Abstract
Northern pig-tailed macaques (NPMs, Macaca leonina) are susceptible to HIV-1 infection largely due to the loss of HIV-1-restricting factor TRIM5α. However, great impediments still exist in the persistent replication of HIV-1 in vivo, suggesting some viral restriction factors are reserved in this host. The APOBEC3 proteins have demonstrated a capacity to restrict HIV-1 replication, but their inhibitory effects in NPMs remain elusive. In this study, we cloned the NPM A3A-A3H genes, and determined by BLAST searching that their coding sequences (CDSs) showed 99% identity to the corresponding counterparts from rhesus and southern pig-tailed macaques. We further analyzed the anti-HIV-1 activities of the A3A-A3H genes, and found that A3G and A3F had the greatest anti-HIV-1 activity compared with that of other members. The results of this study indicate that A3G and A3F might play critical roles in limiting HIV-1 replication in NPMs in vivo. Furthermore, this research provides valuable information for the optimization of monkey models of HIV-1 infection.
Keywords: Macaca leonina, Northern pig-tailed macaques, APOBEC3, HIV-1
INTRODUCTION
The lack of effective HIV-1-infected animal AIDS models hinders our understanding of HIV-1 pathogenesis and the development of AIDS vaccines and drugs (Hatziioannou & Evans, 2012; Zhang et al., 2007). The ideal animal model is one that can be infected by HIV-1 and progress to an AIDS-like disease. However, HIV-1 shows a narrow host range and only infects humans and a handful of nonhuman primates (Kuang et al., 2009). Among them, the pig-tailed macaques (PTMs), which consist of northern pig-tailed macaques (NPMs, Macaca leonina), southern pig-tailed macaques (M. nemestrina) and Mentawai macaques (M. pagensis) (Groves, 2001), are the only Old World monkeys susceptible to HIV-1 infection. To date, however, the mechanism of this susceptibility remains largely unclear.
TRIM5-Cyclophilin A (TRIMCyp) fusion in NPMs, identified in our previous research, might account for the susceptibility of NPMs to HIV-1 infection (Kuang et al., 2009; Liao et al., 2007). Thus, NPMs might present fewer impediments to HIV-1 infection and replication than that of other macaques, such as the widely used rhesus and cynomolgus macaques. In addition, we previously reported on many basic biological parameters of NPMs (Lian et al., 2016; Pang et al., 2013; Zhang et al., 2014, 2016; Zheng et al., 2014; Zhu et al., 2015), which will help in the promotion of NPMs in AIDS and biomedical research.
Though HIV-1 can infect PTMs, considerable obstacles exist in regards to persistent replication in these hosts (Agy et al., 1992; Bosch et al., 2000; Hu, 2005). This has limited the application of the PTM animal model of HIV-1 infection in the field of vaccine and drug testing. Furthermore, although APOBEC3 proteins have the capacity to restrict HIV-1 replication (Jia et al., 2015; Simon et al., 2015; Stavrou & Ross, 2015), it is unclear whether such proteins play an important role in restricting the replication of HIV-1 in NPMs.
The APOBEC3 family, comprised of seven members (APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H) in primates (Prohaska et al., 2014), can inhibit the replication and spread of various retroviruses by inducing C-to-U hypermutation in newly synthesized viral minus DNA, ultimately leading to G-to-A hypermutation in the viral genome. The activity of APOBEC3 proteins, particularly that of APOBEC3G, is inhibited by vif, an accessory protein encoded by lentiviruses. Interestingly, the degradation of APOBEC3 proteins by different vifs shows pronounced species-specificity (Zennou & Bieniasz, 2006), although this is not absolute. The species-specific interaction between vifs and APOBEC3s suggests that APOBEC3s are important obstacles to cross-species transmission of lentiviruses.
The rate of viral hypermutation mediated by APOBEC3s is associated with viral transmission and disease progression. In recent years APOBEC3s have been treated as potential targets of future therapeutic strategies against HIV-1 (Pillai et al., 2012).
Recently, PTMs were shown to develop AIDS following infection with adapted macaque-tropic HIV-1 (Hatziioannou et al., 2014), which is considered a major step forwards in AIDS research. To explore whether APOBEC3 proteins play a role in restricting the replication of HIV-1 in NPMs, we cloned A3A-A3H genes from NPMs and analyzed their anti-HIV-1 activity. The aim of this study was to provide valuable information for optimizing nonhuman primate models for AIDS research.
MATERIALS AND METHODS
Animals, cells and viruses
The NPMs used in this study were obtained from the Kunming Institute of Zoology, Chinese Academy of Sciences (KIZ, CAS), and were maintained in accordance with the regulations and recommendations of the Animal Care Committee of KIZ, CAS, and the Guide to the Care and Use of Experimental Animals, as described previously (Zhang et al., 2014).
Peripheral blood mononuclear cells (PBMCs) in NPM blood samples were isolated using Ficoll density centrifugation (Dai et al., 2013). The isolated PBMCs were cultured for 72 h in RPMI 1640 medium (10% fetal bovine serum (FBS) containing interleukin-2 (IL-2, 50 U/mL) and Con A (1 mg/mL). In addition, 293T cells (Type Culture Collection (TCC), CAS) and TZM-bl cells (Medical Research Council, AIDS Reagent Project, UK) were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS (Lei et al., 2014). HIV-1NL4-3 was made by transfection of 293T cells using Lipofectamine TM 2000 according to the manufacturer’s instructions (Invitrogen Carlsbad, CA). HIV-1NL4-3 proviral plasmids were kindly donated by Prof. Guang-Xia Gao (Institute of Biophysics, CAS, China).
