Abstract
Hematopoietic differentiation is a highly complex process originating from an extraordinary population of cells called long-term repopulating hematopoietic stem cells (LT-HSCs). The unique feature of all stem cells, including HSCs, is their exceptional ability to divide asymmetrically giving rise to two different kinds of offspring. One daughter cell becomes an LT-HSC itself (self-renews) to maintain the LT-HSC pool, whereas the second daughter cell pursues a differentiation fate to ultimately give rise to terminally differentiated mature blood cells (Orkin and Zon, 2008). Quantification of phenotypic LT-HSCs can be performed by multi-color flow cytometry and the gold standard for assessment of LT-HSC self-renewal and function is competitive bone marrow transplantation (Miller et al., 2008). Although these methods are irreplaceable to determine LT-HSC abundance and functionality, they have their disadvantages and limitations. For example, competitive bone marrow transplantation is typically monitored as a function of peripheral blood donor contribution over 12–16 weeks. While reduced peripheral blood donor contribution by itself signifies impairment in the stem/progenitor cells compartment, it cannot unambiguously discriminate between reduced LT-HSC self-renewal, impaired LT-HSC differentiation or compromised progenitor cell differentiation. Here we describe an LT-HSCs methylcellulose colony-forming assay, as a fast complementary in vitro method to directly assess LT-HSC differentiation capacity. As described in Kerenyi et al. (2013), this technique acts as a powerful tool to differentiate between LT-HSC or progenitor cell differentiation defects.
Materials and Reagents
Mice (C57BL/6, 12–16 weeks)
Bone marrow cells from experimental mice
7-amino-actinomycin D (7AAD) (BD, catalog number: 559925)
CD150 Alexa488 antibody (BioLegend, catalog number: 115916)
CD48 APC antibody (eBiosciences, catalog number: 17048182)
cKit APC-efluor 780 antibody (eBiosciences, catalog number: 47117182)
DynaBeads Biotin Binder (Life Technologies, Invitrogen™, catalog number: 11047)
EDTA (0.5 M, pH 8.0) (Boston BioProducts, catalog number: BM-150)
Fetal Bovine Serum (FBS) (Gemini Bio-Products, catalog number: 100106)
Ficoll-Paque Premium (General Electric Company, catalog number: 17544202)
Iscove’s Modified Dulbecco’s Medium (IMDM) (Gibco®, catalog number: 12440053)
L-Glutamine (Gibco®, catalog number: 25030081)
MethoCult GF M3434 methylcellulose-based medium (STEMCELL Technologies, catalog number: 3434)
Mouse Hematopoietic Lineage Biotin Panel (eBiosciences, catalog number: 88777475)
Phosphate buffered saline (PBS) without Ca2+ and Mg2+ (Mediatech, Cellgro®, catalog number: 21031CM)
Penicillin/Streptomycin Solution (Gibco®, catalog number: 15140122)
Purified Anti CD16/CD32 antibody (FC Block) (BD, catalog number: 553142)
Recombinant mouse interferon (IFN) alpha A (R&D Systems, catalog number: 121001)
Sca-1 PE-Cy7 antibody (eBiosciences, catalog number: 25598182)
Streptavidin PerCP (BD, catalog number: 554064)
Staining Medium for FACS (see Recipes)
IMDM for methylcellulose colony assays (see Recipes)
Equipment
Mortar and pestle
Easy Grip 35 mm petri dish (BD Biosciences, Falcon®, catalog number: 351008)
6 well plate
1.5 ml Eppendorf Protein LoBind Tube (Eppendorf, catalog number: 022431081)
1.5 ml Eppendorf Safe-Lock Tubes (Eppendorf, catalog number: 0030120086)
2.0 ml Eppendorf Safe-Lock Tubes (Eppendorf, catalog number: 0030120094)
40 µM BD Cell stainer (BD Biosciences, Falcon®, catalog number: 352340)
15 ml BD Falcon Conical (BD Biosciences, Falcon®, catalog number: 352095)
50 ml BD Falcon Conical (BD Biosciences, Falcon®, catalog number: 1443222)
10 cm BD Falcon Tissue culture dish (BD Biosciences, Falcon®, catalog number: 353003)
5 ml BD Polystyrene Tubes (FACS tubes) (BD Biosciences, Falcon®, catalog number: 352054)
BD PrecisionGlide Needle (16 gauge) (BD Biosciences, Falcon®, catalog number: 305198)
3 ml BD Syringe Luer-Lok Tip (BD Biosciences, Falcon®, catalog number: 309657)
BD FACS Aria I (BD Biosciences, special order product)
Inverted Light Microscope (OLYMPUS, catalog number: CKX31)
DynaMag-2 Magnet (Life Technologies, Invitrogen™, catalog number: 12321D)
Procedure
Day 0
-
1
Thaw sufficient numbers of 3 ml MethoCult GF M3434 aliquots overnight (one 3 ml aliquot per mouse) at 4 °C.
