (SPCSV; genus Crinivirus), it is associated with a severe sweet potato virus disease (SPVD), causing heavy yield losses (Gibson et al., 2004; Tairo et al., 2005). SPVD is particularly important in sub‐Saharan Africa, where sweet potato is the second most important root crop after cassava.
The complete genome sequences of several SPFMV‐group viruses have been determined (Li et al., 2012; Pardina et al., 2012; Sakai et al., 1997; Untiveros et al., 2010; Yamasaki et al., 2010). Although all members of the genus Potyvirus encode a P1 protein, SPFMV‐group viruses encode the largest P1 among currently sequenced potyviruses (Fig. 1a) (Sakai et al., 1997; Valli et al., 2007). P1 is a serine protease that cleaves at its own C‐terminus, but is not known to be involved in other polyprotein‐processing events. Apart from its protease activity, the functions of P1 are not well established. It appears to play a variety of roles in different potyviruses, including enhancement of the RNA silencing suppression (RSS) activity of the helper component proteinase (HCpro), and roles in genome amplification and host specificity (reviewed in Rohožková and Navrátil, 2011; Valli et al., 2007). Although HCpro is generally considered to be the RSS protein in potyviruses (Anandalakshmi et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998), in members of some other potyvirid genera, P1 has been shown to be a potent RSS. In Sweet potato mild mottle virus (SPMMV; genus Ipomovirus), Triticum mosaic virus (genus Poacevirus) and Wheat streak mosaic virus (genus Tritimovirus), P1, but not HCpro, suppresses silencing (Giner et al., 2010; Tatineni et al., 2012; Young et al., 2012). Cassava brown streak virus (genus Ipomovirus) lacks HCpro, and P1 suppresses silencing (Mbanzibwa et al., 2009). Cucumber vein yellowing virus (genus Ipomovirus) also lacks HCpro, but has two P1 serine protease proteins, P1a and P1b, where P1b suppresses silencing (Valli et al., 2006). In Potato virus Y (PVY), P1 may be needed to stabilize the RSS protein HCpro (Tena Fernández et al., 2013). However, macluraviruses lack any P1 protein (Revers and Garcia, 2015).
Figure 1.
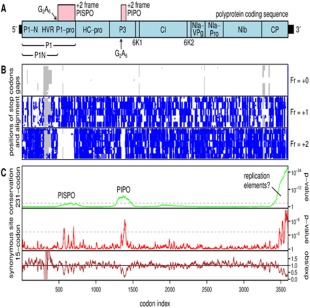
Genome organization of Sweet potato feathery mottle virus (SPFMV)‐group potyviruses. (a) Map of the ∼10.8‐kb genome showing the polyprotein open reading frame (ORF) (light blue) and overlapping pipo and pispo ORFs (pink). The P1‐N and P1‐pro domains and hypervariable region (HVR) are marked. For consistency with previous naming (SPFMV P1‐N and potyvirus P3N‐PIPO), we adopt the convention that ‘P1‐N’ refers to the N‐terminal domain, whereas ‘P1N’ refers to the entire region of P1 encoded upstream of the G2A6 slippage site. (b) Positions of stop codons (blue) in the three forward reading frames, and alignment gaps (grey) in an alignment of 31 SPFMV‐group sequences. (c) Conservation at synonymous sites in the alignment, using a 231‐codon sliding window (green) or a 15‐codon sliding window (red and brown). The upper panels (green, red) depict the probability that the degree of conservation within a given window could be obtained under a null model of neutral evolution at synonymous sites, whereas the bottom panel (brown) depicts the absolute amount of conservation as represented by the ratio of the observed number of substitutions within a given window to the number expected under the null model. The broken grey lines indicate a P = 0.05 threshold after applying a rough correction for multiple testing (namely 3519 codons/15‐ or 231‐codon window size).
The P1 protein of SPFMV‐group viruses appears to contain two distinct protein domains (Fig. 1a): the C‐terminal P1‐pro, which is homologous to the P1 serine protease encoded by all potyviruses, and an N‐terminal extension, designated P1‐N, which is absent from other potyviruses, but bears homology to the N‐terminal region of the P1 protein of the ipomovirus SPMMV (Untiveros et al., 2010; Valli et al., 2007). The P1‐N domain appears to have been transferred between ancestors of the two viruses, presumably during co‐infection of the common host. P1‐N and P1‐pro are separated by a hypervariable region (Untiveros et al., 2010). SPMMV P1‐N is an RSS which functions by binding Argonaute 1 (AGO1) (Giner et al., 2010). This is thought to be mediated via WG/GW motifs, and such motifs are a common feature of AGO‐binding proteins (Azevedo et al., 2010; El‐Shami et al., 2007; Giner et al., 2010; Till et al., 2007). Wild‐type SPFMV contains only one WG/GW motif in P1‐N, which was not found to confer any RSS activity (Szabó et al., 2012), whereas three WG/GW motifs are found in SPMMV P1‐N. Thus, despite the homology, it is unclear whether P1‐N serves a similar RSS function in SPFMV‐group viruses.
It was long believed that the potyvirus gene expression strategy was based on a large, single, open reading frame (ORF) encoding a polyprotein, which is processed to 10 mature proteins by three viral proteinases. However, Chung et al. (2008) discovered that there is another, short ORF in a different reading frame embedded within the region encoding P3. This ORF, designated pipo (‘pretty interesting Potyviridae ORF’), is conserved throughout the Potyviridae family. Polymerase slippage at a highly conserved GA6 sequence at the 5′ end of pipo results in the insertion of an extra A into 1%–2% of transcripts (Olspert et al., 2015; Rodamilans et al., 2015), leading, after translation and proteolytic cleavage, to the production of P3N‐PIPO, a ‘transframe fusion’ of the N‐terminal part of P3 (P3N) with the product (PIPO) of the pipo ORF. An additional, long alternative‐frame ORF in the region encoding P1‐pro was observed in the ‘SPFMV group’ of potyviruses and, in analogy with pipo, was named pispo (‘pretty interesting sweet potato potyvirus ORF’) (Fig. 1a) (Clark et al., 2012). The presence of multiple WG/GW motifs within the translation of pispo was noted by Clark et al. (2012), who suggested a potential role in RSS. Here, we present computational analyses of the pispo ORF that support its coding status, high‐throughput sequencing data demonstrating that pispo can be expressed as part of a transframe protein P1N‐PISPO via polymerase slippage at a conserved G2A6 slippery sequence, and experimental analyses that confirm that P1N‐PISPO is a potent suppressor of RNA silencing.
Results
Comparative genomic analysis of the overlapping pispo ORF
The polyprotein ORFs from the 28 full‐length SPFMV‐group sequences currently available in GenBank, plus three additional sequences determined in this study (see Experimental procedures), were extracted, translated, aligned as amino acid sequences and back‐translated to a nucleotide sequence alignment. Next, the positions of stop codons in the +1 and +2 reading frames relative to the polyprotein ORF, in all 31 sequences, were plotted. This confirmed the conserved presence of a long +2‐frame ORF overlapping the P1 region of the polyprotein ORF (Fig. 1b). The ORF has 230 codons in SPFMV and SPVC, and 228 codons in SPVG and SPV2 (measured from a ubiquitous 5′ G2A6 sequence; see below). The statistical significance of such a long ORF being conserved by chance over such a degree of phylogenetic divergence (estimated >900 phylogenetically independent substitutions within pispo in the SPFMV‐group sequence alignment) was assessed as described previously (Fang et al., 2012). Using this method, the probability of such a long +2 frame ORF occurring by chance at this location is P < 1 × 10−16.
Next, the alignment was analysed for enhanced conservation at polyprotein‐frame synonymous sites (an indicator of overlapping functional elements), as described previously (Firth, 2014). The analysis revealed regions of enhanced synonymous site conservation where the polyprotein ORF is overlapped by the pipo ORF (Chung et al., 2008), and at the 3′ end of the polyprotein ORF, where overlapping replicational RNA elements have been mapped in other potyvirus species (Fig. 1c) (Haldeman‐Cahill et al., 1998). Enhancement of polyprotein‐frame synonymous site conservation within the region overlapped by pispo was modest, suggesting that the PISPO amino acid sequence is not subject to strong purifying selection. Nonetheless, the mean rate of synonymous substitutions in the region of the polyprotein ORF overlapped by pispo was 10% below the genome average, with a corresponding total P value of 0.01 (Fig. 1c).