PCR of APOBEC3 mRNA
Total RNA from PBMCs was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) and then reverse transcribed into cDNA using the PrimeScript® RT reagent kit with gDNA Eraser (Takara, Dalian, China). The primers used were synthesized by Generay Biotech (Shanghai, China), and the sequences and PCR conditions are listed in Table 1. The PCR products were analyzed on a 2% agarose gel, visualized by ethidium bromide staining, and purified using a DNA gel extraction kit (Generay Biotech, Shanghai, China). The purified fragments were cloned into pMD19-T simple vector (Takara, Dalian, China) and finally sequenced (Majorbio, Shanghai, China). Sequences were analyzed by MEGA5.0 software and the NCBI’s online BLAST server (http://www.ncbi.nlm.nih.gov/blast/blast.cgi).
Table 1.
Primer sequences and PCR conditions for APOBEC3 mRNA
| Primers | Primer DNA sequences | PCR conditions |
| NPM-A3A-F | 5'-CAGGAACCGAGAAGAGACAAGCACAT-3' | 94 ℃ 5 min, (94 ℃ 30 sec,59 ℃ 30 sec,72 ℃ 90 sec)×32 cycle,72 ℃ 10 min |
| NPM-A3A-R | 5'-CTGCCTTCCTTAGAGACTGAGGC-3' | |
| NPM-A3B-F | 5'-AGAGCGGGACTGGGACAAG-3' | 94 ℃ 5 min, (94 ℃ 30 sec,59 ℃ 30 sec,72 ℃,2 min) × 32 cycle,72 ℃ 10 min |
| NPM-A3B-R | 5'-TTAGAGACTGAGGCCCATCCTTC-3' | |
| NPM-A3C-F | 5'-ACAAGCGTATCTAAGAGGCCG-3' | 94 ℃ 5 min, (94 ℃ 30 sec,59 ℃ 30 sec,72 ℃ 90 sec)×32 cycle,72 ℃ 10 min |
| NPM-A3C-R | 5'-GGAGGCCCGTGCAGCATG-3' | |
| NPM-A3D-F | 5'-GCGTATCTAAGAGGCTGAACAT-3' | 94 ℃ 5 min, (94 ℃ 30 sec,59 ℃ 30 sec,72 ℃,2 min) × 32 cycle,72 ℃ 10 min |
| NPM-A3D-R | 5'-TTATGTTGGGGAGATGGGAAGAG-3' | |
| NPM-A3F-F | 5'-ACAAAGATCTTAGTCGGGACT-3' | 94 ℃ 5 min, (94 ℃ 30 sec,59 ℃ 30 sec,72 ℃,2 min) × 32 cycle,72 ℃ 10 min |
| NPM-A3F-R | 5'-AGGCTAGAGGAGACAGACCA-3' | |
| NPM-A3G-F | 5'-GTCAGGACTAGCCGGCAAAGGAT-3' | 94 ℃ 5 min, (94 ℃ 30 sec,59 ℃ 30 sec,72 ℃,2 min) × 32 cycle,72 ℃ 10 min |
| NPM-A3G-R | 5'-CTTCCTTAGAGACTGAGGCCCATC-3' | |
| NPM-A3H-R | 5'-GACCAGCAGGCTATGAGGCAA-3' | 94 ℃ 5 min, (94 ℃ 30 sec,59 ℃ 30 sec,72 ℃ 90 sec)×32 cycle,72 ℃ 10 min |
| NPM-A3H-F | 5'-CCAGAAGCACAGATCAGAAACACGAT-3' |
Molecular cloning of NPM A3s
For expression studies, FLAG primers were designed based on the CDSs of the NPM APOBEC3s, with the sequences and PCR conditions listed in Table 2. The flag-APOBEC3s amplicons were cloned into the pcDNA3.1 (+) vector (Invitrogen, Carlsbad, CA) using the XbaI and HindIII restriction sites and then sequenced (Majorbio, Shanghai, China). Finally, the constructs of these recombinant plasmids were verified by restriction mapping of XbaI and HindIII.
Table 2.
Primer sequences and PCR conditions for molecular cloning of NPM A3s
| Primers | Primer DNA sequences | PCR conditions |
| NPM-A3A-CD-F | 5'-gcgaagcttgccaccATGGACGGCAGCCCAGCATCCAG-3' | 94 ℃ 5 min, (94 ℃ 30 sec,58 ℃ 30 sec,72 ℃ 90 sec) ×32 cycle,72 ℃ 10 min |
| NPM-A3A-CD-R | 5'-cctctagaggcTCACTTATCGTCGTCATCCTTGTAATCgccgccGTTTCCCTGATTCTGGAG AATGGCCC-3' | |
| NPM-A3B-CD-F | 5'-gcgaagcttgccaccATGAATCCACAGATCAGAAATCCGATGGAGC-3' | 94 ℃ 5 min, (94 ℃ 30 sec,58 ℃ 30 sec,72 ℃ 2 min) ×32 cycle,72 ℃ 10 min |
| NPM-A3B-CD-R | 5'-cctctagaggcTCACTTATCGTCGTCATCCTTGTAATCgccgccGTTTCCCTGATTCTGGAG AATGGCC-3' | |
| NPM-A3C-CD-F | 5'-gcaagcttgccaccATGAATCCACAGATCAGAAACCCGATGA-3' | 94 ℃ 5 min, (94 ℃ 30 sec,58 ℃ 30 sec,72 ℃ 90 sec) ×32 cycle,72 ℃ 10 min |
| NPM-A3C-CD-R | 5'-cctctagaggcTCACTTATCGTCGTCATCCTTGTAATCgccgccCTGAAGAATCTCCCGTAG GC-3' | |
| NPM-A3D-CD-F | 5'-gcgaagcttgccaccATGAATCCACAGATCAGAAATCCGATGGAGGG-3' | 94 ℃ 5 min, (94 ℃ 30 sec,58 ℃ 30 sec,72 ℃ 2 min) ×32 cycle,72 ℃ 10 min |
| NPM-A3D-CD-R | 5'-cctctagaggcTCACTTATCGTCGTCATCCTTGTAATCgccgccCTGAAGAATCTTCCATAG GCGTCTTTCCAGA-3' | |
| NPM-A3F-CD-F | 5'-gcgaagcttgccaccATGAAGCCTCAGTTCAGAAACACAGT-3' | 94 ℃ 5 min, (94 ℃ 30 sec,58 ℃ 30 sec,72 ℃ 2 min) ×32 cycle,72 ℃ 10 min |
| NPM-A3F-CD-R | 5'-CctctagaggcTCACTTATCGTCGTCATCCTTGTAATCgccgccCTCGAGAATCTCCTGCA GCTTG-3' | |
| NPM-A3G-CD-F | 5'-gcgaagcttgccaccATGAATCCTCAAATCAGAAACATGGTGGA-3' | 94 ℃ 5 min, (94 ℃ 30 sec,58 ℃ 30 sec,72 ℃ 2 min) ×32 cycle,72 ℃ 10 min |
| NPM-A3G-CD-R | 5'-cctctagaggcTCACTTATCGTCGTCATCCTTGTAATCgccgccGTTTCCCTGATTCTGGAG AATGGC-3' | |
| NPM-A3H-CD-F | 5'-gcgaagcttgccaccATGGCTCTGCTAACAGCCAAAACATTC-3' | 94 ℃ 5 min, (94 ℃ 30 sec,58 ℃ 30 sec,72 ℃ 90 sec) ×32 cycle,72 ℃ 10 min |
