Abstract
Rationale
S-nitrosylation (SNO), an oxidative post-translational modification of cysteine residues, responds to changes in the cardiac redox-environment. Classic biotin switch assay and its derivatives are the most common methods used for detecting SNO. In this approach, the labile SNO group is selectively replaced with a single stable tag. To date, a variety of thiol-reactive tags have been introduced. However, these methods have not produced a consistent dataset which suggests an incomplete capture by a single tag and potentially the presence of different cysteine subpopulations.
Objective
To investigate potential labeling bias in the existing methods with a single tag to detect SNO, explore if there are distinct cysteine subpopulations, and then, develop a strategy to maximize the coverage of SNO proteome.
Methods and Results
We obtained SNO-modified cysteine datasets for wild-type and S-nitrosoglutathione reductase (GSNOR) knock-out mouse hearts (GSNOR is a negative regulator of GSNO production) and NO-induced human embryonic kidney cell using two labeling reagents; the cysteine-reactive pyridyldithiol and iodoacetyl based tandem mass tags. Comparison revealed that <30% of the SNO-modified residues were detected by both tags, while the remaining SNO sites were only labeled by one reagent. Characterization of the two distinct subpopulations of SNO residues indicated that pyridyldithiol reagent preferentially labels cysteine residues that are more basic and hydrophobic. Based on this observation, we proposed a parallel dual labeling strategy followed by an optimized proteomics workflow. This enabled the profiling of 493 SNO-sites in GSNOR knock-out hearts.
Conclusions
Using a protocol comprising two tags for dual labeling maximizes overall detection of SNO by reducing the previously unrecognized labeling bias derived from different cysteine subpopulations.
Keywords: S-nitrosylation, nitric oxide, biotin switch assay, tandem mass tag, proteomics, S-nitrosoglutathione reductase, oxidation, redox
Subject terms: Basic Science Research, Functional Genomics, Cell Signaling/Signal Transduction
INTRODUCTION
Nitric oxide (NO) is well known to play important roles in the regulation of redox signaling through cGMP-dependent pathway1. In the past decade, however, it also has been determined to signal through an oxidative post-translational modification of cysteine thiols, S-nitrosylation (SNO, also known as S-nitrosation)1–7. SNO has been proposed to trigger molecular redox-switches which respond to changes in the redox-environment by regulating protein function. This modification has been implicated in a broad range of physiology and pathophysiology in the cardiovascular systems2,8.
This labile cysteine modification is most commonly detected using variations of the biotin switch assay, originally described in 20019. In this approach, unmodified cysteine residues are blocked with a thiol reactive agent, then SNO-modified residues are selectively reduced with ascorbate and labeled with biotin-HPDP, allowing the formerly SNO-modified proteins or peptides to be enriched using affinity chromatography. Since then, various labeling reagents10–19 have been introduced into the biotin switch technique with the underlying assumption that labeling of free cysteine residues following the reduction of SNO groups, is complete. However, we have observed there is very little overlap in the SNO sites obtained from different proteomic switch experiments20; although, at the time, these differences were not recognized, indicating that there could be differences in the chemistry between the various labeling reagents and the individual SNO-modifiable cysteine residues.
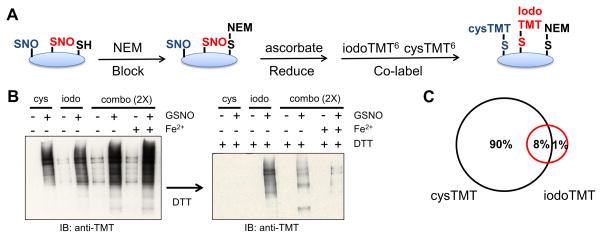
To investigate this potential labeling bias, we compared SNO-modified cysteine datasets equivalently handled but detected using two recently developed isobaric reagents that share same chemical structure except their cysteine reactive group; cysteine-reactive tandem mass tag (cysTMT)18,21,22 and iodoacetyl tandem mass tag (iodoTMT)19,23 (Figure 1). Comparison of these reagents in both cultured cells and cardiac tissues revealed a systemic labeling bias in biotin switch-style assays that, if not addressed, would result in an incomplete assessment of SNO-modifications. These findings also indicate distinct subclasses of reactive SNO residues.
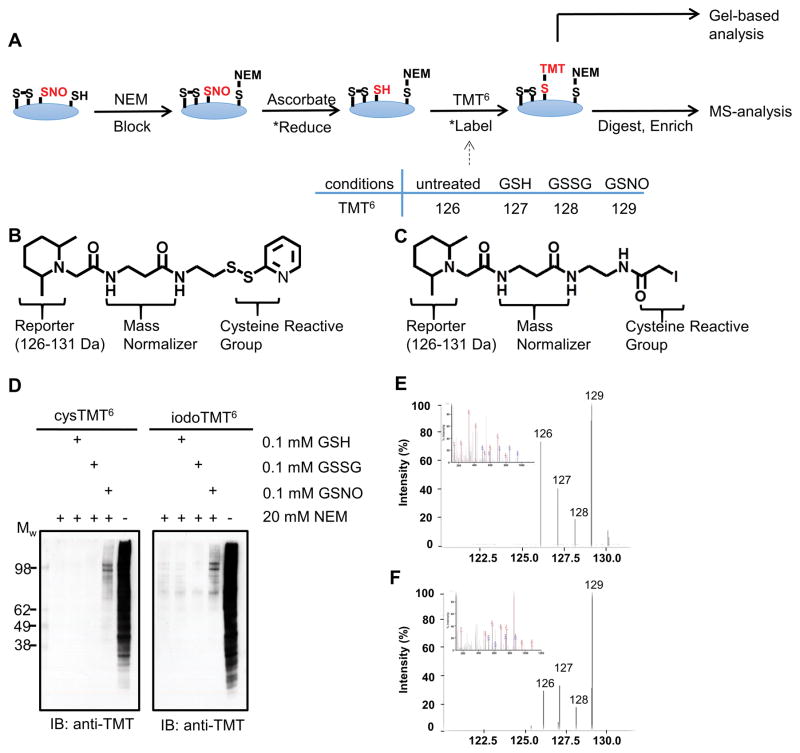
Figure 1. Comparison of the cysTMT6 and iodoTMT6 labeling reagents.
A, Reaction scheme for the TMT switch assay. Asterisks (*) indicate reactions preformed in situ. B and C, Chemical structure of cysTMT6 (B) and IodoTMT6 (C). D, Western blot analysis indicating cysTMT6 and iodoTMT6 labeling. Both TMTs label SNO proteins under NO-inducing conditions. GSH and GSSG were used as negative controls. E and F, MS2 spectra (inset) and reporter ions clusters for the same peptide (PC[TMT]SEETPAISPSK, dUTPase nuclear isoform) identified by cysTMT6 (E) and iodoTMT6 (F). SNO-modification is determined when the GSNO treated reporter ion intensity (129 Da) is at least two-fold greater than greatest control condition: untreated (126 Da), GSH (127 Da) or GSSG (128 Da).
METHODS
An extended methods section is available in the Online Data Supplement.
Cell lysates and heart homogenates preparation
Human embryonic kidney (HEK) 293 cells were lysed in lysis buffer in the dark.
Left ventricles of 8-months-old male C57BL/6 (n=3) and 4 to 5-months-old male/female C57BL/6 (n=3) and S-nitrosoglutathione reductase (GSNOR) knock-out24 (n=4) mice were utilized in this study. Hearts were excised and homogenized on ice in the dark. Protein concentration was determined using the BCA assay (Pierce).
