Abstract
The traditional explanation for the exceptional diversity of herbivorous insects emphasizes host shift as the major driver of speciation. However, phylogenetic studies have often demonstrated widespread host plant conservatism by insect herbivores, calling into question the prevalence of speciation by host shift to distantly related plants. A limitation of previous phylogenetic studies is that host plants were defined at the family or genus level; thus, it was unclear whether host shifts predominate at a finer taxonomic scale. The lack of a statistical approach to test the hypothesis of host‐shift‐driven speciation also hindered studies at the species level. Here, we analyze the radiation of leaf cone moths (Caloptilia) associated with maples (Acer) using a newly developed, phylogeny‐based method that tests the role of host shift in speciation. This method has the advantage of not requiring complete taxon sampling from an entire radiation. Based on 254 host plant records for 14 Caloptilia species collected at 73 sites in Japan, we show that major dietary changes are more concentrated toward the root of the phylogeny, with host shift playing a minor role in recent speciation. We suggest that there may be other roles for host shift in promoting herbivorous insect diversification rather than facilitating speciation per se.
Keywords: Diversification, herbivorous insect, host plant, host shift, speciation
Introduction
Herbivorous insects comprise one of the major components of earth's biodiversity. Because the diversity of herbivorous insects is often correlated with host plant diversity (Lawton and Schroeder 1977; Wiegmann et al. 2002; Janz et al. 2006; Joy and Crespi 2012; Ferrer‐Paris and Sánchez‐Mercado 2013; Isaka and Sato 2015; Lin et al. 2015), the cycle of host plant adaptation and host plant shift is commonly invoked as the major process generating high diversity (Mitter and Brooks 1983; Craig et al. 2001; Wheat et al. 2007; Futuyma and Agrawal 2009; Bennett and O'Grady 2012). For example, a classical study by Farrell (1998) showed that herbivorous insects using angiosperms as hosts are more species rich than those using gymnosperms among the Phytophaga beetles, suggesting that the diversity of angiosperms has facilitated speciation by host shift in the beetles that feed on them. Studies of host races in herbivorous insects showed that specialization to a novel host plant sometimes results in reproductive isolation between insects using different hosts (Feder et al. 1988; Groman and Pellmyr 2000; Hawthorne and Via 2001; Nosil et al. 2002; Thomas et al. 2003; Malausa et al. 2005; Ohshima 2012; Xue et al. 2014), providing a mechanistic explanation of how host shifts may promote speciation. Understanding the role of host plant shifts in generating diversity is thus a current focus in the study of herbivorous insect diversification (Marvaldi et al. 2002; Stireman et al. 2005; Wheat et al. 2007; Winkler et al. 2009; Fordyce 2010; Funk 2010; Matsubayashi et al. 2010; Nyman 2010; Soria‐Carrasco et al. 2014).
However, phylogenetic analyses of herbivorous insect radiation have often demonstrated conservatism in host plant use by herbivorous insects (Crespi et al. 1998; Lopez‐Vaamonde et al. 2003; Wahlberg 2007; Winkler and Mitter 2008; Nyman et al. 2010; Jousselin et al. 2013; Doorenweerd et al. 2015). For example, Nyman et al. (2010) showed that only 20% of the speciation events in nematine sawflies were accompanied by shifts between host plant families, and Doorenweerd et al. (2015) showed that host use was generally conserved at the plant family level, with biogeographic processes playing a greater role in the recent speciation of nepticulid moths. Extreme cases of host plant conservatism are found in gall wasps feeding on oaks (Stone et al. 2009) or micropterigid moths that have radiated on a single liverwort species (Imada et al. 2011). However, many phylogenetic studies that tested for host conservatism defined host plants at the plant family or genus level (Lopez‐Vaamonde et al. 2003; Wahlberg 2007; Nyman et al. 2010; Jousselin et al. 2013; Doorenweerd et al. 2015). The relative importance of host shifts in herbivorous insect speciation should ideally be assessed using species‐level phylogenies with data on all known host associations.
Two major obstacles hamper analysis at the species level. First, because most radiations of herbivorous insect groups occur at the continental scale, it is usually difficult to achieve complete taxon sampling while having host association data for each species. It is therefore not surprising that some of the best‐sampled phylogenies are those for less mobile herbivorous insect groups (e.g., Imada et al. 2011). Second, an appropriate method of analyzing host plant shifts along phylogenies has been lacking. Coding host plant associations at the family or genus level would simplify analysis because methods such as ancestral character state reconstructions are then applicable. However, many herbivorous insects use several closely related plant species (i.e., polyphagy) with varying levels of preference (Smiley 1978; Roininen and Tahvanainen 1989; Thompson 1998; Scheirs et al. 2000; D'Costa et al. 2013; Nakadai and Murakami 2015), which complicates analysis of the ancestral state regarding host use. In addition, individual host plant species cannot be considered as discrete character states because they are phylogenetically nonindependent (Pearse and Altermatt 2013). Ideally, the dissimilarity of host use between a pair of herbivorous insect species should be weighed by the phylogenetic disparity of the host plants.
In this study, we assess the importance of host shifts in the speciation process of herbivorous insects by developing a new method that overcomes these issues. This method focuses on whether host plant shifts are concentrated toward the roots or the tips of the insect phylogenetic tree, while taking into account host plant phylogeny in the calculation of host use dissimilarity between a pair of herbivorous insect species. If most speciation events are associated with host shifts, the level of disparity in host use between a pair of herbivorous insect species will on average be greater for phylogenetically more closely related pairs (Fig. 2A). Alternatively, if most host shifting events occurred during the initial stage of the radiation and more recent speciation events were independent of host shifts, the level of difference in host use would be larger toward the root of the phylogenetic tree (Fig. 2B). We focused on the interaction between a group of leaf cone moths (Caloptilia, Gracillariidae) and their maple hosts (Acer, Sapindaceae). The Caloptilia–Acer interaction is appropriate for testing host‐shift‐driven speciation at fine taxonomic scales because a previous study demonstrated large variation in the pattern of host use among Caloptilia species (Nakadai and Murakami 2015). The genus Acer is one of the most taxonomically diverse groups of trees in the Northern Hemisphere, particularly in the temperate regions of East Asia, eastern North America, and Europe (van Gelderen et al. 1994). The genus comprises 124 species in the Northern Hemisphere, 81% of which are distributed in China, Korea, and Japan (Renner et al. 2007). A previous taxonomic study of Caloptilia identified 11 species associated with Acer in Japan alone, which have high morphological affinity to each other (Kumata 1982). Based on extensive geographic sampling, we establish full host plant records for these 11 species and three newly found ones, and analyze them using the above method to assess the relative importance of host shift in the speciation of Caloptilia moths feeding on Acer trees.
Figure 2.
