Abstract
In recent genome-wide association studies, the extracellular chaperone protein, clusterin, has been identified as a novel risk factor in Alzheimer’s disease (AD). We have examined the interactions between clusterin and the AD-associated amyloid-β1–40 peptide (Aβ1–40) which is prone to aggregate into an ensemble of oligomeric intermediates implicated in both the proliferation of amyloid fibrils and in neuronal toxicity. Using highly sensitive single molecule fluorescence methods, we have found that Aβ1–40 forms a heterogeneous distribution of small oligomers (from dimers to 50mers), all of which interact with clusterin to form long-lived, stable complexes. Consequently, clusterin is able to influence strongly both the aggregation and disaggregation of Aβ1–40 by sequestration of the Aβ oligomers. These results not only elucidate the protective role of clusterin but also provide a molecular basis for the genetic link between clusterin and AD.
Introduction
The aggregation of amyloidogenic peptides and proteins is a highly heterogeneous process as monomers aggregate in a stepwise manner via a series of oligomeric intermediates to form β-sheet-rich fibrillar structures1,2. These oligomers have been implicated not only in the further generation of aggregates but also in the neurotoxicity associated with many neurodegenerative diseases1,3–7. The detection and characterization of the various oligomeric intermediates and their interactions with other proteins has proved to be a difficult task for ensemble-level techniques as a result of the inherent heterogeneity of the species involved. Recently, however, a variety of techniques has been developed to detect and characterize molecular interactions at the level of single molecules8–12. We have applied two forms of such techniques to investigate the aggregation and fibril disaggregation processes of the Alzheimer’s disease (AD)-associated peptide, amyloid-β1–40 (Aβ1–40), and its interactions with clusterin, an extracellular chaperone protein which has been found to be colocalized with senile amyloid plaques and has recently been genetically associated with AD13–16.
The single molecule techniques that we have utilized are able to detect and characterize the ensemble of oligomeric species formed during the self-assembly and disassembly processes of Aβ1–40 and have been implicated in the toxicity associated with AD1,5,6,17. The first of these techniques, confocal two color coincidence detection (cTCCD), uses two spatially overlapped Gaussian laser beams that are combined to create a single confocal volume. cTCCD identifies multimolecular species bearing two different types of fluorophores through the observation of coincident bursts of fluorescence when they pass through this confocal volume; this enables oligomeric species to be detected and analyzed8,10. The number of peptide molecules present in each oligomer, denoted the oligomer size, can be measured by comparing the fluorescence brightness values of the coincident fluorescence bursts to the brightness values of non-coincident (monomeric) bursts. This method is based on the principle that oligomers contain a greater number of molecules, and hence fluorophores, than monomers and therefore will emit proportionally brighter bursts. In this work, we refer to this metric as the “apparent size” of an oligomer since this parameter is determined indirectly from the fluorescence intensity of the species (see Methods, Supplementary Methods and Supplementary Figs. 1–3 for details).
The second technique is a two-color version of total internal reflection fluorescence microscopy (TIRFM), an imaging technique that uses two overlapped lasers of different wavelengths to create dual excitation within the imaging field. Species bearing different fluorophores deposited onto a surface within this field can be sampled, and the distribution of colocalization of fluorescence can be analyzed to give images with detailed information about the composition, size and morphology of the species present. (See Methods, Supplementary Fig. 1, and Supplementary Methods for details of data acquisition and instrumentation).
In this study, we have combined the solution-phase detection by cTCCD with imaging of surface-bound species by TIRFM in order to gain insight not only into the aggregation process of Aβ1–40 but also into the interactions between the AD-linked extracellular chaperone, clusterin, and the various species populated along the Aβ1–40 aggregation pathway.
Results
Characterizing the aggregation of Aβ1–40
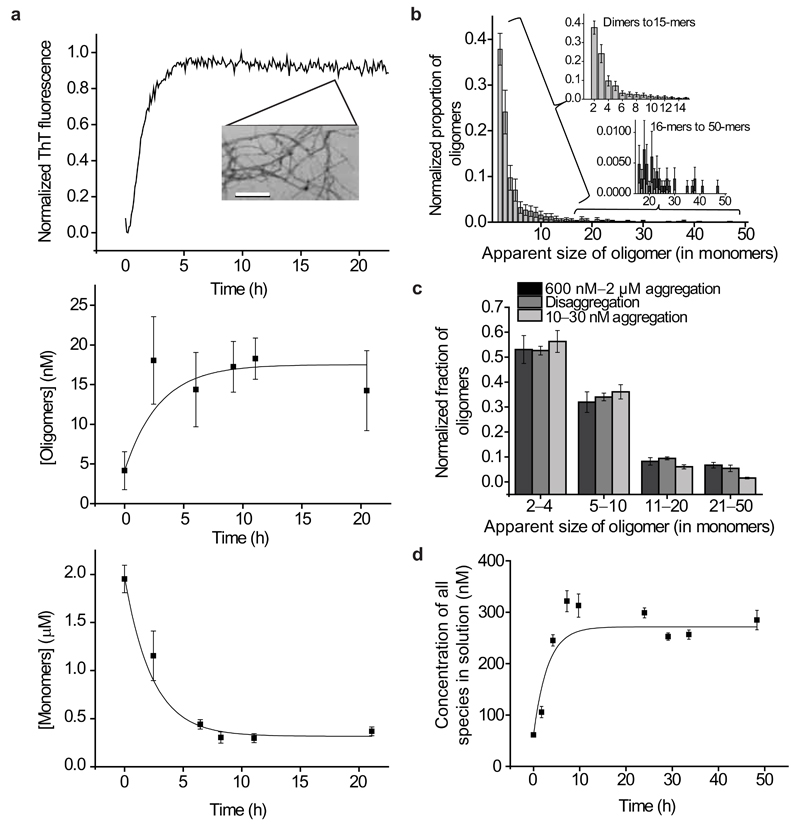
We first used cTCCD to characterize the species formed during the process of Aβ1–40 aggregation and disaggregation in the absence of clusterin using. To study the aggregation process, equimolar quantities of the Aβ1–40 peptide, labeled with either the HiLyteFluor488 fluorophore or the HiLyteFluor647 fluorophore (Anaspec), were mixed and the sample was incubated at pH 7.4 and 37 °C; cTCCD measurements were then made every 2–3 h as the peptides aggregated and eventually formed amyloid fibrils (Fig. 1a). A series of control experiments revealed that the presence of neither fluorophore had any appreciable effect on the bulk aggregation behavior of Aβ1–40, a result in accord with other studies using the same fluorescent peptides18–21 (Supplementary Fig. 2). Analysis of the distribution of oligomers reveals a variety of soluble species, the majority of which are stable for over three hours at the concentrations and temperature required for cTCCD characterization (~25–50 pM, 21 °C). In addition, control experiments reveal that the technique is capable of detecting even small quantities of aggregates and that the species present after dilution to picomolar sample concentrations are highly representative of those present at the higher concentrations under which aggregation readily takes place (600 nM – 2 µM) (see Supplementary Information and Supplementary Fig. 3a,b).
