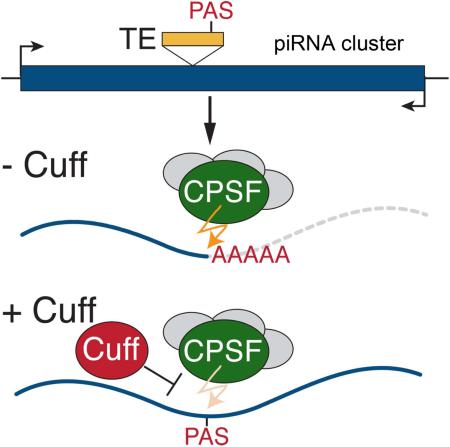
Abstract
Small non-coding RNAs called piRNAs serve as guides for an adaptable immune system that represses transposable elements in germ cells of Metazoa. In Drosophila the RDC complex, composed of Rhino, Deadlock and Cutoff (Cuff) binds chromatin of dual-strand piRNA clusters, special genomic regions, which encode piRNA precursors. The RDC complex is required for transcription of piRNA precursors, though the mechanism by which it licenses transcription remained unknown. Here we show that Cuff prevents premature termination of RNA polymerase II. Cuff prevents cleavage of nascent RNA at poly(A) sites by interfering with recruitment of the cleavage and polyadenylation specificity factor (CPSF) complex. Cuff also protects processed transcripts from degradation by the exonuclease Rat1. Our work reveals a conceptually different mechanism of transcriptional enhancement. In contrast to other factors that regulate termination by binding to specific signals on nascent RNA, the RDC complex inhibits termination in a chromatin-dependent and sequence-independent manner.
Graphical abstract
Introduction
The piRNA pathway functions as an intracellular immune system that recognizes and represses transposable elements (TEs), which are selfish genome invaders that occupy large portions of eukaryotic genomes. Transposon expression is activated in the germline of piRNA pathway mutants leading to accumulation of double-stranded DNA breaks, which are believed to be direct products of transposon mobilization, and ultimately to sterility in flies and mice (Malone and Hannon, 2009; Siomi et al., 2011). Transcripts of active transposons are recognized by complementary 23-30 nucleotide piRNAs that associate with members of the Piwi clade of the Argonaute family proteins. The piRNA-Argonaute complexes ensure post-transcriptional and transcriptional repression of transposable elements. The piRNA pathway is believed to adapt to new transposons by changing the repertoire of piRNAs expressed in germ cells (Aravin et al., 2007). Once acquired, immunity against transposons is transmitted to the progeny.
In Drosophila the most piRNAs are encoded in genomic regions dubbed piRNA clusters (Brennecke et al., 2007). piRNA clusters are transcribed by RNA polymerase II into long non-coding transcripts that are processed into mature 23-30 nucleotide piRNAs in the cytoplasm. Most piRNA clusters in the Drosophila genome are transcribed from both genomic strands and are therefore called dual-strand clusters; the rarer uni-strand clusters are transcribed from one strand.
The adaptation of the piRNA pathway to novel transposons is believed to occur when an active transposon randomly inserts into a piRNA cluster, triggering generation of piRNAs against this element. Indeed, exposure of a naïve genome to P-element led to insertion of this transposon in piRNA clusters (Khurana et al., 2011). Furthermore, insertion of heterologous sequences into piRNA clusters results in generation of new piRNAs that are able to silence expression of these sequences (Muerdter et al., 2012). Thus piRNA clusters are believed to serve as traps for transposon insertions and as a source of piRNAs from any sequence present inside the cluster. The ability to incorporate novel transposon sequences allows piRNA clusters to maintain and update information about genome invaders and to transmit this information to the next generations. Insertions of transposons into piRNA clusters have a potential to interfere with transcription and processing of the long RNA transcribed from these regions. Indeed, transposons carry termination signals that might cause premature termination of transcription. The properties of piRNA clusters that allow them to take advantage of transposon integrations without their transcription being interrupted remain unknown.
Genetic studies, including several whole-genome RNAi screens, identified numerous genes involved in piRNA-mediated repression of transposable elements. Interestingly, a group of proteins composed of the HP1 homologue Rhino (Rhi), the RNA helicase UAP56, and two proteins of unknown function Cutoff (Cuff) and Deadlock (Del) were shown to be essential for piRNA biogenesis from dual-strand, but not uni-strand clusters, indicating that piRNA biogenesis from these two types of clusters is quite different (Chen et al., 2007; Czech et al., 2013; Klattenhoff et al., 2009; Pane et al., 2011; Zhang et al., 2012). Subsequent studies revealed that Rhi, Del, and Cuff form the RDC complex that associates with chromatin of dual-strand, but not uni-strand clusters (Le Thomas et al., 2014; Mohn et al., 2014; Zhang et al., 2014). The chromodomain of Rhi directly binds the H3K9me3 mark, which is enriched in chromatin of dual-strand clusters (Le Thomas et al., 2014; Mohn et al., 2014; Yu et al., 2015). The main function of Rhi seems to be to anchor other components of the RDC complex to chromatin. Del appears to be a scaffold protein bridging Rhi and Cuff, and Cuff is the effector of the molecular function of the RDC complex (Mohn et al., 2014).
Rhi (presumably as a part of RDC complex) is required for transcription of piRNA precursors (Le Thomas et al., 2014; Mohn et al., 2014) and to suppress splicing of piRNA precursor transcripts (Zhang et al., 2014). The molecular mechanism of transcriptional enhancement by RDC is unknown and surprising considering that Rhi is a homolog of HP1, which is a major player in heterochromatin-mediated transcriptional repression. The observation that Cuff is required for read-through transcription and piRNA generation at the hsp70 locus led Mohn and co-authors to hypothesize that the RDC complex licenses transcription by suppressing termination of piRNA precursor transcripts (Mohn et al., 2014). Here, we tested Cuff functions and found that it prevents premature termination of piRNA precursor transcription. Cuff suppresses cleavage of pre-mRNA at canonical poly(A) sites and also suppresses termination of transcripts that have been cleaved at poly(A) sites. Our studies identify a novel mechanism of transcriptional regulation that ensures the function of piRNA clusters as adaptable repositories of information about genome invaders.
RESULTS
Transcription of non-polyadenylated piRNA precursors from dual-strand piRNA clusters is dependent on Cuff
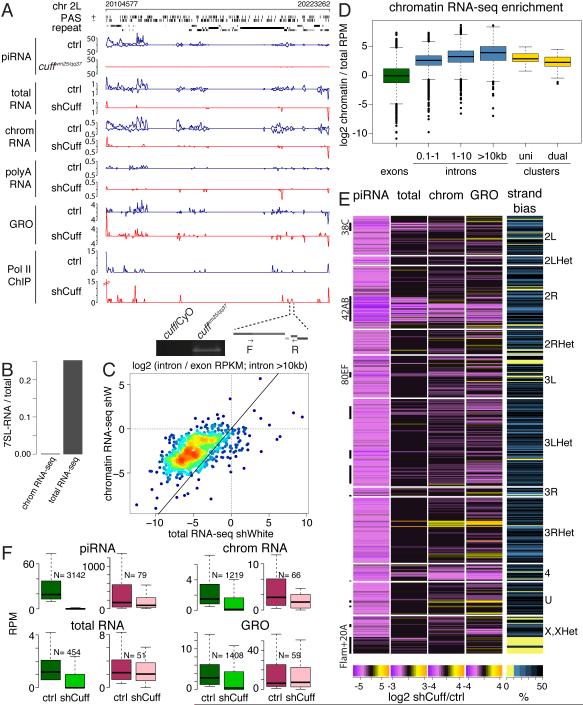
To understand the role of Cuff in piRNA biogenesis we cloned and sequenced libraries of small and long RNAs from ovaries of control and Cuff-depleted flies. cuff was knock-downed in the germline using an shRNA driven by nos-GAL4 (shCuff); shRNA against the white gene, was used as a control (shWhite). As previously reported (Pane et al., 2011), Cuff depletion leads to a dramatic decrease in the amount of mature piRNA generated from dual-strand clusters, such as 42AB and 38C (99% and 98% depletion, respectively), whereas piRNAs from uni-strand clusters were only mildly affected (35% depletion, Fig. 1A and S1A). To comprehensively analyze the effect of Cuff on piRNA generation, we defined 1 kb genomic regions that generate piRNA in ovaries of wild-type flies using a 10 RPKM cutoff. 3142 intervals (~3.1 Mb) generate piRNAs in a Cuff-dependent manner as they show greater than 80% decrease of piRNA upon Cuff knockdown. The majority (83%) of the Cuff-dependent regions generate piRNAs from both genomic strands supporting previous observations that Cuff is specifically required for generation of piRNA from dual-strand clusters (Fig. 1E). 14% of Cuff-dependent regions overlap with the top 10 dual-strand piRNA clusters described previously.