At the 5′ end of the pispo ORF, there is a conserved G2A6 sequence (Figs 2a and S1, see Supporting Information). In SPFMV and SPVC isolates, this takes the form GG_AAA_AAA, where underscores separate polyprotein codons. In SPVG and SPV2 isolates, it takes the form G_GAA_AAA_A, displaced a few codons upstream from the sequence in SPFMV and SPVC (Fig. 2a). A similar highly conserved sequence, G1–2A6–7 (G2A6 in SPFMV‐group viruses), is present at the 5′ end of the pipo ORF in nearly all potyvirids (Chung et al., 2008). The presence of the same motifs at the 5′ end of pispo suggests that pispo is expressed by the same slippage mechanism as pipo. This would result in the production of a P1N‐PISPO fusion protein (Fig. 2b). In the Ruk73 strain of SPFMV, P1 has a predicted mass of 77.1 kDa, whereas P1N‐PISPO has a predicted mass of 75.7 kDa. Analysis of additional sequences with only partial coverage of the polyprotein ORF, but with coverage of the 5′ end of pispo and/or the entire pispo ORF, confirmed the conserved presence of the potential slippage site (14 of 14 sequences) and the conserved presence of the pispo ORF (nine of nine sequences).
Figure 2.

Representative nucleotide and peptide sequences. (a) Nucleotide sequences flanking the proposed slippage sites in representative Sweet potato feathery mottle virus (SPFMV)‐group sequences (GenBank accession numbers shown on left, species on right). Spaces separate polyprotein‐frame codons; pispo is in the +2 frame. The G2A6 sequence, where slippage is proposed to occur, is highlighted in orange. Numbers on the left indicate the genomic coordinates of the first nucleotide in each line. (b) Amino acid sequences of P1 and P1N‐PISPO in the Ruk73 isolate of SPFMV. The amino acids encoded at the slippage site are highlighted in orange. Sequences corresponding to the epitopes of the two polyclonal antibodies (pAbs) are highlighted in pale blue. GW/WG motifs are underlined. (c) Sequences of the epitopes against which the two pAbs were raised. Sequence differences from the Ruk73 isolate are highlighted in red. SPV2, Sweet potato virus 2; SPVC, Sweet potato virus C; SPVG, Sweet potato virus G.
High‐throughput sequencing reveals polymerase slippage at the pispo and pipo G2A6 sites
The occurrence of the G2A6 sequence at the 5′ end of the pispo and pipo ORFs in different phases with respect to the polyprotein reading frame suggests that pispo and pipo are unlikely to be expressed via translational frameshifting, although, in principle, both are compatible with −1 ribosomal frameshifting (reviewed in Firth and Brierley, 2012). Further, using RNA‐folding software, such as alidot (Hofacker et al., 2002) and pknotsRG (Reeder et al., 2007), we failed to predict a convincing 3′ RNA secondary structure at a canonical spacing (i.e. 5–9 nucleotides) downstream of the slippery sites for the stimulation of −1 ribosomal frameshifting. To evaluate whether frameshifting could occur instead during RNA synthesis, we first utilized high‐throughput small RNA sequencing data obtained from SPFMV‐infected sweet potato plants and SPV2‐ and SPVG‐infected Ipomoea setosa plants. At both the pispo and pipo G2A6 sequences, a fraction of small RNA reads contained an additional ‘A’ insertion and, for most samples, insertions at these sites occurred well above background frequencies (Fig. 3; Table 1). In these data, ‘A’ insertions were found to occur at a frequency ranging from 0.9% to 11.4% at the pispo G2A6 site and 0.0% to 1.0% at the pipo G2A6 site, depending on the sample. The 0% values are probably a result of low coverage and the lower rate of insertions at the pipo site relative to the pispo site (see below). In comparison, average single nucleotide insertions at other sites across the genome were estimated to occur at a frequency ranging from 0.001% to 0.01% per nucleotide, depending on the sample (Table 1).
Figure 3.
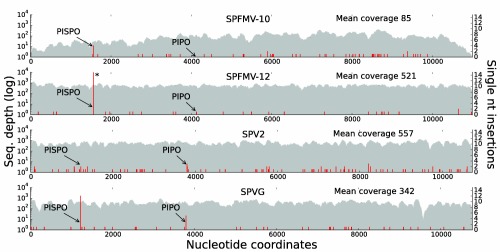
Single nucleotide insertions in short‐RNA sequencing of sweet potato virus‐infected plants. Short‐RNA reads from Sweet potato virus 2 (SPV2), Sweet potato virus G (SPVG) and two Sweet potato feathery mottle virus (SPFMV) [10, sweet potato only infected with SPFMV; 12, sweet potato infected with SPFMV + Sweet potato chlorotic stunt virus (SPCSV)] samples were mapped to the corresponding reference genomes. For each sample, the sequencing depth of fully matched reads is plotted as a 100‐nucleotide moving average in grey (log scale on left axis). The mean coverage is indicated at the top right. Positions of single nucleotide insertions are indicated in red (linear scale on right axis). For each virus, the positions of the G2A6 sequences at the beginning of the pispo and pipo open reading frames (ORFs) are indicated with arrows. An asterisk (*) denotes the off‐scale value of 43 insertions at the pispo G2A6 site for sample SPFMV‐12.
Table 1.
Number of small RNA sequences with (FS) and without (WT) an ‘A’ insertion at the pispo or pipo G2A6 slippage sites.
| Sample | PISPO | PIPO | Background % insertion rate | ||||
|---|---|---|---|---|---|---|---|
| WT | FS | FS % | WT | FS | FS % | ||
| SPFMV‐10* | 31 | 4 | 11.4 | 42 | 0 | 0.0 | 0.0106 |
| SPFMV‐12* | 576 | 43 | 7.0 | 219 | 1 | 0.5 | 0.0013 |
| SPV2 † | 226 | 2 | 0.9 | 291 | 3 | 1.0 | 0.0035 |
| SPVG † | 219 | 12 | 5.2 | 580 | 5 | 0.9 | 0.0023 |
SPCSV, Sweet potato chlorotic stunt virus; SPFMV,Sweet potato feathery mottle virus; SPV2,Sweet potato virus 2; SPVG,Sweet potato virus G.
*10, sweet potato only infected with SPFMV; 12, sweet potato infected with SPFMV + SPCSV.
†Ipomoea setosa infected with SPV2 or SPVG.
Although these results support the interpretation that polymerase slippage occurs at the pispo and pipo G2A6 sites, low coverage, variability between samples and low accuracy of insertion calling as a result of short read lengths mean that the slippage frequency is likely to be poorly estimated by this approach. Therefore, a separate sequencing experiment was conducted in which both the pispo and pipo sites in viral RNA were specifically targeted for amplification and sequencing. This experiment was conducted with RNA extracted from sweet potato or I. nil infected with SPFMV‐Ruk73, and led to sequencing depths ranging from 6.1 to 32.5 million reads for each slippage site region (Fig. 4; Table 2). Single ‘A’ insertions were observed within the pispo G2A6 sequence for 5.0% and 5.5% of sweet potato and I. nil reads, respectively. Within the pipo G2A6 sequence, single ‘A’ insertions were observed for 0.9% and 1.0% of reads, respectively. Control DNAs for the same regions showed no insertions at all in more than eight million reads, indicating that insertions did not occur during amplification, library preparation or sequencing. Parallel work with the Turnip mosaic virus G2A6 pipo slippage site indicated that any insertional slippage occurring during reverse transcription is less than ∼0.05% (Olspert et al., 2015). At a lower level, deletions were also observed to occur (Fig. 4), although these could be an artefact of deletional slippage during reverse transcription (Olspert et al., 2015).
Figure 4.
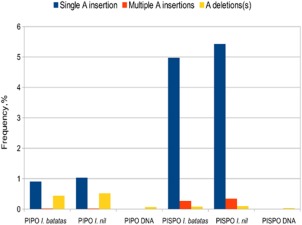
Transcriptional slippage at the pispo and pipo G2A6 sequences in Sweet potato feathery mottle virus (SPFMV)‐infected plants. Results from targeted high‐throughput sequencing of systemically infected Ipomoea batatas and I. nil. The frequencies of transcripts with a single ‘A’ insertion at the G2A6 sequence are shown in blue; the frequencies of transcripts with two or more inserted ‘A’ nucleotides are shown in orange; and the frequencies of transcripts with one or more ‘A’ nucleotides deleted are shown in yellow. Controls from plasmid template were included to assess the variability introduced during amplification and sequencing.
Table 2.
Targeted high‐throughput sequencing of the pispo and pipo G2A6 slippage sites.
| Sample | # reads | Reads with a single ‘A’ inserted (i.e. G2A7) | Reads with >1 ‘A’ inserted | Reads with 1 or more ‘A's deleted | |||
|---|---|---|---|---|---|---|---|
| % | # reads | % | # reads | % | # reads | ||
| PIPO Ipomoea batatas | 9149358 | 0.90 | 82637 | 0.02 | 1717 | 0.44 | 40314 |
| PIPO Ipomoea nil | 37396166 | 1.03 | 385513 | 0.02 | 8711 | 0.52 | 193787 |
| PIPO DNA | 8449879 | 0.00 | 0 | 0.00 | 0 | 0.07 | 5808 |
| PISPO I. batatas | 7298433 | 4.98 | 363120 | 0.27 | 19936* | 0.08 | 5771 |
| PISPO I. nil | 30255818 | 5.45 | 1650021 | 0.31 | 95093 † | 0.10 | 28964 |
| PISPO DNA | 8142625 | 0.00 | 0 | 0.00 | 0 | 0.03 | 2366 |
*74% G2A8, 26% G2A9.