Figure 1.
Bulk and single molecule studies of Aβ1–40. (a) Appearance and disappearance of species populated during the aggregation of Aβ1–40 ( 2 µM at 37 °C with agitation). Fibril formation monitored by thioflavin T (ThT) fluorescence (top). The inset (top) is a transmission electron microscopy (TEM) image of the fibrils present after 24 h of incubation (scale bar is 200 nm). Concentration of soluble oligomers (dimers–50mers) (middle), and concentration of monomeric species (bottom) are both tracked using cTCCD. The data are averaged from multiple experimental repetitions (2 μM Aβ1–40, n=3, error bars are s.e.m.). (b) A representative distribution of apparent sizes of oligomers formed during Aβ1–40 aggregation and disaggregation (error bars are s.d.). Insets are zoomed into regions of dimers–15-mers and 16-mers–50-mers to provide greater detail. (c) A comparison of the distributions of apparent oligomer sizes during aggregation and disaggregation experiments (2 μM Aβ1–40 aggregation, n=3; disaggregation, n=12; 10–30 nM Aβ1–40 aggregation, n=4; error bars are s.e.m.). (d) Time dependence of the concentration of soluble species released from a pellet of fibrils (n=12, error bars are s.e.m.).
We first carried out aggregation experiments at a concentration of Aβ1–40 of 30 nM, a value similar to its typical physiological concentration in cerebrospinal fluid (CSF)22. We observed the formation of 300 pM of oligomers representing approximately 1% of the total number of molecules present. This result confirms previous speculation that oligomeric species can form at Aβ1–40 concentrations similar to those in physiological environments but at which fibril formation can be very slow23. In order to be able to collect data under concentrations where aggregation occurs more rapidly and it where it is possible to perform bulk experiments and single molecule measurements with high signal-to-noise ratios, the concentrations were increased either to 600 nM or to 2 µM for further experiments. These experimental conditions were also selected as they yielded highly reproducible data. The aggregation experiments at these higher concentrations reveal a rise in the population of stable oligomeric species to a peak concentration of ~20 nM during the incubation of 2 μM Aβ1–40 (again representing about 1% of the total molecules) along with a 75% decrease in the monomer concentration during the growth phase of fibril formation (Fig. 1a).
The relative quantities of the different oligomeric forms that could be observed decreased exponentially with increasing apparent size under all conditions examined in this study (dimers were most abundant; the probability of finding a 10-mer relative to a dimer is ~3000:1). Indeed, this distribution of apparent sizes was sustained throughout the progress of the aggregation reaction suggesting that many intermediate species are involved in the growth of amyloid fibrils (Fig. 1b, Supplementary Fig. 3c–e). At these higher concentrations we observe that the distribution of stable oligomer sizes is effectively the same as that formed at a concentration of 30 nM (Fig. 1d). This exponential form of the distribution of apparent oligomer sizes directly observed in our experiments has been predicted by theoretical analysis of aggregation of other protein systems, and is consistent with models of both the bulk kinetics and the molecular processes of amyloid growth7,24.
Characterizing disaggregation of Aβ1–40 fibrils
We then set out to study the species present during the process that is the reverse of aggregation, namely the disaggregation of fibrils. Aβ1–40 fibrils were formed from the monomeric peptides at a concentration of 8 µM, pelleted by centrifugation, washed, and finally re-suspended in buffer in the absence of any additional Aβ1–40. Using TCCD, we monitored the concentration of species released into the buffer from the fibrils at a rate of (9.3 ± 3.1) ×10−5 s−1 (n=12, s.e.m.) (Fig. 1d, see Supplementary Methods for details). These results are similar to previous observations of exchange between amyloid fibrils and soluble proteins25,26. However, the single molecule experiments in the present study additionally reveal that the species released include a small population of stable oligomers as well as monomeric species (Fig. 1c).