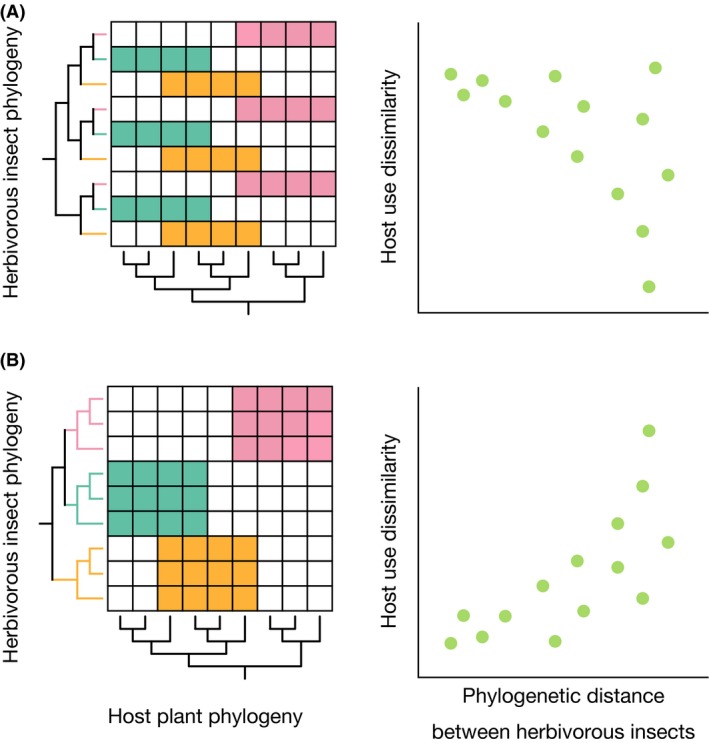
Phylogenetic distributions of host use arising from different speciation modes in herbivorous insects. (A) Distribution of host use on the phylogeny of a hypothetical insect group in which speciation is mainly associated with host shifts. (B) Distribution of host taxa when speciation mainly involves other processes without host shifts.
Materials and Methods
Study material
The genus Caloptilia is globally distributed and includes nearly 300 described species, of which 27 feed on maples (De Prins and De Prins 2015). In Japan, 51 species are described feeding on 21 host plant families, and 11 of them use Acer, which is the most common host plant genus of Japanese Caloptilia (Kumata et al. 2013). The feeding habits of the larvae change dramatically between the early and late developmental stages. Upon hatching, larvae mine the surface layer of the leaf (i.e., leaf miners) until the third instar, then exit the mine, and form the edge of the leaf into a roll within which they feed externally until the final instar (hence the name leaf cone moth) (Kumata et al. 2013). Some species are leaf gallers or blotch miners at the final instar and do not roll leaves. Each species is usually associated with a single plant genus.
Sampling, DNA sequencing, and phylogenetic analyses
We sampled Caloptilia moths that use Acer trees at 73 sites covering a wide geographic range in Japan (Figs. 1, S2) during May–October of 2011–2015. Moths were sampled by searching for larvae in leaf rolls (fourth or fifth instar) or pupae on maple leaves. In total, 254 specimens were obtained, used to delimit species and to establish the host range for each species. Delimitation of species was based on sequences of the mitochondrial cytochrome oxidase subunit I (COI) gene; major divergences in COI sequences clearly corresponded with differences in wing pattern and genital morphology. Species were morphologically identified following Kumata (1982). To further determine whether the Caloptilia species feeding on maples resulted from a single radiation, we additionally sampled 44 Caloptilia species that use nonmaple hosts and six species in closely related genera (Gracillaria, Calybites, and Eucalybites; for details, see Table S1) and reconstructed a species‐level phylogeny of Caloptilia. For the species‐level phylogeny, one representative specimen of each Caloptilia species feeding on maple was included in the analysis. All moth specimens were kept in ethanol prior to DNA extraction.
Figure 1.
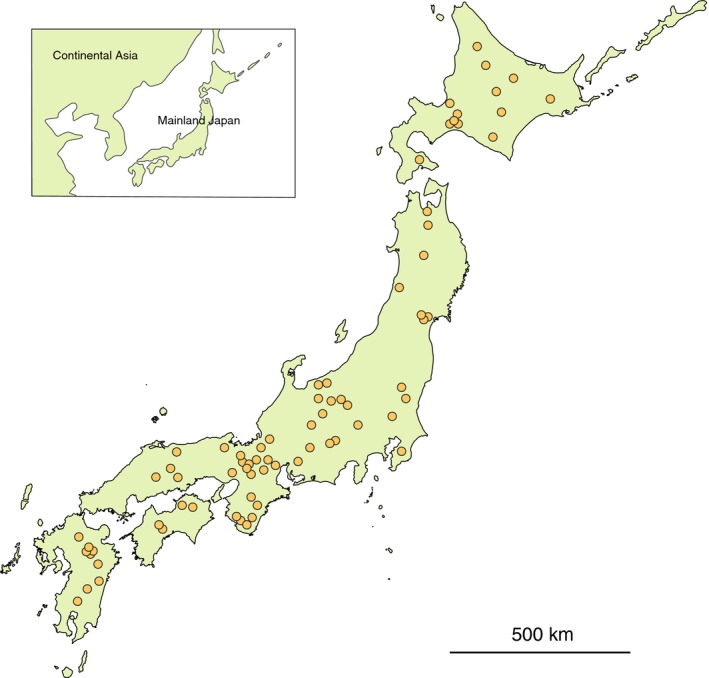
Sampling localities of Caloptilia moths collected from Acer trees in Japan. Sampling information for each species shown in Figure S2.
We extracted genomic DNA using the NucleoSpin Tissue Kit (Macherey‐Nagel, Düren, Germany). The head capsule of the larva or the head, wings, and abdomen of the adult were stored as vouchers. The COI gene was sequenced for all of the 254 moths collected from maples. For the species‐level phylogenetic analysis, we sequenced four genomic regions: COI and the nuclear arginine kinase (ArgK), carbamoyl‐phosphate synthetase 2 (CAD), and elongation factor 1‐alpha (EF‐1α) genes. We designed new primer sets for ArgK, CAD, and EF‐1α (Table S3) based on sequences available for other species of Gracillariidae in the database. The information on existing primer sets for CO1 and EF‐1α is also provided in Table S3. Polymerase chain reaction (PCR) amplifications were carried out under the following conditions: initial denaturation step at 94°C for 5 min; 30 cycles of 94°C for 30 sec, 50°C for 30 sec, and 72°C for 1 min; and a final extension at 72°C for 7 min. Products were sequenced on an ABI 3100 automated sequencer using BigDye chain termination chemistry (Applied Biosystems, Foster City, CA), and obvious sequence errors were manually corrected using MEGA 6.06 (Tamura et al. 2013). Obtained sequences were aligned using Mafft ver. 6.901 (Katoh and Toh 2008) under the default settings. The resulting dataset contained 658, 573, 614, and 541 base pairs of COI, ArgK, CAD, and EF‐1α, respectively. Species‐level phylogenetic trees were constructed using two datasets: (1) an all‐genes dataset (COI + ArgK + CAD + EF‐1α) and (2) a nuclear‐only dataset (ArgK + CAD + EF‐1α). The latter was created because a previous phylogenetic study of Gracillariidae suggested that nuclear genes provide strong phylogenetic signals at the genus and species levels (Kawahara et al. 2011). We reconstructed phylogenetic trees by maximum‐likelihood and Bayesian methods for each dataset. The maximum‐likelihood analysis was performed using RAxML ver. 8.0 (Stamatakis 2014). We conducted 100 replicates of shotgun search for the likelihood ratchet and assessed nodal support using bootstrap analyses with 1000 replications. We also conducted Bayesian phylogenetic analysis using MrBayes5D (Tanabe 2008), a modified version of MrBayes3.1.2 (Ronquist and Huelsenbeck 2003). We used the following settings for the Bayesian analysis: number of Markov chain Monte Carlo generations, five million; sampling frequency, 100; and burn‐in, 5001. The burn‐in size was determined by checking the convergence of log likelihood (ln L) plotted against generation time. In both methods, we used Kakusan4 (Tanabe 2011) to determine appropriate models of sequence evolution under the BIC4 criterion.