Day 1
-
2
Euthanize mice according to the method of choice, e.g., cervical dislocation, CO2.
-
3
Disinfect mice with 70% ethanol. Open skin on top of the femur medially (on the interior side of hindlegs). Using small scissors and forceps, dissect out ilii (hip bones), femora and tibiae from mice and place them in one well of a 6 well plate containing 2 ml ice-cold PBS/2% FBS 2 mM EDTA (= staining medium).
-
4
Isolate bone marrow (BM) using autoclaved mortar and pestle: Put femora, tibiae, iliii of one mouse into mortar and add 5 ml staining medium and grind until bones are not red anymore. Filter cell suspension into a 50 ml conical tube through a 40 µM nylon mesh and use the plunger of a 5 ml syringe to push cells trough the mesh which are held back by bone marrow fat. Repeat at least 2 more times.
-
5
Fill 50 ml conical tube up to 40 ml with staining medium and spin down at 300 × g at 4 °C.
-
6
Resuspend BM cells in 5 ml staining medium.
-
7
Fill 5 ml Ficoll at room temperature (!) into a 15 ml conical tube.
-
8
Slowly overlay 5 ml of the bone marrow cell suspension onto the Ficoll.
-
9
Spin down cells for 15 min at 400 × g at room temperature, with the brake off (or low). This will get rid of the bulk of erythroid and myeloid cells.
-
10Using a pasteur pipette transfer the interface, containing mononuclear cells, into another 15 ml tube.Note: The interface may not appear if Ficoll is not at room temperature or if the centrifuge chamber is chilled.
-
11
Wash the bone marrow mononuclear cells with 15 ml staining medium.
-
12
Spin down cells for 7 min at 300 × g at 4 °C and resuspend again in 10 ml staining medium. At this point a small aliquot can be drawn to enumerate the cell number.
-
13
Spin down cells for 7 min at 300 × g at 4 °C. Aspirate supernatant completely. Resuspend pellet in 50 µl staining buffer.
-
14
Add 1 µl of purified anti CD16/CD32 antibody (FC-Block) to keep FC-receptors from capturing antibodies, which would result in false positive staining. Incubate for 15 min in the fridge.
-
15Add 50 µl of biotinylated lineage marker antibody cocktail (Mouse Hematopoietic Lineage Flow Panel):
- 1× lineage marker cocktail:
- Biotinylated anti-Ter-119 (2 µl/1 × 107 cells)
- Biotinylated anti-Gr-1 (2 µl/1 × 107 cells)
- Biotinylated anti-Mac-1 (2 µl/1 × 107 cells)
- Biotinylated anti-B220 (2 µl/1 × 107 cells)
- Biotinylated anti-CD3 (2 µl/1 × 107 cells)
- Up to 50 µl with staining buffer
-
16
Incubate cells in fridge for 30 min.
-
17
Wash cells once with 5 ml of staining buffer and spin down for 5 min at 300 × g at 4 °C.
-
18
Resuspend cells in 1.5 ml staining medium (in 2 ml Eppendorf tube).
-
19Preparation for DynaBeads Biotin Binder for lineage depletion:
- Resuspend the Dynabeads in the vial.
- Transfer the desired volume of Dynabeads to a 1.5 ml tube.
- Add the same volume of staining buffer.
- Place the tube in a magnet for 3 min and discard the supernatant.
- Repeat steps c and d twice.
- Remove tubes from magnet and resuspend washed Dynabeads in the same volume of staining buffer as the starting volume of beads was.
-
20
Add ~300 µl DynaBeads Biotin Binder (or 4 Dynabeads per target cell) per mouse and incubate cells rotating at 2–8 °C for 30 min.
-
21
Put tubes containing the cells in magnet for 3 min. Transfer supernatant into FACS tube.
-
22
Wash cells once with 10 ml of staining buffer and resuspend in 50 µl staining buffer.
-
23Add 50 µl of HSC staining cocktail and stain 30 min at 4 °C. Flick sample every 10 min to mix.