After 50 h of disaggregation the concentrations of the various species in solution were constant and we could assume that the system had reached equilibrium. We then derived the thermodynamic stabilities of the various species present. The values of ΔGº are −18.2 ± 0.5 kJ mol−1 (s.d.) for dimers and an average of –38.9 ± 2.7 kJ mol−1 (s.d.) for the larger oligomers. These values are in good agreement with previous bulk measurements of the free energy of the formation of amyloid fibrils (–36 kJ mol−1)27. They also indicate that the dimers are inherently less thermodynamically stable than all larger oligomers, and that the stability of the larger oligomers is largely independent of their size (Table 1), findings that are consistent with previous computational analyses28.
| Parameter | Without clusterin (n) | With clusterin (n) |
|---|---|---|
| Rate of monomer and oligomer release (s−1) | (9.3 ± 3.1)×10−5 (12) | (1.7 ± 0.3)×10−5 (8) |
| Rate of clusterin release (s−1) | — | (9.8 ± 0.9)×10−7 (3) |
| Final concentration of all species (nM) | 270 ± 20 (12) | 120 ± 20 (8) |
| Final concentration of oligomeric species (nM) | 0.16 ± 0.06 (12) | 0.42 ± 0.1 (8) |
| Final soluble clusterin concentration (nM) | — | 90 ± 14 (3) |
| Aggregation | ||
| ΔG° (dimer) (kJ mol−1) | −23.0 ± 2.9 (4) | — |
| ΔG° (larger oligomer) (kJ mol−1) | −39.3 ± 3.0 (11) | — |
| Disaggregation | ||
| ΔG° (dimer) (kJ mol−1) | −18.2 ± 0.5 (3) | −25.8 ± 2.6 (4) |
| ΔG° (larger oligomer) (kJ mol−1) | −38.9 ± 2.7 (12) | −43.9 ± 1.0 (12) |
| ΔH° (dimer) (kJ mol−1) | −43.8 ± 24.5 (3) | — |
| ΔH° (larger oligomer) (kJ mol−1) | −29.1 ± 0.3 (12) | — |
| ΔS° dimer (kJ mol−1) | −80.8 ± 83.5 (3) | — |
| ΔS° larger than dimer (kJ mol−1) | 28.3 ± 1.1 (12) | — |
The rates were derived from fitting monomolecular dissociation functions to the plots of species released as a function of time from disaggregation experiments. All thermodynamic values—free energies (ΔG°), enthalpies (ΔH°), and entropies (ΔS°) of formation for various sizes of oligomers—were determined from the steady state apparent size distributions of the various species. Errors for rate values are s.e.m. and errors for thermodynamic values are s.d.
Effect of clusterin on Aβ1–40 aggregation
The observation of stable distributions of oligomers by cTCCD during the aggregation and disaggregation processes of Aβ1–40 provides a framework for the investigation of the manner in which clusterin interacts with such ensembles of species. In a series of experiments, we first used unlabeled clusterin to explore any changes in the Aβ1–40 oligomer distribution resulting from the presence of the chaperone and then AlexaFluor647-labeled clusterin to characterize the specific interactions between clusterin molecules and Aβ1–40 species. Control experiments confirmed that AlexaFluor647-labeled clusterin behaved in a similar manner to its unlabeled counterpart which indicates that fluorescent labeling has no marked effect on the properties of clusterin functionality studied in this work (Supplementary Fig. 4a).
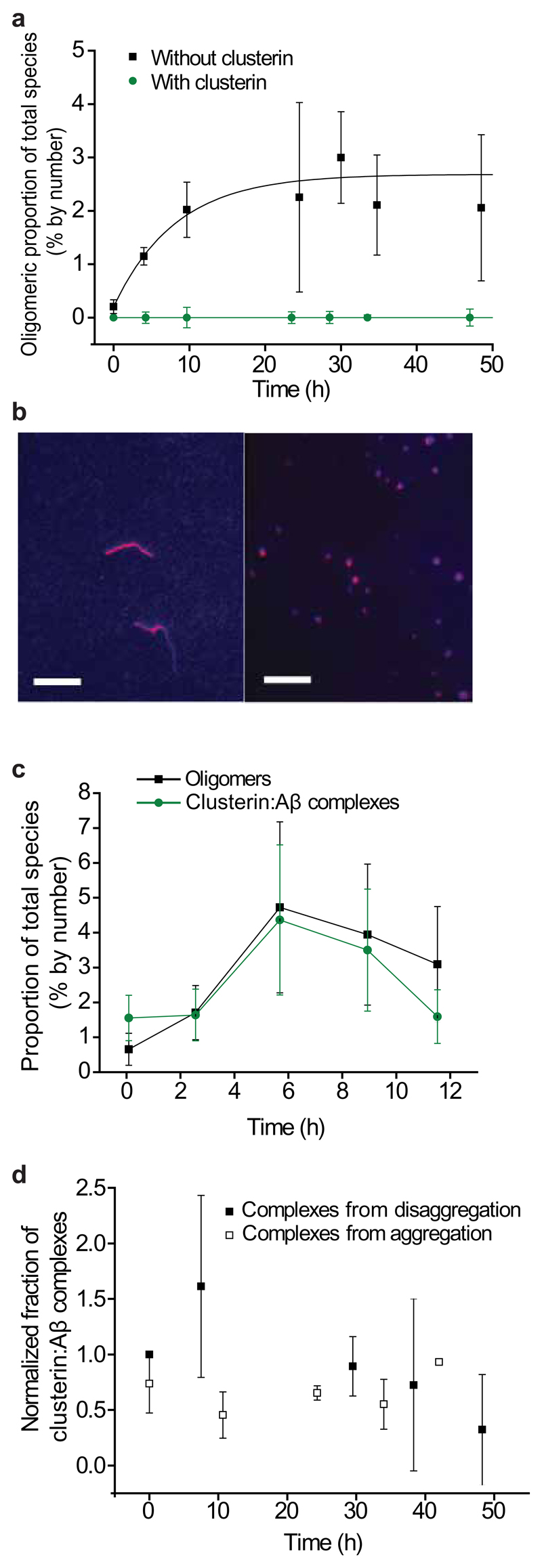
Addition of equimolar quantities of clusterin to monomeric Aβ1–40 prior to the start of the aggregation reaction dramatically inhibits the appearance of small oligomeric species, a finding that is consistent with bulk measurements showing that clusterin strongly inhibits fibril formation (Fig. 2a)29,30. Moreover, when clusterin was added to solutions in which aggregation had progressed to a stage where a population of oligomers is present, further self-association of the peptides was halted (Fig. 2b). Throughout the reaction, the fraction of Aβ1–40 peptides in oligomers corresponded well to the fraction of Aβ1–40 peptides in clusterin:Aβ complexes, indicating that even in the presence of an excess of monomeric Aβ, it is the oligomeric fraction of the Aβ species that binds to clusterin (Fig. 2c). These clusterin:Aβ complexes are very long-lived, persisting for over 50 h following dilution to nanomolar concentrations (Fig. 2d). The binding between clusterin and Aβ oligomers was observed not only at concentrations of 2 µM but also at low concentrations of 100 nM, conditions under which fibrils are not observed to form for days. The clusterin:Aβ complexes formed at these low concentrations were found to persist, after dilution, at picomolar concentrations for over 200 h (Supplementary Fig. 4b–d). Furthermore, the distribution of the sizes of the Aβ1–40 oligomers observed in our experiments was not substantially altered in the presence of clusterin (Fig. 3a, Supplementary Fig. 4c). We therefore conclude that the growth of larger aggregates, and ultimately amyloid fibrils, is inhibited as a result of the very effective sequestration of Aβ oligomers by clusterin molecules.
