Abstract
In the past three years, RNA-guided Cas9 nuclease from the microbial clustered regularly interspaced short palindromic repeats (CRISPR) adaptive immune system has been used to facilitate efficient genome editing in many model and non-model animals. However, its application in nonhuman primates is still at the early stage, though in view of the similarities in anatomy, physiology, behavior and genetics, closely related nonhuman primates serve as optimal models for human biology and disease studies. In this review, we summarize the current proceedings of gene editing using CRISPR/Cas9 in nonhuman primates.
Keywords: CRISPR/Cas9, Non-human primates, Transgene, paCas9, Animal ethics
INTRODUCTION
The emergence of recombinant DNA technology in the 1970s marked the beginning of a new era for biology. With advances in recent years, however, a series of programmable nucleasebased genome editing technologies have been developed, which enable targeted and efficient modification of the genomes of a variety of species. The most rapidly developing genome editing technique is the RNA-guided endonucleases known as Cas9, originally derived from the microbial adaptive immune system CRISPR (clustered regularly interspaced short palindromic repeats), which can target virtually any genomic location of choice (Brouns et al., 2008; Cong et al., 2013). CRISPR nuclease Cas9 is targeted by a short guide RNA, which recognizes the target DNA via Watson-Crick base pairing, and can generate DNA double-strand breaks (DSB) (Nishimasu et al., 2013). Cas9-induced DSBs have been used to introduce non-homologous end joining (NHEJ)-mediated insertiondeletion (indel) mutations as well as to stimulate homologous DNA recombination (HDR) with both double-stranded plasmid DNA and single-stranded oligonucleotide donor templates (Ran et al., 2013).
Compared with mice, nonhuman primates are genetically and phenotypically closer to humans, particularly in regards to anatomy, physiology, cognition and gene sequences (Zhang et al., 2014). They are, therefore, optimal animal models for genetic modification in an attempt to understand human biology, especially in neurobiology and human evolution. At the same time, however, we must address the potential ethical issues involved due to the potential off-target effect when using the CRISPR/Cas9 tool for genome editing.
GENERATION OF GENETICALLY MODIFIED MONKEYS VIA CRISPR/CAS9
In 2001, the first transgenic monkey (rhesus, Macaca mulatta) was born in the USA, thus demonstrating the plausibility of modifying the primate genome (Chan et al., 2001). Until 2013, however, progress in regards to the generation of transgenic monkeys was rather slow. Firstly, compared with the widely used gene editing tools in model species (e.g., mice), few techniques existed for genetically manipulating embryonic stem cells or germline cells of monkeys. Furthermore, it was difficult to use traditional gene targeting technology to establish primate animal models. In 2014, however, thanks to the emergence of the Cas9-RNA-mediated gene targeting technique, Chinese scientists generated the first gene knockout cynomolgus monkey via targeting one-cell embryos using CRISPR/Cas9 (Niu et al., 2014), opening the door to primate genome manipulation. Now, the use of the CRISPR/Cas9 tool to generate genetically modified nonhuman primates has greatly intensified.
In rodents, the CRISPR/Cas9 system has achieved highly-efficient directed modification of the genome and regulation of gene expression (Perez-Pinera et al., 2013; Sander & Joung, 2014; Wang et al., 2013; Yang et al., 2013; Yin et al., 2014). In monkeys, however, genome targeting efficiency is still low. Several gene knockout studies in monkeys have been published, but successful gene replacement in monkeys via the CRISPR/Cas9 system remains elusive, possibly due to the complexity of DNA repair mechanisms in monkeys. In this review, we summarize the previous work done and discuss the existing problems in current primate genome modification and the potential direction for future research.
