Abstract
The ability to modulate stem cell differentiation in a three dimensional (3D) microenvironment for bone tissue engineering in absence of exogenous pharmaceutical agents such as bone morphogenic protein (BMP-2) remains a challenge. In this study, we introduce extracellular matrix (ECM)-mimicking nanocomposite hydrogels to induce osteogenic differentiation of human mesenchymal stem cells (hMSCs) for bone regeneration in absence of any osteoinducting factors. In particular, we have reinforced photocrosslinkable collagen-based matrix (gelatin methacryloyl, GelMA) used disk-shaped nanosilicates (nSi), a new class of two-dimensional (2D) nanomaterials. We show that nanoengineered hydrogels supported migration and proliferation of encapsulated hMSCs, with no signs of cell apoptosis or inflammatory cytokine responses. The addition of nSi significantly enhances osteogenic differentiation of encapsulated hMSCs as evident by the increase in alkaline phosphates (ALP) activity and deposition of biomineralized matrix compared to GelMA without nSi. We also show that microfabricated nanoengineered microgels can be used to pattern and control cellular behaviour. Furthermore, we also show that nanoengineered hydrogel have high biocompatibility as determined by in vivo experiments using immunocompetent rat model. Specifically, the hydrogels showed minimum localized immune responses, indicating it ability for tissue engineering applications. Overall, we showed the ability of nanoengineered hydrogels loaded with 2D nanosilicates for osteogenic differentiation of stem cells in vitro, in absence of any growth factors such as BMP-2. Our in vivo studies show high biocompatibility of nanocomposites and show the potential for growth factor free bone regeneration.
Keywords: nanomaterials, hydrogels, bone regeneration, tissue engineering, nanosilicates
Graphical abstract
We report development of a silicate nanoclay-rich bioactive hydrogel to promote osteogenic differentiation of adult stem cells in a three dimensional microenvironment.
Introduction
Despite decades of intensive research, bone regeneration has been a formidable clinical challenge in orthopedics and associated traumatic injuries.1 This is mainly attributed to the uneven healing and transplant integration to host tissue, potential infection and limited availability with current modes of treatments, such as bone allo- and autografting. These challenges have imposed a strong impetus to explore effective alternative strategies such as bioengineering artificial bone tissue grafts, immobilizing growth factors on biomaterials and implant surfaces to activate the inherent bone regeneration capacity.1, 2 In fact, tissue engineering approach, comprising of biomaterial-based scaffolds and stem cells alone, can be an important tool for better bone treatment.
Recent advancements in biomaterials research have considerably enhanced our understanding and ability to control the stem cell behavior, making them a promising tool in regenerative medicine.3, 4 Hydrogel-based scaffolds, resembling the natural extracellular matrix (ECM) microenvironment, represent a unique platform to exploit the benefits of stem cells for tissue engineering.5, 6 The versatile features of hydrogels ranges from finely tunable mechanical, chemical properties to ease in fabrication of spatially controlled microstructures for tissue engineering applications.7-9 Naturally derived polymers, such as gelatin, have the inherent properties of providing cell binding motifs, biodegradability and ability to be tethered with complex biofunctional molecules.5, 7 Approaches that integrate stem cells with hydrogels for bone tissue regeneration applications offer immense potential to address the problems associated with currently available regeneration therapy methods.10, 11 In addition, culturing stem cells in hydrogel-based 3D environment helps the cells to grow in native-like conditions as well as to maintain their inherent cell properties with respect to expression of cell surface markers, natural RNA expression profiles and proliferation abilities.12 Moreover, growing knowledge to understand the underlying mechanisms and interactions in stem cell-biomaterial constructs has significantly enhanced our abilities to manipulate the stem cell fate.4, 13 Consequently an increasing application of such bioactive hydrogels has garnered considerable interest in the biomaterials and tissue engineering community.
One of the approach to design bioactive hydrogels is to incorporate ceramics nanomaterials, such as hydroxyapatite, carbon nanotubes (CNTs), magnetic nanoparticles (MNPs), calcium phosphate, and bioactive glasses. For example, in a recent work, nanocrystalline hydroxyapatite and single-walled carbon nanotubes were used in combination with hydrogel to impart improved bioactivity and mechanical properties to the nanocomposite hydrogels for bone tissue engineering.14 In another approach, Henstock et al, remotely activated human mesenchymal stem cells (hMSCs) through mechanotransduction in collagen hydrogel using MNPs to provide necessary stimulations for bone regeneration.15 Other nanomaterials recently used to prepare nanocomposite hydrogels for such orthopedic applications include bioprinted ceramic nanoparticles, gold nanoparticles, electrospun organic–inorganic nanofibers, and bioceramics.16-18 All, these studies demonstrate the cooperative effects of nanocomposite hydrogels but in 2D cell culture conditions, most of which are multi-step complex processes. In addition, it is difficult to obtain a uniform dispersion of these nanomaterials within hydrogel network, which results in heterogeneous structures and properties.
Recently, we have introduce a novel 2D nanomaterials known as synthetic nanosilicates to induce osteogenic differentiation of hMSCs in absence of any growth factors.19 In addition, these 2D nanosilicates have high surface-to-volume ratio due to their unique disc-shape morphology that results in enhances interactions with different hydrophilic polymers.20, 21 In spite of the wide range of potential biomedical applications, the role of 2D nanosilicates in tissue engineering is so far limited, as they cannot individually represent the natural 3D cell microenvironment.