Figure 2.
The effects of clusterin on the aggregation of Aβ1–40. (a) Fraction of oligomers detected in solution during the aggregation of Aβ1–40 with and without clusterin (Aβ1–40 and clusterin are both at a concentration of 600 nM, n=3, error bars are s.e.m.). (b) TIRFM image of the species present after 24 h of aggregation of a 2 μM solution of Aβ1–40 without clusterin (left). TIRFM image of a 2 μM solution of Aβ1–40 after 24 h of aggregation but with 2 µM clusterin added 4 h after the start of the reaction, during the fibril growth phase (right). An approximately 50% reduction in the average size of species present is observed in the presence of clusterin (from 1400 ± 200 nm without clusterin to 780 ± 60 nm with clusterin, s.e.m., P-value is 0.01, two-sample independent, two-tailed, t-test). Scale bars are 5 µm. (c) Fractions of species formed during the aggregation of a 2 μM solution that are oligomeric and that are in Aβ:clusterin complexes. (n=3, error bars are s.e.m.). (d) Proportion of Aβ:clusterin complexes persisting at 10–20 nM (total peptide concentration) at 21 °C. Complexes were formed between clusterin and oligomers from both aggregation and disaggregation reactions. For both traces, n=3, error bars are s.e.m. There is no statistically significant change in the proportion of complexes with oligomers formed during either the disaggregation experiment (P-value of 0.77, ANOVA single-factor) or the aggregation experiment (P-value of 0.99, ANOVA single-factor).
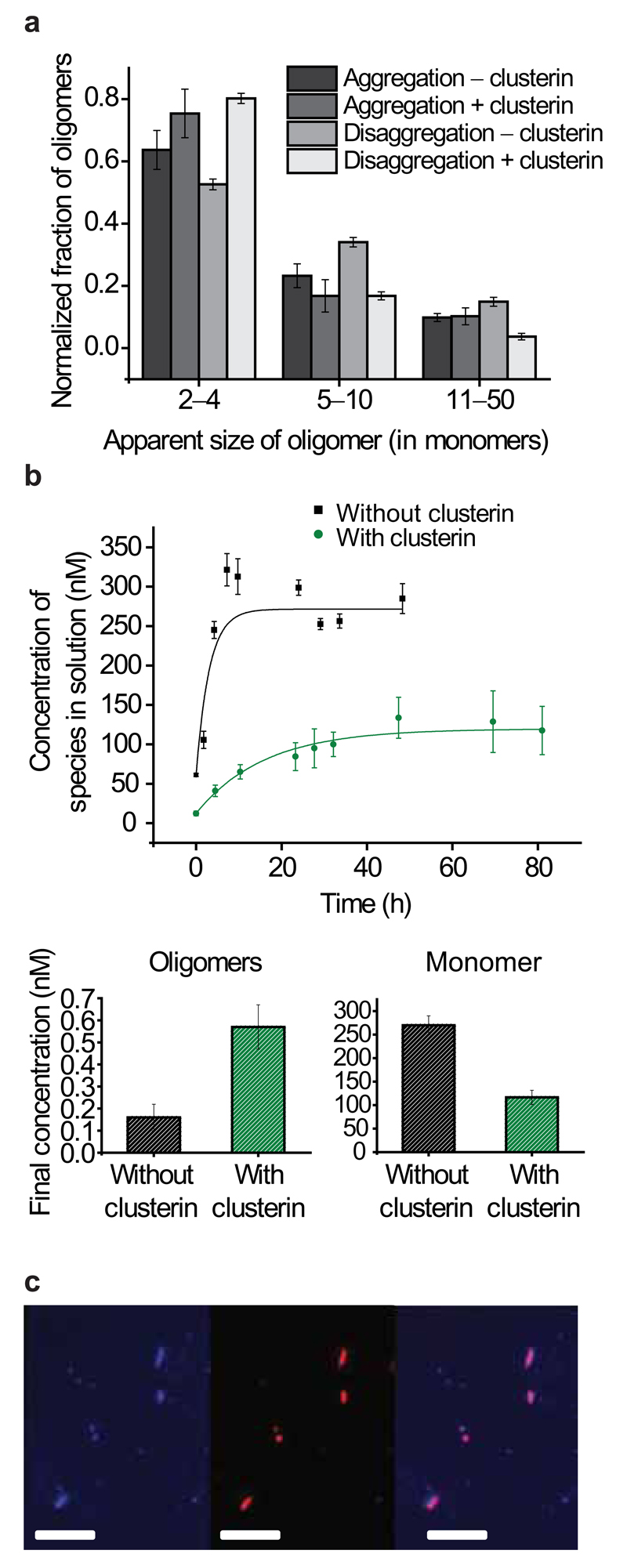
Figure 3.