TMT switch assay with Cys and IodoTMT6
HEK cell lysates were diluted to 0.8 g/l in PEN buffer (PBS pH 7.4, 1 mmol/L EDTA and 0.1 mmol/L neocuproine). For parallel labeling, 400 μg of HEK cell lysate per condition in PEN buffer was treated with 0.1 mmol/L S-nitrosoglutathione (GSNO) or three different control treatments (untreated vehicle, 0.1 mmol/L reduced glutathione (GSH), or 0.1 mmol/L oxidized glutathione (GSSG) for negative controls and untreated vehicle as a positive control) for 20 min at 37 °C (n=4, from separately cultured plates). GSNO was removed using a Zeba desalt spin column (Thermo) equilibrated with PEN pH 7.4 according to the manufacturer’s protocol. The solution of remaining free thiols diluted to 0.5 g/l was blocked with 20 mmol/L of NEM in the presence of 2.5% (w/v) SDS and incubated for 20 min at 50°C. Excess NEM was removed by the PEN pH 8.0 equilibrated Zeba desalt spin column. As a positive control, an additional untreated sample (400 μg) was processed but not blocked with NEM. GSNO-treated and control samples were split for labeling with either cys- or iodoTMT6 (200 μg/each) and half of them (each n=4) were labeled with each sixplex (126–131) of cys- or iodoTMT6, individually (n=4 for cys TMT and n=4 for iodo TMT). For cysTMT6 labeling, samples were diluted to 0.41 g/l in PEN pH 7.4, 1 mmol/L sodium ascorbate, 1 mmol/L CuSO4 and 0.3 mmol/L cysTMT6 (cysTMT0 in the case of the positive control) and incubated for 2 h at 37 °C. For iodoTMT6 labeling, samples were diluted to 0.41 g/l in PEN pH 8.0, 5 mmol/L sodium ascorbate and 0.3 mmol/L iodoTMT6 with equivalent incubation. Excess label was removed by acetone precipitation and the resultant pellets were carefully washed with an additional volume of acetone. Pellets were re-suspended to 1 g/l with PEN and 0.5% of SDS.
Sequential labeling was performed in the iodoTMT6 labeling condition. After acetone precipitation, pellets were re-suspended with cysTMT6 labeling buffer as described and incubated for 2h for the second labeling. For simultaneous labeling, samples were incubated with equal concentration of cysTMT6 and iodoTMT6, 5 mmol/L sodium ascorbate and trace of metals as described in Online Figure VIII.
TMT switch assay with individual recombinant proteins or heart homogenates (450 μg/each) was performed in the same manner as parallel labeling.
TMT switch assay in identical labeling conditions regarding the two labels was performed following the methods for iodoTMT6 as described above (PEN pH 8.0, 5 mmol/L sodium ascorbate and 0.3 mmol/L cys- or iodoTMT6), with recombinant protein, HEK cell lysates, and heart homogenates. In these sets of assays, non-reduced condition (no-sodium ascorbate) was used as a negative control.
All steps were protected from light.
SNO peptides enrichment for Mass Spectrometry (MS) analysis
Elutions from parallel labeling experiments were performed in two ways: Unlabeled peptides were removed by washing with TBS and TMT-labeled peptides were eluted with 500 mmol/L triethylammonium bicarbonate pH 8.5 buffer containing 10 mmol/L N,N-diisopropylethylamine (n=2 in separate 6′plexes for both labels). Or elution was performed as described previously with 50 % (v/v) acetonitrile/0.4 % (v/v) trifluoroacetic acid18 (n=2 in separate 6′plexes for both labels). Both elution methods were applied alternatively in different replicates to increase TMT peptide identification.
Data analysis
Raw MS data was searched using OMSSA (version 2.1.9)25 against the human or mouse Uniprot database. The MS spectra was also searched using the X!Tandem algorithm (version TPP v4.4, rev 1)26. SNO sites were searched based on FASTA sequence in Uniprot using customized code. Quantitative values for the TMT reporter ions were collected with the Libra module of the trans-proteome pipeline using a custom condition file (Online Figure IX). The fold-increase in TMT intensity of GSNO over maximum background signal was indicative of the detected extent of SNO-modification and used as ‘intensity index’. For experiments performed with non-reduced control (no-ascorbate), a site of SNO-modification was determined if the TMT intensity in the ascorbate treated sample was greater than the no-ascorbate sample.
The MS data have been deposited to the ProteomeXchange (http://www.proteomexchange.org) via the PRIDE partner repository27 (PXD000741).
Functional properties analysis
Gene ontology enrichment analysis, molecular function, biological process, and cellular compartment, were executed using cytoscape28 with the Bingo plug-in29. The statistical test employed was the hypergeometric test with a Benjamini & Hochberg false discovery rate correction with a selected significance level of 0.05. The results were compared between groups, and unique ontology categories between the groups were analyzed. Spindle graphs of the enriched gene ontologies were created using a custom script and cytoscape28.
Statistics
Difference of pI was analyzed using a t-test using GraphPad Prism 5. Data in bar graphs represents mean ± SEM. p values less than 0.05 were considered significant.
RESULTS
Detection and site mapping of SNO-modifications using CysTMT or IodoTMT
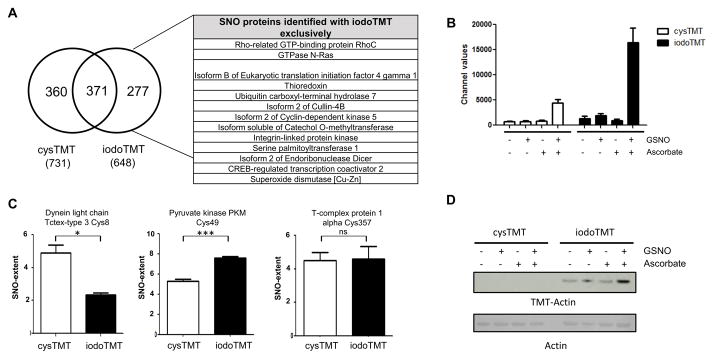
In our initial investigations, we found that although the newly developed iodoTMT6 was effective at labeling cysteine residues, it could not be simply substituted into the previously reported cysTMT6 switch protocol18 for SNO-modifications (Figure 1A; Online Figure IA). The cysTMT6 multiplex reagent shares a similar chemical structure that releases the same reporter ions (Figure 1B and 1C) and both reagents are enriched by the TMT affinity chromatography. However, cysTMT6 has a pyridyldithiol reactive group that forms a reversible disulfide bond with an available cysteine, while iodoTMT6 utilizes an iodoacetyl group to irreversibly alkylate cysteines. IodoTMT6 was recently developed to circumvent the reversible cysTMT6 modification which would allow iodoTMT6 labeled samples to undergo routine reduction and alkylation prior to MS-analysis19. To achieve effective incorporation of iodoTMT6 into the assay, various labeling conditions were evaluated including potential radical initiators or propagators (Online Figure I). Independently optimized protocols (Online Table I) were used to detect SNO-modifications using the iodoTMT6 and cysTMT6 reagents. TMT switch assays were carried out on HEK 293 cell lysates treated with five different conditions; GSNO to induce SNO, three negative control conditions (untreated, GSH, GSSG) and unblocked positive control (labeling of all available cysteine residues). Samples were divided, labeled with cysTMT6 as previously established18 or iodoTMT6 followed by the optimized protocol (see Method Section), in a multiplex format. Then they were subsequently examined by western blot analysis (Figure 1D) or digested, desalted and analyzed by MS (Figure 1E and 1F). Analysis of the complete spectral dataset resulted in the identification of 1008 unique SNO-modified residues on 773 proteins (Online Table II and III). The iodoTMT6 reagent identified 648 SNO-modified cysteine residues (corresponding to 495 proteins), while cysTMT6 identified 731 SNO-modified sites in 557 proteins in the same biological samples, handled equivalently (Figure 2A). Approximately one third of the SNO-modified residues (371 sites) were detected by both of the reagents. 277 SNO-modified cysteine residues were identified exclusively with iodoTMT6 and this included 200 proteins whose SNO sites were only detected using iodoTMT6 (Figure 2A). When the samples were labeled with either cysTMT6 or iodoTMT6 under an identical reducing condition (see Online Method), less than one third of the total SNO-sites were identified by both of the reagents (Online Table IV), while more than 60% of the sites were commonly detected between replicates of the same label (Online Figure II).