†70% G2A8, 27% G2A9, 4% G2A10.
PISPO contains GW/WG motifs
The amino acid sequence of PISPO is less conserved than the amino acid sequence of the part of P1 that it overlaps (Table S1, see Supporting Information), e.g. there is 29%–40% amino acid identity in PISPO, but 48%–67% identity in P1‐pro between potyviruses that encode PISPO. Application of blastp, CS‐Blast, InterProScan, hhpred and phyre‐2 to PISPO revealed no significant homology to previously annotated or characterized proteins, as expected, as PISPO clearly evolved by ‘overprinting’ the ancestral P1‐pro domain (Altschul et al., 1990; Angermüller et al., 2012; Kelley and Sternberg, 2009; Quevillon et al., 2005; Söding et al., 2005). In addition, ∼60% of PISPO was predicted to be disordered according to PONDR‐FIT (Xue et al., 2010) (Fig. S2, see Supporting Information), again typical of many overlapping genes (Rancurel et al., 2009).
Further inspection of the amino acid sequence revealed WG/GW (‘AGO‐hook’) motifs. Such motifs play a key role within the P1‐N domain of SPMMV P1 (Giner et al., 2010) and some other plant viral silencing suppressors (Azevedo et al., 2010; de Ronde et al., 2014). Among 31 sequenced isolates, the PISPO sequences contain between one and three GW/WG motifs, whereas full‐length P1 and full‐length P1N‐PISPO contain between two and four GW/WG motifs (Fig. 5). It should be noted, however, that WG/GW di‐amino acids are not particularly uncommon in generic protein sequences.
Figure 5.
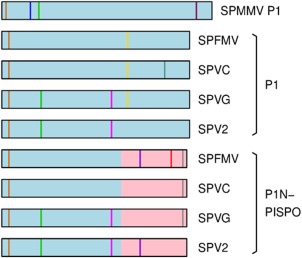
Locations of GW/WG sequences in P1 and P1N‐PISPO proteins of Sweet potato feathery mottle virus (SPFMV)‐group viruses. Schematic diagram of the P1 protein of Sweet potato mild mottle virus (SPMMV) and the P1 and P1N‐PISPO proteins of SPFMV‐group viruses, showing the positions of GW/WG sequences. The P1 sequence is indicated in light blue; the PISPO sequence is indicated in pink. GW/WG sequences are indicated by vertical coloured lines, with homologous instances (by blastp alignment) indicated with the same colour. SPV2, Sweet potato virus 2; SPVC, Sweet potato virus C; SPVG, Sweet potato virus G.
P1 and P1N‐PISPO are RNA silencing suppressors with distinguishable phenotypes
To test whether P1 and/or P1N‐PISPO have RSS activity, we used a standard silencing assay, whereby transgenic Nicotiana benthamiana line 16c plants that constitutively express green fluorescent protein (GFP) were agroinfiltrated with recombinant Agrobacterium tumefaciens strains harbouring binary expression constructs for GFP (pBIN:GFP) and a test protein (e.g. pBIN:P1). Overexpression of GFP mRNA from pBIN:GFP induces sense‐mediated silencing of GFP and results eventually in a red leaf patch under UV light, unless the test protein suppresses silencing, in which case the leaf patch will show enhanced green fluorescence under UV light. We tested various potyvirus proteins in this system (Fig. 6). At 4 days post‐infiltration (dpi), tissues co‐infiltrated for expression of GFP and P1 or P1N‐PISPO showed slightly brighter GFP fluorescence than those infiltrated for expression of SPFMV HCpro, Sweet potato latent virus (SPLV) P1 or a β‐glucuronidase (GUS) control (Fig. 7a), indicating that P1 and P1N‐PISPO have RSS activity. Leaf tissues expressing Potato virus A (PVA) HCpro exhibited the strongest GFP signals (Fig. 7a). These phenotypic differences correlated positively with the accumulation of GFP (Fig. 7b) and GFP mRNA (Fig. 7c), and were negatively correlated with the accumulation of GFP mRNA‐derived small interfering RNA (siRNA) in the leaf tissue (Fig. 7c). The expression of P1 and P1N‐PISPO was verified by western analysis using peptide antibodies (Abs) generated against an epitope (Fig. 2c) common to P1 and P1N‐PISPO (blot ‘P1’ in Fig. 7b) and peptide Abs specific to P1N‐PISPO (Fig. 7b). The observed bands were of slightly different sizes than expected, possibly as a result of post‐translational modifications.
Figure 6.
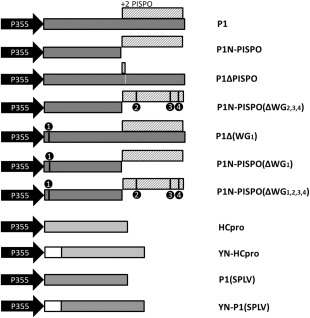
Binary vectors for the expression of viral proteins in leaves by agroinfiltration. Cauliflower mosaic virus 35S promoter‐driven binary vectors were designed to express the following proteins: P1, P1 of Sweet potato feathery mottle virus (SPFMV) [the shaded bar indicates the position of the +2 frameshift pispo open reading frame (ORF) encoding 230 residues]; P1N‐PISPO, engineered transframe sequence for the expression of the N‐proximal part of P1 together with PISPO; P1ΔPISPO, P1 including a stop codon inserted in the +2 frame pispo ORF to ensure that P1N‐PISPO is not produced; P1N‐PISPO(ΔWG2,3,4), P1N‐PISPO mutated at three positions (W514G515, W624G625 and W665G666; designated as WG motifs 2, 3 and 4, respectively) to substitute the tryptophan residues for alanines; P1(ΔWG1), P1 mutated to substitute residue W25 for alanine (WG motif 1); P1N‐PISPO(ΔWG1), P1N‐PISPO with W25 mutated to alanine; P1N‐PISPO(ΔWG1,2,3,4), P1N‐PISPO with all four WG motif tryptophans mutated to alanine; HCpro, HCpro of SPFMV; YN‐HCpro, the N‐proximal part of the yellow fluorescent protein (YN) fused to the N‐terminus of SPFMV HCpro; P1(SPLV), P1 of Sweet potato latent virus; YN‐P1(SPLV), the N‐proximal part of the yellow fluorescent protein (YN) fused to the N‐terminus of SPLV P1.
Figure 7.
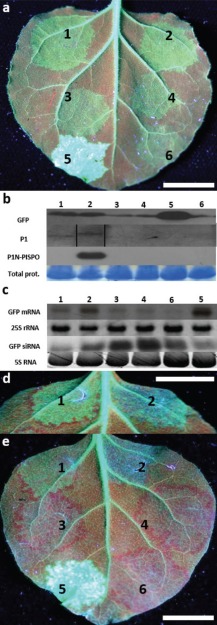
Suppression of sense‐mediated RNAi of green fluorescent protein (GFP) expression by potyviral proteins in leaves of GFP‐transgenic Nicotiana benthamiana line 16c. (a) Silencing of gfp was induced by overexpression of GFP mRNA ‘on the spot’ from a binary vector pBIN:GFP introduced by agroinfiltration, and interference with silencing was tested by co‐introduction of binary vectors for the expression of: (1) Sweet potato feathery mottle virus (SPFMV) P1; (2) SPFMV P1N‐PISPO; (3) SPFMV HCpro; (4) Sweet potato latent virus (SPLV) P1; (5) Potato virus A (PVA) HCpro; or (6) β‐glucuronidase (GUS) (control). The leaves were photographed at 4 days post‐infiltration (dpi) (a), 6 dpi (d) and 8 dpi (e). Scale bars indicate 2 cm. (b) Expressed proteins were detected at 4 dpi using antibodies to GFP, peptide antibodies generated against an epitope common to P1 and P1N‐PISPO, and peptide antibodies specific to the PISPO domain of P1N‐PISPO. In the P1 panel, lane 2 was loaded in a different order in the gel, and spliced electronically afterwards (indicated by black vertical lines), for consistency with other panels. Staining of total proteins by Coomassie blue was used as a loading control (shown for GFP gel). (c) Northern blot analysis for the detection of GFP mRNA and gfp‐derived small interfering (si)RNA in leaf tissues co‐infiltrated with pBIN:GFP and the viral constructs in (A) at 4 dpi; gfp was used as a probe and 28S and 5S ribosomal RNAs (rRNA) were used as loading controls. Note the different order of samples 5 and 6 in panels (b) and (d). (d) The infiltrated leaf tissue expressing P1 was surrounded by a red halo (left), in contrast with the tissue expressing P1N‐PISPO (right), which was most apparent at 6 dpi. (e) Suppression of silencing by P1, P1N‐PISPO and PVA HCpro was indicated by GFP fluorescence in the co‐infiltrated leaf tissue at 8 dpi, whereas no suppression of silencing was observed with SPFMV HCpro and SPLV P1.
