Abstract
Background
Recent studies have provided a detailed census of genes that are mutated in acute myeloid leukemia (AML). Our next challenge is to understand how this genetic diversity defines the pathophysiology of AML and informs clinical practice.
Methods
We enrolled a total of 1540 patients in three prospective trials of intensive therapy. Combining driver mutations in 111 cancer genes with cytogenetic and clinical data, we defined AML genomic subgroups and their relevance to clinical outcomes.
Results
We identified 5234 driver mutations across 76 genes or genomic regions, with 2 or more drivers identified in 86% of the patients. Patterns of co-mutation compartmentalized the cohort into 11 classes, each with distinct diagnostic features and clinical outcomes. In addition to currently defined AML subgroups, three heterogeneous genomic categories emerged: AML with mutations in genes encoding chromatin, RNA-splicing regulators, or both (in 18% of patients); AML with TP53 mutations, chromosomal aneuploidies, or both (in 13%); and, provisionally, AML with IDH2R172 mutations (in 1%). Patients with chromatin–spliceosome and TP53–aneuploidy AML had poor outcomes, with the various class-defining mutations contributing independently and additively to the outcome. In addition to class-defining lesions, other co-occurring driver mutations also had a substantial effect on overall survival. The prognostic effects of individual mutations were often significantly altered by the presence or absence of other driver mutations. Such gene–gene interactions were especially pronounced for NPM1-mutated AML, in which patterns of co-mutation identified groups with a favorable or adverse prognosis. These predictions require validation in prospective clinical trials.
Conclusions
The driver landscape in AML reveals distinct molecular subgroups that reflect discrete paths in the evolution of AML, informing disease classification and prognostic stratification. (Funded by the Wellcome Trust and others; ClinicalTrials.gov number, NCT00146120.)
Acute Myeloid Leukemia (AML) is characterized by clonal expansion of undifferentiated myeloid precursors, resulting in impaired hematopoiesis and bone marrow failure. Although many patients with AML have a response to induction chemotherapy, refractory disease is common, and relapse represents the major cause of treatment failure.1
Cancer develops from somatically acquired driver mutations, which account for the myriad biologic and clinical complexities of the disease. A classification of cancers that is based on causality is likely to be durable, reproducible, and clinically relevant. This is already evident in the case of AML, for which there has been a progressive shift from a morphologic classification scheme to one informed by causative genomic changes.2–4 Systematic studies of the genomic landscape of AML, such as analyses of data from the Cancer Genome Atlas5 (TCGA), have generated a catalogue of leukemia genes that is increasingly comprehensive. It is therefore an opportune time to revisit the possibility of an AML classification scheme that is fully genomic.
With whole-genome sequencing, AML emerges as a complex, dynamic disease.5–8 There are many leukemia genes, most of which are infrequently mutated, and patients typically have more than one driver mutation.5 The disease evolves over time, with multiple competing clones coexisting at any time.5–8 These discoveries are revealing the biologic intricacies of AML, but how they inform clinical practice is unclear.
Here we report a comprehensive study of leukemia genes in three clinical trials of intensive AML treatment, acknowledging that the landscape of AML in older patients may be underrepresented. The structure of driver mutations identifies nonoverlapping subgroups of patients, allowing a fully genomic classification of AML. Beyond class, we explore patterns of gene–gene co-occurrences and investigate how such compound genotypes are related to clinical outcomes.
Methods
Study Participants, Treatment, and Oversight
We obtained samples from patients participating in three prospective multicenter clinical trials of the German–Austrian AML Study Group: AML-HD98A, AML-HD98B, and AMLSG-07-04 (see the Supplementary Appendix, available with the full text of this article at NEJM.org).9–11 In AML-HD98A, patients 18 to 65 years of age received induction chemotherapy consisting of idarubicin, cytarabine, and etoposide (ICE); high-risk patients were offered allogeneic stem-cell transplantation, intermediate-risk patients a stem-cell allograft (if a matched related donor was available) or intensive chemotherapy, and low-risk patients intensive chemotherapy. AMLSG-07-04 had a similar design, but patients 18 to 61 years of age were randomly assigned to receive ICE or ICE plus all-trans retinoic acid (ATRA) as induction therapy. In AML-HD98B, patients 58 to 84 years of age were randomly assigned to induction therapy with ICE or ICE plus ATRA, with further therapy dictated by the response (see Figs. S1 and S2 in the Supplementary Appendix). The median follow-up period was 5.9 years (Table S1 in the Supplementary Appendix). All patients provided written informed consent.
The last two authors guided the analysis and data interpretation.
Genetic Studies
Genetic profiling included cytogenetic analyses and sequencing of 111 genes (Table S2 in the Supplementary Appendix). Sequencing data have been deposited in the European Genome-Phenome Archive (www.ebi.ac.uk/ega) under accession number EGAS00001000275. We based our analysis on variants that we classified as driver mutations, using widely accepted genetic criteria.12 Recurrent fusion genes, aneuploidies, and leukemia gene mutations, including base substitutions and small (<200-bp) insertions or deletions (indels), were all included as drivers.
Statistical Analysis
We used Bayesian Dirichlet processes13 to establish classification rules that partitioned patients into subgroups, minimizing overlap between categories. The Dirichlet process defines an infinite prior distribution for the number and proportions of clusters in a mixture model, fitted with the use of the Markov chain Monte Carlo method.
We modeled overall survival using Cox proportional-hazards methods with the study variables treated as random effects14 (see the Methods section in the Supplementary Appendix). Random effects are modeled as independently distributed normal variables with identical mean and variance across eight variable categories: point mutations (58 variables), fusion genes (8), copy-number alterations (18), clinical variables (11), demographic characteristics (2), treatment (3), nuisance (4; nuisance variables are other or missing variables, such as the trial a patient was enrolled in, the year a patient entered the clinical trial, and whether cytogenetic data were missing), and gene–gene interaction terms, defined as nonadditive effects on survival between two genes when both are mutated (126). We used an expectation-maximization algorithm to estimate the shared means and variances; maximum a posteriori estimates were used for the effects of individual variables.