Recent research in generating gene knockout monkeys has shown considerable progress, with gene knockout efficiency becoming similar to that achieved in mice. However, there are still limitations in regards to monkey species (Guo & Li, 2015). First of all, the genome sequences of published monkey species (e.g., rhesus macaque, cynomolgus monkey and common marmoset) are still of low quality, thereby hindering the design of sgRNAs based on genome sequences. For example, when using genome sequence data from the NCBI or Ensemble databases, -AGG must be selected as the PAM sequence, despite the real read being -ATG, which results in failed sgRNA. Second, nonhuman primates require much more time to reach sexual maturity than that required by mice. For example, rhesus macaques need 4-5 years to obtain sexual maturity. Hence, it is not plausible to use the breeding strategies of mice to generate homozygous genetically modified monkeys. To overcome this problem, researchers have tried to improve efficiency by increasing the concentrations of Cas9 mRNA and sgRNA components when they inject primate embryos. While certain increases in the injection concentration can indeed improve gene targeting efficiency, high concentrations can also produce toxicity, and thus interfere with embryonic development (Table 1). In the DMD gene targeting case, the efficiency of embryos developing into morulae or blastocysts is only 23.9% (Chen et al., 2015).
Table 1.
Reported CRISPR/Cas9 edited genes in monkey embryos
| Targeted gene | Cas9mRNA: sgRNA (ng/μL) | Efficiency in embryos (%) | Morulae and Blastocysts (%) | Off-target mutation | References |
| Nr0b1 | 20: 5 | 26.6 | 15/22(68.1) | No | Niu et al., 2014 |
| Ppar-γ | 46.7 | ||||
| Rag1 | 60 | ||||
| p53 | 20: 5 | 62.5 | 8/12(66.7) | No | Wan et al., 2014 |
| 100: 10 | 100 | 10/15(66.7) | |||
| 200: 10 | 100 | 5/16(31.25) | |||
| DMD | 200: 25 | 46.47 | 34/142(23.9) | No | Chen et al., 2015 |
The efficiency of gene replacement (or knockin) in primate embryos also remains very low. Because primate embryos are very expensive, and difficult to obtain in large numbers, we need to learn from practice with other animal species. To improve the efficiency of gene replacement or gene homologous recombination, a growing number of methods have been developed. Researchers have tried inhibitors of the NHEJ-repair pathway enzyme to inhibit DNA repair in order to, in theory, raise the efficiency of homologous recombination through the HDR-repair pathway. Recently, Dr. Ploegh's team discovered inhibitor Scr7, which significantly improved homologous repair efficiency in mice, exhibiting 19 times greater efficiency compared with that of the control group (Maruyama et al., 2015). Based on the same strategy, researchers have also tested inhibitors that hinder key enzymes of the NHEJ-repair pathway, including KU70, KU80 and DNA ligase Ⅳ, to improve the efficiency of homologous recombination. Furthermore, Dr. Rajewsky's team used shRNA and Ad4s or shRNA, Ad4s and Scr7 to improve the efficiency of homologous recombination (Chu et al., 2015). In monkeys, we tested inhibitor Scr7, and although it inhibited NHEJ repair, we did not observe an increase in homologous recombination for improving replacement efficiency, implying that different enzymes or mechanisms might be involved in embryos of different species.
It was recently reported that small molecules (e.g., L755507 and Brefeldin A) can enhance CRISPR-mediated HDR efficiency, 3-fold for large fragment insertions and 9-fold for point mutations (Yu et al., 2015). As these small molecules are generally non-toxic, they may serve as a better choice for gene replacement manipulation in monkeys.
Alternatively, the concentration of the homologous repair template can be adjusted to achieve an optimal ratio between the repair template and target genomic segment, and therefore improve homologous recombination efficiency. A recent study found that a higher concentration of ssODN remarkably reduced HDR-derived mutations in pig zygotes, suggesting a possible balance for optimal HDR-derived mutations in zygotes between the excessive accessibility to HDR templates and the activities of HDR relative to NHEJ, which are negatively correlated to ssODN concentration (Zhou et al., 2016). In the previous case, the efficiency of homologous recombination achieved 80% following injection of the homologous repair template at a low concentration in the pig embryos. Again, these data suggest between-species differences for improving homologous recombination.