Hypothesis and randomization tests for validation
To test the relative importance of host shift in the speciation process from phylogeny, we assumed two contrasting scenarios (Fig. 2). If most speciation events are associated with host shifts, the dissimilarity in host use will on average be larger for phylogenetically more closely related pairs of Caloptilia moths (Fig. 2A). Conversely, if most speciation events occur during the initial stage of the radiation and more recent speciation events are independent of host shifts, host use dissimilarity will be larger for phylogenetically more distantly related pairs of Caloptilia moths (Fig. 2B). A similar framework was proposed by Nyman et al. (2010), but their method cannot be applied to species‐level analysis. Following Barraclough et al. (1999), we used randomizations to compare the observed pattern of host use to that expected under a null model of no association with cladogenesis. Our null model hypothesized that changes occurred at random and independently across the tree. The statistic used to test the association between phylogenetic distance and the degree of difference in host use is expressed as the sum across all nodes of phylogenetic distance X i multiplied by the degree of host use dissimilarity H i (see the next section for detailed calculation of dissimilarity),
If differences in host use are greater between closely related species, the above statistic is expected to be smaller than that under the null model and vice versa. Thus, we tested for a significant concentration of changes toward either the tips or the root of the tree. A positive sign indicates the concentration of changes toward the tips, whereas a negative sign indicates that more changes occurred toward the root. The null distribution was obtained by randomly shuffling observed changes among branches of the tree and calculating the above statistic in each null trial. The two‐tailed probability of the observed value was calculated based on 10,000 randomizations. A similar randomization method was used by Barraclough et al. (1999) and Sauer and Hausdorf (2009) to study adaptive character evolution in tiger beetles and land snails, respectively.
In addition, we calculated the standardized effect size (SES) as the observed test statistic minus the mean of the null distribution, divided by the standard deviation of the null distribution. This null model approach is commonly used for expressing biological differences regardless of the units of the indices (McCabe et al. 2012).
Indices of dissimilarity in host use
We used both Jaccard (Jaccard 1912; Koleff et al. 2003) and Unifrac (Lozupone and Knight 2005) indices to quantify the degree of difference in host use between a pair of Caloptilia moths feeding on Acer trees. Both indices are commonly used in community ecology for assessing the degree of dissimilarity between two communities (Cavender‐Bares et al. 2009). The Unifrac index is analogous to the Jaccard dissimilarity index, but takes into account phylogenetic information (Lozupone and Knight 2005), which in the present case is the plant phylogeny. The Unifrac index has an advantage over the Jaccard index especially when there is missing information on host association; the latter index assumes an equal weight for all host plant species, whereas the former weighs host plants according to their phylogenetic relatedness and is thus less sensitive to missing data. In this study, we used the phylogeny of 30 Japanese Acer species published by Nakadai et al. (2014). In addition, both Jaccard and Unifrac indices can be partitioned into two components of dissimilarity: turnover and nestedness (Baselga 2010; Leprieur et al. 2012). In community ecology, the turnover of a species assemblage refers to the replacement of some species by others as a consequence of historical events, such as geographic barrier formation or environmental sorting (Baselga 2010). In contrast, the nestedness of a species assemblage occurs when the species composition of sites with a smaller number of the species is a subset of that of species‐rich sites, which reflects a spatial pattern of species loss resulting from dispersal limitation or environmental filtering (Hirao et al. 2015). In our study, the turnover component indicates the degree of nonoverlapping host use, and the nestedness component represents the difference in the degree of specialization between insect species with shared host plants (Fig. 3). All indices were calculated using the “betapart” package (Baselga and Orme 2012) in R ver. 3.2.2 (R Core Team 2015).
Figure 3.
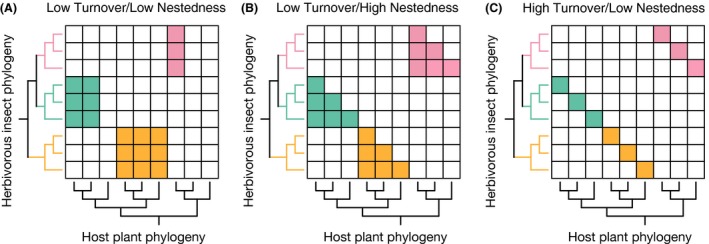
Possible patterns of plant–herbivore association. (A) Low turnover/low nestedness, (B) low turnover/high nestedness, and (C) high turnover/low nestedness. Both Jaccard and Unifrac indices perform similarly in (A) and (B), whereas in (C), the nestedness component of the Unifrac index between a pair of closely related herbivores will be lower than that of the Jaccard index. This is because host use is similar when host phylogeny is taken into account but maximally dissimilar in the absence of host phylogenetic information.
Results
Extensive sampling of Caloptilia moths throughout Japan identified 14 species feeding on maples (Figs. 4, S1), of which three were newly discovered in this study. This represents ca. 40% of the Caloptilia species known to feed on maples (De Prins and De Prins 2015). Most species were widely distributed throughout the range, although some were only found at a limited number of sites (Fig. S2). Some species were apparently specialists on single Acer species (e.g., Caloptilia hidakensis, Caloptilia kurokoi), whereas others were collected from many hosts. Overall, each species uses 1–11 Acer species, with a mean of 3.0 ± 3.0 (Fig. 5).
Figure 4.
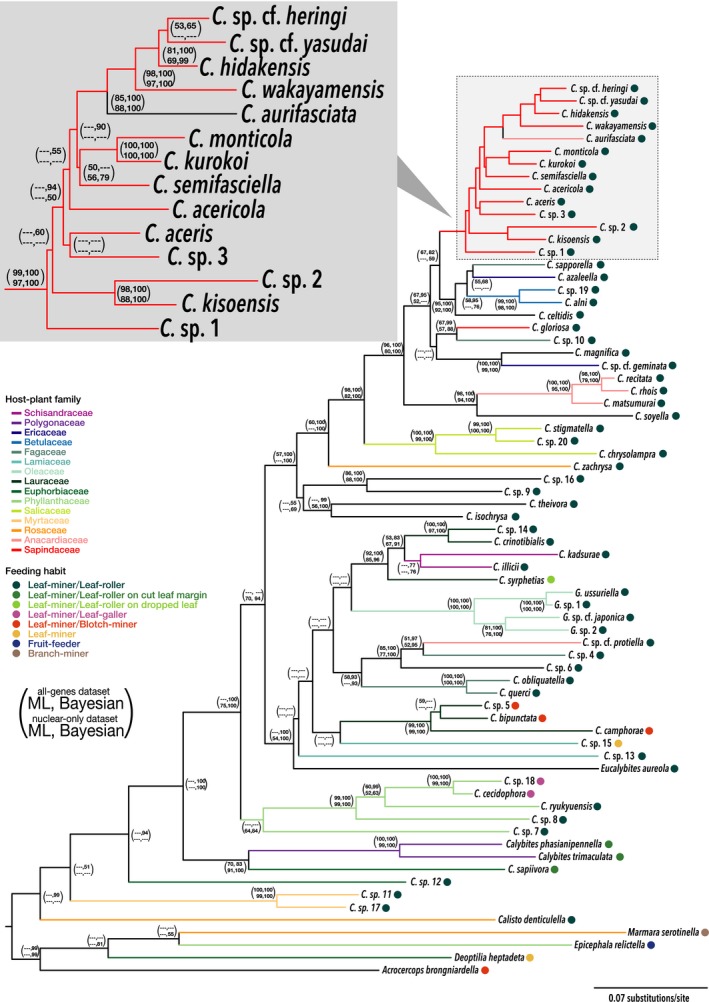
Phylogeny of Caloptilia moths and their related groups. The phylogeny was constructed by maximum‐likelihood method using four genomic regions (COI, ArgK, CAD, and EF‐1α) of 71 species.