| NPM-A3H-CD-R | 5'-cctctagaggcTCACTTATCGTCGTCATCCTTGTAATCgccgccTCTTGAGTTGCGTATTGA CGATGAGGGG-3' |
Western blot analysis
The 293T cells were transfected with pcDNA3.1-npmA3s-Flag (pcDNA3.1-npmA3A-Flag, pcDNA3.1-npmA3B-Flag, pcDNA3.1-npmA3C-Flag, pcDNA3.1-npmA3D-Flag, pcDNA3.1-npmA3F-Flag, pcDNA3.1-npmA3G-Flag, pcDNA3.1-npmA3H-Flag), pcDNA3.1-humanA3G-Flag, and pcDNA3.1-empty vector in 6-well plates with Lipo2000 (Invitrogen, Carlsbad, CA), and were lysed with cell lysis buffer (Beyotime, , Shanghai, China after 48 h transfection. The extracted proteins were denatured in SDS/PAGE loading buffer by boiling for 15 min and separated by SDS/PAGE. The flagtagged proteins were detected using mouse monoclonal antiflag antibody (Abmart, Shanghai, China) and then HRPconjugated secondary antibody, followed by light staining with chemiluminescent detection reagents (Millipore, Bedford, MA).
Co-transfection
The HIV-1NL4-3 proviral plasmid (3 μg) was transiently cotransfected with the NPM A3A-A3G and human-A3G expression plasmid (1 μg) and pcDNA3.1 (+) control plasmid (1 μg) in the 293T cells with Lipo2000, according to the manufacturer’s protocols (Invitrogen, Carlsbad, CA). The cell culture medium was replaced with fresh medium 8 h after transfection. On day 2 after transfection, the cell supernatant containing the virus was collected, filtered, and then treated by DNase I (Takara, Dalian, China) at 37 °C for 1 h to prevent plasmid carryover.
Single-round viral infectivity assays
The amount of viral particles harvested from the co-transfection supernatants was assayed by p24 antigen enzyme-linked immunosorbent assay (ELISA) (ZeptoMetrix Corp., Buffalo, NY, USA). The TZM-bl cells seeded at a density of 1×104 cells per well in 96-well plates were infected with equal amounts of virus. The infection was performed in triplicate for 48 h. After incubation, luminescence was measured by Bright-Glo luciferase assay reagent (Promega, Madison, WI). Statistical differences between mean percentages were compared by Student's t-tests (two-tailed, P < 0.05) using GraphPad Prism software v5.0.
RESULTS
PCR amplification and sequence analysis of NPM A3s
Total RNA was extracted from PBMCs and reverse transcribed into cDNAs, which were used as PCR templates. The sizes of the A3A to A3H amplicons were 665 bp, 1 205 bp, 629 bp, 1 181 bp, 1 178 bp, 1 208 bp, and 689 bp, respectively (Figure 1).
Figure 1.

Analysis of PCR products of NPM A3A-A3H with agarose gel electrophoresis
The purified amplicons were cloned into pMD19-T simple vector and finally sequenced. Sequences were analyzed by MEGA5.0 software and BLAST online. BLAST analysis of the NPM A3A-A3H CDSs showed 94%-99% nucleotide identity with those of rhesus, crab-eating, and southern pig-tailed macaques, and 89%-93% of nucleotides were identical to those of humans. Among them, the identities of NPM cDNA sequences with those of rhesus and southern pig-tailed macaques reached 99% (Table 3). To explore the clustering of NPM A3A-A3H sequences with specific primate lineages, we constructed a phylogenetic tree (Figure 2). Amino acid sequence alignment was subjected to phylogenetic analyses using the neighbor joining method. The results showed that NPM A3A-A3H sequences clustered according to the corresponding A3A-A3H sequences from different primate taxa. These analyses demonstrated that the cloned NPM A3A-A3H gene classifications were correct. GenBank accession numbers of the NPM A3A-A3H CDSs are KX583650, KX583652, KX583655, KX583653, KX583651, KX583654, and KX583656, respectively.
Table 3.
Nucleotide identity of A3A-A3H genes from NPMs compared with other primate species
| Gene | Species | Percentage identified with NPM (%) |
| NPM-A3A | Macaca mulatta | 99 |
| Homo sapiens | 90 | |
| NPM-A3B | Macaca mulatta | 96 |
| Homo sapiens | 92 | |
| NPM-A3C | Macaca mulatta | 98 |
| Macaca fascicularis | 98 | |
| Homo sapiens | 91 | |
| NPM-A3D | Macaca mulatta | 94 |
| Homo sapiens | 90 | |
| NPM-A3F | Macaca nemestrina | 99 |
| Macaca mulatta | 99 | |
| Homo sapiens | 93 | |
| NPM-A3G | Macaca nemestrina | 99 |
| Macaca mulatta | 99 | |
| Macaca fascicularis | 99 | |
| Homo sapiens | 89 | |
| NPM-A3H | Macaca mulatta | 99 |
| Homo sapiens | 91 |
Figure 2.