The CRISPR/Cas9 system consisting of Cas9 nuclease and sgRNA is the most commonly used approach in genome editing due to its high convenience and robust targeting (Doudna & Charpentier, 2014). However, it is still unknown whether sgRNA works more efficiently than dual-crRNA: tracrRNA, especially for the production of knockin animals. The highly efficient generation of knockin mice carrying a functional gene cassette using a cloning-free CRISPR/Cas9 system was achieved by combining the Cas9 protein with chemically synthesized dual-crRNA: tracrRNA, with conventional mRNA pronuclear injection or Cas9 protein injection combined with sgRNA being unsuccessful (Aida et al., 2015). Additionally, chemically modified guide RNAs were found to enhance CRISPR/Cas9 genome editing in human primary cells (Hendel et al., 2015), especially 2'-O-methyl 3' phosphorothioate (MS) or 2'-O-methyl 3' thioPACE (MSP) modified sgRNAs. Besides sgRNA, modified ssODNs have demonstrated superior genome editing efficacy (Renaud et al., 2016), possibly by protecting ssODNs from degradation.
Figure 1.
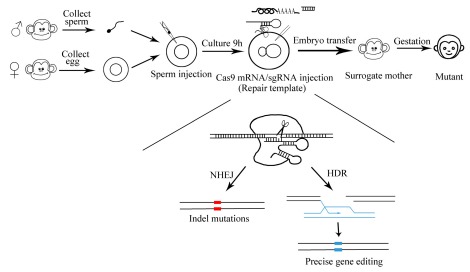
Establishment of nonhuman primate models via CRISPR/Cas9
Figure 2.
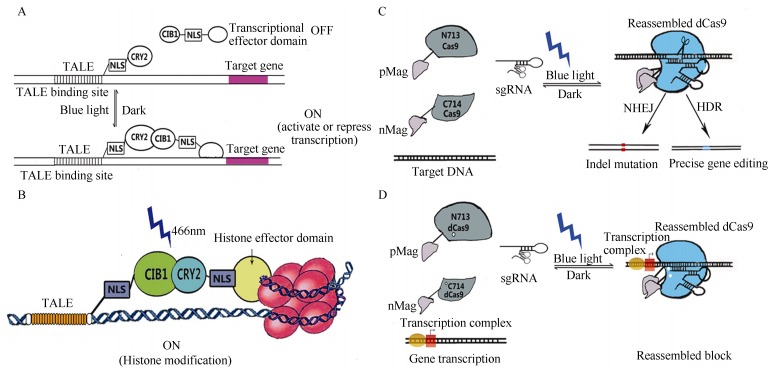
Schematic of the TALEN-LITE system (A, B) and photoactivatable Cas9 (paCas9) (C, D)
Researchers have also optimized the structure of Cas9 to improve knockin efficiency. Dr. Xue and colleagues from Tsinghua University added a "GGSGP" linker and HA tag to the C-terminal of Cas9, which had a profound effect on both editing efficiency and accuracy (Zhao et al., 2016). They also developed a structurally optimized sgRNA, sgRNA(F+E), in which "F" is an A-U base pair flip that can destroy a potential polymerase Ⅲ terminator (UUUU) and "E" is a 5-bp extension of the Cas9-binding hairpin structure that likely improves the assembly of the sgRNA/Cas9 complex. Furthermore, they showed that using sgRNA(F+E) with modified Cas9 led to a 32-fold increase in editing efficiency in the Caenorhabditis elegans genome. Similar strategies may be used to improve the knockin efficiency in monkeys.
Collectively, the newly developed CRISPR/Cas9 systems hold great promise for application in nonhuman primates to generate knockin models of human diseases or to modify specific genes.