Results
Driver-Mutation Landscape of AML
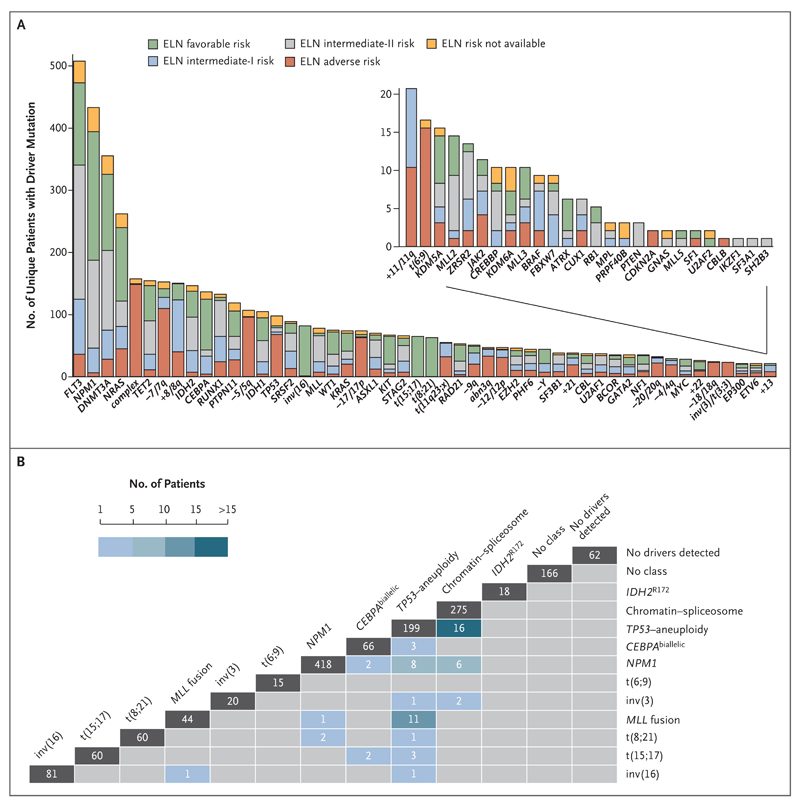
We identified 5234 driver mutations involving 76 genes or regions in 1540 patients (Fig. 1A, and Tables S5 and S6 in the Supplementary Appendix), with mutation frequencies that were consistent with those in previous studies4,5 (Fig. S2c in the Supplementary Appendix). Point mutations accounted for 73% of all drivers (3824 of 5234) (Fig. S2d in the Supplementary Appendix) and were often enriched in patients with AML classified as intermediate risk according to current guidelines.3 We identified at least 1 driver mutation in 1478 of 1540 samples (96%), and 2 or more driver mutations in 86% of samples (Fig. S2e in the Supplementary Appendix). The mutation landscape in this cohort, including novel hotspot mutations in MYC, is described in detail in the Results S1 section in the Supplementary Appendix.
Figure 1. Landscape of Driver Mutations in Acute Myeloid Leukemia (AML).
Panel A shows driver events in 1540 patients with AML. Each bar represents a distinct driver lesion; the lesions include gene mutations, chromosomal aneuploidies, fusion genes, and complex karyotypes. The colors in each bar indicate the molecular risk according to the European LeukemiaNet (ELN) classification. Panel B shows the distribution of samples and overlap (cross-sections) across molecular subgroups (vertical bars). Patients who had no driver mutations and those who had driver mutations but did not meet the criteria for any specific class are also included. The number at the top of each column is the number of patients assigned solely to the designated class; the numbers of patients meeting criteria for two or more classes are shown at the intersection of classes.
Although patterns of co-mutation and mutual exclusivity have been described previously in AML,4,5 our sample size afforded a more comprehensive analysis, which identified many new pairwise gene–gene correlations (see the Results S2 section, Tables S7 and S8, and Fig. S3a in the Supplementary Appendix). We also found surprising differences in patterns of co-mutation for hotspots within genes. NPM1, for example, preferentially associated with NRASG12/13 but not with NRASQ61. Distinct patterns of co-mutation were also observed for IDH2R140 and IDH2R172 and for FLT3 internal tandem duplication (FLT3ITD) and FLT3 tyrosine kinase domain (FLT3TKD) mutations (Fig. S3b). Thus, the functional consequences of hotspot mutations within genes may not be equivalent.
Leukemias often comprise heterogeneous mixtures of subclones,6–8,15 but samples to date have been too small to define common patterns of evolution. We used allele fractions of point mutations to infer clonal relationships5,7,16,17 (see the Results S3 section in the Supplementary Appendix). Mutations in DNMT3A, ASXL1, IDH1/2, and TET2, genes that encode epigenetic modifiers, were often acquired earliest (Fig. S4a in the Supplementary Appendix). These genes are frequently mutated in elderly persons with clonal hematopoiesis and confer an increased risk of hematologic cancers.18–22 In our study population with AML, mutations in these genes were typically in the founding clone and were almost never found in isolation, suggesting that they are not sufficient for overt leukemia (Fig. S4b in the Supplementary Appendix). Mutations in receptor tyrosine kinase–RAS pathway genes typically occurred late23–25 and frequently more than once in the same patient (Fig. S5 in the Supplementary Appendix). NPM1 mutations were usually secondary events, often occurring after DNMT3A, IDH1, or NRAS mutations (Fig. S6 and Table S9 in the Supplementary Appendix). These data suggest that development of AML follows specific and ordered evolutionary trajectories.
Implications of Genomic Structure for Classification of AML
In the 2008 World Health Organization (WHO) classification, molecular groups in adult AML include t(15;17), t(8;21), inv(16)–t(16;16), t(6;9), inv(3)–t(3;3), MLL fusion genes, and provisionally, CEBPA or NPM1 mutations.26 In our cohort, AML in 736 patients (48%) would not be classified according to these genomic lesions, even though 96% of the patients had driver mutations. The characterization of many new leukemia genes, multiple driver mutations per patient, and complex co-mutation patterns prompted us to reevaluate genomic classification of AML from the beginning.
We developed a Bayesian statistical model to compartmentalize AML into mutually exclusive subtypes on the basis of patterns of co-mutation (see the Methods section and Fig. S7 in the Supplementary Appendix). From this model, we defined simple rules to generate 11 subgroups or classes of AML (Table 1, Fig. 1B, and Fig. 2; and Results sections S4 through S7 in the Supplementary Appendix). We found that inv(16), t(15;17), t(8;21), inv(3), t(6;9), and MLL fusions each represent small, individual subgroups (≤5% of the study cohort), confirming the WHO classification. NPM1-mutated AML and CEBPAbiallelic AML were also identified as distinct subgroups. NPM1-mutated AML was the largest class in our cohort (accounting for 27% of the cohort), with 73% of patients (319 of 436) also carrying mutations in DNA methylation or hydroxymethylation genes (DNMT3A, IDH1, IDH2R140, and TET2).