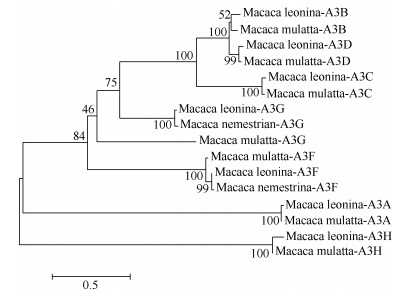
Phylogenetic tree of APOBEC3 proteins
Enzyme digestion and expression product identification of NPM A3A-A3H expression plasmids
Positive colonies containing non-mutated target fragments were picked and identified by double digestion with restriction enzymes HindIII and XbaI. Agarose gel electrophoresis showed a series of DNA bands, which were in accordance with the expected sizes (Figure 3).
Figure 3.

Identification of pcDNA3.1-NPM A3A-A3H by restriction analysis
To verify the constructed pcDNA3.1-NPM-A3s expressed proteins, the recombinant plasmid and an empty pcDNA3.1 vector as a control were transfected into 293T cells. The cells were then subjected to protein detection by Western blot analysis. Protein expressions were detected and the molecular weights of the products were consistent with the expected values (Figure 4).
Figure 4.
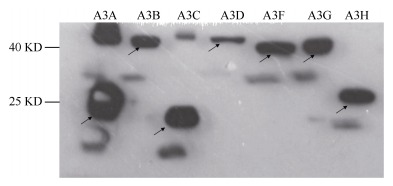
Detection of protein expression in 293T cells by Western blot analysis
In vitro anti-HIV-1 activities of NPM APOBEC3s
To address whether different NPM APOBEC3 isoforms were able to effectively inhibit HIV-1 infectivity, we performed a single-cycle infectivity assay in TZM-bl cells. We observed more efficient inhibition of HIV-1NL4-3 infectivity by NPM A3G and A3F. Furthermore, NPM A3B and A3D showed less efficient inhibition than A3G/F, A3H showed relatively weaker inhibition compared with that of A3B/D, and the HIV-1 produced in the presence of A3A and A3C expression vector did not show any reduction in relative infectivity (Figure 5).
Figure 5.
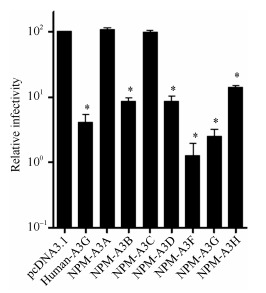
Anti-HIV-1 activities of NPM APOBEC3s
DISCUSSION
NPMs express a TRIM5-Cyclophilin A (TRIMCyp) fusion protein, instead of a TRIM5α protein, which makes them more susceptible to HIV-1 infection than other macaques (Kuang et al., 2009; Liao et al., 2007). However, HIV-1 still encounters considerable obstacles in regards to persistent replication in this host (Agy et al., 1992; Bosch et al., 2000; Hu, 2005), suggesting that other restriction factors act as barriers. In previous studies, APOBEC3s have demonstrated a capacity to restrict HIV-1 replication (Jia et al., 2015; Simon et al., 2015; Stavrou & Ross, 2015). However, it is unclear whether the APOBEC3 proteins of NPMs play an important role in restricting the replication of HIV-1.
In the current study, the NPM A3A-A3H genes were cloned, with BLAST analysis of their CDSs showing 94%-99% nucleotide identity with those of rhesus, crab-eating, and southern pig-tailed macaques, and 89%-93% nucleotide identity with those of humans. Among them, the identities of NPM cDNA sequences with those of rhesus and southern pig-tailed macaques reached 99%. Such a high nucleotide identity suggests that APOBEC3 genes might be important for the species to survive in nature. In addition, compared with rodents, which have only one APOBEC3 gene, primates have as many as seven (Jarmuz et al., 2002; Wedekind et al., 2003; Zhang & Webb, 2004), indicating that APOBEC3 gene expansion has been critical for primate survival during evolution.
About 35-50 million years ago, the dramatic decline in retrotransposon activities in primates might have resulted from the successful expansion of the APOBEC3 gene (Zhang & Webb, 2004). Retrotransposon activities increase genomic instability. Frequent retrotransposition can be detrimental for species living in stable environments and adapting to the environment at the same. However, moderate retrotransposon activities promote gene mutation, which is helpful for a species population to adapt to changing living environments. The APOBEC3 gene family has undergone rapid expansion, from one or two genes in non-primate mammals to at least seven in primates. In the past 30 million years, the primate genome has been infected by many viruses (Belshaw et al., 2004; Wolfe et al., 2004). The rapid evolution of the APOBEC3 genes has kept pace with the rapidly evolving viruses (Sawyer et al., 2004), suggesting that the APOBEC3 gene family plays an important role in restricting virus infection and maintaining genomic stability.
We analyzed the anti-HIV-1 activity of the A3A-A3H genes. NPM A3G/F demonstrated very efficient anti-HIV-1 activity; however, NPM A3A/C exhibited no anti-HIV-1 activity, which is consistent with the anti-HIV-1 activity of APOBEC3s in humans and rhesus macaques (Virgen & Hatziioannou, 2007). Our study indicates that A3G and A3F might play critical roles in restricting HIV-1 replication in NPMs in vivo.
Although results indicated that NPM A3A and A3C exhibited no anti-HIV-1 activity, their antiviral activity against other viruses and important biological roles cannot be ignored. Some studies have suggested that primate A3A limits the replication of the hepatitis B virus (HBV) and human papillomavirus (HPV) (Suspène et al., 2005; Vartanian et al., 2008) and the retrotransposition of Alu elements (Stenglein et al., 2010). In this study, we only determined the anti-HIV-1 activity of NPM APOBEC3s in vitro. Whether NPM A3G and A3F exhibit better anti-HIV-1 activity in vivo needs to be further explored.