Here, we report the development of a nanosilicates-based biodegradable and biocompatible hydrogel platform, which can induce differentiate of encapsulated hMSCs into osteogenic lineage without supplementing any osteoinducting agents such as BMP-2 or dexamethasone. To achieve this, we incorporated nanosilicates in photocrosslinkable GelMA hydrogel. GelMA- nanosilicates hydrogel were then investigated for its cytocompatibility using encapsulated hMSCs by determining production of reactive oxygen species, apoptotic caspase activities and inflammatory responses in order to substantiate its potential in tissue regeneration therapy. For osteoinductivity of nanosilicate in 3D microenvironment, we determined upregulation of alkaline phosphates (ALP) activity and deposition of biomineralized matrix by encapsulated hMSCs. Lastly, in vivo tests in an immunocompetent rat model were performed to determine the biocompatibility of the developed nanocomposite hydrogels at the implant site. This biomimetic nanocomposite hydrogel can provide effective treatment of bone defects by augmenting bone repair and regeneration.
Materials and Methods
Materials
Gelatin type A from porcine skin and methacrylic anhydride were obtained from Sigma-Aldrich and used without further modification. Poly (ethylene glycol) 1000 dimethacrylate (PEGDMA) was procured from Polysciences Inc and Irgacure 2959 photoinitiator for UV curing was obtained from Ciba Speciality Chemicals. Nanosilicate (Laponite XLG) containing SiO2, MgO, Na2O and Li2O were obtained from Southern Clay Products, Inc. (Louisville).
Preparation of photocrosslinkable nanocomposite hydrogels
Photocrosslinkable gelatin (GelMA) was synthesized following previously published studies.22 In brief, gelatin was added at 10% (w/v) in phosphate buffer saline (PBS) and stirred for 1 h at 50 °C until completely dissolved. Methacrylate anhydride (MA) was added drop wise (0.8 mL/g gelatin) to the gelatin solution maintaining the temperature at 50 °C for additional 2 h. The mixture was dialyzed against distilled water (Mw cut-off 12-14 kDa) to remove unreacted components followed by freeze-drying. To prepare nanocomposite hydrogel, 2D nanosilicates at specified amounts (0.01, 0.05, and 0.5 wt%) was first dispersed in a photoinitiator (0.5 % w/v) solution by vigorous vortexing for 15 minutes. This was followed by the addition of lyophilized GelMA foam (7 wt%) followed by vortexing for another 15 minutes to prepare a 7 wt% nSi prepolymer solution. The 7 wt% GelMA prepolymer solutions were prepared similarly without the addition of nanosilicates. To fabricate the hydrogel thin films, 50 μL of the prepolymer solution was added to the space in between the glass slides with 150 μm spacer. It was then exposed to UV at 6.9 mW/cm2 (wavelength 360 - 480 nm) for 20 seconds. The hydrogels on glass slides were then washed in PBS solution to remove unreacted chemical components.
Structural, mechanical and degradation characterization of nanocomposite hydrogels
The hydrogels were imaged under scanning electron microscopy (SEM) for structure and porosity analysis. Porosity measurements of the hydrogel samples were generated by analyzing the SEM images with NIH Image J software. To determine the presence of silicate nanoparticles, Fourier Transform Infrared spectra (FTIR) of the freeze-dried nanocomposite hydrogels were analyzed using a Thermo Scientific Nicolet iS10 instrument. All FTIR experiments were performed at room temperature using the Smart iTR mode utilizing DTGS-KBr detector and KBr beam splitter. Transmittance was measured from 500 to 2100 cm-1 with spectral data spacing of 0.964 cm-1. The mechanical properties of the hydrogels were using a mechanical tester (Instron 5943, Norwood, MA). Cylindrical hydrogel discs (diameter = 7 mm, thickness =2 mm) were fabricated using the conditions explained above and hydrated for 8h in DPBS before a uniaxial mechanical compression test. Compressive load readings versus displacement were obtained using the mechanical tester and a 10 N load cell. Each sample was conditioned to a pre-load of 0.1 N and compressed at a strain rate of 20% per minute to a displacement corresponding to 60% of its thickness. The hydrogels were tested for degradation under enzymatic conditions. The hydrogels were placed in 2.0 U/mL collagenase type II solutions and incubated at 37 °C for 0, 8, 16 and 24 hours. At each time point, the remaining hydrogels were washed with ultrapure water. After washing, the gels were lyophilized. The percent mass degraded was calculated using the weight ratio of dried experimental hydrogel and untreated control hydrogel. Data at each time point was obtained from five samples per group.
hMSC encapsulation in micropatterned nanocomposite hydrogels
Bone marrow-derived hMSCs (Lonza) were cultured in α-MEM growth medium (Gibco, USA), supplemented with 10% of heat-inactivated fetal bovine serum and 1% penicillin/streptomycin (Gibco, USA) at 37 °C with 5% CO2. The cells were cultured until 80% confluency and were used before passage 6 for all experiments. To prepare normal cell-laden nSi hydrogels, hMSCs were trypsinized and suspended in a 7% (w/v) nSi prepolymer solution containing specified concentrations of nSi at a cell density of 3×106 cells/mL This was followed by UV exposure and photocrosslinking to form hydrogels as described above. The fabricated cell-laden hydrogel was then washed in normal media to remove the non-crosslinked components and grown under normal cell culture conditions in an incubator. To perform differentiation studies, the normal media was replaced after 24 hours with osteogenic media which contains normal growth media with 50 μM ascorbic acid phosphate and 10 mM β-glycerophosphate (without any supplementary drugs such as dexamethasone). Media was replaced every alternate day during the experimental period. To prepare micropatterned nSi hydrogels, photomasks with arrays of linear microchannels (100 μm width) were designed using Auto-CAD. A thin layer of PEGDA (20%) was coated on a glass slide to avoid direct cell attachment to the slides. Microchannel units (150 μm height) encapsulating hMSCs were fabricated on PEGDA-coated glass slides similar to the previously described method23 followed by exposure to 7.9 mW/cm2 UV light (360-480 nm) with UV exposure time of 30 seconds. The glass slides containing the microchannels were washed to remove unreacted components and subsequently incubated in normal growth media containing 1% penicillin/streptomycin with standard culture conditions. For differentiation experiments, the media was replaced after 24 hours with osteogenic media without drugs.