Table 1. Proposed Genomic Classification of Acute Myeloid Leukemia (AML).
| Genomic Subgroup | Frequency in the Study Cohort (N = 1540) | Most Frequently Mutated Genes* |
|---|---|---|
| no. of patients (%) | gene (%) | |
| AML with NPM1 mutation | 418 (27) | NPM1 (100), DNMT3A (54), FLT3ITD (39), NRAS (19), TET2 (16), PTPN11 (15) |
| AML with mutated chromatin, RNA-splicing genes, or both† | 275 (18) | RUNX1 (39), MLLPTD (25), SRSF2 (22), DNMT3A (20), ASXL1 (17), STAG2 (16), NRAS (16), TET2 (15), FLT3ITD (15) |
| AML with TP53 mutations, chromosomal aneuploidy, or both‡ | 199 (13) | Complex karyotype (68), −5/5q (47), −7/7q (44), TP53 (44), −17/17p (31), −12/12p (17), +8/8q (16) |
| AML with inv(16)(p13.1q22) or t(16;16)(p13.1;q22); CBFB–MYH11 | 81 (5) | inv(16) (100), NRAS (53), +8/8q (16), +22 (16), KIT (15), FLT3TKD (15) |
| AML with biallelic CEBPA mutations | 66 (4) | CEBPAbiallelic (100), NRAS (30), WT1 (21), GATA2 (20) |
| AML with t(15;17)(q22;q12); PML–RARA | 60 (4) | t(15;17) (100), FLT3ITD (35), WT1 (17) |
| AML with t(8;21)(q22;q22); RUNX1–RUNX1T1 | 60 (4) | t(8;21) (100), KIT (38), −Y (33), −9q (18) |
| AML with MLL fusion genes; t(x;11)(x;q23)§ | 44 (3) | t(x;11q23) (100), NRAS (23) |
| AML with inv(3)(q21q26.2) or t(3;3)(q21;q26.2); GATA2, MECOM(EVI1) | 20 (1) | inv(3) (100), −7 (85), KRAS (30), NRAS (30), PTPN11 (30), ETV6 (15), PHF6 (15), SF3B1 (15) |
| AML with IDH2R172 mutations and no other class-defining lesions | 18 (1) | IDH2R172 (100), DNMT3A (67), +8/8q (17) |
| AML with t(6;9)(p23;q34); DEK–NUP214 | 15 (1) | t(6;9) (100), FLT3ITD (80), KRAS (20) |
| AML with driver mutations but no detected class-defining lesions | 166 (11) | FLT3ITD (39), DNMT3A (16) |
| AML with no detected driver mutations | 62 (4) | |
| AML meeting criteria for ≥2 genomic subgroups | 56 (4) |
Genes with a frequency of 15% or higher are shown in descending order of frequency. Key contributing genes in each class are shown in boldface type.
Classification in this subgroup requires one or more driver mutations in RUNX1, ASXL1, BCOR, STAG2, EZH2, SRSF2, SF3B1, U2AF1, ZRSR2, or MLLPTD. In the presence of other class-defining lesions — namely, inv(16), t(15;17), t(8;21), t(6;9), MLL fusion genes, or complex karyotype or driver mutations in TP53, NPM1, or CEBPAbiallelic — two or more chromatin–spliceosome mutations are required.
Classification in this subgroup requires TP53 mutation, complex karyotype, or in the absence of other class-defining lesions, one or more of the following: −7/7q, −5/5q, −4/4q, −9q, −12/12p, −17/−17p, −18/18q, −20/20q, +11/11q, +13, +21, or +22.
Multiple fusion partners for MLL were found, with the clinical implications depending on the specific fusion partner.
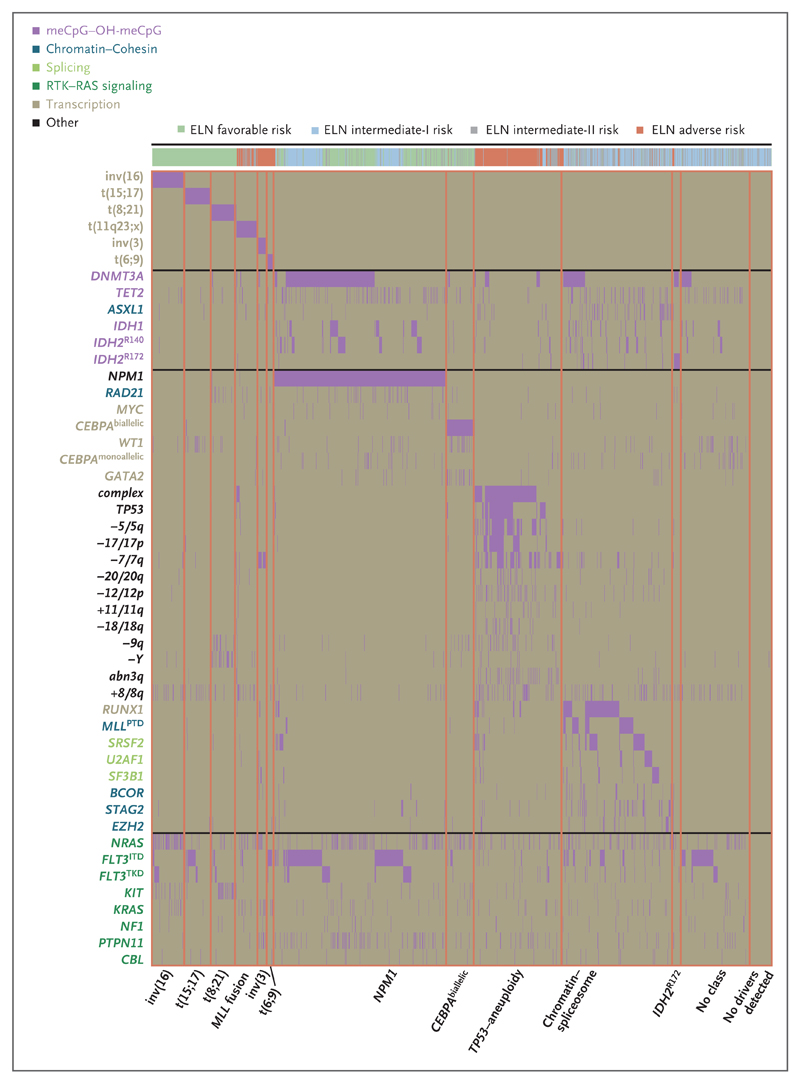
Figure 2. Identification of Molecular Subgroups in AML.