Fig. 1. Cuff deficiency eliminates the majority of transcripts from piRNA clusters.
(A) Cuff depletion leads to diminished piRNA cluster transcription. Shown are profiles of piRNA, total cell and poly(A)-selected RNA-seq, chromatin RNA-seq, global nuclear run-on seq (GRO-seq), and RNA polymerase II ChIP-seq from ovaries of control and cuff depleted flies on the dual-strand piRNA cluster 38C. The distribution of predicted poly(A) signals on plus and minus genomic strands is shown in the poly(A) signal (PAS) tracks. Below is an expansion of the indicated region from which poly-adenylated transcripts detected by RT-PCR using oligo-dT primer are produced in cuffwm25/qq37 flies.
(B) Chromatin-associated RNA sequencing libraries are depleted of signal recognition particle (7SL) RNA.
(C) Chromatin-associated RNAs are enriched in intronic sequences as measured by the ratio of intronic to exonic RPKM signals. Solid line indicates no difference between standard and chromatin RNA-seq.
(D) piRNA cluster transcripts are enriched in chromatin-associated RNA.
(E) Cuff suppresses primary transcripts from Cuff-depended piRNA producing loci. Heatmap shows signal change in Cuff-depleted versus control ovaries for small RNA, total RNA, chromatin-associated RNA and GRO-seq in genomic windows that produce piRNA in a Cuff-dependent manner. Intervals overlapping uni-strand piRNA clusters that are not affected by Cuff depletion are shown for comparison (Flam+20A).
(F) Box plots show the distribution of piRNA, chromatin-associated RNA, total RNA and GRO signals in control and Cuff-depleted ovaries. Green boxes corresponds to all genomic regions that produce piRNA in Cuff-dependent manner; pink boxes correspond to windows overlapping uni-strand piRNA clusters (flamenco and 20A). See also Figure S1.
Long transcripts from both strands of dual-strand piRNA clusters were readily detected in control flies when total RNA was used as a source to prepare the RNA-seq library. However, when libraries were prepared from poly(A)-selected RNA, almost no signal from dual-strand piRNA clusters was detected (Fig. 1A). This suggests that the vast majority of dual-strand piRNA cluster transcripts are not polyadenylated. In agreement with a previous study (Goriaux et al., 2014), we found that uni-strand piRNA clusters produce polyadenylated piRNA precursors, indicating that there is a difference in the mode of transcription between the two types of genomic regions that generate piRNAs (Fig. 1A and S1A). To further explore the nature of piRNA precursors before their processing into mature piRNA, we isolated and sequenced chromatin-associated nascent transcripts (chromatin RNA-seq) from fly ovaries. We first explored the composition of chromatin-associated RNA libraries to validate the isolation of transcripts from this fraction. 7SL RNA, a component of the cytoplasmic SRP is over 300-fold depleted in chromatin-bound RNA compared to total cellular RNA (Fig. 1B). Furthermore, chromatin-bound RNA-seq libraries were strongly enriched in intronic reads from protein-coding pre-mRNAs. Enrichment of intronic reads was particularly strong when genes with long introns were analyzed: for genes with intronic regions longer than 10 kB, the ratio of intron/exon RPKM was increased on average ~7-fold in chromatin-bound compared to total cellular RNA (Fig. 1C). This result indicates that, as expected, the chromatin-bound RNA fraction contains nascent pre-mRNAs. Together, analyses of standard RNA-seq libraries from chromatin-bound RNA indicated specific purification of nuclear transcripts. We found that, similarly to introns of protein-coding genes, transcripts from piRNA clusters are enriched on chromatin (Fig. 1D). Due to enrichment of piRNA precursors in the chromatin fraction, chromatin RNA-seq provides deeper coverage of piRNA precursors than standard RNA-seq libraries prepared from total tissue, where piRNA precursors are not abundant.
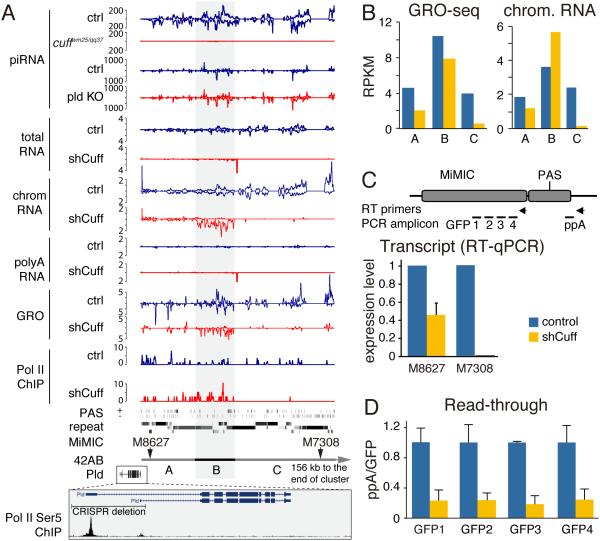
Over 80 % of the genomic regions that require Cuff for piRNA generation show loss of long RNA precursors as measured by both total cell and chromatin RNA-seq. In total RNA the median expression dropped from 1.2 RPKM to zero, while it was reduced ~12-fold in chromatin RNA-seq (Fig. 1E, F). However, loss of long RNA upon Cuff depletion was not universal even within a single piRNA cluster. One example is provided by the major 42AB dual-strand piRNA cluster that spans ~250 kB and has an overall ~10 fold decrease in piRNA precursors in chromatin-associated RNA-seq. Examination of 42AB piRNA precursors showed that the cluster can be divided into three distinct regions (Fig 2A, S2A). The longest (~200 kB) region C shows almost complete elimination (~95%) of transcripts upon Cuff depletion, while transcript levels drops less than 2-fold in the ~24 kB region A (Fig 2A, B). Surprisingly, the ~18 kB region B continues to generate transcripts that are slightly more abundant upon Cuff depletion compared to wild-type (Fig 2A, B). However, Cuff affects transcription pattern even in this region: while in wild-type flies both genomic strands generate transcripts, only one strand is expressed upon Cuff depletion. Remarkably, despite the observation that Cuff-depleted flies have high level of transcripts from region B, almost no piRNA are expressed, indicating that in the absence of Cuff these RNAs are not processed into piRNA. Overall, we conclude that the strong decrease in piRNA precursor abundance is likely a major reason for the lack of mature piRNA in Cuff mutants.
Fig. 2. Cuff suppresses transcription termination at canonical poly(A) sites.
(A) Profiles of different libraries over the 42AB cluster which is further divided into thee regions, A, B and C. Below shown are the locations of two insertions of artificial MiMIC transposons, and the site of CRISPR-mediated deletion of promoters of the Pld gene, which flanks the 42AB cluster. The entire cluster is shown on Figure S2A.