- 1× HSC staining cocktail:
- Streptavidin PerCP (0.125 µg/1 × 107 cells)
- CD150-Alexa488 (0.5 µg/1 × 107 cells)
- CD48-APC (0.125 µg/1 × 107 cells)
- cKit-APC-eFluor780 (0.125 µg/1 × 107 cells)
- Sca1-PE-Cy7 (0.125 µg/1 × 107 cells)
- Biotinylated anti-Ter-119 Ab (2 µl/1 × 107 cells)
- Biotinylated anti-Gr-1 Ab (2 µl/1 × 107 cells)
- Biotinylated anti-Mac-1 Ab (2 µl/1 × 107 cells)
- Biotinylated anti-B220 Ab (2 µl/1 × 107 cells)
- Biotinylated anti-CD3 Ab (2 µl/1 × 107 cells)
- Up to 50 µl with staining buffer
-
24
Wash cells once with 3 ml staining buffer, spin down for 5 min at 300 × g at 4 °C and resuspend in 300 µl staining buffer.
-
25Add 3 µl 7-AAD and incubate 10 min at room temperature.Note: The LT-HSC isolation part of this protocol has been adapted and modified from Ema et al. (2006).
-
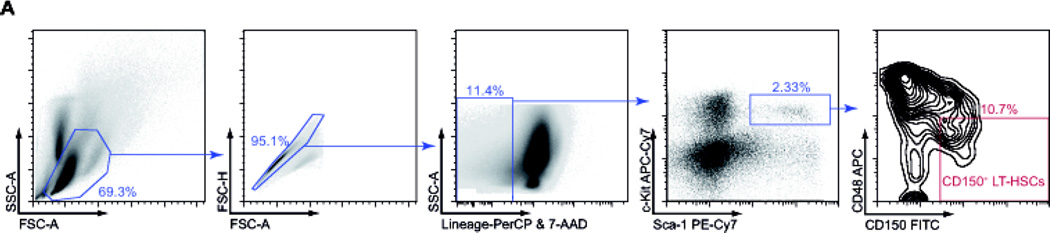
26Sort cells on a BD FACS ARIA I (or comparable Cell Sorter). To achieve over 95% purity of LT-HSCs, perform 2 rounds of sorting! See Figure 1 for gating strategy.
- 1st sort: Sort at least 5000 LT-HSCs (i.e. lineage negative Sca-1+ c-Kit+ CD150+CD48− cells) into 250 µl of IMDM 2% FCS + 1% P/S in a low protein binding 1.5 ml Eppendorf tube. Using a low protein binding tube at this step will maximize your LT-HSC recovery.
- 2nd sort: Sort exactly 1000 LT-HSCs into 500 µl of IMDM 2% FCS + 1% P/S in a low protein binding 1.5 ml Eppendorf tube. You will now have 200 LT-HSCs/100 µl.
Optional: If using Mx1Cre BM you can either delete your floxed allele in vivo with pIpC (not described here) or in vitro using 1,000 U/ml recombinant mouse Interferon alpha (IFNα). In vitro deletion may be desirable in cases in which knock out of your gene of interest results in altered cell surface immunophenotypes.Note: The specific activity of IFNα varies from Lot to Lot. Final volume of IFNα to obtain 3,000 U/3 ml Methocult aliquots will have to be adjusted for every lot. -
27A total of 300 µl volume can be added to 3 ml Methocult M3434 aliquot:
- 60 µl HSCs (from 200 LT-HSCs/100 µl) = about 40 LT-HSCs/ml Methocult M3434
- 30 µl Pen/Strep
- Optional: × µl IFNα (Lot dependent–but a total of 3,000 U)
- Up to 300 µl with IMDM 2% FCS 1% Pen/Strep 1% L-Glut
-
28
Mix well by vortex, and let methylcellulose settle at least for 5 min to allow air bubbles to dissipate.
-
29Plate 2 × 1.1 ml of Methocult M3434 including your LT-HSCs per 35 mm petri dish using a 3 CC syringe with a 16 gauge needle. You now have technical duplicates of your colony assay.Note: You have to use a petri dish not a tissue culture dish!
-
30
Place the two dishes into a 100 mm petri dish. Add a third, uncovered 35 mm dish containing 3 ml of sterile water. Replace lid of 100 mm petri dish.
-
31
Transfer the duplicate samples and your petri dish filled with water into a 10 cm tissue culture grade dish and put the lid on it. This installation will keep your methylcellulose from drying out. Transfer to tissue culture incubator and incubate for 10–12 days at 37 °C.