Figure 2. MS comparison of SNO-detection using cysTMT6 or iodoTMT6 switch reveals incomplete cysteine labeling.
A, Cys- or iodoTMT6 labeled SNO sites from HEK293 cells were analyzed by MS. The table contains a selection of proteins whose cysteine residues were only detected by iodoTMT6. B, MS analysis and quantitation of TMT6 reporter ions for Cys49 of pyruvate kinase PKM demonstrating the labeling bias (n=4/group). C, Bar graphs indicating TMT labeling for a selection of SNO-modified cysteines: Cys8 of dynein light chain Tctex-type 3 (n=3/group), Cys49 of pyruvate kinase PKM (n=4/group), and Cys357 of T-complex protein 1 alpha (n=3/group). *P<0.05, ***P< 0.00001 by t-test. These sites show different preference for either the cys- or iodoTMT6 reagents. D, Western blot analysis of different TMT labeling on recombinant human actin after cys- or iodoTMT6 switch assay. Extent of SNO on actin in HEK293 cell was distinguished by MS analysis (Online Table IV).
Labeling bias in classic biotin switch-style assay reveals two subpopulations of cysteine residues
We have shown that cysTMT6 and iodoTMT6 target different cysteine residues in the same experiment, suggesting the possibility for distinct subclasses of cysteine residues. Our dataset contained numerous examples highlighting the specificity of the labeling bias including Cys8 of dynein light chain Tctex-type 3 which was labeled preferentially by cysTMT6 (Figure 2C). Other sites, such as Cys49 of pyruvate kinase PKM, were preferentially labeled by iodoTMT6 while Cys357 of T-complex protein 1 alpha reacted equivalently to both reagents (Figure 2B and 2C).
This labeling bias was validated by a TMT-switch assay with recombinant proteins. For example, SNO cysteine residues on human actin was only labeled with iodoTMT, while SNO-modified bovine actin was preferentially detected by cysTMT (Figure 2D; Online Figure III).
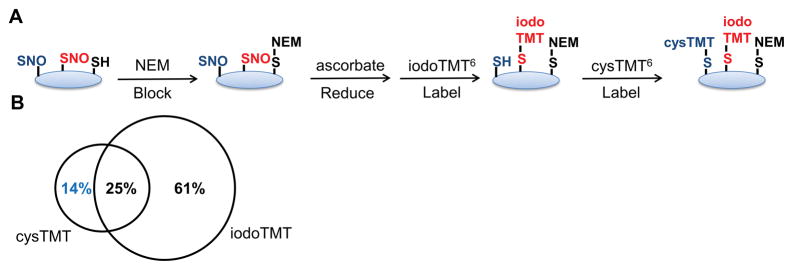
This observation is further supported by a sequential labeling experiment where SNO sites were first labeled with iodoTMT6 followed by cysTMT6 (Figure 3A; Online Table V). MS analysis detected a group of cysTMT6 labeled cysteine residues that remained available throughout the iodoTMT6 labeling (Figure 3B). This finding demonstrates that labeling is not fully saturating and suggests individual cysteine residues preferentially react with different labeling reagents.
Figure 3. Sequential cys- and iodoTMT6 labeling for the TMT-switch assay in HEK cells.
A, Scheme depicting sequential labeling methods. B, MS analysis of the sequential labeling found 14% of the SNO sites were detected by the second label, cysTMT6, indicating incomplete labeling by iodoTMT6 and suggesting distinct subclasses of cysteine residues.
Validation of SNO-labeling bias in mouse hearts
Next, we applied the dual reagent protocol and optimized proteomics workflow for MS (discussed in a later section) to identify SNO modifications in mouse hearts. Left ventricular tissue from intact mouse hearts were homogenized, labeled by either iodo- or cysTMT6 and analyzed through the optimized pipeline as described later. As observed in HEK cell analysis, of the 315 total SNO sites identified, 98 sites were detected with both reagents while 113 sites and 104 sites were identified only with cys or iodoTMT respectively, (Online Table VI). We also performed the switch assay under identical reducing conditions with 4 to 5-month-old wild-type mouse hearts (Online Table VII). More than 70% of TMT-labeled sites were identified with only one tag. Additional 44 sites not identified with cysTMT, were determined with iodoTMT, confirming the labeling bias. SNO of muscle proteins such as cardiac troponin I (cTnI) (Cys81), alpha-actinin-2 (Cys781) and isoform 3 of LIM domain-binding protein 3 (Cys511/Cys514) were exclusively identified via iodoTMT-labeling.
Using our two-label strategy we were able to confirm SNO-modifications previously identified30–32 in untreated cardiac tissue as well as identify several novel modified sites (Selected sites are listed in Table 1). For example, cardiac isoform sarcoplasmic/endoplasmic reticulum calcium ATPase (SERCA2A), a key protein for Ca2+ handling33–35, was identified to be S-nitrosylated in this study. It’s been reported that the SNO increases activity of SERCA2A and thus enhances Ca2+ update into sarcoplasmic reticulum and decreases cytosolic Ca2+ during ischemia/reperfusion, suggesting SNO-SERCA2A as a potential mediator for cardioprotection36. SERCA2A had several SNO-modified cysteine residues previously reported such as Cys364 or Cys47130,31 and also new sites Cys344 and Cys349 were identified in this study, which were exclusively labeled with either of TMT reagents. The other examples, the sites of the muscle proteins descrived above, Cys781 of alpha-actinin-2 and Cys511/Cys514 of isoform 3 of LIM domain-binding protein 3 were newly identified to be S-nitrosylated. Without use of both tags, the detection of SNO-modification would be incomplete. Based on these data, it is clear that the addition of an alternate cysteine reactive tag is required to reduce the labeling bias and thus increase SNO proteome coverage.
Table 1. SNO sites from hearts.
Selected examples of endogenous SNO sites, which were detected from cardiac tissues of wild-type mice.