OFF-TARGET EFFECTS AND MOSAIC MUTATIONS CAUSED BY CSRISP/CAS9
The specificity of CRISPR/Cas9 targeting relies on 20 bp guide RNA and PAM. There are many guide RNA-like sequences in the genome. Consequently, CRISPR/Cas9 may generate a number of nonspecific mutations, leading to off-target effects. Although the published data has not reported detectable offtarget effects in the CRISPR/Cas9 derived gene knockout monkeys, this might be due to the limited number of genes studied. Human tripronuclear zygotes injected with CRISPR/Cas9 showed an off-target effect (Liang et al., 2015), which could confound the phenotypes of founder animals. Hence, many methods have been developed to reduce offtarget effects (Koo et al., 2015).
The off-target effect of Cas9 can be reduced to the detection limit of deep sequencing by choosing unique target sequences in the genome and modifying both guide RNA and Cas9. For example, paired Cas9 nickases, composed of D10A Cas9 and guide RNA, which generate two single-strand breaks (SSBs) or nicks on different DNA strands, are highly specific in human cells, thus avoiding off-target mutations without sacrificing genome-editing efficiency (Cho et al., 2014). sgRNAs with 5'-GG-can greatly reduce the off-target effect. In addition, using truncated guide RNAs can also improve CRISPR/Cas9 specificity (Fu et al., 2014). The off-target effect can be induced by the high concentration of Cas9 and sgRNA, suggesting that an optimal concentration of Cas9 and sgRNA should be considered for reducing off-target effect.
Another important issue with CRISPR/Cas9 is mosaic mutations. The CRISPR/Cas9 system can repeatedly target genes at different stages of embryonic development, which could lead to mosaicism of the introduced mutation (s). It was speculated that mosaicism might result from a prolonged expression of Cas9 mRNA. However, direct injection of the Cas9 protein rather than Cas9 mRNA into cells can also lead to mosaic mutations (Kim et al., 2014; Sung et al., 2014). To understand its mechanism, however, rigorous quantification of mosaicism from one-cell to multiple-cell embryos is required.
PACAS9: CRISPR/CAS9 MEETS OPTOGENETICS
Compared with rodents, nonhuman primates have bigger and more complex brains, and their neurogenesis is more similar to that of humans. For example, during neurogenesis, macaques have an outer subventricular zone (OSVZ) like humans, but rodents do not have this cell layer in their developing brains (Betizeau et al., 2013). Researchers have been hoping to manipulate the brains of living nonhuman primates by changing the expression of certain genes under controlled spatial and temporal conditions in order to observe the brain's neural network connections. The pathogenesis of human brain diseases can be clearly revealed via this approach; however, traditional technology has thus far been unable to complete this task.
In 2013, Dr. Zhang and colleagues from the Broad Institute combined the TALENs gene targeting technology and optogenetics, and developed a new system for manipulating living brain gene expression in mice (Konermann et al., 2013). Furthermore, they also developed light-inducible transcriptional effectors (LITEs), an optogenetic two-hybrid system integrating the customizable TALEN DNA-binding domain with the light-sensitive cryptochrome 2 protein (CRY2) and its interacting partner CIB1 from Arabidopsis thaliana (Kennedy et al., 2010; Liu et al., 2008). LITEs do not require additional exogenous chemical cofactors, and can be easily customized to target and activate many endogenous genomic loci within minutes. This process is also reversible and LITEs can also be packaged into viral vectors and target specific cell populations.
Similarly, Dr. Sato and colleagues from the University of Tokyo attempted to combine the Cas9 protein with CRY2 to modify the genome of living cells or animals; however, they found that Cas9 could not work when combined with CRY2. They next developed the paCas9 system using pairs of photoswitching proteins called Magnets that use electrostatic interactions to join proteins when activated by light (Kawano et al., 2015; Nihongaki et al., 2015a, b). They first created paCas9 by splitting the Cas9 protein into two inactive fragments, and then coupled each fragment with one Magnet protein of a pair. When irradiated with blue light, the Magnets come together so that the split Cas9 fragments merge to reconstitute the nuclease's RNA-guided activity. Importantly, this process is reversible, and when the light is turned off, the paCas9 nuclease splits again, and nuclease activity is halted.