In vitro cytocompatibility studies with GelMA-nSi hydrogels
To determine the in vitro cyototoxicity of different nSi hydrogel formulations (0%, 0.01%, 0.05% and 0.5% nSi in prepolymer solution) on encapsulated hMSCs grown in normal media MTS cell proliferation assay (Promega) was used according to the previous method following the manufacturer's instructions.24 Briefly the cells encapsulated in hydrogels, as described above, were grown in normal media for 24 hours in 24 well plates and absorbance of each formulation was measured at 490nm in a plate reader using MTS assay and percentage cell viabilities were analyzed taking only GelMA group (0% nSi) as 100%. A serum starved group with GelMA hydrogel was used as the control group [Ctrl (0)] for the experiment. The same assay was used to perform 96 hours hMSC proliferation study with micropatterned nSi hydrogels. In addition, Alexa Fluor 488 phalloidin (Life Technologies) staining was done and photographed under a fluorescence microscope to trace the filamentous F-actin cytoskeletons of encapsulated hMSCs within the 3D hydrogel environment.
Caspases are a family of genes which helps to maintain h omeostasis through regulating apoptosis and inflammation. To trace the effect of nSi hydrogels on cell apoptosis, caspase-3/7 activity was measured after 24 hours with Apo-ONE Homogeneous Caspase-3/7 Assay (Promega, WI, USA) as per the manufacturer's instructions using a plate reader (Biotek Synergy 2, Winooski, VT). The obtained results were represented as relative fluorescence units (RFU) from experimental groups normalized to GelMA control group (0% nSi) taken as 1.
Intracellular production of superoxide by hMSCs due to nSi exposure in 3D hydrogels was evaluated using intracellular reactive oxygen species (ROS) assay (Cell Biolabs, Inc) as described elsewhere.21 Briefly, hMSCs were grown in nSi hydrogels for 24 hours in normal growth media and fluorescence signals in each well were quantified with a microplate reader (excitation wavelength 488 nm, emission wavelength 533 nm) and represented as normalized fluorescence intensity values where data from 0.05% nSi group was taken as 1. The fluorescence intensity in each well was directly proportional to the ROS level within the cell. The increase in ROS can be invoked by different stress-related factors, including the interactions of the cells with nanosilicates. To determine the possible inflammatory effects of nSi hydrogels, RAW 264.7 macrophages were seeded on the top of photocrosslinked nSi hydrogels at 2×103 cells/well. As controls, RAW cells were treated with 100 ng/mL of lipopolysacchride (LPS). After 24 hours, ELISA assay (SA Biosciences) was performed according to the manufacturer's protocol. The supernatants from all the experimental nSi hydrogel groups were collected to quantify the secreted cytokines IL-6 and TNF-α.25 Each experiment was performed in triplicates.
In vitro 3D osteogenic differentiation of hMSCs in nanocomposite hydrogels
To quantify the calcium biomineralization by hMSCs in hydrogels, the wet weight of hydrogels were first weighed followed by lyophilization. The lyophilized samples were homogenized in 0.5 M HCl and vortexed overnight at 4 °C. The supernatant was collected for calcium colorimetric assay following the manufacturer's protocol (Sigma Aldrich). The calcium content was quantified as μg per mg of wet weight of hydrogel.26 For histology analysis, cell embedded hydrogels were fixed in 4% paraformaldehyde and washed three times with PBS. The fixed constructs were washed with distilled water to remove residual salts followed by the addition of 2% (wt/v) Alizarin Red S (Sigma Aldrich) solution with pH 4.2. After incubation for 20 minutes, the constructs were thoroughly washed three times with 15 minutes incubation times in a shaker and bright field pictures were taken. For intracellular alkaline phosphatase (ALP) activity study of hMSCs encapsulated in nSi hydrogels, total DNA present in the nSi hydrogels was determined with the PicoGreen dsDNA reagents (Invitrogen).27 In brief, disks were rinsed in PBS and placed in a passive lysis buffer (Promega). After one freeze-thaw cycle, the lysate was sonicated and centrifuged at 8,000 rpm for 10 minutes, and the supernatant was quantified for DNA content. ALP activities of encapsulated hMSCs from different groups were quantified with a p-nitrophenyl phosphate (pNPP) colorimetric assay at 405 nm and normalized to DNA content.
To trace the osteogenic differentiation of hMSCs in hydrogel by immunostaining, the hydrogels were first fixed with formalin. Then the constructs were treated with 0.1% Triton X solution for 10 minutes to permealize the cells, followed by treatment with 10% goat serum blocking solution. The cells were then incubated with primary antibodies: mouse monoclonal anti-human osteocalcin (1:100), mouse monoclonal anti-human osteopontin (1:100) and mouse monoclonal anti-human RunX2 (1:100). The cells were treated with secondary antibody: Alexa Fluor 594 goat anti-mouse (1:500) and counterstained with diamidino-2-phenyindole dilactate (DAPI), for staining nuclei. The stained samples were imaged under fluorescence and confocal microscopes.