Figure 5.
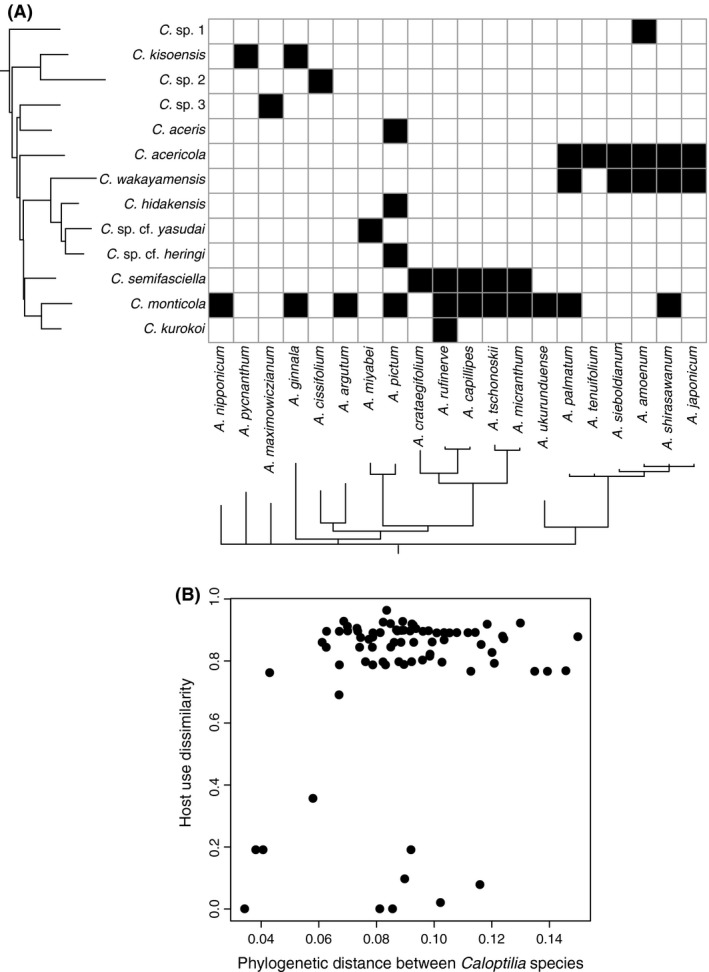
The results of Acer–Caloptilia interactions obtained from wide range sampling in Japan. (A) Phylogram of 13 species of Caloptilia pruned from a phylogeny of this genus and related groups (Table S3) and a phylogram of 20 species of Acer trees pruned from a phylogeny of this genus in Japan (Nakadai et al. 2014). The complete phylogeny of Acer trees was the 50% majority‐rule consensus of trees sampled from the stationary distribution of a Bayesian analysis of four chloroplast DNA loci sampled from 30 species, including some varieties. (B) The plot of phylogenetic distance between Caloptilia moths (all‐genes dataset) versus host use dissimilarity (turnover and nestedness components of the Unifrac index).
Species‐level phylogenetic analyses based on 2386 bp of the combined COI, ArgK, CAD, and EF‐1α dataset produced a well‐resolved phylogeny (Fig. 4). All of the Caloptilia species feeding on Acer were closely related, although they were not monophyletic. One species, Caloptilia gloriosa, was positioned outside of the clade consisting mainly of Acer‐feeding Caloptilia (Fig. 4), and another species, Caloptilia aurifasciata, feeding on Toxicodendron (Anacardiaceae), was embedded within this clade (Fig. 4). We thus focused on the clade containing C. aurifasciata and the 13 species feeding on Acer for the analysis of host shifts. We conducted randomization tests separately for datasets with and without C. aurifasciata. Because information on the phylogenetic distance between Acer and Toxicodendron (the host of C. aurifasciata) was not available, we assumed the maximum turnover (1) and minimum nestedness (0) for the calculation of dissimilarity indices between C. aurifasciata and Acer‐feeding Caloptilia.
The results of randomization tests indicated that the turnover components and the combined turnover and nestedness components of both Jaccard and Unifrac indices are greater between distantly related species than expected under the null model (positive signs in Table 1), although the trend was not significant for the Jaccard index except for the turnover component of the all‐genes dataset. The nestedness component showed negative signs but was not statistically significant (Table 1). These results support the hypothesis of phylogenetic conservatism in host use (Fig. 2B). Inclusion of C. aurifasciata, which feeds on Toxicodendron, did not change the overall pattern but slightly strengthened the trend, with tests using both Jaccard and Unifrac indices becoming significant (Table S4).
Table 1.
Relationships between differences of host use and phylogenetic distance between Caloptilia species feeding on Acer according to randomization tests
| Dataset | Turnover + nestedness | Turnover | Nestedness | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sign | SES | Sign | SES | Sign | SES | |||||
| All‐genes dataset | Jaccard index | + | 1.66 | n.s. | + | 1.95 | * | – | −1.26 | n.s. |
| Unifrac index | + | 2.17 | * | + | 2.16 | * | – | −0.85 | n.s. | |
| Nuclear‐only dataset | Jaccard index | + | 1.90 | n.s. | + | 1.95 | n.s. | – | −1.10 | n.s. |
| Unifrac index | + | 2.72 | ** | + | 2.40 | * | – | −0.60 | n.s. | |
Positive signs of differences in host use with phylogenetic distance suggest that changes are concentrated toward the root and negative signs suggest that changes occur near the tips. Significance level: n.s., P ≥ 0.05; *P < 0.05; **P < 0.01.
The SES values provide a quantitative measure of the strength of association between host use dissimilarity and phylogenetic distance (Table 1). Overall, the values for the turnover component and the combined turnover + nestedness component were greater when host plant phylogeny was taken into account (Unifrac index) than when it was not (Jaccard index).