The rows in the graph represent individual genomic lesions, and the columns represent patients in the study. Vertical purple lines (some of which appear as blocks because of clustering) indicate the presence of a specified driver mutation in a patient. The patients have been ordered by group membership; orange lines demarcate boundaries between classes. OH-meCpG denotes hydroxymethyl CpG.
The second largest subgroup, accounting for 18% of the cohort, was defined by mutations in genes regulating RNA splicing (SRSF2, SF3B1, U2AF1, and ZRSR2), chromatin (ASXL1, STAG2, BCOR, MLLPTD, EZH2, and PHF6), or transcription (RUNX1); we call this the chromatin–spliceosome group. In contrast to the WHO classes of AML, no single genomic lesion defines this group. In our cohort, it could not be further subdivided because of the overlapping patterns of co-mutations among the defining genes (Fig. 2).
Mutations in TP53, complex karyotype alterations, cytogenetically visible copy-number alterations (aneuploidies), or a combination, characterize an additional AML subgroup (accounting for 13% of the cohort). This group emerges because specific chromosomal abnormalities are closely correlated with one another and with TP53 mutations, but both are mutually exclusive with other class-defining lesions. Since this subtype includes patients with TP53 mutations, isolated chromosomal-arm losses or gains, or both, it is broader than previously proposed groups, such as “monosomal karyotype AML” and “complex karyotype AML”2,3,27,28 (see the Results S5 section in the Supplementary Appendix). Patients in this subgroup were older and had fewer RAS-pathway mutations than patients in other subgroups (Fig. 2, and Fig. S3a in the Supplementary Appendix). This may reflect redundancy between RAS-pathway activation by point mutation and loss of RAS regulators by chromosomal aneuploidies.29
Finally, our analysis identified a subgroup of AML with IDH2R172 mutations, accounting for 1% of the cohort. Unlike IDH2R140 mutations, which show strong co-mutation with NPM1 (odds ratio for co-mutation, 3.6; P= 5×10−10), IDH2R172 mutations are mutually exclusive with NPM1 (odds ratio for co-mutation, 0.06; P= 4×10−5) and other class-defining lesions. IDH2R172 AML is associated with gene-expression and DNA-methylation profiles that differ from the profiles for other IDH mutations and that lead to more severe aberrations in metabolic activity,30,31 adding further support for it as a distinct entity.
Under this schema, 1236 of the 1540 patients with driver mutations (80%) were unambiguously classified in a single subgroup, and 56 patients (4%) met criteria for two or more categories (Fig. 1B). These mostly fell in the TP53–aneuploidy and chromatin–spliceosome classes. A total of 166 patients (11%) with driver mutations remained unclassified, potentially harboring mutations in drivers not sequenced here6 or class-defining mutations that were missed. Notably, 105 of 166 unclassified patients had two or more driver mutations, with DNMT3A, TET2, IDH1, FLT3, and NRAS observed most frequently.
We applied the classification scheme developed here to the independent cohort that was evaluated for the Cancer Genome Atlas (TCGA),5 which included many older patients with AML. The absence of overlap among subgroups was replicated, and the relative frequencies were equivalent to those in our cohort (see the Results S8 section in the Supplementary Appendix).
Clinical Implications of Genomic Classification of AML
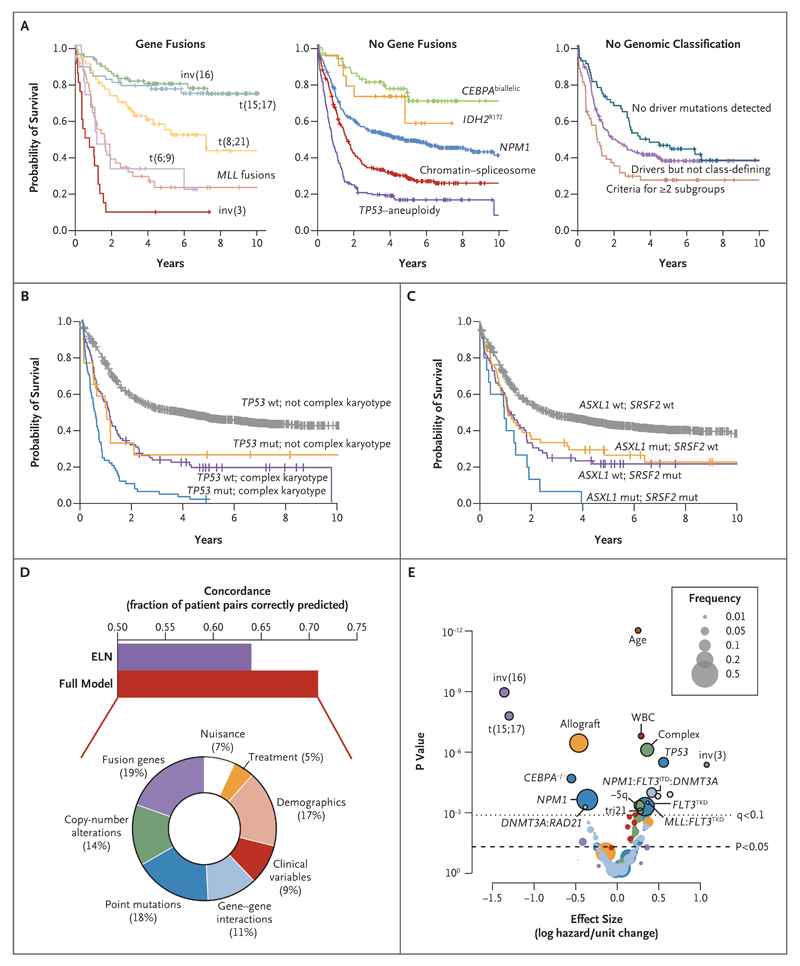
Although a genomic classification does not pre-suppose clinical relevance, its foundation on causal mutations could plausibly provide a bridge from molecular to clinical features of disease. We found considerable differences in clinical presentation and overall survival across the genomic subgroups (Fig. 3A, and Fig. S8 and the Results S5 section in the Supplementary Appendix). For subgroups defined by fusion genes, NPM1 and CEBPAbiallelic mutations, survival curves were as expected. Despite its more inclusive definition, the TP53–aneuploidy subgroup had dismal outcomes, as previously described.3,26,28,32
Figure 3. Molecular Subclassification and Overall Survival.