(B) Cuff depletion leads to reduced transcription of the 42AB cluster. Normalized densities of GRO-seq and chromatin RNA-seq signals are calculated for three regions shown in (A).
(C) Depletion of Cuff has a different effect on two promoterless MiMIC insertions located at different positions in the 42AB cluster. Shown is the amount of MiMIC RNA in control and cuff-depleted (shCuff) ovaries as determined by RT-qPCR. Error bars show standard errors from three biological replicates.
(D) Depletion of Cuff suppresses read-through transcription downstream of the canonical poly(A) site of the M8627 insertion. Amounts of RNA upstream (GFP1-4) and downstream (ppA) of the poly(A) site were quantified by strand-specific RT-qPCR and the ratios of read-through (ppA/GFP1-4) transcripts were plotted. RT primers and qPCR amplicons are shown in panel C. Error bars show standard errors from three biological replicates. See also Figure S2B.
A decrease of piRNA precursors can be caused by rapid post-transcriptional degradation or by inhibition of transcription. To discriminate between these two possibilities, we performed nuclear run-on followed by RT-qPCR and global nuclear run-on (GRO-seq) to measure transcriptional activity in ovaries from control and Cuff-depleted flies. Nuclear run-on and GRO-seq both showed that transcription of dual-strand clusters is decreased in Cuff depleted ovaries (Fig. 1A, E, F, 2A, B, S1B). Cuff depletion did not greatly decrease levels of transcription on the uni-strand piRNA clusters 20A and flamenco (Fig. 1E,F, S1B). These results show that Cuff is essential for transcription of dual-strand, but not uni-strand piRNA clusters.
Analyzing poly(A) selected RNA sequencing data after Cuff knockdown led to a surprising observation: in the Cuff-depleted ovaries, polyadenylated transcripts that were absent in control flies appeared. These transcripts correspond to a few distinct positions along the piRNA clusters (Fig. 1A). A similar phenomenon was also seen in the cuffwm25/qq37 mutant (data not shown). We used independent RT-PCR to confirm the existence of poly(A)+ RNA in the cuffwm25/qq37 flies (Fig. 1A). Cloning and Sanger sequencing of the RT-PCR product confirmed that the poly(A)+ transcripts that were exclusively detected in cuff wm25/qq37 flies span several TE fragments within the piRNA cluster. Nuclear run-on and GRO-seq results show that depletion of Cuff leads to a decrease in transcription from dual-strand clusters (Fig. 1A-C); however, transcription does not stop completely. Instead, certain regions within clusters produce poly(A)+ as well as poly(A)-RNA upon Cuff depletion. These results indicate that cuff deficiency causes a switch from read-through transcription, which generates non-polyadenylated RNAs from the entire length of the piRNA cluster, to formation of a few RNAs that are transcribed from within the cluster.
Cuff suppresses transcription termination at canonical poly(A) sites
Multiple poly(A) sequence motifs (PAS), AAUAAA, are present on both genomic strands of dual-strand clusters and these motifs are often located inside fragments of transposons (Fig. 1A). In wild-type flies, non-polyadenylated RNA transcripts are generated from the entire length of dual-strand clusters despite the presence of these motifs. It is possible that the transcribing polymerase does not recognize these motifs as signals for termination and polyadenylation because additional important sequence elements are not present. Alternatively, as proposed by Mohn an co-authors (Mohn et al., 2014), termination and polyadenylation on even canonical poly(A) sites embedded in dual-strand clusters may be suppressed. To discriminate between these possibilities, we analyzed transcription of transgenes with strong, well-characterized poly(A) signals that have been inserted into piRNA clusters. By analysis of a collection of strains with insertions of the MiMIC transgene, a transgene that carries an SV40 poly(A) signal (Venken et al., 2011), we found two strains that have identical MiMIC insertions located in two different positions inside the dual-strand cluster 42AB (Fig. 2A). The transgenes do not have their own promoters and are transcribed as parts of long transcripts in ovaries of control flies.
Interestingly, in agreement with differential effect of Cuff depletion on transcription inside different regions of the 42AB cluster (Fig. 2A, B), knock-down of cuff also has different effects on transcription of the two MiMIC insertions. Expression of transgene M8627, which is located in region A close to the flank of the cluster, 6.5 kb downstream of the Pld gene, was reduced ~2-fold, whereas expression of transgene M7308, which is located deep inside the cluster in region C (83 kb from the Pld gene) was almost completely eliminated (Fig. 2C). The difference in transgene expression correlates with the efficiency of transcription in these two regions as measured by GRO-seq (Fig. 2A, B). GRO-seq signal was only modestly affected in region A that harbors insertion M8627. In contrast, Cuff depletion led to about a 10-fold decrease in GRO-seq signal in the internal region C that contains M7308. These data confirm that the effect of Cuff on transcription is not uniform along the length of the piRNA cluster.
Taking advantage of the fact that transgene M8627 continues to be transcribed upon Cuff depletion we analyzed the effect of Cuff on termination at the canonical poly(A) site of this transgene. Strand-specific RT-qPCR was used to measure transcript levels at several regions upstream and downstream of the poly(A) signal in ovaries of control and Cuff-depleted flies (Fig. 2C). To determine efficiency of read-through transcription we calculated the ratio of transcript abundance downstream and upstream of the poly(A) cleavage site. Upon Cuff depletion this ratio is decreased ~5-fold indicating that read-through transcription is suppressed (Fig. 2D). Importantly, our results argue against gradual termination as four amplicons upstream of the poly(A) site showed the same ratios suggesting that increase in termination in cuff knock-down occurs at or very close to the poly(A) site. To quantify the efficiency of termination we created qPCR standard curves by amplifying the same fragments from a DNA template and calculating the absolute transcripts levels (Fig. S2B). This analysis showed that in wild-type flies 24% of transcripts do not terminate at the poly(A) site, while Cuff depletion reduced the fraction of read-through transcripts to 5.6%. Thus, the presence of Cuff leads to high levels of transcription through the canonical SV40 poly(A) signal. Together, these data suggest that Cuff promotes transcription of long piRNA precursors by preventing termination.
It is not clear where transcription of piRNA precursors is initiated as, in contrast to uni-strand clusters, dual-strand piRNA clusters, including 42AB, lack obvious promoters marked by characteristic chromatin signatures of high Pol II Ser5 and H3K4me3 peaks (Mohn et al., 2014, our unpublished data). Therefore, it was proposed that transcription of piRNA precursors might be initiated on promoters of flanking protein-coding genes followed by read-through transcription that ignores the genes polyA/termination signal (Mohn et al., 2014). The Cuff-dependent suppression of termination we observed provides support for this hypothesis, suggesting that Cuff might ensure read-through transcription from flanking protein-coding genes towards the cluster. We detected read-through transcripts spanning the 3’UTR of the flanking Pld mRNA and extending a few hundred nucleotides downstream of the poly(A) site into the 42AB cluster by RT-PCR in ovaries (data not shown). Therefore, we decided to test if Pld supplies the promoter for transcription of 42AB piRNA precursors. We deleted a 4.8 kB region that includes two alternative promoters of the Pld gene using CRISPR-mediated genome engineering (Fig. 2A). Though expression of the Pld gene is abolished in flies homozygous for the deletion, expression of piRNA from the 42AB cluster remained essentially unperturbed (Fig. 2A). This result shows that, at least in the case of the 42AB cluster and the Pld gene, the expression of piRNA precursors is not driven by read-through transcription of flanking protein-coding gene. Instead, we propose that suppression of termination plays a role primarily within the clusters by facilitating generation of long read-through transcripts.