Discussion
Application of randomization test in the study of herbivorous insect speciation
In this article, we describe a new method for testing the role of host shift in herbivorous insect speciation. We identified three beneficial features of this method. First, it is less sensitive to incomplete species sampling. It is usually difficult to sample every species for the entire radiation (Lopez‐Vaamonde et al. 2003; Nyman et al. 2006; Agrawal and Fishbein 2008; Stone et al. 2009; Doorenweerd et al. 2015), and conventional methods of analyzing the effects of host shifts on phylogeny (e.g., ancestral character state reconstruction) are sensitive to species sampling. However, because our analysis focuses on whether host use changes are concentrated toward either the root or the tips of the phylogenetic tree, complete sampling is not required as long as species sampling is not biased (e.g., toward species feeding only on a particular species of host).
Second, the method permits analysis of speciation by host shift at a broader geographic scale. In many cases, herbivorous insect species have broader distributions than individual host plant species, so sister herbivore species occurring in allopatry should always use different hosts, even if diet shift was not the major cause of speciation. The use of a dissimilarity index controlling for host phylogeny partly remedies this problem (Pearse and Altermatt 2013; Pearse et al. 2013) because related plant species are generally similar in their traits associated with susceptibility to herbivores (Rasmann and Agrawal 2011; D'Costa et al. 2014; Nakadai and Murakami 2015), and thus host use dissimilarity will consistently be low if no major diet shift has occurred during speciation. Caution is needed in cases where the group of herbivores being studied has extremely high or low host specificity because, in both cases, the method may overestimate host use conservatism.
Finally, calculation of SES allows comparison of trends among different studies (McCabe et al. 2012) because SES is independent of differences in the number of herbivore species included in the dataset. Previous phylogenetic studies assessed the percentage of host shifts between host plant families in each taxonomic group (Lopez‐Vaamonde et al. 2003; Nyman et al. 2010; Doorenweerd et al. 2015), but quantitative comparisons among studies were difficult due to the lack of a standardized measure for comparison.
We note that our method has a link to those developed previously to test the degree of cospeciation between a pair of host and parasite. However, because they are designed to test for cospeciation, they either assume that each parasite is associated with only one host at any given time (Page 1994; Ronquist 1995; Charleston and Robertson 2002; Merkle and Middendorf 2005; Conow et al. 2010) or that host and parasite speciation events are simultaneous in time (Legendre et al. 2002), which are not realistic for many plant–herbivore associations. Recently, Rafferty and Ives (2013) and Hadfield et al. (2014) developed methods that do not require such assumptions and uses GLMM to test for interaction effect of two phylogenies, but the methods are not designed to test the polarity of trait divergence occurring either toward the tips or the root of the phylogeny as in our method.
One weakness of our analysis is that we treated host association based on presence/absence, but in reality, preference levels are not equal for all of the host plant species observed. We could not quantify host preference in this study because it is necessary to standardize both sampling effort and host abundance to obtain a comparative measure of host preference, which was difficult to accomplish at all sampling sites. However, the above‐described method can easily incorporate host preference when such data are available, as dissimilarity measures (Unifrac and Jaccard indices) are also designed for quantitative data. The newly developed method is presently intended for testing host‐shift‐driven speciation in herbivorous insects, but the overall framework is applicable, in principle, to studies of other types of ecological speciation. The source code for running the analysis in R is provided as Data S4. The source code and datasets for running the analysis in R is provided as Data S1–4.
Alternative hypothesis on the speciation process of leaf cone moths feeding on maples
Application of the present method to the 13 species of maple‐feeding leaf cone moths suggested that major dietary changes are concentrated toward the root of the herbivore phylogenetic tree (Table 1). Because the Unifrac index takes into account plant phylogeny whereas the Jaccard index does not, significant positive sign for the Unifrac index and lack of significance for the Jaccard index indicate that the trend exists only when host plant phylogeny is taken into account in the calculation of dissimilarity. Thus, the results indicate that major dietary shifts play a minor role in recent speciation events, but shifts between very closely related hosts may have took place during recent Caloptilia speciation. The addition of C. aurifasciata generally strengthened the trend for both Jaccard and Unifrac indices because C. aurifasciata diverged from all other species toward the root of the tree and has a completely different diet. The Jaccard test, which was only marginally insignificant in the absence of C. aurifasciata, became significant after the inclusion of this species (Table S4).
Although our test indicated that speciation assisted by host shift may be relatively minor in this group, we do not deny the importance of major dietary changes as such events occur in some of the earliest speciation events. Nevertheless, host‐shift‐driven speciation may not be as important as commonly thought in generating the current diversity of Caloptilia. Because our analysis only tests for patterns, the alternative process that drives speciation in Caloptilia cannot be inferred from our data. However, previous studies proposed several possible processes by which herbivorous insects speciate without changing their diet (Imada et al. 2011; Bennett and O'Grady 2012; Yamamoto and Sota 2012; Hamm and Fordyce 2015). For some phytophagous insect groups, allopatric speciation without host shift may be a major factor causing radiation (Nyman et al. 2010; Imada et al. 2011), but in the case of Japanese leaf cone moths, the pattern is unclear based on visual inspection of the current geographic distribution (Fig. S1). Ecological shift within a host plant is also a significant process (Condon and Steck 1997; Cook et al. 2002; Joy and Crespi 2007; Althoff 2014; Mishima et al. 2014). For example, Zhang et al. (2015) demonstrated divergence induced by host plant ages in sympatric sister beetles (Pyrrhalta maculicollis and Pyrrhalta aenescens) feeding on elm. There is clearly a need to sample from a broader geographic area and to collect additional information on microniche divergence among leaf cone moths to fully understand the process underlying their diversification. Adding timeline to the divergence events of both herbivores and host plants should also facilitate the understanding of the role of host shift in herbivore radiation.
Revealing the role of host shifts in herbivorous insect diversification
Our study proposed a method for assessing the relative importance of host shifts in herbivorous insect speciation. This method allows quantitative analysis at a fine taxonomic scale, but because we only applied it to one herbivorous insect group, the application of this method to various herbivorous insect groups will facilitate a more general discussion on herbivorous insect diversification. If host‐shift‐driven speciation turns out to be relatively minor in recent speciation, there may be another role for host shifts in promoting herbivorous insect diversification rather than facilitating speciation per se, such as facilitating the entry into novel niche spaces (Janzen 1968) and the coexistence of already diverged species (Rabosky 2009). Information on the phylogenetic pattern of host use is clearly increasing rapidly, and a standardized method would link studies using different systems and facilitate our understanding of the effects of host shift on herbivorous insect diversity.
Conflict of Interest
None declared.
Data Archiving
Obtained DNA sequences have been deposited in the DDBJ database under accession numbers LC127539–LC128013. Nucleotide alignments will be archived in TreeBase.
Supporting information
Figure S1. Phylogeny of Caloptilia moths feeding on Acer based on mitochondrial COI with information on sampling site.
Figure S2. Distributions of 14 Caloptilia moth species feeding on Acer.
Table S1. Specimen information.
Table S2. DDBJ accession numbers.
Table S3. Primers used in this study.
Table S4. The results of randomization tests including C. aurifasciata that feeds on Toxicodendron.
Data S1. Newick format data containing phylogenetic information of 13 Caloptilia moths feeding on Acer trees.
Data S2. Newick format data containing phylogenetic information of 20 Acer species.
Data S3. CSV format file containing host use information.