The suppression of silencing by P1 and P1N‐PISPO was quite apparent at 6 dpi (Fig. 7d) and at 8 dpi (Fig. 7e), as indicated by GFP fluorescence in the co‐infiltrated leaf tissue, whereas the leaf spots infiltrated for the expression of SPFMV HCpro, SPLV P1 and GUS had lost GFP fluorescence (Fig. 7e). However, P1 and PIN‐PISPO differed in that a red halo developed around the leaf tissue infiltrated for the expression of P1. The red halo was most apparent at 6 dpi (Fig. 7d,e, leaf spot 1), indicating short‐distance movement of the silencing signal. A red halo also developed around the leaf spots infiltrated for the expression of SPFMV HCpro, SPLV P1 and GUS (Fig. 7e). In contrast, only a faint red halo, if any, developed around the leaf tissue infiltrated for the expression of P1N‐PISPO (Fig. 7d,e, leaf spot 2), indicating interference with the short‐distance signalling for silencing.
As no Abs were available for detection of SPFMV HCpro and SPLV P1, these proteins were also expressed tagged with the N‐proximal part (YN) of yellow fluorescent protein (YFP) (Fig. 6) and tested by western blot using anti‐YN Abs. YN‐P1 (SPLV) was detected at 2 dpi (Fig. S3, see Supporting Information), but only occasionally at later time points (3–5 dpi), whereas YN‐HCpro could not be detected at any time point (2–10 dpi) in eight independent experiments.
As it is possible that the P1 construct could also express P1N‐PISPO via translational or transcriptional frameshifting, we generated another P1 expression construct, P1ΔPISPO, in which expression of the pispo ORF was prevented by the insertion of a stop codon (Fig. 6). As with P1 (leaf spot 1; Fig. 8a), P1ΔPISPO (leaf spot 3; Fig. 8a) suppressed GFP silencing and was associated with the formation of a red halo.
Figure 8.
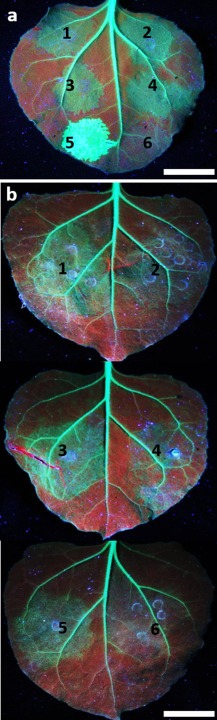
Effects of mutation of the WG motifs in P1 and P1N‐PISPO on the suppression of sense‐mediated RNAi of green fluorescent protein (GFP) expression in leaves of GFP‐transgenic Nicotiana benthamiana line 16c. Silencing of gfp was induced by overexpression of GFP mRNA from a binary vector pBIN:GFP introduced by agroinfiltration, and interference with silencing was tested by co‐introduction of binary vectors for expression of Sweet potato feathery mottle virus (SPFMV) P1, P1N‐PISPO or their mutated forms (see Fig. 3). (a) Suppression of silencing by: (1) P1, (2) P1N‐PISPO, (3) P1ΔPISPO, (4) P1N‐PISPO(ΔWG2,3,4), (5) Potato virus A (PVA) HCpro or (6) β‐glucuronidase (GUS) control at 4 days post‐infiltration (dpi). (b) Half‐agroinfiltrated leaf arrays were used to test the suppression of silencing by: (1) P1, (2) P1(ΔWG1), (3) P1N‐PISPO, (4) P1N‐PISPO(ΔWG1), (5) P1N‐PISPO(ΔWG2,3,4) or (6) P1N‐PISPO(ΔWG1,2,3,4). Scale bars indicate 2 cm.
The WG motif at the N‐terminus of P1 is pivotal for suppression of RNA silencing
The completely conserved WG motif near the N‐terminus of SPFMV‐group P1 (residues 25–26) aligns with the first WG motif in SPMMV P1 (Fig. 5), which has been shown to be required for SPMMV P1 RSS activity (Giner et al., 2010). To test whether this WG motif is also required for the RSS activity of SPFMV P1 and/or P1N‐PISPO, we mutated the tryptophan residue (W25) to alanine and tested the mutated P1 and P1N‐PISPO for RSS activity, as above. In contrast with P1 and P1N‐PISPO (Fig. 8b, leaf spots 1 and 3, respectively), the mutated constructs for P1(ΔWG1) and P1N‐PISPO(ΔWG1) (Fig. 6) were unable to suppress silencing (Fig. 8b, leaf spots 2 and 4, respectively). Similarly, when all four WG motifs were mutated [P1N‐PISPO(ΔWG1,2,3,4)] (Fig. 6), no suppression of silencing was observed (Fig. 8b, leaf spot 6). In contrast, similar mutations introduced in all three WG motifs in the PISPO domain [P1N‐PISPO(ΔWG2,3,4)] (Fig. 6) did not prevent the suppression of silencing (Fig. 8b, leaf spot 5). Taken together, these results indicate that the WG motif near the N‐terminus of P1 is essential for suppression of RNAi by both P1 and P1N‐PISPO.
Discussion
Potyviruses were believed to use a single polyprotein gene expression strategy until the recent discovery of the overlapping pipo ORF (Fig. 1a) (Chung et al., 2008). The pipo ORF has a highly conserved G1–2A6–7 sequence at its 5′ end. Previously, it was proposed that the expression of P3N‐PIPO might depend on translational or transcriptional frameshifting occurring at this site (Chung et al., 2008). However, the true nature of the expression mechanism was not confirmed until recently, when, during the preparation of this article, both we and others observed the production of virus transcripts with single nucleotide insertions at the pipo G1–2A6–7 sequence in various potyviruses, thus allowing the expression of P3N‐PIPO (this study; Olspert et al., 2015; Rodamilans et al., 2015). The ubiquitous presence of a G2A6 sequence at the 5′ end of the pispo ORF, conserved in all SPFMV‐group sequences, suggested that pispo might be expressed via the same polymerase slippage mechanism, leading to a P1N‐PISPO trans‐frame fusion protein. The potential for P1N‐PISPO to be expressed in this way was confirmed by the observation of edited transcripts in our small RNA sequencing data for SPFMV, SPV2 and SPVG, and our targeted high‐throughput sequencing of SPFMV RNA. The latter data also indicate that slippage at the SPFMV pispo slip site (5.0%–5.4%) is significantly more efficient than at the SPFMV pipo slip site (0.9%–1.0%). Whilst this article was in preparation, a similar difference was also observed by others (Rodamilans et al., 2015). The reasons for the differences between the two sites are unknown, but may be a result of the flanking nucleotide context which may affect the stability of the template:nascent RNA duplex adjacent to the slippage site (Hausmann et al., 1999). Although we have currently been unable to detect P1N‐PISPO in infected plants by western analysis (perhaps because of low expression, masking of the Ab epitope and/or the kinetics of virus infection and protein turnover), the presence of edited transcripts implies that P1N‐PISPO can indeed be expressed during virus infection.
P1 is the most variable potyvirus protein in terms of length and sequence (Adams et al., 2005; Valli et al., 2007). The discovery of PISPO adds a new dimension to the functional variability occurring in the P1 region. Unlike PIPO, which is found universally in the Potyviridae family, PISPO is restricted to potyviruses in the SPFMV group. It does not occur in the related SPLV, nor in the ipomovirus SPMMV with which SPFMV‐group viruses share a related P1‐N domain. The PISPO sequence is highly variable (Table S1) (Li et al., 2012) and less conserved than the overlapping region of P1‐pro, in clear contrast with PIPO and the region of P3 that it overlaps (Chung et al., 2008). Differences in the evolutionary plasticity of pipo and pispo may reflect the functional importance of the corresponding transframe proteins. P3N‐PIPO is an essential protein, and appears to be of ancient origin, as attested by its conservation throughout the Potyviridae family, whereas P1N‐PISPO seems to be a more recent evolutionary development and may therefore not yet have acquired a critical role, allowing more sequence plasticity, as has been observed for other recently evolved overlapping viral genes (Sabath et al., 2012). PISPO is predicted to be largely disordered (Fig. S2). The function of such an intrinsically unstructured protein region may be largely independent of most of the primary sequence, allowing for rapid evolution and exploration of sequence space (Gitlin et al., 2014).