Activation of piRNA biogenesis by exposure to homologous piRNAs correlates with increased read-through transcription
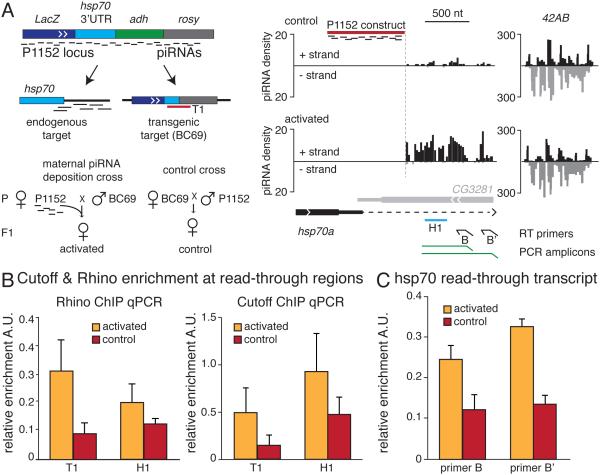
The exposure of a naïve locus to homologous maternally-inherited piRNAs triggers piRNA generation (de Vanssay et al., 2012; Le Thomas et al., 2014). Insertion of the P1152 transgene into a telomeric piRNA cluster leads to production of transgenic piRNAs (Muerdter et al., 2012; Todeschini et al., 2010). When inherited through the maternal germline those transgenic piRNAs trigger piRNA generation from targeted regions, such as other lacZ transgenes as well as the endogenous adh and hsp70 loci (Le Thomas et al., 2014; Muerdter et al., 2012) as shown schematically in Figure 3A. ChIP-qPCR showed that both Cuff and Rhi accumulated on target lacZ and hsp70 loci after piRNA biogenesis was activated as a result of exposure to homologous maternally-inherited piRNA (Fig. 3B). Thus, the presence of Rhi and Cuff proteins on chromatin correlates with piRNA generation from loci exposed to homologous piRNA.
Fig. 3. Exposure of target locus to homologous piRNA enhances read-through transcription.
(A) Exposure to maternally inherited homologous piRNAs activates piRNA biogenesis. The insertion of the P{lArB} construct in a subtelomeric piRNA cluster in P1152 strain produces abundant piRNAs that target homologous loci, the lacZ-containing BC69 transgene and the endogenous hsp70 locus, in trans. piRNA biogenesis from target loci is activated if inducer is inherited from the mother (‘activated’ progeny), but not in control flies produced by the reciprocal cross. The right panel shows the density of piRNAs at the endogenous hsp70 locus in the ovaries of activated and control progenies compared to 42AB cluster.
(B) Rhi and Cuff associate with target loci exposed to homologous piRNA. Binding of Rhi and Cuff to chromatin of the BC69 transgene (T1) and the hsp70 locus (H1) was measured in ovaries of activated and control progenies by ChIP-qPCR using amplicons shown in (A). Error bars represent standard deviations of six technical replicates.
(C) Activation of primary piRNA biogenesis on the hsp70 locus is associated with increased read-through transcription as measured by strand-specific RT-qPCR using primers shown in panel A. Error bars represent the standard deviations of three biological replicates. See also Figure S3.
We used the hsp70 locus as a model to understand how transcription of the target locus changes upon exposure to homologous piRNA. piRNA that seem to be processed from read-through transcripts of the endogenous hsp70Aa gene are generated in wild-type flies without exposure to homologous trans-piRNA. Generation of these piRNA and their precursor read-through transcript of hsp70Aa gene requires Rhi (Mohn et al., 2014) and Cuff (Hur et al., 2016). Exposure to piRNA homologous to the P1152 transgene greatly enhances production of piRNAs from read-through transcripts of the endogenous hsp70Aa gene (Fig. 3A). Activation of piRNA biogenesis from this region might be explained by two, not necessarily mutually exclusive, mechanisms. First, processing of piRNA from the same amount of pre-existing hsp70 read-through transcripts might be enhanced. In this case, activation of piRNA biogenesis should lead to decreased levels of long read-through transcripts, as they will be more efficiently processed into piRNAs. Alternatively, activation of piRNA biogenesis might be caused by an increase in the level of precursor molecules, that is, the long hsp70 read-through transcripts. Strand-specific RT-qPCR showed that exposure to homologous piRNAs led to an increase in the amount of transcripts downstream of the hsp70 gene (Fig. 3C and S3), supporting the second explanation. This result suggests that exposure to homologous piRNAs suppresses termination of transcription through the hsp70 region. This leads to accumulation of read-through hsp70 transcripts that are processed into piRNAs. Thus, activation of piRNA biogenesis as a result of exposure to homologous piRNA correlates with the presence of Cuff and an inhibition of transcription termination.
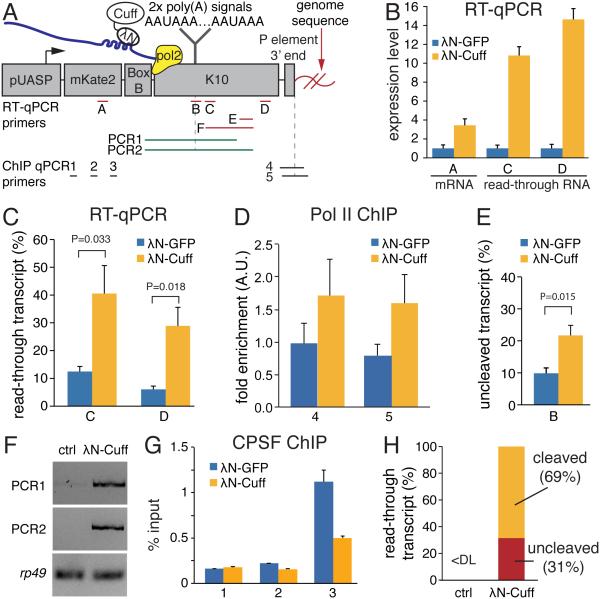
Cuff tethering to a reporter gene suppresses poly(A) site cleavage and termination
Transcription termination is a complex process consisting of two major steps. First, recognition of the poly(A) signal on nascent RNA leads to cleavage by the cleavage and polyadenylation specificity factor (CPSF) complex. Cleavage of the nascent transcript does not lead to immediate transcription termination, in fact the polymerase continues to transcribe a few hundred nucleotides downstream of the poly(A) site before the second step in which the polymerase stops and dissociates from the template DNA (Proudfoot, 2011). To explore the effects of Cuff on transcription and termination, we tethered Cuff to the 3’ UTR of reporter mRNA by fusing it to the RNA-binding domain of λN protein, which recognizes four BoxB hairpins in the reporter (Keryer-Bibens et al., 2008) (Fig. 4A). Tethering of Cuff to the reporter in fly ovaries led to a 3.4-fold increase in the steady-state level of reporter mRNA as measured by RT-qPCR (Fig. 4B, region A in Fig. 4A). Importantly, upon Cuff tethering, amounts of read-through transcripts 132-nt and 805-nt downstream of the cleavage site (regions C and D, respectively, in Fig. 4A) were increased 10.8-fold and 14.6-fold, respectively (Fig. 4B). To quantify the efficiency of termination we normalized the levels of these read-through transcripts to the level of reporter mRNA using a standard curve (Fig. S4). This analysis showed that upon Cuff tethering, the fractions of read-through transcripts increased from 12% to 41% for region C and from 6% to 29% for region D (Fig. 4C). To test if increase in read-through transcription is correlated with decreased termination of RNA polymerase we have measured Pol II occupancy downstream of poly(A) site. Tethering of Cuff leads to ~2-fold increase in Pol II occupancy ~850 bp downstream of poly(A) site as measured by ChIP-qPCR (Fig. 4D). These results indicate that tethering of Cuff to the reporter in fly ovaries leads to defects in RNA polymerase termination and subsequently to read-through-transcription.
Fig. 4. Tethering of Cuff to a heterologous mRNA reporter suppresses termination and poly(A) site cleavage.