In conclusion, A3A-A3H genes from NPMs were cloned, and their CDSs were found to be 99% identical to relevant sequences from rhesus and southern pig-tailed macaques. NPM A3G and A3F showed the greatest anti-HIV-1 activity compared with that of the other members, whereas A3A and A3C exhibited no anti-HIV-1 activity at all. This study indicates that A3G and A3F might play critical roles in limiting HIV-1 replication in NPMs in vivo. Our study provides valuable information for the optimization of monkey models of HIV-1 infection.
Supplementary Table 1.
Full-length coding sequences (CDSs) of NPM A3POBECs
| Gene name | Full-length coding sequences |
| NPM-A3A | 5′-ATGGACGGCAGCCCAGCATCCAGGCCCAGACACTTGATGGATCCAAACACGTTCACTTTCAACTTTAACAATGACCTTTC GGTCCGTGGACGGCACCAGACCTACTTGTGCTACGAGGTGGAGCGCCTGGACAATGGCACCTGGGTCCCGATGGACGAGCGCAGGGGCTTTCTACAAAACAAGGCTAAGAATGTTCCCTGTGGTGATTATGGCTGCCACGTGGAGCTGCGCTTCCTGTGTGAGGTTCCTTCTTGGCAGTTGGACCCGGCCCAGACATACAGGGTCACTTGGTTCATCTCCTGGAGCCCCTGCTTCAGGAGGGGCTGTGCCGGGCAAGTGCGTGCGTTCCTTCAGGAGAACAAACACGTGAGACTGCGCATCTTCGCTGCCCGCATCTATGATTACGACCCCCTGTATCAGGAGGCACTGCGAACGCTGCGGGATGCTGGGGCCCAAGTCTCCATCATGACCTACGAGGAATTTAAGCACTGCTGGGACACCTTTGTGGACCGCCAGGGACGTCCCTTCCAGCCCTGGGATGGACTAGATGAGCACAGCCAAGCCCTGAGTGAGAGGCTTCGGGCCATTCTCCAGAATCAGGGAAACTGA-3′ |
| NPM-A3B | 5′-ATGAATCCACAGATCAGAAATCCGATGGAGCGGATGTATCAACGCACATTCTACTACCACTTTGAAAATAAACCCATCCT CTATGGTCGGAGCTACACCTGGCTGTGCTACGAAGTGAAAATAAGGAAGGACCCCTCAAAGCTCCCTTGGGACACAGGGGTCTTTCGAGGCCAGGTGTATTCCAAGCCTGAGCACCACGCAGAAATGTGCTTCCTCTCTCGGTTCTGTGGCAACCAGCTGCCTGCTTACAAGCGCTTCCAGATCACCTGGTTTGTATCCTGGAACCCCTGCCCGGACTGTGTGGTGAAGGTGATTGAATTCCTGGCTGAGCACCCTAATGTCACCCTGACCATCTCCACCGCCCGCCTCTACTACTACTGGGGTAGAGATTGGCAGAGGGCGCTCTGCAGGCTGCGTCAGGCAGGGGCCCGAGTGAAGATCATGGACTATGAAGAATTTGCATACTGCTGGGAAAACTTTGTGTACAATGAAGATCAGTCATTCATGCCTTGGTACAAATTCGATGACAATTATGCATTCCTGCACCGCATGCTAAAGGAGATTCTCAGACACTTGATGGATCCAGACACGTTCACTTCCAACCTTAACAATGACCTTTCGGTCCGTGGACGGCACCAGACCTACTTGTGCTACGAGGTGGAGCGCCTGGACAATGGCACCTGGGTCCCGATGGACCAGCACTGGGGATTTCTTTGCAACCAGGCTAAGAATGTTCCCCGTGGTGATTACGGCTGCCACGTGGAGCTGTGCTTCCTGGGCAAGGTTCCTTCTTGGCAGTTGGACCCGGCCCAGACGTACAGGGTCACTTGGTTCATCTCCTGGAGCCCCTGCTTCAGCTGGGGCTGTGCCGAGCAAGTGCGTGCGTTCCTTCAGGAGAACAAACACGTGAGACTGCGCATCTTCGCTGCCCGCATCTATGATTACGACCCCCTGTATCAGGAGGCACTGCGAACGCTGCGGGATGCTGGGGCCGAAGTCTCCATCATGACCTACGAGGAATTTGAGTACTGCTGGGACACCTTTGTGGACCGCCAGGGACGTCCCTTCCAGCCCTGGGATGGACTAGATGAGCACAGCCAAGCCCTGAGTGGGAGGCTTCGGGCCATTCTCCAGAATCAGGGAAACTGA-3′ |
| NPM-A3C | 5′-ATGAATCCACAGATCAGAAACCCGATGAAGGCAATGGATCCAGGCACATTCTACTTCCAATTTAAAAACCTATGGGAAG CCAACAATCGGAACGAAACCTGGCTGTGCTTCACCGTGGAAGTTATAAAGCAGTGTTCAACTGTCTCCTGGGAGAGGGGCGTCTTCCGAAACCAGGTCGATCTTGAGACCCATTGTCATGCAGAAAGGTGCTTCCTCTCCTGGTTCTGCGAGGACATACTGTCTCCTAACACAGACTACCAGGTCACCTGGCACACATCTTGGAGCCCTTGCCCAGAGTGTGCCAGGGAGGTGGCCAAGTTCCTGGCCAGGCACAACAATGTGATGCTCACCATCTATACCGCCCGCCTCTACTACTCCCAGTATCCGAATTACCAGCAGGGGCTCCGCAGCCTGAGTGAGAAAGGAGTCTCTGTGAAGATTATGGACTACGAAGATTTTAAATATTGTTGGGAAAAGTTTGTGTACGATGATGGTGAGCCATTCAAGCCTTGGAAGGGACTAAAAACCAGCTTTCGATTTCTGAAAAGACGCCTACGGGAGATTCTTCAGTGA-3′ |
| NPM-A3D | 5′-ATGAATCCACAGATCAGAAATCCGATGGAGGGGATGTATCGACACATATTCAACTACAACTTTGAAAATGAACCCATCCT CTATGGTCGGAGCTACACCTGGCTGTGCTACGAAGTGAAAATAAGGAAGGACCCCTCAAAGCTCCCTTGGTACACAGGGGTCTTTCGAGGCCAGGTGTATTTCCAGCCTCAGTACCACGCAGAAATGTGCTTCCTCTCTTGGTTCTGTGGCAACCAGCTGCCTGCTAACAAGCGCTTCCAGATCACCTGGTTTGTATCCTGGAACCCCTGCCCGGACTGTGTGGCGAAGGTGACCGAATTCCTGGCTGAGCACCCTAATGTCACCCTGACCATCTCTGTCGCCCGCCTCTACTACTACCGGGGCAAAGATTGGCGGAGGGCGCTCCGCAGGCTGCATCAGGCAGGGGCTCGCGTGAAGATCATGGACTATGAAGAATTTGCATACTGCTGGGAAAACTTTGTGTACAATGAAGGTCAGTCATTCATGCCTTGGGACAAATTCGATGACAATTATGCATCCCTGCACTGCAAGCTAAAGGAGATTCTCAGAAACCCGATGAAGGCAATGTATCCACACACATTCTACTTCCACTTTGAAAACCTACAGAAAGCCTATGGTCGGAATGAAACCTGGCTGTGCTTCGCCGTGGAAATTATAAAGCAGCACTCAACTGTCCCCTGGAAGACGGGCGTCTTCCGAAACCAGGTTGATCCTGAGACCCATTGTCATGCAGAAAGGTGCTTCCTCTCTTGGTTCTGCGACAACACACTGTCTCCTAAGAAAAACTACCAGGTCACCCGGTACATATCTTGGAGCCCTTGTCCAGAGTGTGCAGGGGAGGTGGCCGAGTTCCTGGCCACGCACAGCAACGTGAAGCTCACCATCTATACCGCCCGCCTCTACTACTTCTGGGATACAGATTACCAGGAGGGGCTCCGCAGCCTGAGTGAGGAAGGGGCCTCCATGGAGATCATGGGCTACGAAGATTTTAAATATTGTTGGGAAAACTTTGTGTGCGATGATGGTGAGCCATTCAAGCCTTGGAAGGGAATAAACACCAACTTTCGATTTCTGGAAAGACGCCTATGGAAGATTCTTCAGTGA-3′ |