In vivo subcutaneous implantation of GelMA-nSi hydrogels and immnohistological staining
Male Wistar rats (n=4; ∼250 g, Charles River, MA, USA), were housed in animal care facility (Partners Research Building). All experiments were conducted according to the NIH “Guide for the Care and Use of Laboratory Animals”, and approved by the HMA Standing Committee on Animals. Under general anesthesia with inhalative isoflurane and carprofen-based analgesia, the dorsal skins of the rats were incised. Subcutaneous pockets were generated, and the following cylindrical hydrogel disk constructs (diameter = 7 mm, thickness = 2 mm) were inserted: GelMA only control group with no nSi (Ctrl), GelMA loaded with 0.01% nSi (Low), GelMA loaded with 0.05% nSi (Medium) and GelMA loaded with 0.5% nSi (High). Three or fourteen days after wound closure, the animals were euthanized and the hydrogel implants along with the adjacent subcutaneous tissue were explanted. The collected samples were embedded in O.C.T. and frozen at -80 °C. Hematoxylin/eosin (H&E) staining and immunohistology were performed on 6 μm cryosections of the frozen tissues according to standard protocols.28 For immunohistology, cryosections were first fixed in formalin. This was followed by permeabilization with 0.25% Triton-X, blocking and treatment with primary antibodies: mouse monoclonal anti-rat CD 3 (1:100) and rabbit monoclonal anti-rat CD 68 (1:100). The samples were then treated with secondary antibodies labeled with fluorescent molecules: Alexa Fluor 594 goat anti-mouse (1:500) and Alexa Fluor 488 goat anti-rabbit (1:500). Finally, the samples were mounted with embedding medium solution (Vector labs) containing DAPI.
Statistical Analysis
Continuous variables are represented as mean ± standard deviation (SD) from independent experiments as depicted in the figures. Statistical analysis were performed using one-way analysis of variance (ANOVA) followed by Bonferroni's multiple comparison post-hoc test. All statistical analyses were performed with Graphpad Prism Software. A p value less than 0.05 indicates statistical significance, which was displayed as *= p <0.05, **=p<0.01, ***=p<0.001.
Results and Discussion
Characteristics of synthesized GelMA-nSi hydrogel
A range of hydrogels were synthesized by varying the concentrations of nSi [0, 0.01, 0.05, 0.5 % (w/v)] in GelMA as illustrated in Figure 1A. The NS used in the study had a dimension of 20-30 nm in diameter confirmed by Transmission Electron Microscope (Figure S1A). The microstructure examination of GelMA and GelMA-NS indicates the formation of a highly porous and interconnected network without the aggregation of nSi. The SEM micrograph (Fig. 1B) revealed well distributed, smooth polyhedral pores in the crosslinked hydrogel matrix. The porosity of the scaffolds can significantly affect the cellular behaviour both actively and passively by, for example, affecting the rate of diffusion of nutrients and byproducts. We examined the changes in porosity due to the addition of silicates by analyzing the SEM images. Porosity was measured as an index of area occupied by the pores using Image J. Previous reports with silica-based nanoparticles embedded hydrogel matrices suggested a potential decrease in porosity with an increasing concentration of silicates.29 We also noticed a similar trend in the group with the highest concentration of nSi (Fig. 1C). However, the other two groups showed no significant changes in porosity compared to the control. Here, we aimed to dope the hydrogel with a minimum quantity of nSi to achieve stem cell differentiation with limited toxicity or inflammation. Such a concentration of nSi did not drastically alter the physical properties. Moreover, the porosity values of our study correspond well with those present in the literature with high cell viability.30 Additionally, the presence of nSi within the GelMA after the crosslinking process was confirmed via infrared (IR) spectra, as represented in Fig. 1D. The GelMA-nSi sample demonstrated the presence of both nSi (Si-O-Si peak at 960 cm-1) and GelMA (amine peaks at 1500 cm-1 and 1650 cm-1), indicating successful formation of nanocomposite materials.
Figure 1.
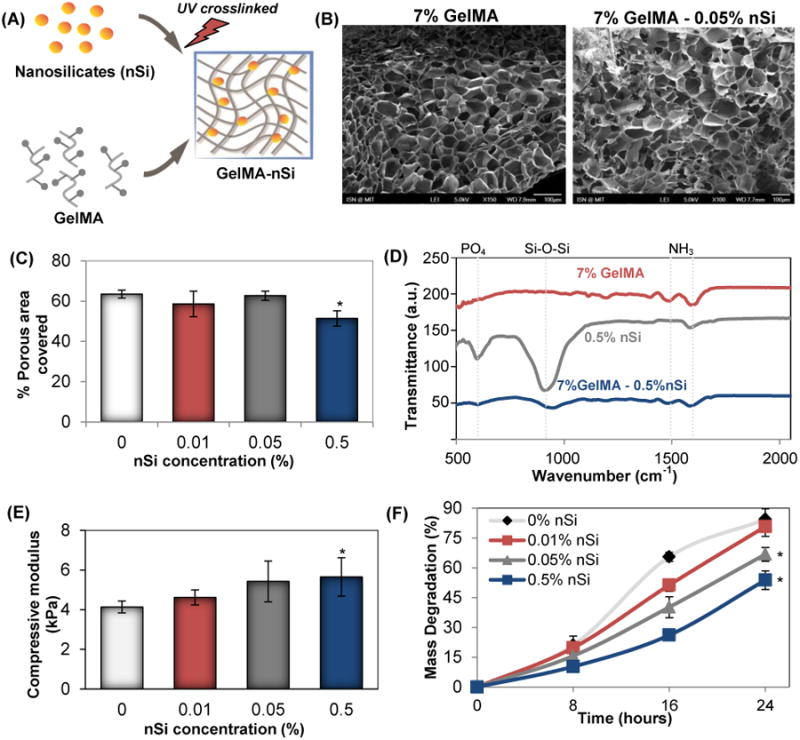
Characterization of photocrosslinked GelMA-nSi hydrogel. (A) Schematic of GelMA-nSi hydrogel prepared by UV crosslinking GelMA prepolymer with NS. (B) SEM images of cross-sections of GelMA and GelMA-nSi hydrogels (Scale: 100μm). (C) Percentage porous area covered was quantified by SEM image analysis of 7% GelMA hydrogels with different concentrations of NS. (D) FTIR spectra of GelMA-nSi after UV crosslinking, confirming the presence of silicate and gelatin components in the nanocomposite construct. (E) Compressive mechanical properties of 7% GelMA hydrogel with different nSi concentrations with respect to compressive modulus. (F) Degradation profiles of 7% GelMA hydrogels with various nSi concentrations when exposed to collagenase for 24h. Data represent Mean ± SD (n=3). *=p< 0.05 compared to control GelMA group (0% nSi).