(A) Schematic diagram of the reporter used to study the effect of Cuff recruitment to RNA. Cuff fused to the λN peptide, which binds BoxB hairpins in reporter RNA, was co-expressed with the reporter in ovaries.
(B) Cuff tethering leads to increased reporter transcript levels. λN-Cuff or control λN-GFP were co-expressed with the reporter in fly ovaries and RNA levels from three regions shown on (A) were quantified by RT-qPCR. Error bars show standard errors of four biological replicates.
(C) Tethering of Cuff increases read-through transcription of the reporter pre-mRNA as measured by the fraction of read-through transcripts relative to total reporter RNA. Error bars are standard errors from four biological replicates. P values were calculated by t-test. See also Figure S4.
(D) Tethering of Cuff suppresses Pol II termination as measured by Pol II ChIP-qPCR downstream of the poly(A) signals (fragments 4 and 5 on panel A). Error bars show standard errors of two biological replicates.
(E) Tethering of Cuff inhibits cleavage at poly(A) sites. Shown is the fraction of unprocessed transcripts calculated as described on Figure S5. Error bars are standard errors from four biological replicates. P was calculated by t-test.
(F) Unprocessed reporter transcripts that span the poly(A) site are detected by RT-PCR upon λN-Cuff tethering (amplified regions are shown in panel A).
(G) Tethering of Cuff suppresses binding of CPSF complex as measured by Cpsf73 ChIP-qPCR on different portions of the reporter shown on (A). Error bars show standard errors of four biological replicates.
(H) The majority of read-through transcripts formed upon Cuff tethering are cleaved near poly(A) sites. The fractions of cleaved and unprocessed RNA in two biological replicas were determined using standard curves shown on Figure S4 and S5.
The increase in read-through transcription upon Cuff tethering might be caused by inefficient pre-mRNA cleavage at the poly(A) site or by failure of the polymerase to stop after cleavage or both. To explore the effect of Cuff on pre-mRNA cleavage we used RT-qPCR to quantify amounts of transcripts that are not cleaved at either of the two poly(A) cleavage sites in the K10 3’ UTR of the reporter (region B in Fig. 4A). This analysis showed that Cuff tethering led to an increase in the fraction of non-cleaved transcripts from 10% to 22% (Fig. 4E). An even more dramatic change in the efficiency of pre-mRNA cleavage was seen when strand-specific long RT-PCR was used to detect uncleaved reporter transcripts (regions PCR1 and PCR2 on Fig. 4A). Upon Cuff tethering abundant non-cleaved reporter transcripts were readily detected; no such transcripts were seen in ovaries of control experiments (Fig. 4F). These data suggest that recruitment of Cuff to nascent transcripts suppresses the cleavage of pre-mRNA at poly(A) sites. To determine the molecular mechanism of suppression of pre-mRNA cleavage by Cuff, we measured the association of FLAG-tagged Cpsf73, the catalytic subunit of the CPSF complex, with the reporter. ChIP-qPCR shows that Cpsf73 is enriched on chromatin of the reporter locus close to the poly(A) signal (region 3), while less Cpsf73 is present at the upstream open reading frame sequences (regions 1 and 2) (Fig. 4G). Importantly, tethering of Cuff decreases the amount of Cpsf73 associated with the reporter locus more than 2-fold (Fig. 4G), despite the increase in transcript abundance. This result indicates that Cuff interferes with recruitment of the CPSF complex, which might result in suppressed cleavage at the poly(A) site.
To explore whether, in addition to suppression of the poly(A) site cleavage, Cuff enhances read-through transcription by another mechanism, we determined the fractions of cleaved and uncleaved RNA. For this, we used an RT-qPCR strategy to determine the absolute amount of all read-through transcripts (region E in Fig. 4A) as well as the amount of uncleaved read-through RNA (region F in Fig. 4A) using standard curves (Fig. S5). This analysis showed that there was dramatically more read-through transcription upon Cuff tethering than was observed in the absence of Cuff (Fig. 4H). Importantly, 69% of read-through transcripts formed upon Cuff tethering were in fact cleaved at the poly(A) site region; only 31% are unprocessed long transcripts that include the portion of mRNA upstream of the cleavage site. Therefore, Cuff recruitment to reporter pre-mRNA promotes read-through transcription downstream of poly(A) sites even if cleavage at the poly(A) site does occur.
Together our data show that Cuff recruitment to heterologous reporter mRNA enhances read-through transcription by two mechanisms. First, it suppresses pre-mRNA cleavage at canonical poly(A) sites; this generates long unprocessed read-through transcripts. Second, it prevents termination of transcription by RNA polymerase II even if cleavage at the poly(A) site does occur. The latter process leads to generation of non-coding transcripts that contain the region downstream of the poly(A) site.
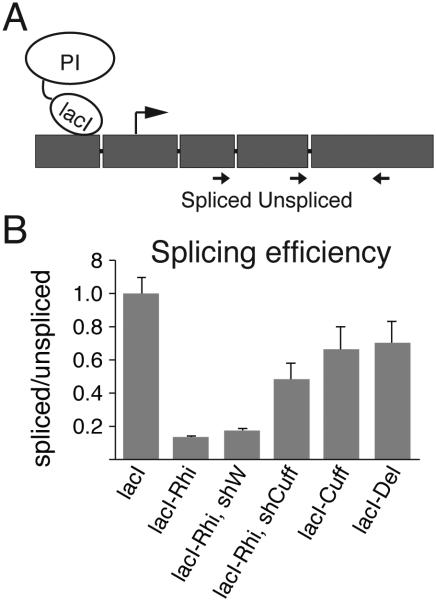
Rhino, which forms a complex with Cuff and Deadlock (Del) was previously implicated in suppression of splicing and it was proposed that Cuff might be responsible for the effect (Zhang et al., 2014). We used a previously described intron-containing reporter that contains lacO-binding sites upstream of the promoter (Fig. 5A) to determine if Cuff plays a role in Rhino-mediated suppression of splicing. First, we have confirmed the previously published observation (Zhang et al., 2014) that recruitment of Rhi upstream of the reporter promoter leads to 6-fold inhibition of splicing (Fig. 5B). LacO/LacI-mediated recruitment of Cuff as well as Del to the reporter also led to inhibition of splicing, though the effect was rather mild compared to Rhi. Importantly, knockdown of Cuff led to partial suppression of splicing inhibition caused by Rhino recruitment. Our results show that Cuff is indeed necessary for Rhi-mediated splicing inhibition indicating that Cuff can influence multiple aspects of early pre-mRNA processing including splicing, poly(A) site cleavage and termination.
Fig. 5. Cuff is required for Rhi-mediated suppression of splicing.
(A) Schematic diagram of the intron-containing reporter used to study the effect of Cuff recruitment on splicing.
(B) Cuff is required for Rhi-mediated suppression of splicing. The splicing efficiency was measured by the ratio of spliced to unspliced transcripts.
Cuff stabilizes non-capped transcripts formed by cleavage at poly(A) sites
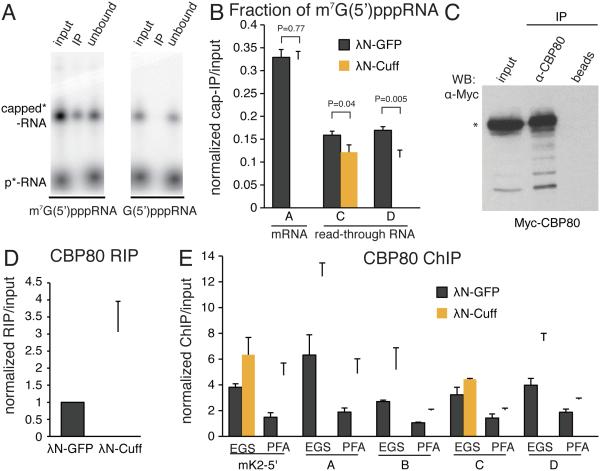
Read-through transcripts that were cleaved at poly(A) sites should have 5’ ends that are distinct in their chemical structure from 5’ ends of pre-mRNA. The 5’ ends of pre-mRNAs are rapidly modified by addition of the m7G(5')ppp cap structure, which then associates with the nuclear CBC. In contrast, the 5’ end of the downstream pre-mRNA fragment produced by CPSF cleavage is not modified by addition of a cap and instead carries a monophosphate group.