The effects of clusterin on the disaggregation of Aβ1–40 fibrils. (a) Distributions of apparent sizes of oligomers formed during aggregation and disaggregation reactions with and without clusterin. (Aggregation without clusterin, n=2, error bars are range; aggregation with clusterin, n=3; disaggregation without clusterin, n=10; disaggregation with clusterin, n=3; error bars are s.e.m.). (b) Time dependence of the release of soluble species during the disaggregation experiments in the presence and absence of clusterin (top), increased oligomer concentration in the presence of clusterin in the concentration plateau region (significant with a P-value of 0.002) (bottom left) and decreased monomer concentration in the presence of clusterin, in the concentration plateau region (significant with a P-value of 0.0003) (bottom right). Both correlations were analyzed using a two-sample independent, two tailed t-test, n=8, and error bars are s.e.m. (c) TIRFM imaging of HiLyte488Fluor-labeled Aβ1–40 fibrils incubated overnight at room temperature with AlexaFluor647-labeled clusterin. Aβ1–40 fluorescence only (left), clusterin fluorescence only (middle), and colocalization of the two species (right). Scale bars are 5 μm.
Effect of clusterin on Aβ1–40 fibril disaggregation
Studies of Aβ1–40 fibril disaggregation in the presence of clusterin reveal that oligomers released from the fibrils bind to clusterin molecules resulting in an increase in the total oligomer population compared to that observed in the absence of clusterin (Fig. 3b). The observation of a population of oligomers resulting from fibril disaggregation is consistent with the observation of oligomeric “halos” around Aβ plaques that have been observed in a mouse model of AD and are linked to neuronal toxicity31. This result is also in line with studies suggesting clusterin increases the observable oligomer population, perhaps due to similar stabilization effects of clusterin on Aβ oligomers32. As found under aggregation conditions, the complexes between clusterin and the Aβ oligomers observed during disaggregation are very long-lived, persisting at nanomolar concentrations for over 50 h (Fig. 2d). In each of these complexes, the ratio of clusterin:Aβ had a median value of 1.2 ± 1.1 (IQR) suggesting that there is a direct correlation between the number of clusterin molecules and the number of Aβ peptides in a complex (Supplementary Fig. 5). In addition, the presence of clusterin bound to the Aβ fibrils is found to decrease both the rate and the extent of fibril disaggregation by inhibiting dissociation into monomers (the rate changes to (17 ± 2.7) × 10−6 s−1, n=8, s.e.m. with clusterin from (9.3 ± 3.1) × 10−5 s−1, n=12, s.e.m. without clusterin) (Fig. 3b,c, Supplementary Fig. 6). Similar binding of chaperones to fibrils and inhibition of their disaggregation has been previously observed in experiments with other systems33–35. The relative change in monomer and oligomer populations is reflected in an increase in the magnitude of the free energies of formation of dimers (from −18.2 ± 0.5 to −25.8 ± 2.6 kJ mol−1, both s.d.) and of larger oligomers (from −38.9 ± 2.7 to −43.9 ± 1.0 kJ mol−1 both s.d.) when clusterin is present (Table 1). In all aggregation and disaggregation experiments, regardless of the presence or absence of clusterin, the distribution of oligomeric species followed an approximately exponential decrease with apparent size (Fig. 3a).
Discussion
The present studies reveal that under the experimental conditions used in this work, clusterin binds and sequesters oligomers formed during both the aggregation of Aβ1–40 monomers and the disaggregation processes of Aβ1–40 fibrils thereby inhibiting the further growth or dissociation of these oligomers. In the CSF, where aggregation is likely to be of particular significance, clusterin is present at concentrations between 20 and 60 nM36, levels in excess of those of Aβ1–40 (between 1 and 10 nM)22, and a concentration range in which our data indicate that stable oligomers of Aβ1–40 can be formed. Clusterin is, therefore, present under normal conditions at levels appropriate for prevention of the formation or growth of oligomers and their consequent toxicity29. Additionally, the persistence times measured here for complexes between clusterin and Aβ1–40 oligomers (from 50 h to greater than 200 h) are long enough to allow for crucial in vivo processes such as Aβ1–40 production (~15 h), Aβ1–40 clearance (~14 h), and astrocytic endocytosis (between 12–24 h)37–39. This is particularly interesting in the context that previous studies have correlated binding of Aβ species by clusterin to the in vivo degradation of the peptides29,40,41.
Our single molecule experiments show that the extracellular chaperone clusterin binds to all members of the ensemble of soluble oligomers found to be present during the aggregation and disaggregation of Aβ1–40 in our experiments. In this way, clusterin is able to prevent further growth and proliferation of aggregated species and in vivo, is likely to sequester the potentially toxic oligomeric species until they can be processed and degraded38,41,42. The methodology described in this study that has enabled these data to be obtained should be readily applicable to different aggregation conditions and to a wide range of disease-associated peptides and proteins and therefore represents a powerful means of probing, at the level of individual molecules, the seminal link between protein misfolding, aggregation and disease1.