| NPM-A3F | 5′-ATGAAGCCTCAGTTCAGAAACACAGTGGAGCGAATGTATCGAGGCACATTCTTCTACAACTTTAATAACAGACCCATCCT TTCTCGTCGGAATACCGTCTGGCTGTGCTACGAAGTGAAAACAAGGGGCCCCTCAATGCCCACTTGGGGCACAAAGATCTTTCGAGGCCAGGTGTGTTTCGAGCCTCAGTACCACGCAGAAATGTGCTTCCTCTCTCGATTCTGTGGCAACCAGCTGCCTGCTTACAAGCGCTTCCAGATCACCTGGTTTGTATCCTGGACCCCCTGCCCAGACTGTGTGGCGAAGGTGGCCGAATTCCTAGCTGAGCACCCTAATGTCACCCTGACCATCTCTGCCGCCCGCCTCTACTACTACTGGGAAACAGATTATCGCAGGGCGCTCTGCAGGCTGCGTCAGGCAGGGGCCCGAGTGAAGATCATGGACTATGAAGAATTTGCATACTGCTGGGAAAACTTTGTGTACAATGAAGATCAGTCATTCATGCCTTGGGACAAATTCGATGACAATTATGCATTCCTGCACCACAAGCTAAAGGAGATTCTCAGAAACCCGATGGAGGCAACGTATCCACACATATTCTACTTCCACTTTAAAAACCTACGGAAAGCCTATGGTCGGAACGAAACCTGGCTGTGCTTCACCATGGAAATTATAAAGCAGCACTCAACTGTCTCCTGGGAGACGGGCGTCTTCCGAAACCAGGTCGATCCTGAGAGCCGTTGTCATGCAGAAAGGTGCTTCCTCTCCTGGTTCTGCGAGGACATACTGTCTCCTAACACAGAGTACCAGGTCACCTGGTACACATCTTGGAGCCCTTGCCTAGATTGTGCAGGGGAGGTGGCCGAGTTCCTGGCCAGGCACAGTAATGTGAAGCTCGCCATCTTTGCCGCCCGCCTCTACTACTTCTGGGATACACATTACCAGCAGGGGCTCCGCAGCCTGAGTGAGAAAGGGGCCTCCGTGGAGATCATGGGCTACAAAGATTTTAAATATTGTTGGGAAAACTTTGTGTACAATGGTGATGAGCCGTTCAAGCCTTGGAAGGGACTAAAATACAACTTTCTATTCCTGGACAGCAAGCTGCAGGAGATTCTCGAGTGA-3′ |
| NPM-A3G | 5′-ATGAATCCTCAAATCAGAAACATGGTGGAGCCAATGGATCCACGCACATTCGTCTCCAACTTCAATAACAGACCTATCCT TTCTGGTCTGAATACCGTCTGGCTGTGCTGCGAAGTGAAAACAAAGGACCCTTCGGGGCCCCCTTTGGACGCAAAGATCTTTCAAGGCAAGGTGTACTCCAAGGCTAAGTACCACCCAGAGATGAGATTCCTCCGCTGGTTCCGCGAGTGGAGGCAGCTGCATCATGACCAGGAGTACAAGGTCACCTGGTACGTATCCTGGAGCCCCTGCACAAGGTGTGCAAACAGTGTGGCCACGTTCCTGGCCAAGGACCCGAAGGTTACCCTGACCATCTTTGTTGCCCGCCTCTACTACTTCTGGAAACCAGATTACCAGCAGGCGCTTCGCATCCTGTGTCAGAAAAGAGGCGGTCTGCATGCCACCATGAAGATCATGAATTATAACGAATTTCAAGACTGTTGGAACAAGTTCGTGGACGGCCGAGGAAAGCCATTTAAGCCTTGGAACAATCTGCCTAAACATTATACATTACTGCAGGCCACGCTGGGGGAGCTTCTCAGACATTTGATGGATCCAGGCACGTTCACTTCCAACTTTAACAATAAACCTTGGGTCAGTGGACAGCATGAGACTTACCTGTGTTACAAGGTGGAGCGCCTGCACAACGACACCTGGGTCCCGCTGAACCAGCACAGGGGCTTTCTACGCAACCAGGCTCCAAATATACACGGTTTCCCTAAAGGCCGCCATGCAGAGCTGTGCTTCCTGGACCTGATTCCCTTTTGGAAGCTGGATGGCCAGCAATACAGGGTTACCTGCTTCACCTCCTGGAGCCCCTGCTTTAGCTGTGCCCAGGAAATGGCTAAATTCATTTCAAATAACGAACACGTGAGCTTGTGCATCTTCGCTGCCCGCATCTATGATGATCAAGGAAGATATCAGGAGGGGCTGCGCACCTTGCACAGGGATGGGGCCAAAATTGCAATGATGAACTACAGTGAATTTGAGTACTGCTGGGACACCTTTGTGGACCGCCAGGGACGTCCCTTCCAGCCCTGGGATGGACTAGATGAGCACAGCCAAGCCCTGAGTGGAAGGCTTCGGGCCATTCTCCAGAATCAGGGAAACTGA-3′ |
| NPM-A3H | 5′-ATGGCTCTGCTAACAGCCAAAACATTCAGCTTACAGTTTAACAACAAGCGCCGTGTCAACAAGCCTTACTACCTGAGGA AGGCCCTCTTGTGTTACCAGCTGACGCCGCAGAATGGCTCCACGCCTACCAGAGGCCACTTAATAAACAAGAAAGAGGACCATGCAGAAATTCGCTTTATTAACGAGATCAAGTCCATGGGACTGGACGAAACCCAGTGCTACCAAGTCACCTGTTACCTCACGTGGAGCCCCTGTCCCTCCTGTGCCGGGGAGCTGGTTGACTTCATCAAGGCTCACCGCCATCTGAACCTGCGCATCTTCGCCTCCCGCCTGTACTACCACTGGCGCCCGAACTATCAGGAGGGGCTGCTGCTTCTGTGTGGATCCCAGGTCCCAGTGGAGGTCATGGGCCTCCCAGAGTTTACTGACTGCTGGGAAAACTTTGTGGACCACAAGGAACCGCCTTCCTTCAACCCCTCTGAGAAGTTAGAGGAGCTAGATAAAAACAGCCAAGCCATAAAGCGACGGCTTGAAAGGATAAAGTCCCGGAGTGTGGATGTTTTAGAGAATGGCTTAAGAAGTTTGCAGCTTGGACCCGTAACCCCCTCATCGTCAATACGCAACTCAAGATGA-3′ |
ACKNOWLEDGMENTS
We thank Prof. Guang-Xia Gao (Institute of Biophysics, CAS) for providing the wild-type HIV-1NL4-3 proviral plasmid. We also thank Dr. Muhammad Shahzad for language editing. We acknowledge the MRC AIDS Reagent Project for providing TZM-bl and the Kunming Primate Research Center of the Chinese Academy of Sciences for providing the macaques in this study.
Funding Statement