Data S4. Text file containing the command for running the randomization analysis in R using Data S1‐3.
Acknowledgments
We thank K. Mochizuki, T. Hirano, S. Furukawa, W. Toki, I. Ohshima, O. Kishida, K. Hashimoto, K. Watanabe, M. Kobayashi, T. Iwasaki, and M. Tsujimoto for sampling and field assistance; staff members of Teshio Experimental Forest, Uryu Experimental Forest, Tomakomai Experimental Forest, Wakayama Experimental Forest, Ashiu Forest Research Station, and Shiiba Research Forest. We also thank two anonymous reviewers and the associated editors for comments that improved the manuscript. This work was supported by a grant from the JSPS KAKENHI Grant Number 26650165, 15H04421 and Grant‐in‐Aid for JSPS Fellows Grant Number 15J00601, and partly supported by the Joint Usage/Research Center, Field Science Center for Northern Biosphere, Hokkaido University.
References
- Agrawal, A. A. , and Fishbein M.. 2008. Phylogenetic escalation and decline of plant defense strategies. Proc. Natl Acad. Sci. USA 105:10057–10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althoff, D. M. 2014. Shift in egg‐laying strategy to avoid plant defense leads to reproductive isolation in mutualistic and cheating yucca moths. Evolution 68:301–307. [DOI] [PubMed] [Google Scholar]
- Barraclough, T. G. , Segraves K. A., Hogan J. E., and Vogler A. P.. 1999. Testing whether ecological factors promote cladogenesis in a group of tiger beetles (Coleoptera: Cicindelidae). Proc. R. Soc. Lond. B Biol. Sci. 266:1061–1067. [Google Scholar]
- Baselga, A. 2010. Partitioning the turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 19:134–143. [Google Scholar]
- Baselga, A. , and Orme C. D. L.. 2012. betapart: an R package for the study of beta diversity. Methods Ecol. Evol. 3:808–812. [Google Scholar]
- Bennett, G. M. , and O'Grady P. M.. 2012. Host‐plants shape insect diversity: phylogeny, origin, and species diversity of native Hawaiian leafhoppers (Cicadellidae: Nesophrosyne). Mol. Phylogenet. Evol. 65:705–717. [DOI] [PubMed] [Google Scholar]
- Cavender‐Bares, J. , Kozak K. H., Fine P. V. A., and Kembel S. W.. 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett. 12:693–715. [DOI] [PubMed] [Google Scholar]
- Charleston, M. A. , and Robertson D. L.. 2002. Preferential host switching by primate lentiviruses can account for phylogenetic similarity with the primate phylogeny. Syst. Biol. 51:528–535. [DOI] [PubMed] [Google Scholar]
- Condon, M. A. , and Steck G. J.. 1997. Evolution of host use in fruit flies of the genus Blepharoneura (Diptera: Tephritidae): cryptic species on sexually dimorphic host plants. Biol. J. Linn. Soc. 60:443–466. [Google Scholar]
- Conow, C. , Fielder D., Ovadia Y., and Libeskind‐Hadas R.. 2010. Jane: a new tool for the cophylogeny reconstruction problem. Algorithms Mol. Biol. 5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, J. M. , Rokas A., Pagel M., and Stone G. N.. 2002. Evolutionary shifts between host oak sections and host‐plant organs in Andricus gall wasps. Evolution 56:1821–1830. [DOI] [PubMed] [Google Scholar]
- Craig, T. , Horner J., and Itami J.. 2001. Genetics, experience, and host‐plant preference in Eurosta solidaginis: implications for host shifts and speciation. Evolution 55:773–782. [DOI] [PubMed] [Google Scholar]
- Crespi, B. J. , Carmean D. A., Mound L. A., Worobey M., and Morris D.. 1998. Phylogenetics of social behavior in Australian gall‐forming thrips: evidence from mitochondrial DNA sequence, adult morphology and behavior, and gall morphology. Mol. Phylogenet. Evol. 9:163–180. [DOI] [PubMed] [Google Scholar]
- D'Costa, L. , Koricheva J., Straw N., and Simmonds M. S. J.. 2013. Oviposition patterns and larval damage by the invasive horse‐chestnut leaf miner Cameraria ohridella on different species of Aesculus . Ecol. Entomol. 38:456–462. [Google Scholar]
- D'Costa, L. , Simmonds M. S. J., Straw N., Castagneyrol B., and Koricheva J.. 2014. Leaf traits influencing oviposition preference and larval performance of Cameraria ohridella on native and novel host plants. Entomol. Exp. Appl. 152:157–164. [Google Scholar]
- De Prins, J. , and De Prins W.. 2015. Global taxonomic database of Gracillariidae (Lepidoptera). Available at http://www.gracillariidae.net. (accessed 6 February 2016).
- Doorenweerd, C. , van Nieukerken E. J., and Menken S. B. J.. 2015. A global phylogeny of leafmining Ectoedemia moths (Lepidoptera: Nepticulidae): exploring host plant family shifts and allopatry as drivers of speciation. PLoS One 10:e0119586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell, B. D. D. 1998. “Inordinate fondness” explained: why are there so many beetles? Science 281:555–559. [DOI] [PubMed] [Google Scholar]
- Feder, J. L. , Chilcote C. A., and Bush G. L.. 1988. Genetic differentiation between sympatric host races of the apple maggot fly Rhagoletis pomonella . Nature 336:61–64. [Google Scholar]
- Ferrer‐Paris, J. , and Sánchez‐Mercado A.. 2013. Congruence and diversity of butterfly‐host plant associations at higher taxonomic levels. PLoS One 8:e63570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordyce, J. A. 2010. Host shifts and evolutionary radiations of butterflies. Proc. R. Soc. Lond. B Biol. Sci. 277:3735–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk, D. J. 2010. Does strong selection promote host specialisation and ecological speciation in insect herbivores? Evidence from Neochlamisus leaf beetles. Ecol. Entomol. 35:41–53. [Google Scholar]
- Futuyma, D. J. , and Agrawal A. A.. 2009. Macroevolution and the biological diversity of plants and herbivores. Proc. Natl Acad. Sci. USA 106:18054–18061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen, D. M. , de Jong P. C., and Oterdoom H. J.. 1994. Maples of the world. Timber Press, Portland, OR. [Google Scholar]
- Groman, J. D. , and Pellmyr O.. 2000. Rapid evolution and specialization following host colonization in a yucca moth. J. Evol. Biol. 13:223–236. [Google Scholar]
- Hadfield, J. D. , Krasnov B. R., Poulin R., and Nakagawa S.. 2014. A tale of two phylogenies: comparative analyses of ecological interactions. Am. Nat. 183:174–187. [DOI] [PubMed] [Google Scholar]
- Hamm, C. A. , and Fordyce J. A.. 2015. Patterns of host plant utilization and diversification in the brush‐footed butterflies. Evolution 69:589–601. [DOI] [PubMed] [Google Scholar]
- Hawthorne, D. J. , and Via S.. 2001. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412:904–907. [DOI] [PubMed] [Google Scholar]
- Hirao, T. , Kubota Y., and Murakami M.. 2015. Geographical patterns of butterfly species diversity in the subtropical Ryukyu Islands: the importance of a unidirectional filter between two source islands. J. Biogeogr. 42:1418–1430. [Google Scholar]