In the particular case of the Aβ peptide discussed in this work, the ability of clusterin to sequester misfolded and potentially toxic oligomers provides a molecular basis for the recently identified genetic association between clusterin and AD13,14. Indeed, any perturbations that result in reduced clusterin levels, or in a reduced ability of clusterin to form stable and long-lived complexes with Aβ oligomers, are likely to increase the susceptibility of the individual concerned to the development of AD13,14.
Online Methods
cTCCD instrumentation
The instrumentation used in this study has been described by Orte and coworkers10. Gaussian laser beams of two colors (a tuned diode laser emitting at 488 nm (Spectra Physics, Cyan CDRH) and a helium-neon laser emitting at 633 nm (Melles Griot, 25LHP151) were overlapped and directed into a confocal microscope (Nikon Eclipse TE2000-U) to achieve dual-excitation in the confocal volume. The beams then pass through an oil-immersion objective (Appochromat 60x, NA 1.40, Nikon) and into the sample with a final beam diameter of approximately 260 nm. The laser powers used were 60 ± 5 μW for the 633 nm laser and 50 ± 5 μW for the 488 nm laser and are low enough that photobleaching of the fluorophores during their time in the probe volume was not appreciable8. During measurements, 200 μL of sample was placed on a 1x1 cm area of a BSA-coated glass coverslip (VWR, 631-1339). Then the beam was focused on a confocal plane that is 10 μm into the measurement solution, above the coverslip surface.
The fluorescence photons emitted from the sample were collected through the objective, passed through a 50 μm pinhole (Melles Griot), separated into the two different detection channels (dichroic 585DRLP Omega Filters), and directed to separate avalanche photodiode detectors (APDs) (SPCM-AQR-14, Perkin-Elmer Optoelectronics). The APDs feed photon count data in 1 ms bins into multichannel scalar cards on a personal computer (MCS-PCS, Ortec) which were saved for analysis in 600–800, 8000 ms frame units. During each measurement, the microscope stage was scanned using two orthogonal DC motors (M-112.1DG, Physik Instrumente, Karlsruhe, Germany) across the confocal volume at a constant rate of 200 µm s−1. See Supplementary Methods for further instrumentation details.
TIRFM instrumentation
Imaging was performed using total internal reflection fluorescence microscopy (TIRFM) which aligns the outputs from a HeNe laser operating at 633 nm (25LHP991230, Melles Griot) and a diode laser operating at 488 nm (PC13589, Spectra Physics) and directs them down the edge of a TIRF objective (60x Plan Apo TIRF, NA 1.45, Nikon) mounted on a Nikon TE200 microscope. Fluorescence collected by the same objective was separated from the returning TIR beam by a dichroic (FF500646, Laser2000), split into blue and red components (585 DXLR, Omega Optical) and filtered using Dual-View (Optical Insights) mounted filters. The images were simultaneously recorded on an EMCCD (Cascade II: 512 Princeton instruments,) whereby the EMCCD was split so that each color was recorded on one half of the EMCCD, operating at −70 °C. Data were acquired 7.9 frames s-1 using Metamorph software. See Supplementary Methods for further instrumentation details.
Aβ peptide preparation
Monomeric solutions of HiLyteFluor488 and HiLyteFluor647-labeled Aβ1–40 (Anaspec) were prepared by dissolving the lyophilized peptide in pH 12 NaOH followed by sonication over ice for 30 min (Bandelin Sonorex) and subsequent flash freezing into aliquots43. Prior to each of the incubations, aliquots of each peptide were brought to pH 7.4 by diluting into SSPE buffer (150 mM NaCl, 10 mM Na2H2PO4 x H2O, 10 mM Na2EDTA, pH 7.4) to the desired aggregation reaction concentration and placed under relevant conditions (e.g. 37 °C, agitation). The concentration for each labeled peptide was measured before mixing using cTCCD.
Preparation and labeling of clusterin
Clusterin was extracted from human serum from Wollongong Hospital (Wollongong, NSW, Australia), as described previously44 and labeled using the AlexaFluor647 dye with a maleimide linker (Invitrogen). See Supplementary Methods for further details.
cTCCD and TIRFM data acquisition
For the aggregation experiments, Aβ1–40 (30 nM to 2 μM) was incubated in SSPE buffer (defined above) at 37 °C with agitation (200 rpm on a rotary shaker). Samples were taken every 2–3 hours and analyzed using cTCCD. For experiments with clusterin, the chaperone was added at a 1:1 molar ratio.
For disaggregation experiments Aβ1–40 fibrils were first prepared by a static incubation of an 8 μM solution of Aβ1–40 for 72 h at 37 °C. Pelleting and washing were carried out by centrifugation at ~10,000×g for 30 min followed by two identical washing steps involving re-suspension of the pellet in SSPE buffer and additional centrifugation at ~10,000×g for 2 min; finally the pellet was re-suspended in SSPE buffer. The soluble species in the solution were monitored and analyzed using cTCCD. For experiments with chaperones, clusterin and Aβ fibrils were incubated together for 12–16 h at 21 °C and any clusterin that was not bound to fibrils was removed via centrifugation (leaving 100–200 nM clusterin bound to fibrils).
See Supplementary Information for further details.
Quantification of oligomer distributions
The extent of oligomerization of a given sample was tracked by separating bursts coincident in both blue and red detection channels from non-coincident bursts. Quantification of oligomer distributions for cTCCD measurements was performed by analyzing the brightness of the coincident, oligomeric bursts. The brightness in the blue (488 nm) detection channel was corrected to account for changes in emission due to FRET between the HiLyteFluor488-labeled and HiLyteFluor647-labeled peptides within an oligomer (see Supplementary Methods for details). Then the FRET corrected blue channel brightness was scaled by the average burst brightness for a blue monomer to yield an average number of blue Aβ molecules in each oligomer. These data were then scaled by a factor of two to account for the fact that the blue-labeled peptides comprise only half of the molecules in the aggregating solutions. We term this number the “apparent size”. Uncertainties in its measurement arise from factors including the Gaussian nature of the probe volume illumination, corrections for the error associated with assuming oligomers have equal numbers of red and blue peptides, as well as the inherent variation in the number of photons emitted by a single fluorophore. Both Aβ oligomers and clusterin:Aβ complexes were treated similarly with respect to effects of FRET.