The spatiotemporal and reversible properties of paCas9 are well suited for the dissection of causal gene functions in diverse biological processes and for medical applications, such as in vivo and ex vivo gene therapies. Furthermore, paCas9 also has the potential of reducing off-target indel frequencies in Cas9-based genome editing. There have been several studies showing that transient introduction of a Cas9: sgRNA complex prepared in vitro can improve the specificity of genome editing (Kim et al., 2014; Ramakrishna et al., 2014; Zuris et al., 2015). Because paCas9 can be switched off by stopping light irradiation, optically controlling the duration of paCas9 activation would contribute to reducing off-target effects. Some scientists have also combined dCas9, an epigenetic effector, with paCas9 to expand the application of this system, making it more useful in studies of the brain.
ETHICAL CONSIDERATIONS IN GENETIC MODIFICATION OF NONHUMAN PRIMATES
The CRISPR/Cas9 technology has gained considerable attention recently due to debate among scientists about the possibility of genetically modifying the human germ line and the ethical implications of doing so (Caplan et al., 2015; Holdren et al., 2015; Lanphier et al., 2015). Researchers in China have used CRISPR/Cas9 to alter a gene in a human embryo that can cause a blood disorder when mutated. The US National Institutes of Health (NIH), however, has reaffirmed its ban on research that involves genetic editing of human embryos due to their concern about the safety of the technique and the ethical implications of altering genes that will be passed to future generations. It has been pointed out that there are few clinical situations in which gene editing would be the only way to prevent the passage of a genetic disease from parent to child. Instead, parents with a genetic disease could create embryos in vitro and screen them for the presence of the defect gene.
Similarly, we must consider the potential ethical issues when we generate genetically modified nonhuman primates using CRISPR/Cas9. The use of animals in research must be justified in terms of the value of the research for understanding fundamental biological processes and ameliorating devastating human diseases. Scientists must consider whether experimental alternatives exist and whether the species used are appropriate to the specific problem being studied (Blakemore et al., 2012; Belmonte et al., 2015). For example, the genetic and mechanistic determination of certain human neurological and psychiatric diseases might be better approximated by nonhuman primate models.
Bearing these considerations in mind, we see the weight of the argument in favor of moving forward on transgenic nonhuman primate disease models with due care, responsibility and transparency.
CONCLUSIONS
Nonhuman primates and humans share many anatomical, perceptual, cognitive and behavioral traits. Recent advances in CRISPR/Cas9 gene-editing techniques have made it possible to create genetically modified nonhuman primates, opening up new and exciting ways to gain insight into the primate brain. However, many problems remain to be solved. The efficiency of homozygous knockout and homologous recombination is still low, and off-target effects and mosaic mutations caused by the CRISPR/Cas9 system need to be improved. In future studies, researchers should attempt to understand the potentially primate-specific aspects to optimize the gene targeting system. China has rich nonhuman primate resources, which could allow for the full use of the latest technological advancements. China is positioning itself as a world leader in primate research, with a new national monkey facility currently being constructed at the Kunming Institute of Zoology, Chinese Academy of Sciences, and similar projects being launched in Shanghai, Beijing, Guangzhou and Shenzhen (Cyranoski, 2016).