Although it is clear that PISPO is biologically relevant, as an ORF of this length would not be conserved by chance (P < 1 × 10−16) over the given phylogenetic divergence, at present there is no infectious clone of SPFMV, and so it is not possible to verify the functional relevance of P1N‐PISPO during infection. However, given the importance of WG/GW domains in other silencing suppressors (Giner et al., 2010; de Ronde et al., 2014), the evaluation of P1N‐PISPO for RSS activity was an obvious course. Surprisingly, we found that both P1N‐PISPO and P1 are RNA silencing suppressors in SPFMV. Intrinsic silencing suppression has not been described previously for P1 proteins of potyviruses (Rajamäki et al., 2005; Tena Fernández et al., 2013), nor was it found herein for P1 of SPLV (which clusters as an immediate outgroup to the SPFMV group). Most recent reports assign potyvirus P1 a role for increasing the accumulation of HCpro and consequently enhancing its silencing activity (Tena Fernández et al., 2013). We also showed that P1 and P1N‐PISPO seem to have different silencing activities in SPFMV, a strategy described for different RSS proteins of other viruses as well (Lu et al., 2004; Senshu et al., 2011). P1 operates only at a single‐cell or local level, as evidenced by the formation of the red halo around the patch (Fig. 7), as also reported for SPMMV P1 and a few other virus silencing suppressors (Baumberger et al., 2007; Giner et al., 2010). The effect of P1 on silencing was a result of the sole action of this protein, and participation of traces of P1N‐PISPO can be discarded as the phenotype was identical to P1ΔPISPO (Fig. 8), in which PISPO is made untranslatable by introducing a stop codon. However, P1N‐PISPO transiently inhibited RNA silencing at both local and short‐range levels, as reported for suppressors that sequester siRNA, including PVA HCpro and tombusvirus P19 (Lakatos et al., 2004; Shiboleth et al., 2007). The elimination of the three PISPO WG/GW motifs did not remove the suppressor capacity of P1N‐PISPO (Fig. 8). However, the elimination of the single WG near the N‐terminus of P1‐N completely eliminated RSS activity of both P1 and P1N‐PISPO. Thus, the differing C‐termini of P1 and P1N‐PISPO modulate the RSS activity resident within P1‐N.
The results of our experiments differ from those of a previous study in which P1 of SPFMV was reported not to suppress RNA silencing (Szabó et al., 2012), probably because a different experimental system and SPFMV isolate were used.
The functional characterization of P1 and P1N‐PISPO requires further experimental work, in particular during SPCSV synergisms. SPCSV RNase3 protein mediates these synergisms through the suppression of antiviral defence in sweet potato plants, allowing potyviruses to enhance titres (Cuellar et al., 2009). Nevertheless, symptom severity, even among SPFMV‐group viruses, varies greatly (Gutierrez et al., 2003; Kokkinos and Clark, 2006; Untiveros et al., 2007), and this is not necessarily related to the enhancement of viral replication (Kokkinos and Clark, 2006). This suggests that additional viral determinants may be required for the development of unusually severe disease in sweet potato plants co‐infected with SPFMV and SPCSV. Such determinants might include P1 and P1N‐PISPO. Therefore, the results of this study may have important implications for understanding the devastating synergistic diseases caused by these viruses in sweet potato.
Experimental Procedures
Viruses
SPFMV isolate Ruk73 from Uganda (Tugume et al., 2010) was used to generate all the SPFMV‐derived expression vectors. In addition, a Taiwanese isolate of SPLV (Wang et al., 2013), available from the International Potato Center (CIP) reference virus collection, was used to generate the corresponding P1 expression construct. SPFMV‐Piu3 (Kreuze et al., 2009), and SPVG‐LSU1 and SPV2‐LSU2 (kindly provided by Dr Christopher A. Clark, Louisiana State University, Baton Rouge, LA, USA), available from the CIP reference isolate collection, were used for small RNA sequencing.
Sequencing of SPVG and SPV2 by assembly of siRNAs
siRNA sequencing (provider: Fasteris Life Sciences SA, Plan‐les‐Ouates, Switzerland) and assembly were performed on leaf samples from I. setosa plants infected with SPVG‐LSU1 or SPV2‐LSU2, as described by Kreuze et al. (2009). Original siRNA reads can be found at: https://research.cip.cgiar.org/confluence/display/cpx/CIP.sweetpotato.2015. The full genome sequences of SPVG (10 771 nucleotides) and SPV2 (10 732 nucleotides) were assembled at an average sequencing depth of 342‐ and 557‐fold, respectively, and deposited in GenBank (Table S2, see Supporting Information).
Computational analysis
The complete genomes of SPFMV‐Ruk73, SPVG‐LSU1 and SPV2‐LSU2 were determined in this study (see above). Additional SPFMV‐group nucleotide sequences in GenBank were identified using tblastn (Altschul et al., 1990) (see Table S2 for accession numbers). Coding sequences were extracted, translated, aligned and back‐translated to produce nucleotide sequence alignments using EMBOSS and ClustalW (Larkin et al., 2007; Rice et al., 2000). Synonymous site conservation was calculated as described previously (Firth, 2014). The probability of the conserved absence of stop codons in the pispo ORF occurring by chance (i.e. if pispo was actually non‐coding) was assessed via P1‐frame alignment codon column shuffling, as described previously (Fang et al., 2012).
Plasmids
A schematic diagram of the vectors used in this study is displayed in Fig. 6. Briefly, DNA fragments containing the complete coding regions of SPFMV P1 and HCpro (isolate Ruk73), and SPLV P1 (KC443039.1), were amplified by polymerase chain reaction (PCR) using Phusion high‐fidelity DNA polymerase (Thermo Scientific, Waltham, MA, USA) and cloned into the pBIN61 binary vector using XbaI/XmaI restriction sites. The sequence encoding the YFP N‐proximal fragment (YN) was added to the 3′ end of SPFMV HCpro and SPLV P1 using a similar approach to that described previously (Haikonen et al., 2013). Appropriate primers were used to introduce an AUG start or a UAA stop codon for the cloned regions where required. Thus, binary vectors containing wild‐type viral proteins included pBIN61:SPFMV‐P1, pBIN61:YN‐SPFMV‐HCpro and pBIN61:YN‐SPLV‐P1.
In addition, SPFMV P1 (hereafter called P1) was cloned into the expression vector pQE 30 (Qiagen, Venlo, the Netherlands) using HindIII/BamHI restriction sites, generating plasmid pQE30:P1. From this plasmid, other constructs were developed, including P1N‐PISPO, P1ΔPISPO, P1N‐PISPO(ΔWG2,3,4), P1(ΔWG1), P1N‐PISPO(ΔWG1) and P1N‐PISPO(ΔWG1,2,3,4), using appropriate primers and the Site Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). The transframe P1N‐PISPO construct contains a 1‐nucleotide ‘A’ insertion at site 1464 (i.e. G2A6 to G2A7 mutation), resulting in a frame change and the production of a P1N‐PISPO protein identical to that expected from polymerase slippage. In addition, two silent ‘A’ to ‘G’ mutations were introduced in P1N‐PISPO at genomic sites 1460 and 1463 within the G2A7 sequence (Lys 448 and 449; GG_AAA_AAA_A to GG_AAG_AAG_A) in order to prevent translational or transcriptional slippage occurring on the run of seven 'A's. P1ΔPISPO is a modified version of P1 including a synonymous mutation (GCA to GCT) at serine‐456 in the polyprotein frame, resulting in a +2 frame stop codon that prevents any accidental expression of P1N‐PISPO from this construct. P1N‐PISPO(ΔWG2,3,4) has the tryptophan residue in each of the three WG motifs in P1N‐PISPO replaced with alanine. PCR‐derived DNA fragments from the mutant versions were subsequently cloned into pBIN61, generating the corresponding plasmids pBIN61:P1N‐PISPO, pBIN61:P1ΔPISPO, pBIN61:P1N‐PISPO(ΔWG2,3,4), pBIN61:P1(ΔWG1), pBIN61:P1N‐PISPO(ΔWG1) and pBIN61:P1N‐PISPO(ΔWG1,2,3,4). All plasmids were verified by sequencing. The primers used are shown in Table S3 (see Supporting Information). Additional binary vectors pAHCpro, p35S‐GUS‐INT and pAGFP, used in the present study for the expression of PVA HCpro, GUS‐INT and GFP, respectively, have been described previously (Kreuze et al., 2005).