| Protein Group Description | Site | Labeled by | |
|---|---|---|---|
| Aconitate hydratase, mitochondrial | 592 | iodo | |
| Actin, alpha cardiac muscle 1 | 219 | cys | |
| Actin, alpha cardiac muscle 1 | 259 | cys | |
| Acyl-coenzyme A thioesterase 2, mitochondrial | 55 | iodo | |
| ADP/ATP translocase 1 | 257 | cys | iodo |
| Aspartate aminotransferase, cytoplasmic | 160 | iodo | |
| ATP synthase subunit d, mitochondrial | 101 | cys | iodo |
| Creatine kinase S-type, mitochondrial | 180 | cys | |
| Cytochrome b-c1 complex subunit 2, mitochondrial | 192 | cys | iodo |
| Dihydrolipoyl dehydrogenase, mitochondrial | 484 | iodo | |
| Elongation factor 2 | 693 | cys | |
| Heat shock cognate 71 kDa protein | 17 | iodo | |
| Isoform M1 of Pyruvate kinase PKM | 474 | iodo | |
| Malate dehydrogenase, mitochondrial | 89 | cys | iodo |
| Myosin-6 | 1750 | cys | iodo |
| Protein kinase C and casein kinase II substrate protein 3 | 402 | cys | |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | 536 | cys | |
| Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | 245 | cys | |
| Troponin I, cardiac muscle | 81 | iodo | |
| Ubiquitin-conjugating enzyme E2 D3 | 85 | cys | iodo |
Proteomic profiling of SNO in GSNOR knock-out mouse
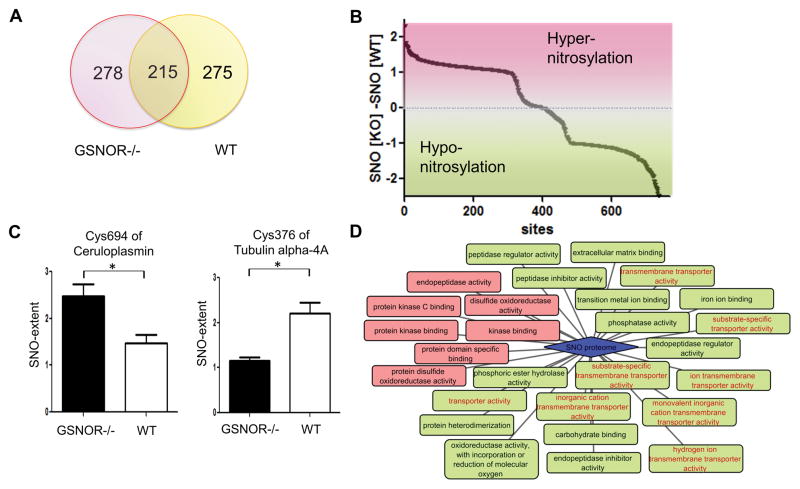
GSNOR is a negative regulator of GSNO in the cell and is thought to govern the SNO level37,38. It has also been identified as a key regulator of cardiac function37,38. Therefore, the GSNOR knock-out mouse model was chosen to perform a high-throughput quantitative analysis of SNO-modification in mouse hearts and also to highlight the labeling bias across different pools of modified cysteines. By applying the dual-reagent protocol, SNO sites in GSNOR knock-out hearts were profiled for the first time; a total of 493 SNO-sites were identified with either or both of the reagents (Online Table VIII) and a set of the sites partially overlapped with the sites of wild-type (Figure 4A). Of these, SNO sites of widely-studied cardiac proteins39 in the GSNOR knock-out hearts were determined.
Figure 4. Analysis of SNO-proteome of GSNOR knock-out hearts.
A, Cys- or iodoTMT6 labeled SNO sites from GSNOR knock-out and wild-type mouse hearts were compared. B, A global view of hyper or hypo-nitrosylation of cysteine residues in GSNOR knock-out hearts. 345 sites were hyper-nitrosylated and 365 sites were hypo-nitrosylated compared to WT. 34 sites out of range (−2~+2) were not indicated in this graph (provided in Online Table VII and VIII). C, Examples of hyper-nitrosylated site (left, n=3/group) and hypo-nitrosylated site labeled with cysTMT6 in GSNOR knock-out (right, n=3 GSNOR−/−, n=2 WT), from the pool of GSNOR knock-out and wild-type’s sites. *P<0.05 by t-test. D, Molecular functional analysis of SNO-proteome of the two subpopulations in GSNOR knock-out heart: hyper-nitrosylated group (red) and hypo-nitrosylated group (green). Proteins that were clustered to unique categories.
cTnI is the inhibitory subunit of the troponin complex which regulates muscle contraction40, and is extensively regulated by phosphorylation41. cTnI fragments exist as a complex with troponin C (cTnC), which is the Ca2+-binding subunit. The binding of Ca2+ to regulatory site of TnC induces a conformational change, which blocks the action of cTnI and generates muscle contraction40,41. Although the functional roles of SNO in those proteins are not well characterized, we site-mapped Cys81 and Cys98 of cTnI and Cys35 of cTnC as S-nitrosylated in GSNOR knock-out mice. In addition, eight SNO sites of SERCA2A and one site of ryanodine receptor 2, one of the Ca2+ handling proteins39, were found to be nitrosylated in this model. The SNO proteomes in WT and the GRNOR knock-out were further compared based on gene ontology enrichment (Online Table XV).
Interestingly, the magnitudes of SNO-shift in GSNOR knock-out hearts varied across sites, as some were hyper-nitrosylated while others were hypo-nitrosylated compared to wild-type. As an example, Cys694 of ceruloplasmin displayed a SNO-increase, while Cys376 of tubulin alpha-4A and Cys349 of cardiac isoform SERCA2A were less nitrosylated in GSNOR knock-out (Figure 4B). 345 sites were hyper-nitrosylated and 365 sites were less nitrosylated in the GSNOR-deficient model (Figure 4C). These two hyper-SNO and hypo-SNO groups were further analyzed based on gene ontology enrichment. Proteins with hypo-nitrosylated sites in GSNOR knock-out were clustered to unique categories, including proteins that have channel or transporter activity or proteins located in extracellular region (Figure 4D and Online Figure IV).
The labeling bias in GSNOR knock-out hearts was observed to be dominant with cysTMT-labeling over iodoTMT. Presumably, the biased preference to one tag occurs by specific cellular chemical environments, which may be different across various biological samples.
Chemical properties of the two subpopulations
To further investigate the potential subclasses of SNO sites detected by the different reagents, we analyzed the physicochemical properties of the cys- and iodoTMT6 specific residues in the HEK and mouse hearts datasets. We hypothesized that a cysteine’s reactivity to a particular TMT labeling reagent is dictated by its surrounding chemical environment. Analogous to the selectivity of SNO-formation42–45, the chemical properties of neighboring amino acids may affect the likelihood that each cysteine residue will react preferentially with either reagent, while the protein’s structure determines accessibility. Although these properties were not enough to represent all elements for the environment of a certain reactive cysteine residue, they provide insight into the physicochemical effect.
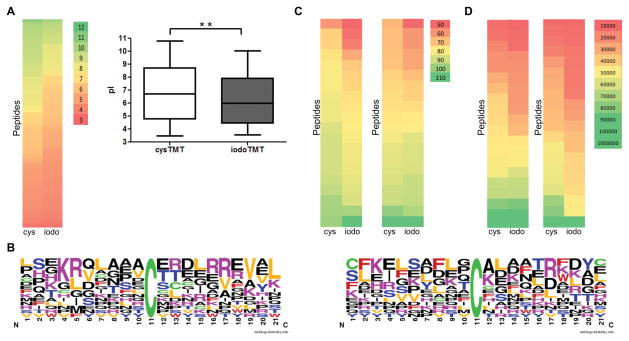
All amino acid residues flanking each side of the cysteine residues that were specifically reactive to either cys- or iodoTMT6 from mouse hearts and HEK cell were compared. It revealed that cysTMT6 specific sites from hearts had a higher proportion of high theoretical pI overall compared to the iodoTMT6 sites (Figure 5A; Online Table IX). The overall distributions for the pI were slightly different between the two reagents groups from HEK cell (Online Figure V; Online Table IX). The top fifteen reactive (or intense) cysteine residues unique to each reagent were compared; the cysTMT6 specific sites were found to have a higher proportion of neighboring positively charged residues (arginine/lysine) compared to the iodoTMT6 sites (Figure 5B; Online Figure VB; Online Table IX).
Figure 5. Physiochemical properties analysis of two subpopulations.