Funding Statement
the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13010000) and the National Natural Science Foundation of China (31130051)
REFERENCES
- 1. Aida T, Chiyo K, Usami T, Ishikubo H, Imahashi R, Wada Y, Tanaka KF, Sakuma T, Yamamoto T, Tanaka K. 2015. Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biology, 16 87- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Belmonte JCI, Callaway EM, Caddick SJ, Churchland P, Feng G, Homanics GE, Lee KF, Leopold DA, Miller CT, Mitchell JF, Mitalipov S, Moutri AR, Movshon JA, Okano H, Reynolds JH, Ringach D, Sejnowski TJ, Silva AC, Strick PL, Wu J, Zhang F. 2015. Brains, genes, and primates. Neuron, 86 (3): 617- 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Ménard A, Afanassieff M, Huissoud C, Douglas RJ, Kennedy H, Dehay C. 2013. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron, 80 (2): 442- 457. [DOI] [PubMed] [Google Scholar]
- 4. Blakemore C, Clark JM, Nevalainen T, Oberdorfer M, Sussman A. 2012. Implementing the 3Rs in neuroscience research: a reasoned approach. Neuron, 75 (6): 948- 950. [DOI] [PubMed] [Google Scholar]
- 5. Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science, 321 (5891): 960- 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caplan AL, Parent B, Shen M, Plunkett C. 2015. No time to waste-the ethical challenges created by CRISPR: CRISPR/Cas, being an efficient, simple, and cheap technology to edit the genome of any organism, raises many ethical and regulatory issues beyond the use to manipulate human germ line cells. EMBO Reports, 16 (11): 1421- 1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan AWS, Chong KY, Martinovich C, Simerly C, Schatten G. 2001. Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science, 291 (5502): 309- 312. [DOI] [PubMed] [Google Scholar]
- 8. Chen YC, Zheng YH, Kang Y, Yang WL, Niu YY, Guo XY, Tu ZC, Si CY, Wang H, Xing RX, Pu XQ, Yang SH, Li SH, Ji WZ, Li XJ. 2015. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Human Molecular Genetics, 24 (13): 3764- 3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. 2014. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Research, 24 (1): 132- 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R. 2015. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nature Biotechnology, 33 (5): 543- 548. [DOI] [PubMed] [Google Scholar]
- 11. Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science, 339 (6121): 819- 823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cyranoski D. 2016. Monkey kingdom-China is positioning itself as a world leader in primate research. Nature, 532 (7599): 300- 302. [DOI] [PubMed] [Google Scholar]
- 13. Doudna JA, Charpentier E. 2014. The new frontier of genome engineering with CRISPR-Cas9. Science, 346 (6213): 1258096- [DOI] [PubMed] [Google Scholar]
- 14. Fu YF, Sander JD, Reyon D, Cascio VM, Joung JK. 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature Biotechnology, 32 (3): 279- 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo XY, Li XJ. 2015. Targeted genome editing in primate embryos. Cell Research, 25 (7): 767- 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, Bacchetta R, Tsalenko A, Dellinger D, Bruhn L, Porteus MH. 2015. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nature Biotechnology, 33 (9): 985- 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holdren JP, Shelanski H, Vetter D, Goldfuss C. 2015. Improving transparency and ensuring continued safety in biotechnology. Office of Science and Technology Policy.
- 18. Kawano F, Suzuki H, Furuya A, Sato M. 2015. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nature Communications, 6 6256- [DOI] [PubMed] [Google Scholar]
- 19. Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. 2010. Rapid blue-light-mediated induction of protein interactions in living cells. Nature Methods, 7 (12): 973- 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim S, Kim D, Cho SW, Kim J, Kim JS. 2014. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Research, 24 (6): 1012- 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. 2013. Optical control of mammalian endogenous transcription and epigenetic states. Nature, 500 (7463): 472- 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koo T, Lee J, Kim JS. 2015. Measuring and reducing off-target activities of programmable nucleases including CRISPR-Cas9. Molecules and Cells, 38 (6): 475- 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lanphier E, Urnov F, Haecker SE, Werner M, Smolenski J. 2015. Don't edit the human germ line. Nature, 519 (7544): 410- 411. [DOI] [PubMed] [Google Scholar]