Agroinfiltration assays
Agroinfiltration was carried out as described in Johansen and Carrington (2001). Agrobacterium tumefaciens C58C1 strains carrying the indicated plasmids were combined before infiltration. The different Agrobacterium cultures were diluted with infiltration medium so that equal final optical densities were reached before combining cultures in a 1 : 1 ratio for infiltration. Transgenic N. benthamiana line 16c, which expresses GFP, was used for infiltration (seeds kindly provided by Professor D. Baulcombe, Department of Plant Sciences, University of Cambridge, Cambridge, UK). Co‐infiltration was carried out with two A. tumefaciens strains expressing GFP and P1, P1N‐PISPO, SPFMV HCpro, SPLV P1, PVA HCpro, GUS, P1ΔPISPO, P1N‐PISPO(ΔWG2,3,4), P1(ΔWG1), P1N‐PISPO(ΔWG1) or P1N‐PISPO(ΔWG1,2,3,4). GFP fluorescence was monitored daily by epi‐illumination with a hand‐held UV source (Blackray UVP 100 AP, Thermo Scientific). Photographs were taken with a digital camera, and images were prepared with Photoshop software (Adobe, San Jose, CA, USA). Samples were collected at different time points to ensure the detection of the relevant proteins.
Antibodies and western blotting
Polyclonal Abs to predicted antigens within PISPO and P1N (present in both P1 and P1N‐PISPO) were prepared by GenScript (Piscataway, NJ, USA). Anti‐P1N was raised against peptide sequence KTQTPGEVHHKMVR. Anti‐PISPO was raised against peptide sequence RAHASSPENSLQKG. Rabbits were injected with one of the two peptides, and Abs were affinity purified from immune sera. The detection of GFP YN‐tagged proteins and GFP was achieved using GFP‐specific polyclonal rabbit antiserum (Invitrogen, Carlsbad, CA, USA). Total proteins were extracted from leaves as follows: 20 mg of leaf tissue were homogenized in liquid nitrogen and resuspended in 80 μL of 2 × extraction buffer [0.1 m Tris‐HCl, 20% glycerol, 10% (v/v) 2β‐mercaptoethanol, 4% (w/v) sodium dodecyl sulfate (SDS), 0.2% bromophenol blue], boiled for 5 min and clarified by centrifugation. When needed, proteins were concentrated from larger amounts of plant material using acetone precipitation. Proteins were analysed on a 12% SDS‐polyacrylamide gel by electrophoresis and electroblotted onto a Hybond‐P polyvinylidene fluoride membrane (GE Healthcare, Chalfont St Giles, Buckinghamshire, UK). Membranes were blocked in 50 mL of Tris‐HCl (TBS) with 0.1% Tween 20 and 5% skimmed milk powder, and then probed with the corresponding Abs (1 : 5000 P1, PISPO) at 4°C overnight. Following washing with 1 × TBS containing 0.1% Tween 20, the membrane was incubated with polyclonal donkey anti‐rabbit immunoglobulins (IgGs) conjugated with horseradish peroxidase (HRP) (GE Healthcare). Proteins were detected with the enhanced chemiluminescence method using Super Signal West Femto chemiluminescent substrate (Thermo Scientific), and visualized by exposure to photographic film (Kodak, Rochester, NY, USA).
RNA analysis
The accumulation of GFP mRNA and GFP mRNA‐derived siRNA was evaluated by northern blot analysis in leaf tissues agroinfiltrated for co‐expression of GFP and P1, P1N‐PISPO, SPFMV HCpro, SPLV P1, PVA HCpro or GUS. Total RNA was extracted with TRIzol LS Reagent (Invitrogen) according to the manufacturer's recommendations and low‐molecular‐weight (LMW) and high‐molecular‐weight (HMW) RNA fractions were separated as described in Cuellar et al. (2009). LMW RNA (15 μg/lane) and HMW RNA (10 μg/lane) were separated by electrophoresis in a denaturing 15% polyacrylamide gel and 1.2% (w/v) agarose gel, respectively, both containing formaldehyde. The two gels were transferred to a nylon membrane (GE Healthcare) and cross‐linked chemically with 1‐ethyl‐3‐(3‐dimethylaminopropyl)carbodi‐imide (Pall and Hamilton, 2008) or by UV light (Sambrook and Russell, 2001), respectively. A GFP mRNA‐specific radioactive RNA probe was synthesized, fractionated with carbonate hydrolysis and used to probe the membranes for GFP mRNA and GFP mRNA‐derived siRNA, as described previously (Cuellar et al., 2009). Signals were detected by autoradiography on an X‐ray film (Kodak).
High‐throughput sequencing
Targeted high‐throughput sequencing was performed on total RNA extracted from SPFMV‐Ruk73‐infected sweet potato or I. nil plants, as described in Olspert et al. (2015), except that reverse transcription‐polymerase chain reaction (RT‐PCR) was performed with primers to amplify the regions surrounding the putative slippage sites for pispo (target site, excluding primer and adaptor sequences: ACTCGTTTGGAAAAAACTGGAC, corresponding to SPFMV‐Ruk73 nucleotides 1448–1469) and pipo (target site, excluding primer and adaptor sequences: CTCATGGAAAAAATTTGGGAT, corresponding to SPFMV‐Ruk73 nucleotides 4017–4037)
High‐throughput sequencing of small RNA was performed on RNA extracted from infected sweet potato (SPFMV‐Piu3) or I. setosa (SPVG‐LSU1 and SPV2‐LSU2) plants as described above. Short‐RNA reads, both forward and reverse sense, were mapped to the respective reference genomes using BWA (Li and Durbin, 2009), and positions of insertions were determined from the BWA CIGAR and TAG report fields. Initial background single nucleotide insertion rates were calculated by dividing the observed number of insertions (excluding those at the pispo and pipo G2A6 sites) by the product of the mean read depth and genome length. As a result of the short length of reads (21–24 nucleotides) and BWA not reporting insertions or deletions (indels) that occur within 5 nucleotides of the termini of reads, these initial values were adjusted for the potentially unreported indels by multiplying by the correction factor ∑i Pi i/(i – 10), where i ranges over the read lengths (i.e. 21–24 nucleotides), Pi is the fraction of mapped reads that have length i, and i/(i – 10) is the reciprocal of the proportion of insertions occurring within a read of length i that would be detected by BWA (i.e. are not within 5 nucleotides of the read termini).
Accession numbers
GenBank sequence accession numbers are provided in Table S2.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Predicted pispo G2A6 slippage sites. Predicted pispo G2A6 slippage sites (orange highlighted) in all available full‐length Sweet potato feathery mottle virus (SPFMV)‐group sequences. SPV2, Sweet potato virus 2; SPVC, Sweet potato virus C; SPVG, Sweet potato virus G.
Fig. S2 PONDR® disorder predictions for P1 and P1N‐PISPO. Common regions predicted to be unstructured or disordered include a small region of ∼25 residues within the N‐terminus of P1N, and the hypervariable region (residues 200–300). In P1, a third region of predicted disorder occurs around residues 459–522, whereas, in P1N‐PISPO, a third region of predicted disorder encompasses almost the entire PISPO domain starting around residue 444. Predicted non‐disordered domains have scores no higher than 0.5.
Fig. S3 Expression and detection of YN‐P1(SPLV) in agroinfiltrated leaf tissue of green fluorescent protein (GFP)‐transgenic Nicotiana benthamiana line 16c. (a) Neither (1) YN‐P1(SPLV) (left half of the leaf) nor (2) YN‐HCpro (right half of the leaf) of Sweet potato feathery mottle virus (SPFMV) suppressed gfp silencing and no enhancement of GFP fluorescence was observed at 4 days post‐infiltration (dpi). The infiltrated tissue was surrounded by a red edge indicating short‐distance movement of the silencing signal. Scale bar indicates 2 cm. (b) Expressed YN‐P1(SPLV) protein was detected at 2 dpi using anti‐YN antibodies, but not at 3, 4 or 5 dpi. The upper band indicated with an arrow (2d) corresponds to the expected size (72 kDa) of YN‐P1(SPLV). The small arrow on the left indicates the position of the 70‐kDa protein marker. Staining of total proteins by Coomassie blue was used as a loading control.
Table S1 Mean amino acid inter‐ and intra‐species identities. Identities (%) calculated for the PISPO (italics; upper left) and P1‐pro (roman; lower right) domains.
Table S2 Virus species and accession number for sequences used for the computational analysis of the pispo open reading frame (ORF). Sequences of Sweet potato virus G (SPVG)‐LSU1 and Sweet potato virus 2 (SPV2)‐LSU2 were determined in this study, and partial sequences with coverage of pispo also included are indicated in a separate column.
Table S3 Primer sequences used in this study.
Acknowledgements
Segundo Fuentes is acknowledged for technical assistance. Work in AEF's laboratory was funded by grants from the Wellcome Trust (088789 and 106207) and the Biotechnology and Biological Sciences Research Council (BBSRC) (BB/J007072/1 and BB/J015652/1). Work at the University of Helsinki was funded by the Academy of Finland (1134335 and 1276136), and work at the International Potato Center (CIP) was funded by the Consultative Group for International Agricultural Research (CGIAR) consortium research programme for roots, tubers and bananas.