Panel A shows Kaplan–Meier curves for overall survival among patients in the 11 genomically defined subgroups and patients who did not have a straightforward classification. Panel B shows Kaplan–Meier curves for overall survival with the additive and independent prognostic effects of TP53 mutation and complex karyotype (TP53 mutation, 17 patients; complex karyotype, 89; and TP53 mutation with complex karyotype, 70). Panel C shows Kaplan–Meier curves for overall survival with the additive and independent prognostic effects of ASXL1 and SRSF2 mutations (ASXL1 mutation, 55 patients; SRSF2 mutation, 74; and ASXL1 and SRSF2 mutations, 15). In Panels B and C, wt denotes wild type, and mut mutation. The bar chart in Panel D shows concordance estimates for overall survival with the use of variables selected in our model (concordance, approximately 71%) as compared with a model using variables considered in the ELN guidelines (concordance, approximately 64%). The doughnut chart shows the relative proportion of explained variance in overall survival in the full model that is accounted for by different categories of predictor variables. Clinical variables are performance status, splenomegaly, bone marrow blasts, and blood counts. Demographic variables are age and sex. Nuisance variables are other variables (e.g., which trial a patient was enrolled in, what year a patient entered the clinical trial, and whether cytogenetic data were missing). The volcano plot in Panel E shows the incremental contribution to the effect size (expressed as the logarithmic hazard on the x axis; positive values indicate a worsening effect) versus P values (expressed on an inverted logarithmic scale on the y axis) for each of the 228 variables included in the random-effects model. The circles above the dotted line represent 18 variables with a q value of less than 0.1; the size of each circle corresponds to the frequency of the variable, as indicated in the box. The incremental contribution of age is shown for every 10 years of age, and the incremental contribution of the white-cell count (WBC) is shown for each increase of 1×109 cells per liter. The colors of the circles correspond to the colors shown in the doughnut chart in Panel D.
As compared with other groups, patients in the chromatin–spliceosome group were older, with lower white-cell and blast counts, lower rates of response to induction chemotherapy, higher relapse rates, and a poor long-term clinical outlook (Fig. S9 in the Supplementary Appendix). Under current guidelines,3 84% of patients in this subgroup (232 of 275) would be classified as being at intermediate risk, whereas their outcomes are in fact similar to those for patients in subgroups of AML with adverse outcomes, such as patients with MLL fusion genes (except for MLLT3–MLL fusion) or t(6;9) (Fig. 3A). In this group, 9% of patients had antecedent chronic myeloid disorders,33 and 91% of the patients in this subgroup had a diagnosis of de novo AML (Fig. S10 in the Supplementary Appendix). Evaluation of the morphologic features in bone marrow specimens from 1064 patients in the cohort showed dysplastic features in 139 patients, 55 of whom were molecularly assigned to the chromatin–spliceosome group (Fig. S11 in the Supplementary Appendix). The same chromatin and splicing factors are also frequently mutated in high-risk myeloproliferative neoplasms34 and myelodysplastic syndromes (MDS),17,35 suggesting that this subgroup could transcend conventional diagnostic boundaries36 between acute and high-risk chronic myeloid disorders.
Although the number of patients in the IDH2R172 subgroup was small, the long-term outcomes in this group were broadly similar to those in patients with NPM1-mutated AML (Fig. 3A).37 Patients in whom no driver mutations were detected had lower blast and whitecell counts and better outcomes (Fig. 3A, and Fig. S12a in the Supplementary Appendix).
Influence of Co-occurring Mutations on Clinical Outcomes
Overall survival was correlated with the number of driver mutations (Fig. S12b in the Supplementary Appendix), independent of age and the whitecell count (P= 8×10−12). One possible explanation for this finding is that driver mutations other than class-defining lesions influence clinical outcomes. For example, despite the frequent cooccurrence of a TP53 mutation and a complex karyotype, they were correlated independently and additively with survival in our cohort (Fig. 3B). Similarly, mutations in chromatin, splicing, and transcriptional regulators are frequently associated with low survival rates, and co-mutation among these genes typically results in even lower survival rates (Fig. 3C, and Fig. S13 in the Supplementary Appendix).
We developed multivariate models to explore the relative contributions of genetic, clinical, and diagnostic variables to overall survival. Using the full model, we could correctly rank approximately 71% of patients for overall survival (vs. 64% with models using only variables in the European LeukemiaNet criteria) (Fig. 3D). Genomic features were the most powerful predictors, accounting for about two thirds of explained variation, with the other third contributed by demographic, clinical, and treatment variables (Fig. 3D). Among genomic factors, fusion genes, copy-number alterations, and point mutations were broadly equivalent. These overall findings were replicated in the TCGA cohort of patients with AML5 (see the Results S8 section and Fig. S14 in the Supplementary Appendix).
Although a number of genomic variants are significant predictors of overall survival (P≤0.01) (Table 2 and Fig. 3E), many more genes show a somewhat weaker correlation with outcome (Table S10 in the Supplementary Appendix). The prognostic effects of class-defining lesions have mostly been described before, but we note the independent deleterious effects of TP53 mutations and the chromatin–spliceosome genes, such as SRSF2 and ASXL1. BRAF mutations are independently associated with a worse prognosis (P=0.009, q=0.06), and BRAF inhibitors might be a useful therapeutic option for patients in this subgroup.