A, The theoretical pIs of 113 exclusively cysTMT6-labeled residues and 104 iodoTMT6-specific residues from mouse hearts were analyzed and illustrated using a bar graph and a color scale from low (red) to high (green) pI (left, cysTMT6; right, iodoTMT6). Higher proportion of low pI was observed in iodoTMT6 specific group. (n=3/group). **P<0.01. B, The frequency of amino acids surrounding the cysteine residues of two groups were analyzed (left, cysTMT6; right, iodoTMT6). Valine/leucine and positively charged lysine/arginine were more frequently observed in cysTMT6 specific group. Reactive cysteine is highlighted in green at position 11. C, Aliphatic index for proteins (whose cysteine residues were reactive, see Online Table X) were computed based on the amino acid sequence and compared between protein groups which were detected with only one TMT reagent. First color scale represents aliphatic index of proteins from HEK cell (left, cysTMT6; right, iodoTMT6) and second color scale displays it from mouse hearts. Overall, cysTMT6 group represented more distribution of higher aliphatic characters. D, Molecular weight of the same proteins for (C) was analyzed and represented in the same way. IodoTMT6 group had higher proportion of low molecular weight proteins compared to cysTMT6 group.
Next, more global characteristics were investigated for the proteins containing the most reactive (or intense) cysteine that were exclusively labeled by one or the other TMT reagent (Figure 5C and 5D; Online Table X). Analysis of aliphatic index revealed that proteins uniquely targeted by cysTMT6 had a higher number of aliphatic amino acid residues (alanine, valine, isoleucine, and leucine) than those by iodoTMT6 (Figure 5C). Analysis of molecular weight revealed iodoTMT6 favored proteins with lower molecular weight than cysTMT6 group (Figure 5D). The analysis with datasets from samples labeled under identical conditions with control groups represented corresponding trends (Online Figure VI). This suggests a synergic effect between basic sequences and a more local/global hydrophobic structure may produce conditions more favorable to cysTMT6 than iodoTMT6. The iodoTMT6 reagent may be more accessible and reactive with cysteine residues located within smaller proteins, local regions with lower pKa or more global polar character of the intact protein, in both HEK cells and cardiac tissue. Although a specific mechanism could not be determined, our findings demonstrate that the reagents can influence selectivity for labeling specific classes of cysteine residues, presenting at least two classes of SNO-modifiable cysteine residues.
Further optimization of experimental strategies to Maximize SNO-detection
To expand the SNO proteome coverage and ensure maximal capture of both subclasses of modified sites, different experimental configurations using both cys- and iodoTMT6 were evaluated: sequential, simultaneous and parallel. Sequential labeling starting with either cys- or iodoTMT6 followed by the other second reagent was determined not to be efficient (Figure 3B; Online Figure VII; Online Table V). There was reduced TMT-intensity for the second reagent along with an increased non-specific labeling compared to the single TMT labeling experiments (Online Figure VII). This loss of reactivity is likely due to oxidation that accumulates during the additional labeling step. For simultaneous labeling experiments, the reaction conditions needed for efficient iodoTMT6 labeling was not compatible with those required for optimal cysTMT6 labeling (Figure 7A; Online Figure VIII). When adapting the cysTMT6 labeling conditions to be compatible with iodoTMT6, the simultaneous labeling was found to be less intense as sum of each single labeling due to poor iodoTMT6 labeling (Figure 7B and 7C; Online Table XI). We found that labeling with cys- and iodoTMT6 in parallel had the greatest efficiency to detect SNO sites as it allows preservation of the optimal buffer and reaction conditions for both reagents. Using the individually optimized conditions in parallel for each reagent, we were able to identify ~700 SNO sites with each labeling reagent, which is more than two-fold increase compared to previous cysTMT6 study using similar conditions18.
Figure 7. Simultaneous cys- and iodoTMT6 labeling for the TMT-switch assay in HEK cells.
C, Scheme of simultaneous labeling protocol. D, The efficiency of iodoTMT6 labeling was assessed for simultaneous labeling. Non-reduced samples are shown in the left and DTT-treated samples are shown in the right. DTT treatment following a co-reagent labeling protocol would remove cysTMT6 (disulfide linked) but not iodoTMT6 (irreversibly alkylated) revealing the extent to iodoTMT6 labeling. IodoTMT6 was found to be less efficient in simultaneous labeling (indicated as combo (2x)) compared to the single labeling. Total labeled SNO in simultaneous labeling approach was shown to increase than either cys- or iodoTMT6 single labeling when the concentration of TMT was double (left) but the efficiency of individual labels decreased than under the optimized conditions for each label, due to poor iodoTMT6-labeling in both iodoTMT6-compatible/incompatible buffers (right). E, Summary of an MS-analysis from a simultaneous labeling experiment demonstrating poor iodoTMT6 labeling efficiency. The majority of the detected TMT-peptides were labeled with cysTMT6.
The improvement in individual reagent’s SNO-identification over the previous study can be attributed to two additional features in the method; the use of OMSSA as a search engine for assignment of the MS spectra (Online Table XII) and the application of new enrichment protocol using a ‘TMT analogue’ for elution (Online Table XIII). With MS-based analysis, a total of 1008 SNO-modified residues were identified, through OMSSA-search (Figure 2A). 731 SNO sites in 557 proteins and 648 sites corresponding to 495 proteins were identified with cys- and iodoTMT6 labeling, respectively (371 sites were detected by both of the reagents). When X!Tandem search engine was applied to the same mass spectra, 315 sites from 271 proteins were found to be SNO with cysTMT6 labeling and 338 SNO sites corresponding to 274 proteins were identified with iodoTMT6 labeling (159 sites detected by both TMTs). Overall, OMSSA identified higher number of SNO sites compared with X!Tandem and had greater than 80% agreement with the X!Tandem dataset (Online Table XII).
Unlike biotin-based labeling, whose labels are removed by reducing agent at enrichment step before MS analysis9,14, the elution protocol of TMT preserves the tag during MS analysis. Previously, TMT-tagged peptides were eluted from immune-affinity chromatography by a low pH buffer to denature the interaction between the resin and TMT peptides18. In a new elution protocol, we introduced a small ‘TMT analogue’, which interacts with affinity resin to compete the bound TMT peptides. As shown in Online Table XIII, the number of SNO-peptides when using the ‘TMT analogue’ was increased ~38 % over the low pH elution. We applied the new elution protocol in addition to the previous low pH elution method in this study to maximize coverage.
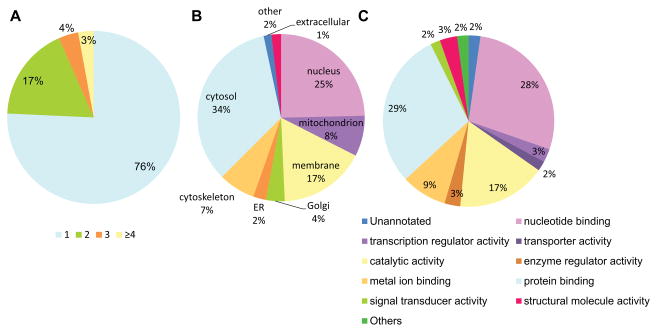
Further investigations indicated the majority of the SNO-modified proteins with a single SNO site and the subcellular locations and molecular functions of them, whose distributions are similar to those of other studies with a single tag (Figure 6; Online Table XIV)18,20. Based on this, our increased number of total SNO proteins was not derived from the specific detection of a certain functional class of proteins (e.g. localized exclusively) and our method increased the depth of proteome coverage.
Figure 6. Analysis of total SNO proteome.