Note Added in Proof
After this manuscript was accepted a complementary study by Mingot et al. was accepted in Journal of Virology (doi:10.1128/JVI.02360‐15; posted online). The study by Mingot et al. largely corroborates our findings and confirms the existence of PISPO expression in infected plants by mass spectrometry.
NCBI GenBank.
References
- Adams, M. , Antoniw, J. and Fauquet, C. (2005) Molecular criteria for genus and species discrimination within the family Potyviridae. Arch. Virol. 150, 459–479. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi, R. , Pruss, G.J. , Ge, X. , Marathe, R. , Mallory, A.C. , Smith, T.H. and Vance, V.B. (1998) A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA, 95, 13 079–13 084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermüller, C. , Biegert, A. and Söding, J. (2012) Discriminative modelling of context‐specific amino acid substitution probabilities. Bioinformatics, 28, 3240–3247. [DOI] [PubMed] [Google Scholar]
- Azevedo, J. , Garcia, D. , Pontier, D. , Ohnesorge, S. , Yu, A. , Garcia, S. , Braun, L. , Bergdoll, M. , Hakimi, M.A. and Lagrange, T. (2010) Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen‐encoded GW repeat protein. Genes Dev. 24, 904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger, N. , Tsai, C.‐H. , Lie, M. , Havecker, E. and Baulcombe, D.C. (2007) The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17, 1609–1614. [DOI] [PubMed] [Google Scholar]
- Brigneti, G. , Voinnet, O. , Li, W.X. , Ji, L.H. , Ding, S.W. and Baulcombe, D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana . EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chung, B.Y.‐W. , Miller, W.A. , Atkins, J.F. and Firth, A.E. (2008) An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA, 105, 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, C.A. , Davis, J.A. , Abad, J.A. , Cuellar, W.J. , Fuentes, S. , Kreuze, J.F. , Gibson, R.W. , Mukasa, S.B. , Tugume, A.K. and Tairo, F.D. (2012) Sweetpotato viruses: 15 years of progress on understanding and managing complex diseases. Plant Dis. 96, 168–185. [DOI] [PubMed] [Google Scholar]
- Cuellar, W.J. , Kreuze, J.F. , Rajamäki, M.‐L. , Cruzado, K.R. , Untiveros, M. and Valkonen, J.P. (2009) Elimination of antiviral defense by viral RNase III. Proc. Natl. Acad. Sci. USA, 106, 10 354–10 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Shami, M. , Pontier, D. , Lahmy, S. , Braun, L. , Picart, C. , Vega, D. , Hakimi, M.‐A. , Jacobsen, S.E. , Cooke, R. and Lagrange, T. (2007) Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE‐binding platforms in RNAi‐related components. Genes Dev. 21, 2539–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y. , Treffers, E.E. , Li, Y. , Tas, A. , Sun, Z. , van der Meer, Y. , de Ru, A.H. , van Veelen, P.A. , Atkins, J.F. and Snijder, E.J. (2012) Efficient −2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc. Natl. Acad. Sci. USA, 109, E2920–E2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth, A.E. (2014) Mapping overlapping functional elements embedded within the protein‐coding regions of RNA viruses. Nucleic Acids Res. 42, 12 425–12 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth, A.E. and Brierley, I. (2012) Non‐canonical translation in RNA viruses. J. Gen. Virol. 93, 1385–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, R.W. , Aritua, V. , Byamukama, E. , Mpembe, I. and Kayongo, J. (2004) Control strategies for sweet potato virus disease in Africa. Virus Res. 100, 115–122. [DOI] [PubMed] [Google Scholar]
- Giner, A. , Lakatos, L. , García‐Chapa, M. , López‐Moya, J.J. and Burgyán, J. (2010) Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 6, e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin, L. , Hagai, T. , LaBarbera, A. , Solovey, M. and Andino, R. (2014) Rapid evolution of virus sequences in intrinsically disordered protein regions. PLoS Pathog. 10, e1004529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, D. , Fuentes, S. and Salazar, L. (2003) Sweetpotato virus disease (SPVD): distribution, incidence, and effect on sweetpotato yield in Peru. Plant Dis. 87, 297–302. [DOI] [PubMed] [Google Scholar]
- Haikonen, T. , Rajamäki, M.‐L. and Valkonen, J.P. (2013) Interaction of the microtubule‐associated host protein HIP2 with viral helper component proteinase is important in infection with Potato virus A. Mol. Plant–Microbe Interact. 26, 734–744. [DOI] [PubMed] [Google Scholar]
- Haldeman‐Cahill, R. , Daròs, J.‐A. and Carrington, J.C. (1998) Secondary structures in the capsid protein coding sequence and 3′ nontranslated region involved in amplification of the tobacco etch virus genome. J. Virol. 72, 4072–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann, S. , Garcin, D. , Morel, A.‐S. and Kolakofsky, D. (1999) Two nucleotides immediately upstream of the essential A6G3 slippery sequence modulate the pattern of G insertions during Sendai virus mRNA editing. J. Virol. 73, 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker, I.L. , Fekete, M. and Stadler, P.F. (2002) Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 319, 1059–1066. [DOI] [PubMed] [Google Scholar]
- Johansen, L.K. and Carrington, J.C. (2001) Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium‐mediated transient expression system. Plant Physiol. 126, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D. and Carrington, J.C. (1998) A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell, 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A. and Sternberg, M.J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. [DOI] [PubMed] [Google Scholar]
- Kokkinos, C. and Clark, C. (2006) Interactions among Sweet potato chlorotic stunt virus and different potyviruses and potyvirus strains infecting sweetpotato in the United States. Plant Dis. 90, 1347–1352. [DOI] [PubMed] [Google Scholar]
- Kreuze, J.F. , Savenkov, E.I. , Cuellar, W. , Li, X. and Valkonen, J.P. (2005) Viral class 1 RNase III involved in suppression of RNA silencing. J. Virol. 79, 7227–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuze, J.F. , Perez, A. , Untiveros, M. , Quispe, D. , Fuentes, S. , Barker, I. and Simon, R. (2009) Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: a generic method for diagnosis, discovery and sequencing of viruses. Virology, 388, 1–7. [DOI] [PubMed] [Google Scholar]
- Lakatos, L. , Szittya, G. , Silhavy, D. and Burgyán, J. (2004) Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23, 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N. , Chenna, R. , McGettigan, P.A. , McWilliam, H. , Valentin, F. , Wallace, I.M. , Wilm, A. and Lopez, R. (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li, F. , Xu, D. , Abad, J. and Li, R. (2012) Phylogenetic relationships of closely related potyviruses infecting sweet potato determined by genomic characterization of Sweet potato virus G and Sweet potato virus 2. Virus Genes, 45, 118–125. [DOI] [PubMed] [Google Scholar]
- Li, H. and Durbin, R. (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Folimonov, A. , Shintaku, M. , Li, W.‐X. , Falk, B.W. , Dawson, W.O. and Ding, S.‐W. (2004) Three distinct suppressors of RNA silencing encoded by a 20‐kb viral RNA genome. Proc. Natl. Acad. Sci. USA, 101, 15 742–15 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbanzibwa, D.R. , Tian, Y. , Mukasa, S.B. and Valkonen, J.P. (2009) Cassava brown streak virus (Potyviridae) encodes a putative Maf/HAM1 pyrophosphatase implicated in reduction of mutations and a P1 proteinase that suppresses RNA silencing but contains no HC‐Pro. J. Virol. 83, 6934–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olspert, A. , Chung, B.Y.W. , Atkins, J.F. , Carr, J.P. and Firth, A.E. (2015) Transcriptional slippage in the positive‐sense RNA virus family Potyviridae. EMBO Rep. 16, 995–1004. e201540509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall, G.S. and Hamilton, A.J. (2008) Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 3, 1077–1084. [DOI] [PubMed] [Google Scholar]
- Pardina, P.R. , Bejerman, N. , Luque, A.V. and Di Feo, L. (2012) Complete nucleotide sequence of an Argentinean isolate of sweet potato virus G. Virus Genes, 45, 593–595. [DOI] [PubMed] [Google Scholar]
- Quevillon, E. , Silventoinen, V. , Pillai, S. , Harte, N. , Mulder, N. , Apweiler, R. and Lopez, R. (2005) InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamäki, M.‐L. , Kelloniemi, J. , Alminaite, A. , Kekarainen, T. , Rabenstein, F. and Valkonen, J.P. (2005) A novel insertion site inside the potyvirus P1 cistron allows expression of heterologous proteins and suggests some P1 functions. Virology, 342, 88–101. [DOI] [PubMed] [Google Scholar]