Most of the studies involving hydrogel-nanoparticle composites have shown a change in the mechanical properties of the hydrogel matrix upon incorporation of the nanoparticles.30, 31 Here, the non-covalent interactions between the heterogeneously distributed charges of discotic nSi and hydrogel matrices at the interface30-32 lead to an increasing trend in elastic modulus with increased nSi concentrations. However, a significant increase in the elastic modulus was noticed only in the group with 0.5% nSi (Fig. 1E), which can be attributed to the possible reinforcement of GelMA by the incorporated nSi.32 Interactions between GelMA and nSi also changed the reaction kinetics of the hydrogel as evidenced by the results of our degradation tests (Fig. 1F). The dry weights of the composite materials were measured for the groups at 0, 8, 16 and 24 hours after the treatment with collagenase enzyme. The groups with 0.05% and 0.5% (w/v) of nSi showed a significantly reduced rate of degradation compared to the other two groups after 24 hours of reaction with collagenase. Such an effect could be due to the physical masking of the sites of action for enzymatic reaction by nSi.33, 34 Similar degradation trends were also reported earlier in collagen-bioactive glass nanocomposite hydrogels as compared to only collagen.35
In vitro cytocompatibility studies with GelMA-NS hydrogels
Investigating the cytocompatibility of the developed hydrogel is critical to consolidate its application for stem cell-based bone tissue engineering. In vitro cytocompatibility of the developed GelMA-nSi hydrogel was analyzed by studying encapsulated cell spreading, viability, apoptosis and stress behaviour. The hMSC encapsulated GelMA group was considered as the positive control and a serum starved group was taken as the negative control. Figure 2A explains the process associated with the encapsulation of hMSCs in the composite hydrogel. After 4 days of culture in normal growth media, the morphology and spreading of cells in the 3D hybrid matrix was analysed by staining for F-actin filaments (Fig. 2B). Uniform spreading of cells were visualized in all groups except for the group with 0.5% NS. This group also showed a lower number of cells and cell spreading compared to the other groups (Fig. 2B). To confirm this observation, we looked at the change in cell metabolic activities of different groups grown in normal growth media with respect to the controls using MTS assay (Fig. 2C). A noticeable decrease was seen only in the group with the highest concentration of nSi. The other two groups with nSi (0.01 and 0.05%) exhibited no significant difference in viability with respect to the positive control. A decrease in metabolic activity in the group with the highest nSi concentration (0.5% nSi) can probably be attributed to the high concentration of nSi itself while a resultant reduced porosity could be an obvious factor that hindered the cell spreading (Fig. 2B). To trace the cell apoptosis, caspase 3/7 activities of encapsulated cells were evaluated (Fig. 2D). Elevated activities of caspase 3/7 were present with the hydrogel loaded with 0.5% nSi, suggesting a higher rate of apoptosis in this group. Reduced activities of caspase (3/7) at lower concentrations of nSi could well be considered as a promising sign of in vitro cytocompatibility. As expected, the serum starved Ctrl (0) group demonstrated significantly high apoptosis. Additionally, we confirmed the biocompatibility of the generated hydrogels by examining the levels of reactive oxygen species (ROS) and pro-inflammatory cytokines (Fig. S2A & S2B). Overproduction of ROS is a well-known indicator of oxidative stress, which leads to cell damage. Prior studies with silicate nanoparticles have employed analysis of ROS levels to identify the threshold concentrations of nanoparticles, that induce oxidative stress.21 No apparent overproduction of ROS was noted in the groups with low concentration of nSi (Fig. S2B). However, in accordance with the already discussed results, 0.5% nSi group induced an up regulation of ROS. Finally, to investigate if the hydrogel constructs can invoke any adverse immune reactions we looked at the in vitro release of IL-6 and TNF-α pro-inflammatory cytokines from macrophages when the latter were cultured on the different hydrogel substrates. For this experiment, LPS-induced inflammation was used as a positive control. The change in cytokine expression profiles helped us to further validate the cytocompatibility of the GelMA-nSi hydrogels with different nSi concentrations (Fig. S2B). The results showed no significant differences in cytokine profile with only GelMA and lower concentrations of nSi, while 0.5% nSi group showed significant pro-inflammatory cytokine productions. These findings together confirmed the in vitro cytocompatibility of GelMA-nSi hydrogels for use in 3D hMSC culture.
Figure 2.