Statistical analysis
Statistical tests were performed to determine the significance of changes in distributions of the apparent sizes of oligomers as well as the persistence of oligomeric species. In order to compare two values, two-sample, two-tailed, independent t-tests were used. For comparison of multiple values, ANOVA one-factor tests were used.
Supplementary Material
Supplementary Information is linked to the online version of the paper.
Acknowledgements
P.N. is supported by the Marshall Aid Commemoration Commission (Marshall Scholarship), and the National Science Foundation (Graduate Research Fellowship). A.O. is supported by an ERG grant from the EU FP7. R.W.C. acknowledges a Research Fellowship from Christ’s College. B.B. is supported by an Alzheimer’s Research Trust Fellowship. S.H. is supported by a BBSRC Fellowship. K.A.G. is supported by fellowships from the EPSRC and Studienstiftung des deutschen Volkes. S.M. is supported by a Royal Society Dorothy Hodgkin Fellowship. M.R.W. acknowledges the support of the Australian Research Council (DP0773555 and DP0984341). D.K. and C.M.D. are supported by the Wellcome Trust and D.K. by the Augustus Newman Foundation. Additionally, we would like to thank James McColl and Paul Dunne for advice on use of the TIRFM instrument, Sarah Shammas and Francisco Newby for help with peptide and protein preparation, and Thomas Jahn, Nunilo Cremades, Leila Luheshi, Tuomas Knowles, Sam Cohen, Allen Chen, and Janet Kumita for discussions.
Footnotes
Author Contributions
P.N., A.O., S.M., M.R.W., C.M.D. and D.K. designed the experiments. P.N performed the cTCCD experiments. P.N., A.O. and R.W.C. refined analysis methods. A.O. and R.W.C. developed instrumentation. R.W.C. wrote the analysis software and designed, built, and calibrated the scanning stage used for cTCCD experiments. P.N. and B.B. performed the bulk scale experiments. P.N and K.A.G. performed the TIRFM experiments. P.N., B.B, K.A.G. and A.O. analyzed the data. S.H. labeled the clusterin that was purified and provided by M.R.W. All authors discussed and interpreted results and contributed to the writing of the manuscript.
The authors declare no competing financial interests.
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Bucciantini M, et al. Prefibrillar amyloid protein aggregates share common features of cytotoxicity. J Biol Chem. 2004;279:31374–31382. doi: 10.1074/jbc.M400348200. [DOI] [PubMed] [Google Scholar]
- 3.Cleary JP, et al. Natural oligomers of the amyloid-β protein specifically disrupt cognitive function. Nat Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 4.Lesne S, et al. A specific amyloid-β protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 5.Selkoe DJ. Soluble oligomers of the amyloid-β protein impair synaptic plasticity and behavior. Behav Brain Res. 2008;192:106–113. doi: 10.1016/j.bbr.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 7.Knowles TPJ, et al. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Ying L, Green JJ, Balasubramanian S, Klenerman D. Ultrasensitive coincidence fluorescence detection of single DNA molecules. Anal Chem. 2003;75:1664–1670. doi: 10.1021/ac026367z. [DOI] [PubMed] [Google Scholar]
- 9.Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2:764–774. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- 10.Orte A, et al. Direct characterization of amyloidogenic oligomers by single-molecule fluorescence. Proc Natl Acad Sci U S A. 2008;105:14424–14429. doi: 10.1073/pnas.0803086105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunne PD, et al. DySCo: quantitating associations of membrane proteins using two-color single-molecule tracking. Biophys J. 2009;97:L5–L7. doi: 10.1016/j.bpj.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiou A, et al. Probing neuroserpin polymerization and interaction with amyloid-β peptides using single molecule fluorescence. Biophys J. 2009;97:2306–2315. doi: 10.1016/j.bpj.2009.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert J-C, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 14.Harold D, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kida E, Choi-Miura N-H, Wisniewski KE. Deposition of apolipoproteins E and J in senile plaques is topographically determined in both Alzheimer's disease and Down's syndrome brain. Brain Res. 1995;685:211–216. doi: 10.1016/0006-8993(95)00482-6. [DOI] [PubMed] [Google Scholar]
- 16.Choi-Miura NH, et al. SP-40,40 is a constituent of Alzheimer's amyloid. Acta Neuropathol (Berl) 1992;83:260–264. doi: 10.1007/BF00296787. [DOI] [PubMed] [Google Scholar]
- 17.Luheshi LM, et al. Systematic in vivo analysis of the intrinsic determinants of amyloid-β pathogenicity. PLoS Biol. 2007;5:e290. doi: 10.1371/journal.pbio.0050290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hickman SE, Allison EK, El Khoury J. Microglial dysfunction and defective β-amyloid clearance pathways in aging Alzheimer's disease mice. The Journal of Neuroscience. 2008;28:8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schauerte JA, et al. Simultaneous single-molecule fluorescence and conductivity studies reveal distinct classes of Aβ species on lipid bilayers. Biochemistry (Mosc) 2010;49:3031–3039. doi: 10.1021/bi901444w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding H, Wong PT, Lee EL, Gafni A, Steel DG. Determination of the oligomer size of amyloidogenic protein β-amyloid(1–40) by single-molecule spectroscopy. Biophys J. 2009;97:912–921. doi: 10.1016/j.bpj.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson RD, Schauerte JA, Wisser KC, Gafni A, Steel DG. Direct observation of single amyloid-β(1–40) oligomers on live cells: binding and growth at physiological concentrations. PLoS ONE. 2011;6:e23970. doi: 10.1371/journal.pone.0023970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta PD, et al. Plasma and cerebrospinal fluid levels of amyloid-β proteins 1–40 and 1–42 in Alzheimer disease. Arch Neurol. 2000;57:100–105. doi: 10.1001/archneur.57.1.100. [DOI] [PubMed] [Google Scholar]