- Rancurel, C. , Khosravi, M. , Dunker, A.K. , Romero, P.R. and Karlin, D. (2009) Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J. Virol. 83, 10 719–10 736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder, J. , Steffen, P. and Giegerich, R. (2007) pknotsRG: RNA pseudoknot folding including near‐optimal structures and sliding windows. Nucleic Acids Res. 35, W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revers, F. and García, J.A. (2015) Chapter Three‐Molecular Biology of Potyviruses. Advances in virus research 92, 101–199. [DOI] [PubMed] [Google Scholar]
- Rice, P. , Longden, I. and Bleasby, A. (2000) EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Rodamilans, B. , Valli, A. , Mingot, A. , San León, D. , Baulcombe, D. , López‐Moya, J.J. and García, J.A. (2015) RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the Potyviridae family. J. Virol. 89, 6965–6967. 10.1128/JVI.00337‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohožková, J. and Navrátil, M. (2011) P1 peptidase—a mysterious protein of family Potyviridae. J. Biosci. 36, 189–200. [DOI] [PubMed] [Google Scholar]
- de Ronde, D. , Pasquier, A. , Ying, S. , Butterbach, P. , Lohuis, D. and Kormelink, R. (2014) Analysis of Tomato spotted wilt virus NSs protein indicates the importance of the N‐terminal domain for avirulence and RNA silencing suppression. Mol. Plant Pathol. 15, 185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath, N. , Wagner, A. and Karlin, D. (2012) Evolution of viral proteins originated de novo by overprinting. Mol. Biol. Evol. 29, 3767–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, J. , Mori, M. , Morishita, T. , Tanaka, M. , Hanada, K. , Usugi, T. and Nishiguchi, M. (1997) Complete nucleotide sequence and genome organization of sweet potato feathery mottle virus (S strain) genomic RNA: the large coding region of the P1 gene. Arch. Virol. 142, 1553–1562. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Senshu, H. , Yamaji, Y. , Minato, N. , Shiraishi, T. , Maejima, K. , Hashimoto, M. , Miura, C. , Neriya, Y. and Namba, S. (2011) A dual strategy for the suppression of host antiviral silencing: two distinct suppressors for viral replication and viral movement encoded by potato virus M. J. Virol. 85, 10 269–10 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiboleth, Y.M. , Haronsky, E. , Leibman, D. , Arazi, T. , Wassenegger, M. , Whitham, S.A. , Gaba, V. and Gal‐On, A. (2007) The conserved FRNK box in HC‐Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J. Virol. 81, 13 135–13 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding, J. , Biegert, A. and Lupas, A.N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó, E.Z. , Manczinger, M. , Göblös, A. , Kemény, L. and Lakatos, L. (2012) Switching on RNA silencing suppressor activity by restoring argonaute binding to a viral protein. J. Virol. 86, 8324–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tairo, F. , Mukasa, S.B. , Jones, R.A. , Kullaya, A. , Rubaihayo, P.R. and Valkonen, J. (2005) Unravelling the genetic diversity of the three main viruses involved in sweet potato virus disease (SPVD), and its practical implications. Mol. Plant Pathol. 6, 199–211. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. , Qu, F. , Li, R. , Morris, T.J. and French, R. (2012) Triticum mosaic poacevirus enlists P1 rather than HC‐Pro to suppress RNA silencing‐mediated host defense. Virology, 433, 104–115. [DOI] [PubMed] [Google Scholar]
- Tena Fernández, F. , González, I. , Doblas, P. , Rodríguez, C. , Sahana, N. , Kaur, H. , Tenllado, F. , Praveen, S. and Canto, T. (2013) The influence of cis‐acting P1 protein and translational elements on the expression of Potato virus Y helper‐component proteinase (HCPro) in heterologous systems and its suppression of silencing activity. Mol. Plant Pathol. 14, 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till, S. , Lejeune, E. , Thermann, R. , Bortfeld, M. , Hothorn, M. , Enderle, D. , Heinrich, C. , Hentze, M.W. and Ladurner, A.G. (2007) A conserved motif in Argonaute‐interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 14, 897–903. [DOI] [PubMed] [Google Scholar]
- Tugume, A.K. , Mukasa, S.B. , Kalkkinen, N. and Valkonen, J.P. (2010) Recombination and selection pressure in the ipomovirus Sweet potato mild mottle virus (Potyviridae) in wild species and cultivated sweetpotato in the centre of evolution in East Africa. J. Gen. Virol. 91, 1092–1108. [DOI] [PubMed] [Google Scholar]
- Untiveros, M. , Fuentes, S. and Salazar, L.F. (2007) Synergistic interaction of Sweet potato chlorotic stunt virus (Crinivirus) with carla‐, cucumo‐, ipomo‐, and potyviruses infecting sweet potato. Plant Dis. 91, 669–676. [DOI] [PubMed] [Google Scholar]
- Untiveros, M. , Fuentes, S. and Kreuze, J. (2008) Molecular variability of sweet potato feathery mottle virus and other potyviruses infecting sweet potato in Peru. Arch. Virol. 153, 473–483. [DOI] [PubMed] [Google Scholar]
- Untiveros, M. , Quispe, D. and Kreuze, J. (2010) Analysis of complete genomic sequences of isolates of the Sweet potato feathery mottle virus strains C and EA: molecular evidence for two distinct potyvirus species and two P1 protein domains. Arch. Virol. 155, 2059–2063. [DOI] [PubMed] [Google Scholar]
- Valli, A. , Martín‐Hernández, A.M. , López‐Moya, J.J. and García, J.A. (2006) RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro. J. Virol. 80, 10 055–10 063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli, A. , López‐Moya, J.J. and García, J.A. (2007) Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. J. Gen. Virol. 88, 1016–1028. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Abad, J. , Fuentes, S. and Li, R. (2013) Complete genome sequence of the original Taiwanese isolate of sweet potato latent virus and its relationship to other potyviruses infecting sweet potato. Arch. Virol. 158, 2189–2192. [DOI] [PubMed] [Google Scholar]
- Xue, B. , Dunbrack, R.L. , Williams, R.W. , Dunker, A.K. and Uversky, V.N. (2010) PONDR‐FIT: a meta‐predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta, 1804, 996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki, S. , Sakai, J. , Fuji, S. , Kamisoyama, S. , Emoto, K. , Ohshima, K. and Hanada, K. (2010) Comparisons among isolates of Sweet potato feathery mottle virus using complete genomic RNA sequences. Arch. Virol. 155, 795–800. [DOI] [PubMed] [Google Scholar]
- Young, B.A. , Stenger, D.C. , Qu, F. , Morris, T.J. , Tatineni, S. and French, R. (2012) Tritimovirus P1 functions as a suppressor of RNA silencing and an enhancer of disease symptoms. Virus Res. 163, 672–677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Predicted pispo G2A6 slippage sites. Predicted pispo G2A6 slippage sites (orange highlighted) in all available full‐length Sweet potato feathery mottle virus (SPFMV)‐group sequences. SPV2, Sweet potato virus 2; SPVC, Sweet potato virus C; SPVG, Sweet potato virus G.
Fig. S2 PONDR® disorder predictions for P1 and P1N‐PISPO. Common regions predicted to be unstructured or disordered include a small region of ∼25 residues within the N‐terminus of P1N, and the hypervariable region (residues 200–300). In P1, a third region of predicted disorder occurs around residues 459–522, whereas, in P1N‐PISPO, a third region of predicted disorder encompasses almost the entire PISPO domain starting around residue 444. Predicted non‐disordered domains have scores no higher than 0.5.
Fig. S3 Expression and detection of YN‐P1(SPLV) in agroinfiltrated leaf tissue of green fluorescent protein (GFP)‐transgenic Nicotiana benthamiana line 16c. (a) Neither (1) YN‐P1(SPLV) (left half of the leaf) nor (2) YN‐HCpro (right half of the leaf) of Sweet potato feathery mottle virus (SPFMV) suppressed gfp silencing and no enhancement of GFP fluorescence was observed at 4 days post‐infiltration (dpi). The infiltrated tissue was surrounded by a red edge indicating short‐distance movement of the silencing signal. Scale bar indicates 2 cm. (b) Expressed YN‐P1(SPLV) protein was detected at 2 dpi using anti‐YN antibodies, but not at 3, 4 or 5 dpi. The upper band indicated with an arrow (2d) corresponds to the expected size (72 kDa) of YN‐P1(SPLV). The small arrow on the left indicates the position of the 70‐kDa protein marker. Staining of total proteins by Coomassie blue was used as a loading control.
Table S1 Mean amino acid inter‐ and intra‐species identities. Identities (%) calculated for the PISPO (italics; upper left) and P1‐pro (roman; lower right) domains.
Table S2 Virus species and accession number for sequences used for the computational analysis of the pispo open reading frame (ORF). Sequences of Sweet potato virus G (SPVG)‐LSU1 and Sweet potato virus 2 (SPV2)‐LSU2 were determined in this study, and partial sequences with coverage of pispo also included are indicated in a separate column.
Table S3 Primer sequences used in this study.