Table 2. Driver Mutations with the Strongest Effect on Overall Survival and Other Class-Defining Lesions in AML.*.
| Variable | Frequency in Study Cohort (N = 1540) | Hazard Ratio for Death (95% CI) | P Value | q Value | Hazard Ratio for Interaction Terms | |
|---|---|---|---|---|---|---|
| no. of patients (%) | ||||||
| Main effects | ||||||
| inv(3), GATA2, MECOM(EVI1) | 23 (1) | 2.9 (1.8–4.7) | 9×10−6 | 0.0003 | ||
| TP53 | 98 (6) | 1.7 (1.4–2.2) | 7×10−6 | 0.0002 | ||
| Complex karyotype | 159 (10) | 1.4 (1.2–1.7) | 2×10−6 | 7×10−5 | ||
| BRAF | 9 (1) | 1.4 (1.1–1.8) | 0.009 | 0.06 | ||
| SRSF2 | 89 (6) | 1.4 (1.1–1.7) | 0.003 | 0.03 | ||
| FLT3ITD | 341 (22) | 1.4 (1.2–1.7) | 0.0008 | 0.01 | ||
| +21 | 39 (3) | 1.3 (1.1–1.6) | 0.001 | 0.02 | ||
| −5/5q | 107 (7) | 1.3 (1.1–1.5) | 0.0007 | 0.01 | ||
| −17/17p | 74 (5) | 1.3 (1.1–1.5) | 0.003 | 0.03 | ||
| +13 | 21 (1) | 1.3 (1.1–1.5) | 0.004 | 0.03 | ||
| −7 | 88 (6) | 1.3 (1.1–1.5) | 0.003 | 0.03 | ||
| −9q† | 53 (3) | 1.2 (1.1–1.5) | 0.01 | 0.08 | ||
| +22† | 26 (2) | 1.2 (1.1–1.4) | 0.008 | 0.06 | ||
| NPM1 | 436 (28) | 0.7 (0.6–0.9) | 0.0004 | 0.007 | ||
| CEBPAbiallelic | 73 (5) | 0.6 (0.4–0.7) | 4×10−5 | 0.001 | ||
| t(15;17), PML–RARA | 65 (4) | 0.3 (0.2–0.4) | 5×10−8 | 4×10−6 | ||
| inv(16), CBFB–MYH11 | 82 (5) | 0.3 (0.2–0.4) | 4×10−9 | 4×10−7 | ||
| Gene–gene interactions | ||||||
| NPM1–FLT3ITD–DNMT3A | 93 (6) | 1.5 (1.2–1.9) | 0.0002 | 0.004 | 1.1 for NPM1–FLT3ITD, 1.0 for DNMT3A–NPM1, 1.2 for DNMT3A–FLT3, 1.1 for DNMT3A | |
| MLLPTD–FLT3TKD | 10 (1) | 1.4 (1.2–1.8) | 0.0005 | 0.008 | 1.2 for MLLPTD, 1.1 for FLT3TKD | |
| DNMT3A–IDH2R140 | 47 (3) | 1.4 (1.1–1.8) | 0.007 | 0.05 | 1.1 for DNMT3A, 1.0 for IDH2R140 | |
| STAG2–IDH2R140 | 11 (1) | 0.8 (0.6–0.9) | 0.01 | 0.09 | 0.9 for STAG2, 1.0 for IDH2R140 | |
| NPM1–FLT3TKD | 53 (3) | 0.7 (0.6–0.9) | 0.009 | 0.06 | 0.7 for NPM1, 1.1 for FLT3TKD | |
| DNMT3A–RAD21 | 19 (1) | 0.7 (0.5–0.9) | 0.0008 | 0.01 | 1.1 for DNMT3A, 1.0 for RAD21 | |
| Other class-defining lesions | ||||||
| t(x;11), not MLLT3–MLL | 37 (2) | 1.4 (1.0–2.1) | 0.06 | 0.2 | ||
| ASXL1 | 70 (5) | 1.3 (1.0–1.6) | 0.04 | 0.2 | ||
| ZRSR2 | 13 (1) | 1.3 (1.0–1.7) | 0.04 | 0.2 | ||
| RUNX1 | 133 (9) | 1.1 (0.9–1.3) | 0.5 | 0.8 | ||
| t(9;11), MLLT3–MLL | 18 (1) | 0.8 (0.4–1.4) | 0.5 | 0.7 | ||
| IDH2R172 | 39 (3) | 0.8 (0.6–1.0) | 0.07 | 0.2 | ||
| t(8;21), RUNX1–RUNX1T1 | 63 (4) | 0.7 (0.4–1.0) | 0.03 | 0.2 | ||
The strongest effect on survival was defined as q<0.1. The effects reported in this table do not take into account the maintenance of favorable outcomes in the context of core-binding leukemias. We did not include interactions between fusion genes and copy-number alterations in this analysis, because they are mutually exclusive for the most part and we wanted to limit the number of variables in the model. See Table S10 in the Supplementary Appendix for the full model.
A +22 lesion frequently co-occurs with inv(16) and is associated with an especially favorable outcome; –9q frequently co-occurs with t(8;21) but does not appear to affect outcomes.
Influence of Complex Gene Interactions on Survival
The prognostic effects of TP53 mutations and complex karyotype (Fig. 3B) and of ASXL1 and SRSF2 mutations (Fig. 3C) are examples of additive associations — that is, the deleterious effect of each lesion remains unchanged whether or not another is present, with co-occurrence indicating a particularly dismal prognosis. We found that 11% of explained variation in survival in the cohort could be attributed to gene–gene interactions (Fig. 3D and Table 2), in which the prognostic effect of one gene is significantly altered if another gene is co-mutated. This suggests that the clinical effect of some driver mutations is modified by the wider genomic context in which they occur.