A total of 1008 SNO proteins detected by TMT reagents were analyzed. A, The number of SNO-modified sites per protein. The majority of the 1008 SNO-modified proteins had a single SNO site while 24% of them had two or more SNO-modified cysteine residues. B and C, Subcellular locations and molecular functions of the proteins were analyzed based on Gene Ontology: Subcellular location of SNO-modified proteins (B) and functional specificity of the SNO-modified proteins (C). These features showed similar distributions to those of other studies for the identification of SNO cysteine20 or data from our previous study with single cysTMT18 (data not shown). This indicates our increased number of SNO proteins is derived from overall proteome, not from a certain class of proteins (e.g. localized or functionalized exclusively); further supporting that SNO is able to impact a broad number of sub-proteomes and various functional classes of proteins.
DISCUSSION
Significant roles of SNO have been proposed in cardiac physiology and pathophysiology. The modification is thought to affect protein function directly by increasing or inhibiting the activity or shield the thiol groups on critical cysteine residues from further oxidation during severe oxidative stress condition such as myocardial ischemia and reperfusion8. It is expected that there are other SNO proteins for cardioprotection not identified yet, especially if they are low-abundant1. In addition, disruption of SNO in myocytes has been recognized as a critical effect in failing hearts46. For example, Cys294 of ATP synthase α was found less S-nitrosylated in dyssynchronous heart failure hearts than in cardiac resynchronization therapy hearts47. Another example, hyponitrosylation of ryanodine receptor 2 due to imbalance redox, was observed in heart failure and suggested to contribute to diastolic Ca2+ leak48. However, molecular mechanisms and potential therapeutic strategies by SNO in numerous cardiovascular diseases remain incompletely understood.
In the aid of better selection among the many existing SNO-labeling reagents such as biotin-HPDP9, ICAT10,17, SNOSID11, SNOCAP13, SNO-RAC14, d5-NEM16, cysTMT18 and iodoTMT19, sets of proteins from two different approaches have been compared49. We have analyzed the datasets studied with three prevalent tags and different experimental protocols, but found little overlaps between them20. Thus, in this study we used same samples with controlled setting where the two labeling regents were identical except for their thiol-reactive groups as thioether or alkyl iodide, which are also common in most of other labeling tags. In addition, we analyzed datasets at an amino acid site level for a more accurate comparison. It has been a fundamental assumption of previous studies that these tags and all reactions proceeded to completion and thus, all ascorbate-exposed cysteine were labeled. However, the result of our study demonstrated ‘unsaturation’ of labeling by an individual label which has underestimated the number of SNO-modified site. In light of this work, it is important to find a method that allows capture of more sites. Our bioinformatics approach to extract the position of each site allowed site-identification for the corresponding peptides. This has allowed removing peptides’ redundancy resulted from mis-cleavage of peptides, producing a more accurate comparison between the pools of cys- and iodoTMT modified sites. More importantly, this enabled site-level quantitation, which provided better accuracy over the traditional peptide-level quantitation. Finally, this site-identification has made it possible to extract flanking amino acid sequences of cysteine, which was used for the analysis of consensus sequence and chemical properties.
This study discovered that there are at least two unique populations of SNO-modifiable cysteine residues with HEK and cardiac muscles that have distinct reactivity to two chemically-classified tags. Based on this, it is clear that the use of a single tag strategy to detect SNO-modifications will introduce a labeling bias. This could affect the interpretation of data and reduce the probability of identification of key modification of proteins since the reactivity of specific set of cysteine residues in a certain environment currently cannot be predicted. To date, all studies on SNO proteomics in the cardiovascular field used only a one tag, which has led to a presumption that there is only a single SNO-reactive population. Thus using both reagents will be required to ensure the detection and quantification of two subpopulations of SNO-modifiable cysteine with different chemical characteristics. Based on this observation, our optimized parallel workflow is a generalizable method and can be applied to other proteomic investigations using a thiol-labeling switch strategy.
Using the dual labeling strategy, we performed a proteomic profiling of SNO in GSNOR knock-out mouse hearts. Previously, most studies have targeted specific SNO-modifications. For example, SNO of β-arrestin2 promoted protein-protein interactions with clathrin in GSNOR knock-out mouse spleen50,51. However, few studies have investigated a global change of the entire SNO proteome in a GSNOR-deficient model38,52. Based on the function of GSNOR, levels of SNO in the knock-out models were expected to increase overall but it wasn’t clear if this would be true across all modified cysteines. Analysis of the nearly 500 SNO-modified cysteine residues revealed that while the majority of sites had increased levels of modifications, more than one third were reduced. This implies that SNO regulation is a complex process and other factors are involved in determining this redox-modification status. Our bioinformatics analysis with the hypo- and hyper-SNO sites suggests that protein functions or subcellular locations may be one of these factors. A set of proteins which contain hypo-SNO sites was uniquely clustered as ‘transporter’ proteins by gene ontology enrichment analysis and this set of proteins was mainly located in membrane or mitochondrion and has metal or nucleotide binding regions.
In the study, we determined that iodoTMT6 didn’t label SNO in the previously reported cysTMT-switch protocol. To achieve effective incorporation of iodoTMT into this assay, various labeling conditions were tested and, although we were unable to determine the exact factors that inhibited labeling of iodoTMT6 compared with cysTMT6, our studies successfully optimized conditions for effective iodoTMT6 labeling. One critical factor was the use of copper during labeling, which has been reported to accelerate ascorbate-dependent reduction of SNO without effecting specificity53. However, for iodo-based TMT switch protocols with NO-inducing conditions, copper over a certain range inhibited iodoTMT6 labeling, while enhancing cysTMT6 labeling (Online Figure I). The alkyl halide moiety in iodoTMT6 has been indicated to be labile during radical-generating conditions54 and labeling was recovered in the presence of 1 mmol/L copper by the addition of glycerol, a known radical scavenger55,56 in this study (Online Figure I). There are several candidates for a radical initiator or propagator in the TMT switch protocol. Some metals have been indicated as a source of radicals similar to those generated in the Fenton-like reaction57 and it was reported hydroxyl radicals generated by Cu2+/H2O2 were stimulated specifically in HEPES buffer58,59. This could explain the different labeling efficiency observed when labeling was carried out in PBS or HEPES buffer. The TMT labeling step is coupled with the reduction of the SNO group by ascorbate, which has been shown to generate hydroxyl radical in copper contaminated water60, suggesting the formation of free radicals that could inhibit labeling.
In conclusion, the parallel combination of both the cysTMT6 and iodoTMT6 increased the coverage of SNO-modifications identified; however, comparison of the individual reagent datasets indicated a clear bias in cysteine labeling between the two reagents. Each of the TMT reagents have thiol-reactive groups commonly used in the biotin switch-style assays but have never been rigorously compared. The pyridyldithiol and iodoacetyl reactive groups in the different TMT reagents were found to label distinct but partially overlapping subclasses of cysteine residues. Our findings demonstrate an important but overlooked aspect of labeling in the classic switch assay, which if accounted for, can expand the accessible cysteine proteome.
Supplementary Material
Novelty and Significance.
What Is Known?
S-nitrosylation (SNO) is a post-translational modification that serves as a key regulator of protein activity in cellular redox status and has many roles in cardiovascular disease, yet the identification and quantification of SNO -modified residues remain limited.
The biotin-switch technique, which first reduces SNO with ascorbate and then chemically labels newly exposed cysteine residues with a stable tag, has been the most general strategy in this field.
What New Information Does This Article Contribute?
When using two chemically different mass spectrometry-based tags to label the ascorbate-exposed cysteine residues, there are subpopulations of SNO-modified amino acid residues which are preferentially detected by one or the other of the tags, indicating that there has been an unrecognized labeling bias in the common biotin-switch assays.