Figure 1. Gating strategy for the isolation of CD150+ LT-HSCs.
A Bone marrow cells were stained with indicated antibodies and sorted by flow cytometry to isolate CD150+ LT-HSCs.
Day 10–12
-
32
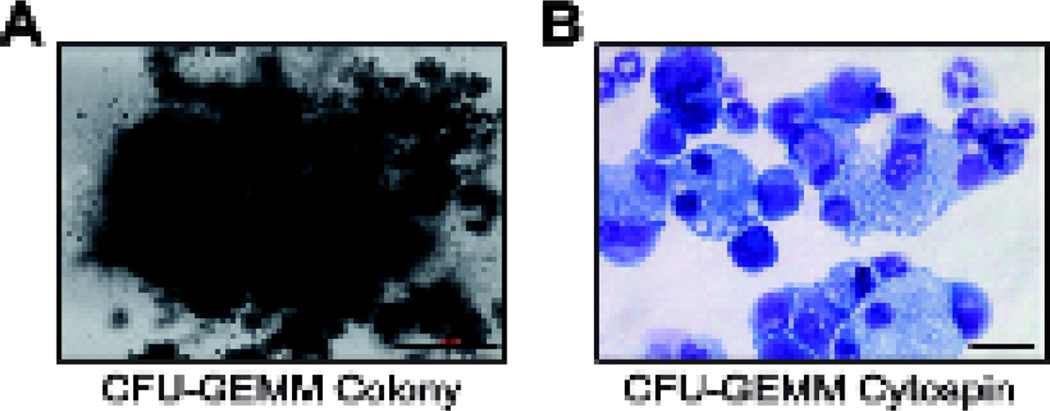
Using an inverted light microscope, count and score the number of colonies according to the instructions found in Miller et al. (2008). You should largely obtain large multipotential CFU-GEMM (Colony Forming Unit–granulocyte, erythrocyte, monocyte, megakaryocyte) colonies (Figure 2A). When seeding 40 wild type LT-HSCs we typically observe ~20 CFU-GEMM colonies. If desired, cells can be harvested from methylcellulose and either subjected to cytospin analysis to determine cell morphologies (Figure 2B) or to flow cytometry, to obtain quantitative cell type distribution enumeration.
Figure 2. Microphotographs of CFU-GEMM Colonies.
A. Microphotograph if a single CFU-GEMM colony (scale bar = 500 µM). B. Microphotograph of a cytospun CFU-GEMM colony stained with hematoxylin and eosin (scale bar = 40 µM).
Recipes
- Staining Medium for FACS (for 500 ml)
- 488 ml PBS (without Ca2+ or Mg2+)
- 10 ml FBS (final concentration: 2% FBS)
- 2 ml 0.5 M EDTA pH 8 (final concentration: 2 mM EDTA)
- IMDM for methylcellulose colony assays (for 500 ml)
- 480 ml IMDM
- 10 ml FBS (final concentration: 2% FBS)
- 5 ml Pen/Strep (final concentration: 1% Pen/Strep)
- 5 ml L-Glutamine (final concentration: 1% L-Glut)
Acknowledgments
This protocol was originally described in and adapted from Kerenyi et al. (2013). This work was supported by the NIH National Heart, Lung and Blood Institute, R01HL075735 and Austrian Science Fund (FWF), J 2948-B19. I would like to thank my mentor Stuart Orkin for giving me the opportunity to conceive, design and author this protocol.
References
- 1.Ema H, Morita Y, Yamazaki S, Matsubara A, Seita J, Tadokoro Y, Kondo H, Takano H, Nakauchi H. Adult mouse hematopoietic stem cells: purification and single-cell assays. Nat Protoc. 2006;1(6):2979–2987. doi: 10.1038/nprot.2006.447. [DOI] [PubMed] [Google Scholar]
- 2.Kerenyi MA, Shao Z, Hsu YJ, Guo G, Luc S, O'Brien K, Fujiwara Y, Peng C, Nguyen M, Orkin SH. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. Elife. 2013;2:e00633. doi: 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller CL, Dykstra B, Eaves CJ. Characterization of mouse hematopoietic stem and progenitor cells. Curr Protoc Immunol. 2008;Chapter 22(Unit 22B):22. doi: 10.1002/0471142735.im22b02s80. [DOI] [PubMed] [Google Scholar]
- 4.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]