We explored whether Cuff tethering to a reporter affects the fraction of non-capped RNA and binding of CBC to its 5’ end. Capped RNA can be isolated using an anti-trimethylguanosine antibody (Chang et al., 2012). Analysis of precipitation of in vitro synthesized oligoribonulceotides that have a complete m7G(5')ppp cap, incomplete G(5')ppp cap, or a monophosphate group at their 5’ ends demonstrated the specificity of the antibody for completely capped RNA (Fig. 6A). Next, we immunoprecipitated RNA from fly ovaries using anti-cap antibodies and determined the fraction of co-purified reporter RNA in control flies and flies that express Cuff fused to λN protein, which recognizes four BoxB hairpins in the reporter. Our analysis indicated that, with or without expression of the Cuff-λN fusion, similar fractions of the upstream mRNA (fragment A) were capped, whereas significant fractions of read-through transcripts (fragments C and D) were not capped (Fig. 6B). Importantly, upon Cuff tethering the fraction of non-capped read-through transcripts increased slightly but significantly (p-values are 0.04 and 0.005 for fragments C and D, respectively). Read-through transcripts that are cleaved at upstream poly(A) sites have monophosphate at their 5’ end instead of a cap. Our result suggests that Cuff stabilizes cleaved non-capped transcripts and corroborates our finding that Cuff promotes read-through transcription even if cleavage at poly(A) sites does occur.
Fig. 6. Cuff stabilizes non-capped transcripts formed by cleavage at poly(A) sites.
(A) Anti-trimethylguanosine antibody specifically precipitates fully capped RNA. Radiolabeled RNAs with different groups at the 5’ end were precipitated with anti-trimethylguanosine antibody and bound (IP) and unbound fractions resolved on PAAG.
(B) Tethering of Cuff increases the fraction of non-capped read-through reporter transcripts. Capped RNA was immunoprecipitated from total ovarian RNA of flies that express the 4BoxB reporter shown on Fig 4A. Error bars show standard errors from six biological replicates. P values were calculated by t-test.
(C) Anti-CBP80 antibody specifically binds to Drosophila CBP80 protein. See also Figure S6.
(D) Tethering of Cuff increases association of CBP80 with reporter RNA. Ovarian lysates from flies expressing the 4BoxB reporter were used for RNA immunoprecipitation (RIP) with anti-CBP80 antibody. Error bars are standard deviations from two biological replicates.
(E) Tethering of Cuff increases association of CBP80 with chromatin of the reporter locus. ChIP using paraformaldehyde (PFA) or ethylene glycol bis(succinimidyl succinate) (EGS) crosslinkers were used to determine association of CBP80 with several regions of the reporter shown in Fig. 4A. Error bars show standard errors from two technical replicates.
The nuclear cap-binding complex (CBC) binds cap after it is formed on nascent RNA. The absence of CBC at the 5’ end of an mRNA might lead to termination defects, as CBC is required for efficient 3’ end processing (Gonatopoulos-Pournatzis and Cowling, 2014). To quantify association of the reporter mRNA with CBC we generated and tested synthetic antibodies that recognize the CBP80 subunit of the complex (Fig. 6C and S6). Next, we immunoprecipitated RNA (RIP) from fly ovaries using anti-CBP80 antibodies and determined the fraction of co-purified reporter RNA. Surprisingly, we found that the fraction of reporter mRNA that associated with CBC increased upon Cuff tethering (Fig. 6D). Increased association of CBC with chromatin at the reporter locus was also observed by ChIP-qPCR (Fig. 6E). Taken together our results indicate that displacement of CBC from the 5’ end of the mRNA in the piRNA reporter is not responsible for enhanced read-through transcription observed upon Cuff recruitment.
Depletion of the Rat1 exonuclease suppresses effects of cuff mutation
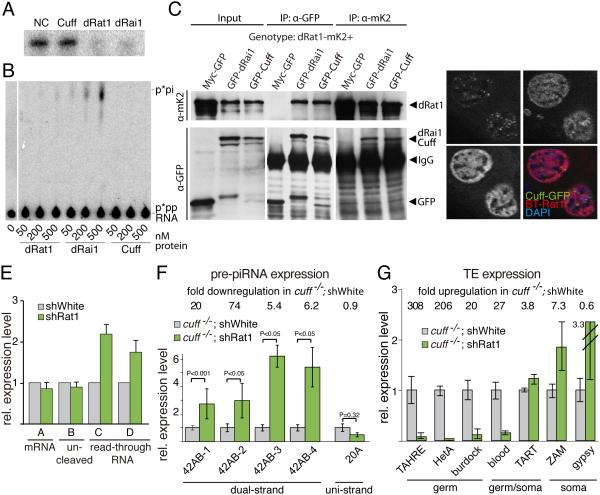
piRNA precursors might be a substrate for degradation by nuclear exonucleases. In eukaryotes, the conserved 5’-3’ exonuclease Rat1/Xrn2 specifically recognizes and degrades RNA with monophosphorylated 5’ ends in the nucleus. Exonuclease activity of Rat1 was shown to be important for RNA polymerase II termination through degradation of nascent RNA downstream of poly(A) site cleavage (Kim et al., 2004; West et al., 2004). In yeast, the exonuclease activity of Rat1 is enhanced by its binding to Rai1 (Xiang et al., 2009; Xue et al., 2000). Rai1 also has intrinsic pyrophosphohydrolase and decapping activities and its paralog in yeast Dxo1 has intrinsic exonuclease activity (Chang et al., 2012; Xiang et al., 2009). Yeast Rai1 and Dxo1 belong to an evolutionary conserved protein family that includes the mammalian Dom3Z, which possesses 5’-3’ exonuclease and pyrophosphohydrolase activities and plays a role in RNA quality control in the nucleus (Jiao et al., 2013). The fly genome encodes two proteins that belong to the Rai1/Dxo1/Dom3Z protein family: one is Cuff, and the other is encoded by the previously uncharacterized CG9125 gene.
As the biochemical activity of Drosophila Rat1 and Rai1 homologs had not been characterized, we expressed them in E. coli. Purified Rat1 effectively degraded 5’-monophosphorylated RNA in vitro (Fig. 7A). The protein product of CC9125 has 5’-3’ exonuclease and pyrophosphohydrolase activities in vitro indicating that it is a functional homolog of Rai1/Dxo1/Dom3Z (Fig. 7A, B). Therefore we refer to it as dRai1. In contrast to dRai1/CC9125, Cuff showed no enzymatic activities under identical conditions. These results are consistent with sequence and structure analyses which indicate that residues responsible for activity are conserved in dRai1/CC9125, but mutated in Cuff rendering it catalytically inactive (Le Thomas et al., 2014; Mohn et al., 2014; Zhang et al., 2014).
Fig. 7. Depletion of Rat1 exonuclease suppresses effects of the cuff mutation.
(A) Drosophila Rat1 and dRai1 have exonuclease activity toward 5’-monophosphate RNA. After incubation with the purified proteins radiolabeled RNA was resolved on a denaturing PAGE. NC: no protein control.
(B) dRai1 but not Cuff has pyrophosphohydrolase activity. See Suppl. methods for details.
(C) Drosophila dRai1 and Cuff physically interacts with Rat1 as measured by Co-IP/Western of tagged proteins.