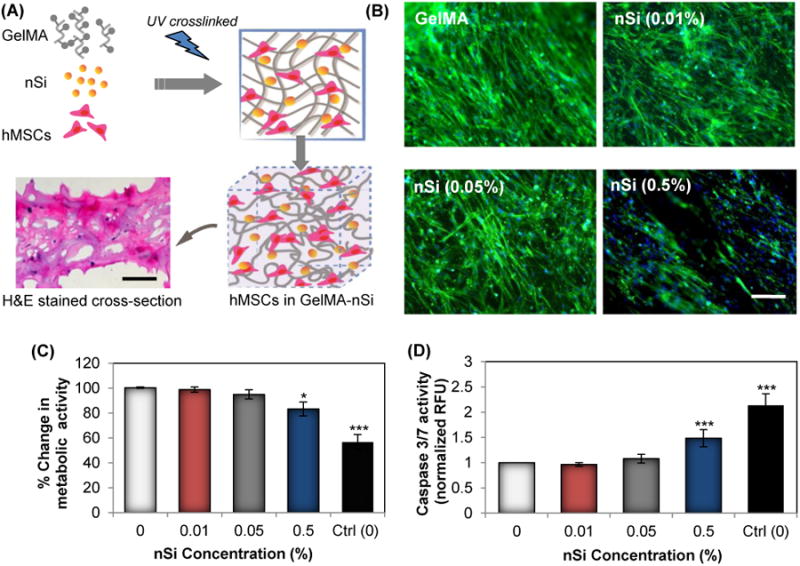
In vitro cell behavior of hMSCs encapsulated in GelMA-nSi nanocomposite hydrogel. (A) Schematic for photo-encapsulation hMSCs in GelMA-nSi hydrogel consisting of nanosilicates and gelatin matrix. Lower left panel microphotograph represents H&E stained cross-section of 7% GelMA hydrogels with 0.05% nSi with dark blue spots showing encapsulated cell nuclei after a day in culture. Scale bar: 200μm. (B) Fluorescence images of F-actin cytoskeleton stained cells (in green) with nuclei (in blue) grown in 3D 7% GelMA hydrogel constructs with different nSi concentrations for 4 days in normal growth media. F-actin cytoskeleton is an indicator and biophysical regulator of cell shape and morphology. F-actin staining confirmed the normal cell architectures in all the experimental groups, except the 0.5% nSi group where proper cell migration and normal cytoskeleton structures were limited. Scale bar: 100μm. (C) Metabolic activity of encapsulated cells in 7% GelMA hydrogels with various nSi concentrations was measured by MTS assay after 24h to determine the cytotoxic effects on the formulated hydrogels. (D) Caspase 3/7 activity assay was used to quantify the apoptotic effects of different nSi concentrations on encapsulated cells. Data represent Mean ± SD (n=3). *=p< 0.05 & ***=p< 0.001 compared to control GelMA group (0% nSi).
Effect of GelMA-nSi hydrogel on in vitro differentiation of encapsulated hMSCs
Hallmarks to detect terminally differentiated osteoblasts from multipotent hMSCs include expression of early markers like ALP and calcium deposition.36 This is normally followed by the production of bone specific proteins for matrix remodelling and finally mineralization. The GelMA-nSi hydrogel was designed to represent a biomimetic niche with an expectation to induce osteogenic differentiation of encapsulated hMSCs. For the differentiation experiments, we selected GelMA-nSi groups with insignificant in vitro cytotoxicity. ALP expressions of encapsulated hMSCs were investigated after 21 days of culture in GelMA-nSi hydrogels (Fig. 3B). Addition of 0.01% and 0.05% of nSi showed a considerable enhancement in the production of ALP compared to the matrix with only GelMA (Fig. 3B). Mineralization of the nanocomposite hydrogel was assessed by Alizarin red staining and determination of calcium levels (Fig. 3C & 3D). Alizarin red staining indicated a proportional rise in mineralization with respect to the concentration of nSi (Fig. 3C & S1). The calcium levels in the groups with 0.01% and 0.05% nSi were significantly elevated (Fig. 3D). This further supports the trend being established through our observation that nSi at particular concentrations were able to trigger osteogenic differentiation of hMSC in 3D matrix. Among various concentrations of nSi, 0.05% (w/v) demonstrated strongest bioactivity and cytocompatibility for in vitro culture. This was further demonstrated through immunostaining for RunX2, a major transcription factor exclusively expressed in mineralized tissues. RunX2 is involved in the upregulation of osteo-related proteins such as osteopontin and osteocalcin that are responsible for bone formation and remodelling.36 Immunostaining for RunX2, osteopontin and osteocalcin, showed a robust expression of these markers in the presence of silicates in 0.05% GelMA-nSi hydrogel as compared to only GelMA (Fig. 3E). A confocal micrograph of osteocalcin-stained hMSCs encapsulated in GelMA-nSi (0.05%) hydrogel confirmed the uniformity of osteogenic differentiation of hMSCs throughout the thickness (150μm) of the 3D construct (Fig. 3F). Overall, the addition of nSi into the 3D hydrogel matrix introduced bioactivity and facilitated osteogenic differentiation of hMSCs without the need of supplemental drugs and growth factors. This is particularly advantageous for eventual clinical realization where delivery and maintenance of the biofunctionality of growth factors is a concern. In contrary to other methods, this strategy also alleviates the need for chemically conjugating therapeutic molecules, such as BMP-2 growth factor, to the hydrogel matrices for bone therapy applications.37
Figure 3.