An optimized proteomic mass spectrometry-based workflow using two (chemically different) tags ensures the detection and quantification of the two subpopulations of SNO-modifiable cysteine with different chemical characteristics on the technical side.
This labeling strategy enables a profiling of nearly 500 SNO-sites in cardiac tissue of GSNOR knock-out mouse, which is an interesting model for studying redox-regulation, and reveals that SNO was reduced on one third of the sites while being increased in the majority.
Investigating SNO-shift is important because SNO is a protein post-translational modification which regulates protein activity and responds to cellular redox status. Therefore, the technique to detect SNO has been a long interest in cardiovascular research and classic biotin-switch style technique has been a widely used method for this. This biotin-switch style techniques shared by a variety of labeling strategies, assumes that the labeling tag captures all available SNO. However, we show that this is not the case and the labeling bias, which has been unrecognized, exists in the commonly used biotin-switch style assays, allowing to detect only subsets of the SNO-sites. By more accurate amino acid site-level comparison between SNO-sites detected by two chemically different tags, we provide the evidence that there are different subpopulations of SNO-modifiable cysteine residues which can only (or preferentially) be detected by one labeling reagent. Collectively, we optimize a proteomic workflow using chemically different tags to maximize coverage of the SNO-proteome of complex samples such as mouse hearts. We describe an important method that may have a large impact on research practice of all future SNO-based studies, the interpretation of existing SNO-data, and our understanding of redox regulation in cardiovascular studies in an unbiased manner.
Acknowledgments
The authors would like to thank Raghothama Chaerkady, Robert O’Meally, Ronald J. Holewinski and Irina Tchernyshyov for assistance with mass spectrometry. We also thank Rosa Viner for discussions regarding TMT-MS data and Gautam Saxena for search engines.
SOURCES OF FUNDING
This work was supported by National Heart, Lung, and Blood Institute grants NHLBI-HV-05(2), PO1HL77189-01 and 1RO1HL101235-01 to J.V.E. American Heart Association Pre-Doctoral Fellowship 0815145E to C.I.M, 15PRE25570009 to H.S.C.
Nonstandard Abbreviations and Acronyms
- cysTMT
cysteine-reactive tandem mass tag
- GSH
reduced glutathione
- GSNO
S-nitrosoglutathione
- GSNOR GSSG
S-nitrosoglutathione reductase oxidized glutathione
- HEK
human embryonic kidney
- iodoTMT
iodoacetyl tandem mass tag
- MS
mass spectrometry
- NEM
N-ethylmaleimide
- NO
nitric oxide
- pI
isoelectric point
- SERCA
Sarcoplasmic/endoplasmic reticulum Ca2+ ATPase
- SNO
S-nitrosylation, S-nitrosation
- TEAB
triethylammonium bicarbonate
Footnotes
DISCLOSURES
None.
References
- 1.Sun J, Murphy E. Protein S-nitrosylation and cardioprotection. Circ Res. 2010;106:285–296. doi: 10.1161/CIRCRESAHA.109.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maron BA, Tang S-S, Loscalzo J. S-nitrosothiols and the S-nitrosoproteome of the cardiovascular system. Antioxid Redox Signal. 2013;18:270–287. doi: 10.1089/ars.2012.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gould N, Doulias P-T, Tenopoulou M, Raju K, Ischiropoulos H. Regulation of protein function and signaling by reversible cysteine S-nitrosylation. J Biol Chem. 2013;288:26473–26479. doi: 10.1074/jbc.R113.460261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulman IH, Hare JM. Regulation of cardiovascular cellular processes by S-nitrosylation. Biochim Biophys Acta. 2012;1820:752–762. doi: 10.1016/j.bbagen.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hess DT, Matsumoto A, Kim S-O, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 6.Martínez-Ruiz A, Lamas S. S-nitrosylation: a potential new paradigm in signal transduction. Cardiovasc Res. 2004;62:43–52. doi: 10.1016/j.cardiores.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Lane P, Hao G, Gross SS. S-nitrosylation is emerging as a specific and fundamental posttranslational protein modification: head-to-head comparison with O-phosphorylation. Sci STKE. 2001;86:re1. doi: 10.1126/stke.2001.86.re1. [DOI] [PubMed] [Google Scholar]
- 8.Murphy E, Kohr M, Sun J, Nguyen T, Steenbergen C. S-nitrosylation: a radical way to protect the heart. J Mol Cell Cardiol. 2012;52:568–577. doi: 10.1016/j.yjmcc.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;86:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 10.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 11.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camerini S, Polci ML, Restuccia U, Usuelli V, Malgaroli A, Bachi A. A novel approach to identify proteins modified by nitric oxide: the HIS-TAG switch method. J Proteome Res. 2007;6:3224–3231. doi: 10.1021/pr0701456. [DOI] [PubMed] [Google Scholar]
- 13.Paige JS, Xu G, Stancevic B, Jaffrey SR. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang B, Chen C. Detection of Protein S-Nitrosation using Irreversible Biotinylation Procedures (IBP) Free Radic Biol Med. 2010;49:447–456. doi: 10.1016/j.freeradbiomed.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Sinha V, Wijewickrama GT, Chandrasena REP, Xu H, Edirisinghe PD, Schiefer IT, Thatcher GRJ. Proteomic and mass spectroscopic quantitation of protein S-nitrosation differentiates NO-donors. ACS Chem Biol. 2010;5:667–680. doi: 10.1021/cb100054m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Huang B, Zhou X, Chen C. Quantitative proteomic analysis of S-nitrosated proteins in diabetic mouse liver with ICAT switch method. Protein Cell. 2010;1:675–687. doi: 10.1007/s13238-010-0087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray CI, Uhrigshardt H, O’Meally RN, Cole RN, Van Eyk JE. Identification and quantification of S-nitrosylation by cysteine reactive tandem mass tag switch assay. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.013441. M111.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan K-T, Chen Y-Y, Pu T-H, Chao Y-S, Yang C-Y, Bomgarden RD, Rogers JC, Meng T-C, Khoo K-H. Mass Spectrometry-Based Quantitative Proteomics for Dissecting Multiplexed Redox Cysteine Modifications in Nitric Oxide-Protected Cardiomyocyte Under Hypoxia. Antioxid Redox Signal. 2014;20:1365–1381. doi: 10.1089/ars.2013.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung HS, Wang S-B, Venkatraman V, Murray CI, Van Eyk JE. Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system. Circ Res. 2013;112:382–392. doi: 10.1161/CIRCRESAHA.112.268680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohr MJ, Aponte A, Sun J, Gucek M, Steenbergen C, Murphy E. Measurement of S-nitrosylation occupancy in the myocardium with cysteine-reactive tandem mass tags: short communication. Circ Res. 2012;111:1308–1312. doi: 10.1161/CIRCRESAHA.112.271320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker J, Zhu N, Zhu M, Chen S. Profiling thiol redox proteome using isotope tagging mass spectrometry. J Vis Exp. 2012;61:e3766. doi: 10.3791/3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu Z, Meng F, Bomgarden RD, Viner RI, Li J, Rogers JC, Cheng J, Greenlief CM, Cui J, Lubahn DB, Sun GY, Gu Z. Proteomic Quantification and Site-Mapping of S-Nitrosylated Proteins Using Isobaric iodoTMT Reagents. J Proteome Res. 2014;13:3200–3211. doi: 10.1021/pr401179v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L, Yan Y, Zeng M, Zhang J, Hanes MA, Ahearn G, McMahon TJ, Dickfeld T, Marshall HE, Que LG, Stamler JS. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell. 2004;116:617–628. doi: 10.1016/s0092-8674(04)00131-x. [DOI] [PubMed] [Google Scholar]