(D) Cuff and Rat1 are localize in nuclei. Immunostaining of GFP-tagged Cuff and BioTAP-tagged Rat1 proteins in ovaries. DAPI staining was used to locate nuclei.
(E) Inhibition of expression of rat1 increases the levels of read-through transcripts downstream of the canonical poly(A) site as measured by RT-qPCR on reporter shown in Fig. 4A. Error bars show standard errors for three biological replicates.
(F-G) Inhibition of rat1 in germline suppresses the phenotype of cuffwm25/qq37 mutation. The plots show the change in expression of piRNA cluster transcripts (F) and transposable elements (G) measured by RT-qPCR in ovaries of flies of specified genotype. Error bars show standard error among four (F) or three (G) biological replicates. P-values are calculated by t-test.
Reciprocal co-immunoprecipitations showed that dRai1 and Cuff form a complex with Rat1 (Fig. 7C). Tagged Rat1 expressed in Drosophila ovary localizes exclusively in the nucleus in a broad pattern that overlaps with chromatin staining (Fig. 7D). The localization pattern of Rat1 is different from Cuff, which localize to distinct nuclear foci. To test whether Rat1 participates in transcription termination in Drosophila, we inhibited its expression in germ cells using RNAi and measured the levels of read-through transcripts downstream of the poly(A) site of our reporter. The levels of read-through transcripts were increased ~2-fold compared to control (Fig. 7E, regions C and D), whereas no change was seen in levels of the mRNA or uncleaved transcripts (regions A and B, respectively). Therefore, Drosophila Rat1 has a conserved function in RNA degradation and promotion of termination. Taken together, biochemical experiments and structure analysis indicate that Rat1 and dRai1 degrade RNA in the nucleus, while Cuff, despite structural similarity with dRai1, does not have the same catalytic activities.
piRNA precursor abundance is severely reduced in Cuff mutants (Fig. 1) (Pane et al., 2011). If Cuff protects piRNA precursor transcripts from degradation by Rat1 nuclease then depletion of Rat1 should suppress the effect of cuff mutation on the abundance of piRNA precursors. To test for a genetic interaction between cuff and rat1 we combined cuff mutation with knockdown of rat1 expression by shRNA in germline and quantified expression of piRNA cluster transcripts by RT-qPCR. As expected from previous observations (Pane et al., 2011) and our RNA-seq data, we found strong depletion of transcripts from dual-strand, but not uni-strand, clusters in ovaries of cuffwm25/qq37 flies. Importantly, inhibition of rat1 expression suppressed the effect of the cuff mutation and partly restored the levels of piRNA transcripts (Fig. 7F). This result shows that a large fraction of nuclear piRNA precursors has mono-P 5’end and that their protection by Cuff from Rat1 is essential to maintain proper piRNA precursor level.
The role of Rat1 in the piRNA pathway can be ultimately revealed by studying its effect on transposon repression. In ovaries of cuffwm25/qq37 flies, there was a dramatic derepression of transposons in the germline, with the steady-state level of HetA and TAHRE mRNA increasing more than 200-fold in ovaries of cuffwm25/qq37 flies compared to wild-type controls (Fig. 7G). As expected, the gypsy and ZAM retrotransposons, which are expressed in somatic follicular cells of the ovary remained repressed in cuffwm25/qq37 flies. Inhibition of rat1 expression using RNAi strongly suppressed the effect of cuff mutation, as HetA levels were reduced 45-fold in ovaries of cuffwm25/qq37; rat1-KD flies compared to flies with the cuffwm25/qq37 mutation alone. A similar, effect was observed for TAHRE, Burdock and Blood transposons. This result indicates that the exonuclease Rat1 plays an essential role in piRNA-mediated transposon silencing. Partial suppression of the cuff phenotype by rat1 depletion shows that one - but not the only - function of Cuff is to protect piRNA precursors from degradation by Rat1. Our data identify Rat1 as a new player in the piRNA biogenesis pathway that is involved in piRNA precursor turnover.
Discussion
Previous studies showed that Rhi and Cuff are required for piRNA biogenesis from dual-strand, but not uni-strand, piRNA clusters in Drosophila germ cells (Klattenhoff et al., 2009; Pane et al., 2011). These proteins, together with Del, form the RDC complex that binds to chromatin of dual-strand clusters (Le Thomas et al., 2014; Mohn et al., 2014; Zhang et al., 2014). Rhi, probably together with other RDC components, suppresses splicing of reporter pre-mRNA (Zhang et al., 2014) and is likely required for piRNA precursor transcription (Le Thomas et al., 2014; Mohn et al., 2014), however, the mechanism of action of the RDC complex remained poorly understood. Here we explored the function of the Cuff protein and showed that it is necessary to generate nuclear piRNA precursors by enhancing transcription of many piRNA producing regions.
How does Cuff enhance transcription of piRNA precursors? Several lines of evidence suggest that Cuff functions to prevent premature termination of transcription of the long piRNA precursor transcripts. First, the canonical poly(A) signals positioned inside dual-strand piRNA clusters are ignored by RNA polymerase II in wild-type flies, but lead to termination upon Cuff depletion. Second, the exposure of a locus to homologous piRNA, which triggers piRNA biogenesis, correlated with binding of Cuff and an increase in read-through transcription. Third, Cuff tethering to an mRNA reporter led to decreased binding of CPSF and to suppression of transcription termination. Though our data suggest that Cuff prevents termination inside dual-strand piRNA clusters, we found no evidence that suppression of termination causes read-through transcription from flanking protein-coding genes into piRNA clusters.
How does Cuff suppress termination of transcription? The termination of transcription by RNA polymerase II is a complex process composed of two linked steps: cleavage of pre-mRNA at poly(A) site by the CPSF complex, followed by actual transcription termination a few hundred nucleotides downstream. Analysis of suppression of termination using tethering of Cuff to a reporter indicates that Cuff suppresses termination at two distinct steps. First, Cuff inhibits association of CPSF complex with nascent RNA and the cleavage at poly(A) site. However, significant fraction of read-through transcripts formed upon Cuff tethering were cleaved suggesting that Cuff ensures that transcription continues even if the nascent transcripts were processed. Our results suggest that Cuff protects cleaved RNA from degradation by the Rat1 exonuclease, therefore, preventing ‘torpedo’-style termination of cleaved transcripts. How Cuff protects RNA from Rat1 remains to be investigated in the future. As Cuff is homologous to the Rai1/Dom3Z proteins, which have a binding pocket for 5’ end of RNA, Cuff might directly bind and occlude 5’ ends, preventing recognition by Rat1 (Fig. S7) (Mohn et al., 2014; Zhang et al., 2014). Unfortunately, our attempts to determine the specificity of Cuff binding to RNAs with different 5’ ends using in vitro crosslinking failed to generate consistent results.
In addition to suppression of termination, protection of piRNA precursors by Cuff is likely to be important for the post-transcriptional fate of these transcripts. An obvious possibility is that Cuff prevents degradation of piRNA precursors during their transport from the nucleus to the cytoplasm. We recently showed that Cuff is required for loading of the conserved TREX complex onto nascent piRNA precursors (Hur et al., 2016). Protection of non-capped transcripts by Cuff can also explain why RDC suppresses splicing of piRNA precursors (Fig. 5)(Zhang et al., 2014). As binding of the nuclear cap-binding complex to pre-mRNA enhances splicing, stabilization of non-capped RNA will increase the fraction of non-spliced transcripts.