This work was supported by the National Special Science Research Program of China (2012CBA01305), National Natural Science Foundation of China (81172876; 81471620; 81273251; 81571606; U0832601), National Science and Technology Major Project (2014ZX10005-002-006), Knowledge Innovation Program of CAS (KJZD-EW-L10-02) and Yunnan Applicative and Basic Research Program (2014FB181)
REFERENCES
- 1. Agy MB, Frumkin LR, Corey L, Coombs RW, Wolinsky SM, Koehler J, Morton WR, Katze MG. 1992. Infection of Macaca nemestrina by human immunodeficiency virus type-1. Science, 257 (5066): 103- 106. [DOI] [PubMed] [Google Scholar]
- 2. Belshaw R, Pereira V, Katzourakis A, Talbot G, Pačes J, Burt A, Tristem M. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proceedings of the National Academy of Sciences of the United States of America, 101 (14): 4894- 4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bosch ML, Schmidt A, Chen JL, Florey MJ, Agy M, Morton WR. 2000. Enhanced replication of HIV-1 in vivo in pigtailed macaques (Macaca nemestrina). Journal of Medical Primatology, 29 (3-4): 107- 113. [DOI] [PubMed] [Google Scholar]
- 4. Dai ZX, Zhang GH, Zhang XH, Zheng YT. 2013. Identification and characterization of a novel splice variant of rhesus macaque MHC IA. Molecular Immunology, 53 (3): 206- 213. [DOI] [PubMed] [Google Scholar]
- 5. Groves CP. 2001. Primate Taxonomy. Washington DC:Smithsonian Institution Press, 222- 224. [Google Scholar]
- 6. Hatziioannou T, Evans DT. 2012. Animal models for HIV/AIDS research. Nature Reviews Microbiology, 10 (12): 852- 867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hatziioannou T, Del Prete GQ, Keele BF, Estes JD, Mcnatt MW, Bitzegeio J, Raymond A, Rodriguez A, Schmidt F, Mac Trubey C, Smedley J, Piatak M Jr, Kewalramani VN, Lifson JD, Bieniasz PD. 2014. HIV-1-induced AIDS in monkeys. Science, 344 (6190): 1401- 1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu SL. 2005. Non-human primate models for AIDS vaccine research. Current Drug Targets-Infectious Disorders, 5 (2): 193- 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics, 79 (3): 285- 296. [DOI] [PubMed] [Google Scholar]
- 10. Jia XF, Zhao Q, Xiong Y. 2015. HIV suppression by host restriction factors and viral immune evasion. Current Opinion in Structural Biology, 31 106- 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuang YQ, Tang X, Liu FL, Jiang XL, Zhang YP, Gao G, Zheng YT. 2009. Genotyping of TRIM5 locus in northern pig-tailed macaques (Macaca leonina), a primate species susceptible to Human Immunodeficiency Virus type 1 infection. Retrovirology, 6 58- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lei AH, Zhang GH, Tian RR, Zhu JW, Zheng HY, Pang W, Zheng YT. 2014. Replication potentials of HIV-1/HSIV in PBMCs from northern pig-tailed macaque (Macaca leonina). Zoological Research, 35 (3): 186- 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lian XD, Zhang XH, Dai ZX, Zheng YT. 2016. Cloning, sequencing, and polymorphism analysis of novel classical MHC class I alleles in northern pig-tailed macaques (Macaca leonina). Immunogenetics, 68 (4): 261- 274. [DOI] [PubMed] [Google Scholar]
- 14. Liao CH, Kuang YQ, Liu HL, Zheng YT, Su B. 2007. A novel fusion gene, TRIM5-Cyclophilin A in the pig-tailed macaque determines its susceptibility to HIV-1 infection. AIDS, 21 (Suppl 8): S19- S26. [DOI] [PubMed] [Google Scholar]
- 15. Pang W, Lü LB, Wang Y, Li G, Huang DT, Lei AH, Zhang GH, Zheng YT. 2013. Measurement and analysis of hematology and blood chemistry parameters in northern pig-tailed macaques (Macaca leonina). Zoological Research, 34 (2): 89- 96. [DOI] [PubMed] [Google Scholar]