- 25.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 26.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinforma. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 27.Vizcaíno JA, Côté RG, Csordas A, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–9. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang P-L, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 30.Kohr MJ, Aponte AM, Sun J, Wang G, Murphy E, Gucek M, Steenbergen C. Characterization of potential S-nitrosylation sites in the myocardium. Am J Physiol Heart Circ Physiol. 2011;300:H1327–1335. doi: 10.1152/ajpheart.00997.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohr MJ, Aponte A, Sun J, Gucek M, Steenbergen C, Murphy E. Measurement of S-Nitrosylation Occupancy in the Myocardium With Cysteine-Reactive Tandem Mass Tags. Circ Res. 2012;111:1308–1312. doi: 10.1161/CIRCRESAHA.112.271320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray CI, Kane LA, Uhrigshardt H, Wang S-B, Van Eyk JE. Site-mapping of in vitro S-nitrosation in cardiac mitochondria: implications for cardioprotection. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.004721. M110.004721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 34.Meyer M, Schillinger W, Pieske B, Holubarsch C, Heilmann C, Posival H, Kuwajima G, Mikoshiba K, Just H, Hasenfuss G. Alterations of Sarcoplasmic Reticulum Proteins in Failing Human Dilated Cardiomyopathy. Circulation. 1995;92:778–784. doi: 10.1161/01.cir.92.4.778. [DOI] [PubMed] [Google Scholar]
- 35.Miyamoto MI, del Monte F, Schmidt U, DiSalvo TS, Kang ZB, Matsui T, Guerrero JL, Gwathmey JK, Rosenzweig A, Hajjar RJ. Adenoviral gene transfer of SERCA2a improves left-ventricular function in aortic-banded rats in transition to heart failure. Proc Natl Acad Sci. 2000;97:793–798. doi: 10.1073/pnas.97.2.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun J, Morgan M, Shen R-F, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res. 2007;101:1155–1163. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 37.Que LG, Liu L, Yan Y, Whitehead GS, Gavett SH, Schwartz DA, Stamler JS. Protection from experimental asthma by an endogenous bronchodilator. Science. 2005;308:1618–1621. doi: 10.1126/science.1108228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beigi F, Gonzalez DR, Minhas KM, Sun Q-A, Foster MW, Khan SA, Treuer AV, Dulce RA, Harrison RW, Saraiva RM, Premer C, Schulman IH, Stamler JS, Hare JM. Dynamic denitrosylation via S-nitrosoglutathione reductase regulates cardiovascular function. Proc Natl Acad Sci U S A. 2012;109:4314–4319. doi: 10.1073/pnas.1113319109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Locatelli J, de Assis LVM, Isoldi MC. Calcium handling proteins: structure, function, and modulation by exercise. Heart Fail Rev. 2014;19:207–225. doi: 10.1007/s10741-013-9373-z. [DOI] [PubMed] [Google Scholar]
- 40.Yar S, Monasky MM, Solaro RJ. Maladaptive modifications in myofilament proteins and triggers in the progression to heart failure and sudden death. Pflugers Arch. 2014;466:1189–1197. doi: 10.1007/s00424-014-1457-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirk JA, Zhang P, Murphy AM, Van Eyk JE. Troponin I alterations detected by multiple-reaction monitoring: how might this impact the study of heart failure? Expert Rev Proteomics. 2013;10:5–8. doi: 10.1586/epr.12.77. [DOI] [PubMed] [Google Scholar]
- 42.Smith BC, Marletta MA. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr Opin Chem Biol. 2012;16:498–506. doi: 10.1016/j.cbpa.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marino SM, Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hess DT, Matsumoto A, Kim S-O, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 45.Doulias P-T, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci U S A. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haldar SM, Stamler JS. S-nitrosylation: integrator of cardiovascular performance and oxygen delivery. J Clin Invest. 2013;123:101–110. doi: 10.1172/JCI62854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S-B, Foster DB, Rucker J, O’Rourke B, Kass DA, Van Eyk JE. Redox regulation of mitochondrial ATP synthase: implications for cardiac resynchronization therapy. Circ Res. 2011;109:750–757. doi: 10.1161/CIRCRESAHA.111.246124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez DR, Treuer AV, Castellanos J, Dulce RA, Hare JM. Impaired S-nitrosylation of the ryanodine receptor caused by xanthine oxidase activity contributes to calcium leak in heart failure. J Biol Chem. 2010;285:28938–28945. doi: 10.1074/jbc.M110.154948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu C, Hu J, Liu T, Ago T, Sadoshima J, Li H. Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J Proteome Res. 2008;7:3789–3802. doi: 10.1021/pr800233r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozawa K, Whalen EJ, Nelson CD, Mu Y, Hess DT, Lefkowitz RJ, Stamler JS. S-nitrosylation of beta-arrestin regulates beta-adrenergic receptor trafficking. Mol Cell. 2008;31:395–405. doi: 10.1016/j.molcel.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whalen EJ, Foster MW, Matsumoto A, Ozawa K, Violin JD, Que LG, Nelson CD, Benhar M, Keys JR, Rockman HA, Koch WJ, Daaka Y, Lefkowitz RJ, Stamler JS. Regulation of beta-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 52.Foster MW, Yang Z, Gooden DM, Thompson JW, Ball CH, Turner ME, Hou Y, Pi J, Moseley MA, Que LG. Proteomic characterization of the cellular response to nitrosative stress mediated by s-nitrosoglutathione reductase inhibition. J Proteome Res. 2012;11:2480–2491. doi: 10.1021/pr201180m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Kettenhofen NJ, Shiva S, Hogg N, Gladwin MT. Copper dependence of the biotin switch assay: modified assay for measuring cellular and blood nitrosated proteins. Free Radic Biol Med. 2008;44:1362–1372. doi: 10.1016/j.freeradbiomed.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cole SJ, Kirwan JN, Roberts BP, Willis CR. Radical chain reduction of alkyl halides, dialkyl sulphides and O-alkyl S-methyl dithiocarbonates to alkanes by trialkylsilanes. J Chem Soc, Perkin Trans. 1991;1:103–112. [Google Scholar]
- 55.Rosenblum WI, El-Sabban F. Dimethyl sulfoxide (DMSO) and glycerol, hydroxyl radical scavengers, impair platelet aggregation within and eliminate the accompanying vasodilation of, injured mouse pial arterioles. Stroke. 1982;13:35–39. doi: 10.1161/01.str.13.1.35. [DOI] [PubMed] [Google Scholar]
- 56.Novogrodsky A, Ravid A, Rubin AL, Stenzel KH. Hydroxyl radical scavengers inhibit lymphocyte mitogenesis. Proc Natl Acad Sci U S A. 1982;79:1171–1174. doi: 10.1073/pnas.79.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valko M, Morris H, Cronin MTD. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 58.Grady JK, Chasteen ND, Harris DC. Radicals from “Good’s” buffers. Anal Biochem. 1988;173:111–115. doi: 10.1016/0003-2697(88)90167-4. [DOI] [PubMed] [Google Scholar]
- 59.Simpson JA, Cheeseman KH, Smith SE, Dean RT. Free-radical generation by copper ions and hydrogen peroxide. Stimulation by Hepes buffer. Biochem J. 1988;254:519–523. doi: 10.1042/bj2540519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asplund KUM, Jansson PJ, Lindqvist C, Nordström T. Measurement of ascorbic acid (vitamin C) induced hydroxyl radical generation in household drinking water. Free Radic Res. 2002;36:1271–1276. doi: 10.1080/1071576021000036425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.