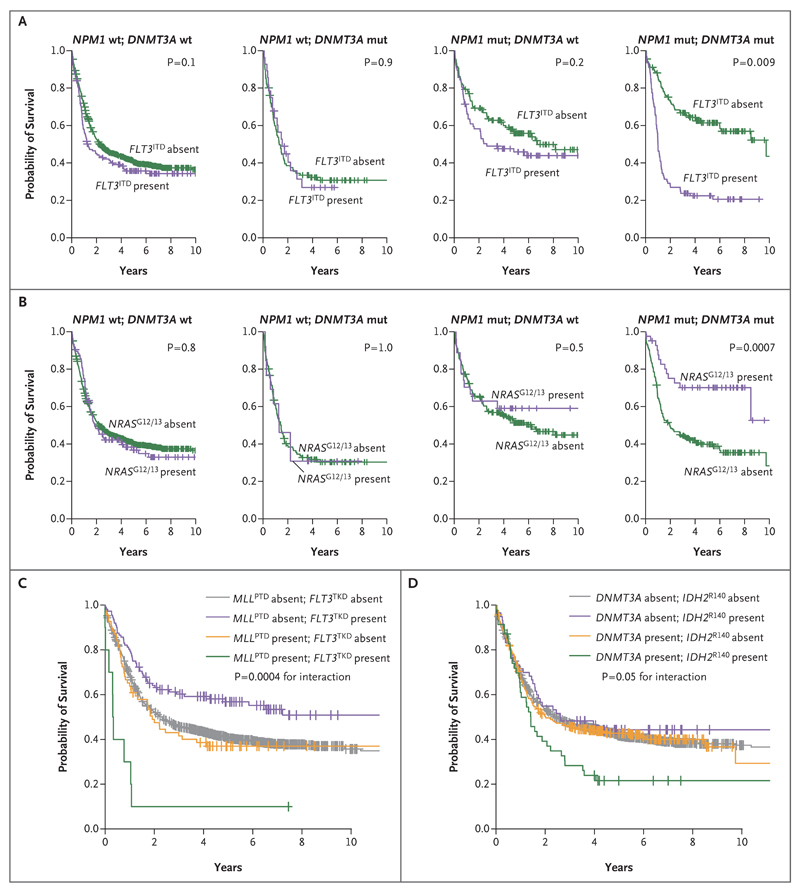
In our data set, this was exemplified by a three-way interaction among NPM1, DNMT3A, and FLT3ITD. This combined genotype represented the most frequent three-gene co-occurrence in our cohort, identified in 93 of the 1540 patients (6%) (P<0.0001). We found that the deleterious effect of FLT3ITD was most clinically relevant in patients with concomitant NPM1 and DNMT3A mutations (P=0.009 for three-way interaction in the univariate analysis, q= 0.004 in the multivariate analysis) (Fig. 4A). When present with either NPM1 or DNMT3A or with neither of these other genes, the effect of FLT3ITD on survival was considerably less pronounced. This observation held true regardless of the ratio of mutant to wild-type FLT3ITD (Fig. S15 in the Supplementary Appendix).
Figure 4. Influence of Gene–Gene Interactions on Overall Survival.
Panel A shows Kaplan–Meier curves for overall survival according to the presence or absence of FLT3ITD. The deleterious prognostic effect of FLT3ITD was significantly greater when both DNMT3A and NPM1 were mutated, as shown in the graph at the right (P = 0.009 for three-way interaction in the univariate analysis; q= 0.004 in the multivariate analysis with correction for multiple hypothesis testing). A total of 28 patients had both DNMT3A and FLT3ITD, 77 had both NPM1 and FLT3ITD, and 93 had all three mutations. Panel B shows Kaplan–Meier curves for overall survival according to the presence or absence of NRAS codon 12/13 mutation. The prognostic effect of NRAS codon 12/13 mutation was significantly greater when both DNMT3A and NPM1 carried the driver mutation (in 45 patients), as shown in the graph at the right (P = 0.0007 for three-way interaction in the univariate analysis). Panel C shows Kaplan–Meier curves for overall survival according to the presence or absence of MLLPTD and FLT3TKD (P = 0.0004 for gene–gene interaction in the univariate analysis; q = 0.008 in the multivariate analysis with correction for multiple hypothesis testing). A total of 69 patients had MLLPTD, 112 had FLT3TKD, and 10 had both. Panel D shows Kaplan–Meier curves for overall survival according to the presence or absence of driver mutations in DNMT3A, IDH2R140, or both (P = 0.05 for gene–gene interaction in the univariate analysis; q= 0.05 in the multivariate analysis with correction for multiple hypothesis testing). A total of 338 patients had DNMT3A, 20 had IDH2R140, and 19 had both.
In contrast, the NPM1–DNMT3A–NRASG12/13 genotype carried an unexpectedly benign prognosis in our cohort (P= 0.04 for three-way interaction) (Fig. 4B). Previous reports have suggested that NPM1–NRAS is a favorable association,38 something we find specific to NPM1–DNMT3A– NRASG12/13. In this cohort, outcomes for patients in the NPM1 subgroup were strongly dictated by the mutation context in which NPM1 mutations occurred (i.e., NRAS, IDH, PTPN11, FLT3, and chromatin–spliceosome mutations) (Fig. S15 in the Supplementary Appendix).
Other significant gene–gene interactions were found. In particular, the prognosis was significantly poorer than expected for the co-occurrence of FLT3TKD mutations with partial tandem duplications of MLL (q= 0.008) and for the co-occurrence of DNMT3A with IDH2R140 (q= 0.05) (Fig. 4C and 4D and Table 2).
Discussion
There is a distinction between a system for classifying patients with AML for diagnostic purposes and a system for predicting the outcome of AML. The former should be stable and durable, based on fixed biologic properties of the disease; hence, the focus on causative driver mutations. The latter should be flexible and adaptable, evolving to reflect advances in treatment and often being influenced by changes in disease surveillance.39 For example, the advent of effective FLT3 and RAS-pathway kinase inhibitors will lead to changes in outcome predictions for patients with these mutations but will not fundamentally alter the fact that these variants co-occur with class-defining mutations such as t(15;17), t(6;9), and NPM1.
It is somewhat counterintuitive that NPM1 emerges as a separate entity, since it appears to be a late driver mutation, is almost never found in isolation, and is associated with a clinical course that is modified depending on the specific co-occurring mutations. Nonetheless, among all the mutations with which it co-occurs, it is the one that most clearly occupies its own distinct niche. Why it occurs late is not clear — it may have transformative effects only in the context of an epigenetic landscape already shaped by initiating mutations such as DNMT3A, TET2, or IDH1/2. NPM1 mutations are not the only route of transformation these initiating lesions can follow: when JAK2 mutations occur, myeloproliferative neoplasms result, whereas co-mutation with SF3B1 leads to MDS with ring sideroblasts.
Beyond the existing WHO subgroups, three genomic categories of AML emerge from our analysis: chromatin–spliceosome, TP53–aneuploidy, and provisionally, IDH2R172 mutations. The chromatin–spliceosome category represents the second largest subgroup of patients with AML, even in this intensively treated, relatively young cohort. In contrast to classes defined by specific fusion genes, no single gene defines this group. We observed intersecting patterns of mutated chromatin and RNA-splicing regulators, suggesting a plasticity of paths of disease evolution in this subgroup, yet these genes showed minimal overlap with other class-defining lesions. Patients in the chromatin–spliceosome subgroup were, on average, older and had lower blast counts and higher rates of antecedent MDS or dysplasia-related morphologic features than patients in other subgroups. This overlap should not be overstated, however — although 20% of patients had a preceding myeloid disorder or evidence of dysplasia as defined by the WHO, 80% did not have such dysplastic features and presented with intermediate-risk, de novo AML.