- Imada, Y. , Kawakita A., and Kato M.. 2011. Allopatric distribution and diversification without niche shift in a bryophyte‐feeding basal moth lineage (Lepidoptera: Micropterigidae). Proc. R. Soc. Lond. B Biol. Sci. 278:3026–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaka, Y. , and Sato T.. 2015. Species richness of sawfly‐host plant associations at higher taxonomic levels. Entomol. Res. 45:294–304. [Google Scholar]
- Jaccard, P. 1912. The distribution of the flora in the alpine zone. New Phytol. 11:37–50. [Google Scholar]
- Janz, N. , Nylin S., and Wahlberg N.. 2006. Diversity begets diversity: host expansions and the diversification of plant‐feeding insects. BMC Evol. Biol. 6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen, D. 1968. Host plants as islands in evolutionary and contemporary time. Am. Nat. 102:592–595. [Google Scholar]
- Jousselin, E. , Cruaud A., Genson G., Chevenet F., Foottit R. G., and Cœur d'acier A.. 2013. Is ecological speciation a major trend in aphids? Insights from a molecular phylogeny of the conifer‐feeding genus Cinara . Front. Zool. 10:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy, J. B. , and Crespi B. J.. 2007. Adaptive radiation of gall‐inducing insects within a single host‐plant species. Evolution 61:784–795. [DOI] [PubMed] [Google Scholar]
- Joy, J. B. , and Crespi B. J.. 2012. Island phytophagy: explaining the remarkable diversity of plant feeding insects. Proc. R. Soc. Lond. B Biol. Sci. 279:3250–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. , and Toh H.. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9:286–298. [DOI] [PubMed] [Google Scholar]
- Kawahara, A. Y. , Ohshima I., Kawakita A., Regier J. C., Mitter C., Cummings M. P., et al. 2011. Increased gene sampling strengthens support for higher‐level groups within leaf‐mining moths and relatives (Lepidoptera: Gracillariidae). BMC Evol. Biol. 11:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleff, P. , Gaston K. J., and Lennon J. J.. 2003. Measuring beta diversity for presence‐absence data. J. Anim. Ecol. 72:367–382. [Google Scholar]
- Kumata, T. 1982. A taxonomic revision of the Gracillaria group occurring in Japan (Lepidoptera: Gracillariidae). Insecta Matsumurana 26:1–186. [Google Scholar]
- Kumata, T. , Kobayashi S., and Hirowatari T.. 2013. Gracillariidae Pp. 91–155 in Nasu Y., Hirowatari T. and Kisida Y., eds. The standard of moths in Japan IV. Gakken Education Publishing, Tokyo, Japan: (in Japanese). [Google Scholar]
- Lawton, J. H. , and Schroeder D.. 1977. Effects of plant type, size of geographical range and taxonomic isolation on number of insect species associated with British plants. Nature 265:137–140. [Google Scholar]
- Legendre, P. , Desdevises Y., and Bazin E.. 2002. A statistical test for host–parasite coevolution. Syst. Boil. 51:217–234. [DOI] [PubMed] [Google Scholar]
- Leprieur, F. , Albouy C., De Bortoli J., Cowman P. F., Bellwood D. R., and Mouillot D.. 2012. Quantifying phylogenetic beta diversity: distinguishing between “true” turnover of lineages and phylogenetic diversity gradients. PLoS One 7:e42760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.‐P. , Cook D. H., Gullan P. J., and Cook L. G.. 2015. Does host‐plant diversity explain species richness in insects? A test using Coccidae (Hemiptera). Ecol. Entomol. 40:299–306. [Google Scholar]
- Lopez‐Vaamonde, C. , Godfray H. C. J., and Cook J. M.. 2003. Evolutionary dynamics of host‐plant use in a genus of leaf‐mining moths. Evolution 57:1804–1821. [DOI] [PubMed] [Google Scholar]
- Lozupone, C. , and Knight R.. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malausa, T. , Bethenod M.‐T., Bontemps A., Bourguet D., Cornuet J.‐M., and Ponsard S.. 2005. Assortative mating in sympatric host races of the European corn borer. Science 308:258–260. [DOI] [PubMed] [Google Scholar]
- Marvaldi, A. E. , Sequeira A. S., O'Brien C. W., and Farrell B. D.. 2002. Molecular and morphological phylogenetics of weevils (Coleoptera, Curculionoidea): do niche shifts accompany diversification? Syst. Biol. 51:761–785. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, K. W. , Ohshima I., and Nosil P.. 2010. Ecological speciation in phytophagous insects. Entomol. Exp. Appl. 134:1–27. [Google Scholar]
- McCabe, D. J. , Hayes‐Pontius E. M., Canepa A., Berry K. S., and Levine B. C.. 2012. Measuring standardized effect size improves interpretation of biomonitoring studies and facilitates meta‐analysis. Freshw. Sci. 31:800–812. [Google Scholar]
- Merkle, D. , and Middendorf M.. 2005. Reconstruction of the cophylogenetic history of related phylogenetic trees with divergence timing information. Theory Biosci. 123:277–299. [DOI] [PubMed] [Google Scholar]
- Mishima, M. , Sato S., Tsuda K., and Yukawa J.. 2014. Sexual isolation between two known intraspecific populations of Hartigiola (Diptera: Cecidomyiidae) that induce leaf galls on upper and lower surfaces of Fagus crenata (Fagales: Fagaceae), indicating possible diversification into sibling species. Ann. Entomol. Soc. Am. 107:789–798. [Google Scholar]
- Mitter, C. , and Brooks D. R.. 1983. Phylogenetic aspects of coevolution Pp. 65–98 in Futuyma D. M. and Slatkin M., eds. Coevolution. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Nakadai, R. , and Murakami M.. 2015. Patterns of host utilisation by herbivore assemblages of the genus Caloptilia (Lepidoptera; Gracillariidae) on congeneric maple tree (Acer) species. Ecol. Entomol. 40:14–21. [Google Scholar]
- Nakadai, R. , Murakami M., and Hirao T.. 2014. Effects of phylogeny, leaf traits, and the altitudinal distribution of host plants on herbivore assemblages on congeneric Acer species. Oecologia 175:1237–1245. [DOI] [PubMed] [Google Scholar]
- Nosil, P. , Crespi B. J., and Sandoval C. P.. 2002. Host‐plant adaptation drives the parallel evolution of reproductive isolation. Nature 417:440–443. [DOI] [PubMed] [Google Scholar]
- Nyman, T. 2010. To speciate, or not to speciate? Resource heterogeneity, the subjectivity of similarity, and the macroevolutionary consequences of niche‐width shifts in plant‐feeding insects. Biol. Rev. Camb. Philos. Soc. 85:393–411. [DOI] [PubMed] [Google Scholar]
- Nyman, T. , Zinovjev A. G., Vikberg V., and Farrell B. D.. 2006. Molecular phylogeny of the sawfly subfamily Nematinae (Hymenoptera: Tenthredinidae). Syst. Entomol. 31:569–583. [Google Scholar]
- Nyman, T. , Vikberg V., Smith D. R., and Boevé J.‐L.. 2010. How common is ecological speciation in plant‐feeding insects? A “higher” Nematinae perspective. BMC Evol. Biol. 10:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima, I. 2012. Genetic mechanisms preventing the fusion of ecotypes even in the face of gene flow. Sci. Rep. 2:506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R. D. M. 1994. Parallel phylogenies: reconstructing the history of host‐parasite assemblages. Cladistics 10:155–173. [Google Scholar]