- 23.Garzon-Rodriguez W, Sepulveda-Becerra M, Milton S, Glabe CG. Soluble amyloid Aβ-(1– 40) exists as a stable dimer at low concentrations. J Biol Chem. 1997;272:21037–21044. doi: 10.1074/jbc.272.34.21037. [DOI] [PubMed] [Google Scholar]
- 24.Oosawa F. Size distribution of protein polymers. J Theor Biol. 1970;27:69–86. doi: 10.1016/0022-5193(70)90129-3. [DOI] [PubMed] [Google Scholar]
- 25.Carulla N, et al. Molecular recycling within amyloid fibrils. Nature. 2005;436:554–558. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- 26.Sánchez L, et al. Aβ40 and Aβ42 amyloid fibrils exhibit distinct molecular recycling properties. J Am Chem Soc. 2011;133:6505–6508. doi: 10.1021/ja1117123. [DOI] [PubMed] [Google Scholar]
- 27.O'Nuallain B, Shivaprasad S, Kheterpal I, Wetzel R. Thermodynamics of Aβ(1–40) amyloid fibril elongation. Biochemistry (Mosc) 2005;44:12709–12718. doi: 10.1021/bi050927h. [DOI] [PubMed] [Google Scholar]
- 28.Auer S, Dobson CM, Vendruscolo M. Characterization of the nucleation barriers for protein aggregation and amyloid formation. HFSP journal. 2007;1:137–46. doi: 10.2976/1.2760023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yerbury JJ, et al. The extracellular chaperone clusterin influences amyloid formation and toxicity by interacting with prefibrillar structures. FASEB J. 2007;21:2312–2322. doi: 10.1096/fj.06-7986com. [DOI] [PubMed] [Google Scholar]
- 30.Matsubara E, Frangione B, Ghiso J. Characterization of apolipoprotein J-Alzheimer's Aβ interaction. J Biol Chem. 1995;270:7563–7567. doi: 10.1074/jbc.270.13.7563. [DOI] [PubMed] [Google Scholar]
- 31.Koffie RM, et al. Oligomeric amyloid β associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc Natl Acad Sci U S A. 2009;106:4012–4017. doi: 10.1073/pnas.0811698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert MP, et al. Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. Proceedings of the National Academy of Sciences. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knowles TPJ, et al. Kinetics and thermodynamics of amyloid formation from direct measurements of fluctuations in fibril mass. Proceedings of the National Academy of Sciences. 2007;104:10016–10021. doi: 10.1073/pnas.0610659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raman B, et al. αB-crystallin, a small heat-shock protein, prevents the amyloid fibril growth of an amyloid β-peptide and β 2-microglobulin. Biochem J. 2005;392:573–581. doi: 10.1042/BJ20050339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shammas SL, et al. Binding of the molecular chaperone αB-crystallin to Aβ amyloid fibrils inhibits fibril elongation. Biophys J. 2011 doi: 10.1016/j.bpj.2011.07.056. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson MR, Yerbury JJ, Poon S. Extracellular chaperones and amyloids. In: Asea AAA, Brown IR, editors. Heat Shock Proteins and the Brain: Implications for Neurodegenerative Diseases and Neuroprotection. Vol. 3. Springer; Netherlands: 2008. pp. 283–315. [Google Scholar]
- 37.Bateman RJ, et al. Human amyloid-β synthesis and clearance rates as measured in cerebrospinal fluid in vivo. Nat Med. 2006;12:856–861. doi: 10.1038/nm1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen HM, Veerhuis R, Holmqvist B, Janciauskiene S. Binding and uptake of Aβ1–42 by primary human astrocytes in vitro . Glia. 2009;57:978–988. doi: 10.1002/glia.20822. [DOI] [PubMed] [Google Scholar]
- 39.Mawuenyega KG, et al. Decreased clearance of CNS β-amyloid in Alzheimer’s disease. Science. 2010;330:1774. doi: 10.1126/science.1197623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nuutinen T, et al. Amyloid-β 1–42 induced endocytosis and clusterin/apoJ protein accumulation in cultured human astrocytes. Neurochem Int. 2007;50:540–547. doi: 10.1016/j.neuint.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Wyatt AR, et al. Clusterin facilitates in vivo clearance of extracellular misfolded proteins. Cellular and Molecular Life Sciences. 2011 doi: 10.1007/s00018-011-0684-8. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luheshi LM, et al. Sequestration of the Aβ peptide prevents toxicity and promotes degradation in vivo. PLoS Biol. 2010;8:e1000334. doi: 10.1371/journal.pbio.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teplow DB, Kheterpal I, Wetzel R. Preparation of amyloid-β protein for structural and functional studies. Methods Enzymol. 2006;413:20–33. doi: 10.1016/S0076-6879(06)13002-5. [DOI] [PubMed] [Google Scholar]
- 44.Wilson MR, Easterbrook-Smith SB. Clusterin binds by a multivalent mechanism to the Fc and Fab regions of IgG. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1992;1159:319–326. doi: 10.1016/0167-4838(92)90062-i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.