The function of Cuff in suppression of termination helps to answer the puzzling question of why the RDC complex is required for piRNA biogenesis exclusively from dual-strand clusters. The insertions of heterologous sequences into piRNA clusters triggers generation of new piRNAs that repress homologous transcripts (Muerdter et al., 2012). This process is believed to be essential for the operation of piRNA-mediated repression as it ensures adaptability to new genome invaders: insertions of new transposons into piRNA clusters will lead to acquisition of heritable immunity against these transposons (Aravin et al., 2007; Khurana et al., 2011). New transposon insertions introduce their own regulatory signals including poly(A)/termination sequences. In the absence of a special mechanism to prevent it, termination signals encountered by the transcribing RNA polymerase would cause termination and collapse of piRNA biogenesis from downstream sequences. We propose that Cuff enables efficient acquisition of immunity to new transposons upon insertion into dual-strand clusters in random orientations. In agreement with this hypothesis, an analog of the RDC complex is absent in mouse, which lacks extended dual-strand piRNA clusters.
Beyond the role of Cuff in piRNA-guided transposon repression, our study illuminates a novel mode of transcriptional regulation in eukaryotes. Most known transcriptional regulators activate or repress transcription at the initiation step. In contrast, Cuff controls efficiency of termination. Furthermore, in contrast to other factors that bind specific sequence signals in nascent RNA, Cuff is recruited to chromatin as part of the RDC complex (Le Thomas et al., 2014; Mohn et al., 2014; Zhang et al., 2014). Regulation of gene expression by control of termination is a well-characterized mechanism in prokaryotes (Nudler and Gottesman, 2002), and recently has gained attention in eukaryotes as well. In the future it will be interesting to test if chromatin structure modulates termination of transcription of protein-coding genes.
Materials and Methods
Cuff tethering to reporter
Transgenic flies with insertions of λN-Cuff (UASp-λN-GFP-Cuff) and BoxB (UASp-mKate2-4xBoxB-K10) constructs were generated by P-element integration. λN-GFP (pUASp-λN-GFP-eGFP) was used as a control. The expression of the constructs was driven by maternal alpha-tubulin67C-Gal4. See the Supplemental Experimental Procedures for description of other stocks.
RNA-seq and small RNA-seq libraries
Total ovarian RNA was either depleted of ribosomal RNA or was poly(A) selected. Isolation of nascent transcripts was performed as previously reported by Khodor and colleagues (Khodor et al., 2011), with some modifications. A detailed protocol is provided in the Supplemental Experimental Procedures. RNA-seq libraries were made using the NEBNext Ultra Directional RNA Library Prep Kit and sequenced on the Illumina HiSeq 2500 platform. Bioinformatic analysis is described in the Supplemental Experimental Procedures. Small RNA-seq data from cuff mutants were previously published (Le Thomas et al., 2014).
Nuclear run-on and GRO-seq
Nuclear run-on experiments were performed on ovarian nuclei isolated from flies that express shRNA against cuff driven by Nos-Gal4. The nuclear run-on was carried out as previously described (Shpiz et al., 2011) with modifications. A detailed protocol is provided in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgements
We thank members of the Aravin lab for discussion. We thank Igor Antoshechkin (Caltech) for help with sequencing and Sergei Manakov for processing the RNA-seq data. E. S. is supported by a PhD fellowship of the Boehringer Ingelheim Fonds. This work was supported by grants from the National Institutes of Health (R01 GM097363, and DP2 OD007371A) and by the the Searle Scholar and the Packard Fellowship Awards to A.A.A., Ellison Medical Foundation award to K.F.T., grants from the Canadian Institutes of Health Research (MOP-14409) and the Natural Sciences and Engineering Research Council of Canada (RGPIN-201) to H.D.L., a grant from the Natural Sciences and Engineering Research Council of Canada (RGPIN-435985) to C.A.S.,and by funds from the Abby Rockefeller Mauze Trust and Maloris and STARR Foundations to D.J.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Experiments were designed and performed by Y.C (Fig. 1-2, 4, 6-7), E.S. (Fig. 1-3), Y.L. (Fig. 5-6), A.L (Pld promoter deletion and piRNA analysis, Fig. 2) with help from E.R. and S.V. M.N. performed all bioinformatic analysis. S.L. and D.J.P modeled the Cuff structure. J.D.L., C.A.S. and H.D.L. generated the CBP80 antibody. A.A.A. and K.F.T. aided in study design and data interpretation. Y.C. and A.A.A. wrote the paper with input from E.S., M.N., Y.L. and K.F.T.
Accession codes
High-throughput sequencing data for RNA-seq and GRO-seq experiments are available through Gene Expression Omnibus (accession no. GSE81090).
References
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Chang JH, Jiao X, Chiba K, Oh C, Martin CE, Kiledjian M, Tong L. Dxo1 is a new type of eukaryotic enzyme with both decapping and 5'-3' exoribonuclease activity. Nat Struct Mol Biol. 2012;19:1011–1017. doi: 10.1038/nsmb.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schupbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Current biology : CB. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Preall JB, McGinn J, Hannon GJ. A Transcriptome-wide RNAi Screen in the Drosophila Ovary Reveals Factors of the Germline piRNA Pathway. Mol Cell. 2013;50:749–761. doi: 10.1016/j.molcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vanssay A, Bouge AL, Boivin A, Hermant C, Teysset L, Delmarre V, Antoniewski C, Ronsseray S. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490:112–115. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- Gonatopoulos-Pournatzis T, Cowling VH. Cap-binding complex (CBC) Biochem J. 2014;457:231–242. doi: 10.1042/BJ20131214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriaux C, Desset S, Renaud Y, Vaury C, Brasset E. Transcriptional properties and splicing of the flamenco piRNA cluster. EMBO Rep. 2014;15:411–418. doi: 10.1002/embr.201337898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Luo Y, Moon S, Ninova M, Marinov GK, Chung YD, Aravin AA. Splicing-independent loading of TREX on nascent RNA is required for efficient expression of dual-strand piRNA clusters in Drosophila. Genes Dev. 2016;30:840–855. doi: 10.1101/gad.276030.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Chang JH, Kilic T, Tong L, Kiledjian M. A mammalian pre-mRNA 5' end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol Cell. 2013;50:104–115. doi: 10.1016/j.molcel.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer-Bibens C, Barreau C, Osborne HB. Tethering of proteins to RNAs by bacteriophage proteins. Biol Cell. 2008;100:125–138. doi: 10.1042/BC20070067. [DOI] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT, 2nd, Rosbash M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011;25:2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147:1551–1563. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Stuwe E, Li S, Du J, Marinov G, Rozhkov N, Chen YC, Luo Y, Sachidanandam R, Toth KF, et al. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 2014;28:1667–1680. doi: 10.1101/gad.245514.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157:1364–1379. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- Muerdter F, Olovnikov I, Molaro A, Rozhkov NV, Czech B, Gordon A, Hannon GJ, Aravin AA. Production of artificial piRNAs in flies and mice. RNA. 2012;18:42–52. doi: 10.1261/rna.029769.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- Pane A, Jiang P, Zhao DY, Singh M, Schupbach T. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J. 2011 doi: 10.1038/emboj.2011.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, Kalmykova A. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Todeschini AL, Teysset L, Delmarre V, Ronsseray S. The epigenetic trans-silencing effect in Drosophila involves maternally-transmitted small RNAs whose production depends on the piRNA pathway and HP1. PLoS One. 2010;5:e11032. doi: 10.1371/journal.pone.0011032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Proudfoot NJ. Human 5' --> 3' exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L. Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Bai X, Lee I, Kallstrom G, Ho J, Brown J, Stevens A, Johnson AW. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol Cell Biol. 2000;20:4006–4015. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Cassani M, Wang M, Liu M, Ma J, Li G, Zhang Z, Huang Y. Structural insights into Rhino-mediated germline piRNA cluster formation. Cell Res. 2015;25:525–528. doi: 10.1038/cr.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, Vreven T, Zamore PD, Weng Z, Theurkauf WE. The HP1 Homolog Rhino Anchors a Nuclear Complex that Suppresses piRNA Precursor Splicing. Cell. 2014;157:1353–1363. doi: 10.1016/j.cell.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.