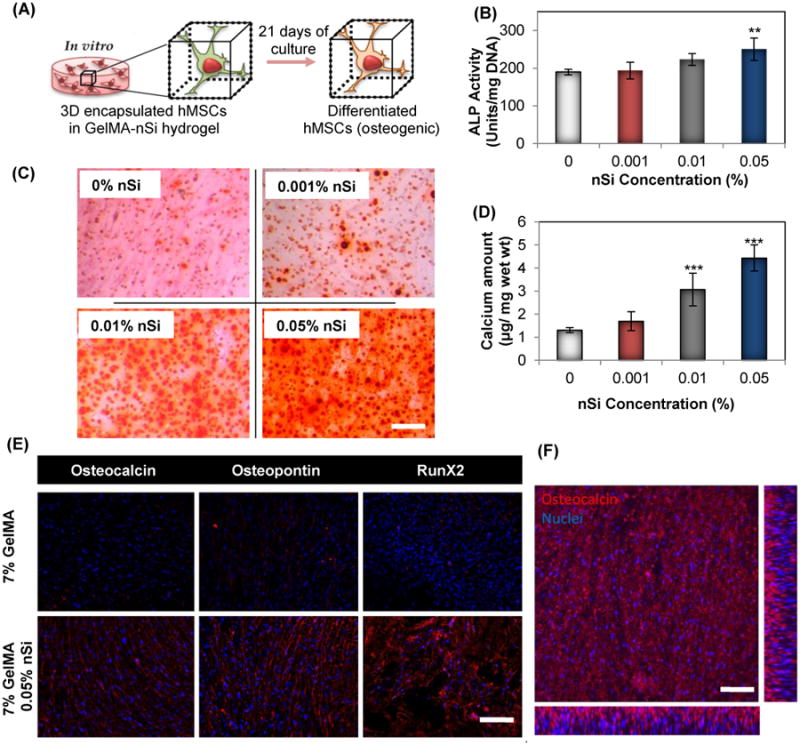
Effect of GelMA-nSi hydrogel on differentiation potential of encapsulated hMSCs. (A) Schematic of hMSCs differentiated to osteogenic lineage when grown in 3D GelMA-nSi hydrogel environment for 21 days. Quantification of osteogenic differentiation of hMSCs within hydrogels was performed by (B) ALP activity, (C) alizarin red staining and (D) calcium quantification in 7% GelMA hydrogels with varying concentration of NS after Day 21 of culture in osteoconductive media with no supplementary growth factors or drugs. (E) Expression of three osteogenic markers - osteocalcin, osteopontin and Runt-related transcription factor 2 (RunX2) by the encapsulated cells were detected by immunostaining and fluorescence microscopy. Osteogenic markers were stained in red, while cell nuclei in blue. (F) Confocal laser microscopy image of hMSCs in GelMA-nSi (0.05%) hydrogel expressing osteocalcin, immunostained in red. The picture represents a single z-plane image and x-z and y-z cross-sectional images of the encapsulated cells in hydrogel. Scale bars: 100μm. Data represent Mean ± SD (n=3). *=p< 0.05, **=p< 0.01 & ***=p< 0.001 compared to control GelMA only group (0% nSi).
The ability to generate patterns of cells at microscale through precise engineering of cell culture substrates such as hydrogels has led to several applications in the field of tissue engineering and regenerative medicine.5 The controlled microenvironment with specific substrate features can impart additional attributes to the developed tissue construct by manipulating the cell behaviour and functionalities. Such micropatterning technologies also help to engineer explicit physiological and pathophysiological conditions for various applications.38, 39 Reports have demonstrated the amenability of gelatin derived hydrogels to be patterned with versatile features using multiple conventional photopatterning and 3D printing techniques.5, 23 This is particularly important as it allows us to mimic the collagen matrix architecture of bone which is a uniaxial arrangement. However, the addition of nanoclay-based materials to such hydrogel matrices can hinder the resolution and stability of patterned structures for bone tissue engineering applications. To study this, the hMSC encapsulated composite GelMA-nSi hydrogels were subjected to a photopatterning technique using a photomask. The overall scheme involved in photopatterning the composite hydrogel has been illustrated in Fig. S3A. Patterning of nSi embedded GelMA hydrogel did not show any sign of loss in resolution compared to the widely established control GelMA. Also, the structural integrity was maintained during the culture period of 21 days (Fig. S3B). The spreading and cytocompatibility of cells in the patterned constructs was monitored for a period of 72 hours using live/dead staining. This indicated partial alignment of cells in the patterned constructs along the axis of the patterns and also expressed high viability (Fig. S3B). In addition, the metabolic activities of the different groups did not show any significant differences in 72 hours compared to the control groups (Fig. S3C). High surface area of the hydrogels due to patterning may have also contributed to the maintenance of the high viability. Finally, the staining for osteogenic markers - ALP and osteocalcin in the patterned construct demonstrated similar results as that of the unpatterned constructs. This confirmed that the micropatterning technique did not adversely affect the differentiation ability of hMSCs, and that the presence of nSi in the GelMA matrix consistently boosted the osteogenic protein expression of hMSCs encapsulated in a 3D microenvironment (Fig. S3D).
In vivo biocompatibility of GelMA-nSi hydrogel
To understand the potential of the developed hydrogel for diverse clinical applications, such as implantation in order to support bone repair in vivo, it is important to study the in vivo biocompatibility of developed GelMA-nSi hydrogels with different nSi percentages (0.01%, 0.05% and 0.5%). We investigated the change in host tissue reactions post subcutaneous implantation of hydrogels in immunocompetent rats. The implanted hydrogels were retrieved on day 3 and day 14 to examine the host immune reactions to the implanted hydrogels with different formulations. Fig. 4A shows the process of subcutaneous implantation of hydrogel constructs and its subsequent integration into the host tissue after 14 days. This was further confirmed by H&E staining in both Ctrl GelMA only and 0.05% nSi groups (Fig. 4B). However, the group with 0.5% nSi showed enhanced cellular infiltration around the implants. Derived from these findings and supported by in vitro results, we suspected a strong presence of immune cells in the group with the highest concentration of nSi.