- Pearse, I. S. , and Altermatt F.. 2013. Predicting novel trophic interactions in a non‐native world. Ecol. Lett. 16:1088–1094. [DOI] [PubMed] [Google Scholar]
- Pearse, I. S. , Harris D. J., Karban R., and Sih A.. 2013. Predicting novel herbivore‐plant interactions. Oikos 122:1554–1564. [Google Scholar]
- R Core Team . 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: Available at https://www.R-project.org/. [Google Scholar]
- Rabosky, D. L. 2009. Ecological limits and diversification rate: alternative paradigms to explain the variation in species richness among clades and regions. Ecol. Lett. 12:735–743. [DOI] [PubMed] [Google Scholar]
- Rafferty, N. E. , and Ives A. R.. 2013. Phylogenetic trait‐based analyses of ecological networks. Ecology 94:2321–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmann, S. , and Agrawal A. A.. 2011. Evolution of specialization: a phylogenetic study of host range in the red milkweed beetle (Tetraopes tetraophthalmus). Am. Nat. 177:728–737. [DOI] [PubMed] [Google Scholar]
- Renner, S. S. , Beenken L., Grimm G. W., Kocyan A., and Ricklefs R. E.. 2007. The evolution of dioecy, heterodichogamy, and labile sex expression in Acer . Evolution 61:2701–2719. [DOI] [PubMed] [Google Scholar]
- Roininen, H. , and Tahvanainen J.. 1989. Host selection and larval performance of two willow‐feeding sawflies. Ecology 70:129–136. [Google Scholar]
- Ronquist, F. 1995. Reconstructing the history of host‐parasite associations using generalised parsimony. Cladistics 11:73–89. [DOI] [PubMed] [Google Scholar]
- Ronquist, F. , and Huelsenbeck J. P.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Sauer, J. , and Hausdorf B.. 2009. Sexual selection is involved in speciation in a land snail radiation on Crete. Evolution 63:2535–2546. [DOI] [PubMed] [Google Scholar]
- Scheirs, J. , Bruyn L. D., and Verhagen R.. 2000. Optimization of adult performance determines host choice in a grass miner. Proc. R. Soc. Lond. B Biol. Sci. 267:2065–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley, J. 1978. Plant Chemistry and the evolution of host specificity: new evidence from Heliconius and Passiflora . Science 201:745–747. [DOI] [PubMed] [Google Scholar]
- Soria‐Carrasco, V. , Gompert Z., Comeault A. A., Farkas T. E., Parchman T. L., Johnston J. S., et al. 2014. Stick insect genomes reveal natural selection's role in parallel speciation. Science 344:738–742. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. 2014. RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stireman, J. O. , Nason J. D., and Heard S. B.. 2005. Host‐associated genetic differentiation in phytophagous insects: general phenomenon or isolated exceptions? Evidence from a goldenrod‐insect community. Evolution 59:2573–2587. [PubMed] [Google Scholar]
- Stone, G. N. , Hernandez‐Lopez A., Nicholls J. A., di Pierro E., Pujade‐Villar J., Melika G., et al. 2009. Extreme host plant conservatism during at least 20 million years of host plant pursuit by oak gallwasps. Evolution 63:854–869. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher G., Peterson D., Filipski A., and Kumar S.. 2013. MEGA 6: molecular evolutionary genetic analysis version 6.0. Mol. Biol. Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, A. S. 2008. MrBayes5D. Available at http://www.fifthdimension.jp/. (accessed on 9 December 2015).
- Tanabe, A. S. 2011. Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol. Ecol. Resour. 11:914–921. [DOI] [PubMed] [Google Scholar]
- Thomas, Y. , Bethenod M.‐T., Pelozuelo L., Frérot B., and Bourguet D.. 2003. Genetic isolation between two sympatric host‐plant races of the European corn borer, Ostrinia nubilalis Hübner. I. Sex pheromone, moth emergence timing, and parasitism. Evolution 57:261–273. [DOI] [PubMed] [Google Scholar]
- Thompson, J. N. 1998. The evolution of diet breadth: monophagy and polyphagy in swallowtail butterflies. J. Evol. Biol. 11:563–578. [Google Scholar]
- Wahlberg, N. 2007. The phylogenetics and biochemistry of host‐plant specialization in Melitaeine butterflies (Lepidoptera: Nymphalidae). Evolution 55:522–537. [DOI] [PubMed] [Google Scholar]
- Wheat, C. W. , Vogel H., Wittstock U., Braby M. F., Underwood D., and Mitchell‐Olds T.. 2007. The genetic basis of a plant‐insect coevolutionary key innovation. Proc. Natl Acad. Sci. USA 104:20427–20431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegmann, B. M. , Regier J. C., and Mitter C.. 2002. Combined molecular and morphological evidence on the phylogeny of the earliest lepidopteran lineages. Zool. Scr. 31:67–81. [Google Scholar]
- Winkler, I. S. , and Mitter C.. 2008. Specialization, speciation and radiation: the evolutionary biology of herbivorous insects Pp. 240–263 in Tilmon K. J., ed. The evolutionary biology of herbivorous insects. Univ. California Press, Berkeley, CA. [Google Scholar]
- Winkler, I. S. , Mitter C., and Scheffer S. J.. 2009. Repeated climate‐linked host shifts have promoted diversification in a temperate clade of leaf‐mining flies. Proc. Natl Acad. Sci. USA 106:18103–18108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, H.‐J. , Li W.‐Z., and Yang X.‐K.. 2014. Assortative mating between two sympatric closely‐related specialists: inferred from molecular phylogenetic analysis and behavioral data. Sci. Rep. 4:18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S. , and Sota T.. 2012. Parallel allochronic divergence in a winter moth due to disruption of reproductive period by winter harshness. Mol. Ecol. 21:174–183. [DOI] [PubMed] [Google Scholar]
- Zhang, B. , Segraves K. A., Xue H.‐J., Nie R.‐E., Li W.‐Z., and Yang X.‐K.. 2015. Adaptation to different host plant ages facilitates insect divergence without a host shift. Proc. R. Soc. Lond. B Biol. Sci. 282:20151649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogeny of Caloptilia moths feeding on Acer based on mitochondrial COI with information on sampling site.
Figure S2. Distributions of 14 Caloptilia moth species feeding on Acer.
Table S1. Specimen information.
Table S2. DDBJ accession numbers.
Table S3. Primers used in this study.
Table S4. The results of randomization tests including C. aurifasciata that feeds on Toxicodendron.
Data S1. Newick format data containing phylogenetic information of 13 Caloptilia moths feeding on Acer trees.
Data S2. Newick format data containing phylogenetic information of 20 Acer species.
Data S3. CSV format file containing host use information.
Data S4. Text file containing the command for running the randomization analysis in R using Data S1‐3.