The most widely accepted classification and prognostic schemes for AML include cytogenetic lesions together with NPM1, FLT3ITD, and CEBPA.3,26 In the short term, TP53, SRSF2, ASXL1, DNMT3A, and IDH2 should be considered for incorporation into prognostic guidelines because they are common and exert a strong influence on clinical outcomes. For AML classification, evaluation of splicing-factor genes RUNX1, ASXL1, and MLLPTD at diagnosis would identify patients in the chromatin–spliceosome group.
In conclusion, we analyzed somatic driver mutations retrospectively in more than 1500 patients with AML, generated a new genetic approach to disease classification with prognostic implications, and obtained similar results for an independent data set from TCGA. Prospective clinical studies are needed for further validation of this schema.
Supplementary Material
Acknowledgments
Supported by grants from the Wellcome Trust (077012/Z/05/Z), Bundesministerium für Bildung und Forschung (01GI9981 and 01KG0605), Deutsche Krebshilfe (DKH, 109675), and Deutsche Forschungsgemeinschaft (DFG) for projects B3 and B4 of Sonderforschungsbereich (SFB) 1074. Dr. Papaemmanuil is a Josie Robertson Investigator and the recipient of a European Hematology Association early career fellowship, Dr. Campbell is the recipient of a Wellcome Trust Senior Clinical Research Fellowship (WT088340MA), and Drs. Bullinger and Heuser are the recipients of grants from the DFG (BU 1339/8-1 and HE 5240/6-1). Drs. Potter and Greaves were supported in part by grants from the Kay Kendall Leukaemia Fund (KKL688) and Wellcome Trust (105104/Z/14/Z). The German-Austrian AML Study Group (AMLSG) treatment trials were supported in part by Amgen.
We thank Daniela Weber for managing clinical data, Veronica Teleanu for assisting with cytogenetic data classification, Dr. Sabine Kayser for assisting with morphologic evaluation, and all the members of the AMLSG (see the Supplementary Appendix for a list of participating institutions and investigators) for participating in this study and providing samples from patients.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–52. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Grimwade D, Hills RK, Moorman AV, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 3.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–74. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 4.Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–89. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–78. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter MJ, Shen D, Ding L, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–8. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–33. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlenk RF, Döhner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–18. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 10.Schlenk RF, Fröhling S, Hartmann F, et al. Phase III study of all-trans retinoic acid in previously untreated patients 61 years or older with acute myeloid leukemia. Leukemia. 2004;18:1798–803. doi: 10.1038/sj.leu.2403528. [DOI] [PubMed] [Google Scholar]
- 11.Schlenk RF, Dohner K, Mack S, et al. Prospective evaluation of allogeneic hematopoietic stem-cell transplantation from matched related and matched unrelated donors in younger adults with high-risk acute myeloid leukemia: German-Austrian trial AMLHD98A. J Clin Oncol. 2010;28:4642–8. doi: 10.1200/JCO.2010.28.6856. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–18. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunson DB. Nonparametric Bayes applications to biostatistics. In: Hjort NL, Holmes C, Müller P, Walker SG, editors. Bayesian nonparametrics. Cambridge, United Kingdom: Cambridge University Press; 2010. p. 35. [Google Scholar]
- 14.Therneau TM, Grambsch PM, Pankratz VS. Penalized survival models and frailty. J Comput Graph Stat. 2003;12:156–75. [Google Scholar]
- 15.Anderson K, Lutz C, van Delft FW, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–61. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 16.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–27. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKerrell T, Park N, Moreno T, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10:1239–45. doi: 10.1016/j.celrep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–8. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busque L, Patel JP, Figueroa ME, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44:1179–81. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–98. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paschka P, Du J, Schlenk RF, et al. Secondary genetic lesions in acute myeloid leukemia with inv(16) or t(16;16): a study of the German-Austrian AML Study Group (AMLSG) Blood. 2013;121:170–7. doi: 10.1182/blood-2012-05-431486. [DOI] [PubMed] [Google Scholar]
- 24.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A. 2014;111:2548–53. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gröschel S, Sanders MA, Hoogenboezem R, et al. Mutational spectrum of myeloid malignancies with inv(3)/t(3;3) reveals a predominant involvement of RAS/RTK signaling pathways. Blood. 2015;125:133–9. doi: 10.1182/blood-2014-07-591461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–51. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 27.Grimwade D, Walker H, Oliver F, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. Blood. 1998;92:2322–3. [PubMed] [Google Scholar]
- 28.Breems DA, Van Putten WL, De Greef GE, et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J Clin Oncol. 2008;26:4791–7. doi: 10.1200/JCO.2008.16.0259. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Z, Chen CC, Rillahan CD, et al. Cooperative loss of RAS feedback regulation drives myeloid leukemogenesis. Nat Genet. 2015;47:539–43. doi: 10.1038/ng.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C, Liu Y, Lu C, et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev. 2013;27:1974–85. doi: 10.1101/gad.226613.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–55. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rücker FG, Schlenk RF, Bullinger L, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119(9):2114–21. doi: 10.1182/blood-2011-08-375758. [DOI] [PubMed] [Google Scholar]
- 33.Lindsley RC, Mar BG, Mazzola E, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–76. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vannucchi AM, Lasho TL, Guglielmelli P, et al. Mutations and prognosis in primary myelofibrosis. Leukemia. 2013;27:1861–9. doi: 10.1038/leu.2013.119. [DOI] [PubMed] [Google Scholar]
- 35.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–7. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taskesen E, Havermans M, van Lom K, et al. Two splice-factor mutant leukemia subgroups uncovered at the boundaries of MDS and AML using combined gene expression and DNA-methylation profiling. Blood. 2014;123:3327–35. doi: 10.1182/blood-2013-07-512855. [DOI] [PubMed] [Google Scholar]
- 37.Ward PS, Lu C, Cross JR, et al. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem. 2013;288:3804–15. doi: 10.1074/jbc.M112.435495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bacher U, Haferlach T, Schoch C, Kern W, Schnittger S. Implications of NRAS mutations in AML: a study of 2502 patients. Blood. 2006;107:3847–53. doi: 10.1182/blood-2005-08-3522. [DOI] [PubMed] [Google Scholar]
- 39.Klco JM, Miller CA, Griffith M, et al. Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA. 2015;314:811–22. doi: 10.1001/jama.2015.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.