- 24. Liang PP, Xu YW, Zhang XY, Ding CH, Huang R, Zhang Z, Lv J, Xie XW, Chen YX, Li YJ, Sun Y, Bai YF, Zhou SY, Ma WB, Zhou CQ, Huang JJ. 2015. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein & Cell, 6 (5): 363- 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu HT, Yu XH, Li KW, Klejnot J, Yang HY, Lisiero D, Lin CT. 2008. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science, 322 (5907): 1535- 1539. [DOI] [PubMed] [Google Scholar]
- 26. Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. 2015. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nature Biotechnology, 33 (5): 538- 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nihongaki Y, Kawano F, Nakajima T, Sato M. 2015. a. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nature Biotechnology, 33 (7): 755- 760. [DOI] [PubMed] [Google Scholar]
- 28. Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M. 2015. b. CRISPR-Cas9-based photoactivatable transcription system. Chemistry & Biology, 22 (2): 169- 174. [DOI] [PubMed] [Google Scholar]
- 29. Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell, 156 (5): 935- 949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Niu YY, Shen B, Cui YQ, Chen YC, Wang JY, Wang L, Kang Y, Zhao XY, Si W, Li W, Xiang AP, Zhou JK, Guo XJ, Bi Y, Si CY, Hu B, Dong GY, Wang H, Zhou ZM, Li TQ, Tan T, Pu XQ, Wang F, Ji SH, Zhou Q, Huang XX, Ji WZ, Sha JH. 2014. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell, 156 (4): 836- 843. [DOI] [PubMed] [Google Scholar]
- 31. Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. 2013. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature Methods, 10 (10): 973- 976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. 2014. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Research, 24 (6): 1020- 1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nature Protocols, 8 (11): 2281- 2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Renaud JB, Boix C, Charpentier M, De Cian A, Cochennec J, Duvernois-Berthet E, Perrouault L, Tesson L, Edouard J, Thinard R, Cherifi Y, Menoret S, Fontaniere S, de Crozé N, Fraichard A, Sohm F, Anegon I, Concordet JP, Giovannangeli C. 2016. Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR-Cas9 nucleases. Cell Reports, 14 (9): 2263- 2272. [DOI] [PubMed] [Google Scholar]
- 35. Sander JD, Joung JK. 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnology, 32 (4): 347- 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sung YH, Kim JM, Kim HT, Lee J, Jeon J, Jin Y, Choi JH, Ban YH, Ha SJ, Kim CH, Lee HW, Kim JS. 2014. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Research, 24 (1): 125- 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wan HF, Feng CJ, Teng F, Yang SH, Hu BY, Niu YY, Xiang AP, Fang WZ, Ji WZ, Li W, Zhao XY, Zhou Q. 2015. One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Research, 25 (2): 258- 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang HY, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell, 153 (4): 910- 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang H, Wang HY, Shivalila CS, Cheng AW, Shi LY, Jaenisch R. 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell, 154 (6): 1370- 1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yin H, Xue W, Chen SD, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. 2014. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nature Biotechnology, 32 (6): 551- 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu C, Liu YX, Ma TH, Liu K, Xu SH, Zhang Y, Liu HL, Russa ML, Xie M, Ding S, Qi LS. 2015. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell, 16 (2): 142- 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang XL, Pang W, Hu XT, Li JL, Yao YG, Zheng YT. 2014. Experimental primates and non-human primate (NHP) models of human diseases in China: current status and progress. Zoological Research, 35 (6): 447- 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao P, Zhang Z, Lv XY, Zhao X, Suehiro SJ, Jiang YN, Wang XQ, Mitani SH, Gong HP, Xue D. 2016. One-step homozygosity in precise gene editing by an improved CRISPR/Cas9 system. Cell Research, 26 (5): 633- 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou XY, Wang LL, Du Y, Xie F, Li L, Liu Y, Liu CH, Wang SQ, Zhang SB, Huang XX, Wang Y, Wei H. 2016. Efficient generation of gene modified pigs harboring precise orthologous human mutation via CRISPR/Cas9-induced homology-directed repair in zygotes. Human Mutation, 37 (1): 110- 118. [DOI] [PubMed] [Google Scholar]
- 45. Zuris JA, Thompson DB, Shu YL, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. 2015. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nature Biotechnology, 33 (1): 73- 80. [DOI] [PMC free article] [PubMed] [Google Scholar]