- 16. Pillai SK, Abdel-Mohsen M, Guatelli J, Skasko M, Monto A, Fujimoto K, Yukl S, Greene WC, Kovari H, Rauch A, Fellay J, Battegay M, Hirschel B, Witteck A, Bernasconi E, Ledergerber B, Gunthard HF, Wong JK. 2012. Role of retroviral restriction factors in the interferon-alpha-mediated suppression of HIV-1 in vivo. Proceedings of the National Academy of Sciences of the United States of America, 109 (8): 3035- 3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prohaska KM, Bennett RP, Salter JD, Smith HC. 2014. The multifaceted roles of RNA binding in APOBEC cytidine deaminase functions. Wiley Interdisciplinary Reviews:RNA, 5 (4): 493- 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sawyer SL, Emerman M, Malik HS. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biology, 2 (9): e275- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simon V, Bloch N, Landau NR. 2015. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nature Immunology, 16 (6): 546- 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stavrou S, Ross SR. 2015. APOBEC3 proteins in viral immunity. The Journal of Immunology, 195 (10): 4565- 4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. 2010. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nature Structural & Molecular Biology, 17 (2): 222- 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suspène R, Guétard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP. 2005. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America, 102 (23): 8321- 8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vartanian JP, Guetard D, Henry M, Wain-Hobson S. 2008. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science, 320 (5873): 230- 233. [DOI] [PubMed] [Google Scholar]
- 24. Virgen CA, Hatziioannou T. 2007. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. Journal of Virology, 81 (24): 13932- 13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wedekind JE, Dance GS, Sowden MP, Smith HC. 2003. Messenger RNA editing in mammals:new members of the APOBEC family seeking roles in the family business. Trends in Genetics, 19 (4): 207- 216. [DOI] [PubMed] [Google Scholar]
- 26. Wolfe ND, Switzer WM, Carr JK, Bhullar VB, Shanmugam V, Tamoufe U, Prosser AT, Torimiro JN, Wright A, Mpoudi-Ngole E, Mccutchan FE, Birx DL, Folks TM, Burke DS, Heneine W. 2004. Naturally acquired simian retrovirus infections in central African hunters. The Lancet, 363 (9413): 932- 937. [DOI] [PubMed] [Google Scholar]
- 27. Zennou V, Bieniasz PD. 2006. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology, 349 (1): 31- 40. [DOI] [PubMed] [Google Scholar]
- 28. Zhang GH, Li MH, Zheng YT. 2007. Application of AIDS macaque animal model in HIV vaccine research. Zoological Research, 28 (5): 556- 562. [Google Scholar]
- 29. Zhang JZ, Webb DM. 2004. Rapid evolution of primate antiviral enzyme APOBEC3G. Human Molecular Genetics, 13 (16): 1785- 1791. [DOI] [PubMed] [Google Scholar]
- 30. Zhang MX, Zheng HY, Jiang J, Pang W, Zhang GH, Zheng YT. 2016. Viral seroprevalence in northern pig-tailed macaques (Macaca leonina) derived from Ho Chi Minh City, Vietnam. Primates, [DOI] [PubMed] [Google Scholar]
- 31. Zhang XL, Pang W, Deng DY, Lv LB, Feng Y, Zheng YT. 2014. Analysis of immunoglobulin, complements and CRP levels in serum of captive northern pig-tailed macaques (Macaca leonina). Zoological Research, 35 (3): 196- 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng HY, Zhang MX, Zhang LT, Zhang XL, Pang W, Lyu LB, Zheng YT. 2014. Flow cytometric characterizations of leukocyte subpopulations in the peripheral blood of northern pig-tailed macaques (Macaca leonina). Zoological Research, 35 (6): 465- 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu L, Lei AH, Zheng HY, Lyu LB, Zhang ZG, Zheng YT. 2015. Longitudinal analysis reveals characteristically high proportions of bacterial vaginosis-associated bacteria and temporal variability of vaginal microbiota in northern pig-tailed macaques (Macaca leonina). Zoological Research, 36 (5): 285- 298. [DOI] [PMC free article] [PubMed] [Google Scholar]