Figure 4.
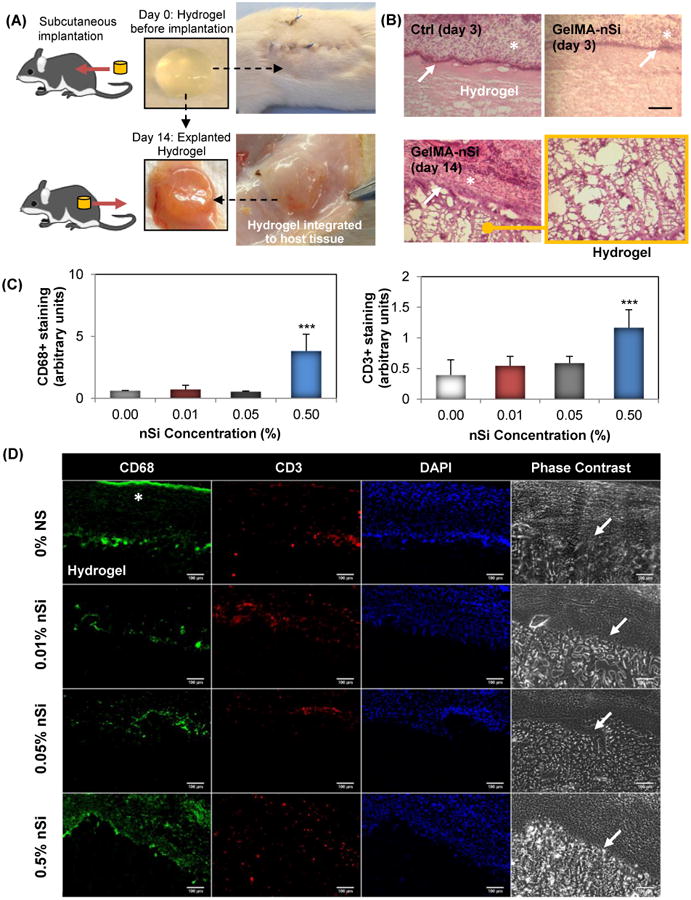
In vivo biocompatibility of the GelMA-nSi hydrogels. (A) Hydrogel constructs with different formulations (0%, 0.01%, 0.05%, 0.5% nSi) were implanted subcutaneously in immunocompetent rat model and collected after 3 and 14 days to study the inflammatory responses. (B) Histological results with H&E staining of Ctrl 0% and 0.05% nSi constructs with 7% GelMA showed no signs of inflammation in the adjacent tissue region. * indicates the host tissue regions and white arrows show the tight interface between intact hydrogel and tissues. A magnified image of the hydrogel region confirms there was no infiltration of macrophages or neutrophils into the hydrogel region. This was further confirmed by (C) immunohistochemistry analysis for CD68 and CD3 inflammatory markers in the hydrogel/tissue interface region where (D) represents the microphotographs of different groups at day 14 post implantation immunostained with CD68 (in green) and CD3 (in red), nuclei (DAPI, in blue) and corresponding phase contrast images. Scale bar: 100μm. The results demonstrate that the hydrogels remained firmly attached to the tissue surface without invasion by foreign macrophage and T-cells in Ctrl 0%, 0.01% and 0.05% nSi groups. However, 7% GelMA 0.5% nSi group showed marked increase in macrophage levels compared to the other groups. Data represent Mean ± SD (n=3). ***=p< 0.001 compared to control GelMA group (0% nSi).
Under a clinical setting, the elicitation of immune reactions to a newly developed material for medical use is of paramount importance. Thus we further assessed the harvested tissue/hydrogel samples for the existence of macrophages and T-lymphocytes by staining for CD68 and CD3, respectively (Fig. 4C & 4D). Fluorescence images from immunostaining were subjected to semi-quantitative analysis using Image J (Fig. 4C). The results were similar to what we concluded from in vitro cytocompatibility tests where groups with 0.01% and 0.05 % nSi showed no noticeable differences in inflammatory responses compared to the Ctrl group, while group with 0.5% nSi demonstrated a significantly elevated number of inflammatory cell infiltration. Such responses could be cited to the elevated levels of pro-inflammatory cytokines and free radicals with respect to increasing concentrations of nSi (Fig. 2E&F). These in vivo results, in addition to the in vitro findings, support the hypothesis that such gelatin-based nanocomposite hydrogels can be used as an in vivo implant for diverse biomedical applications such as bone tissue engineering and regenerative medicine.40-42 In an interesting recent work, Neffe et al reported the development of a lysine-functionalized, biocompatible, porous gelatin graft for in vivo bone regeneration in critical size rat bone defect model.40 A comparative study analysis of developed GelMA-nSi and other nanocomposite hydrogels used for in vitro and in vivo bone regeneration applications has been summarized in Table S1. Together, these findings suggest the immense potential of gelatin to develop mechanically tunable, strong interconnected porous nanocomposite hydrogel networks which can guide stem cell based osteogenic differentiation. This will be particularly useful to treat non-load-bearing orthotopic sites such as in cranial defects. However, further studies with clinically relevant bone defect animal models, such as murine models with critical-size calvarial defects, are needed to fully understand the long term safety and efficacy of the developed GelMA-nSi hydrogel.
Conclusion
Overall, we have demonstrated that photocrosslinkable gelatin hydrogels loaded with an optimal concentration of osteoinductive silicate nanoclay, can promote the survival, migration, proliferation and in situ differentiation of encapsulated human stem cells without the addition of growth factors. The study confirmed in vitro osteogenic differentiation of encapsulated hMSCs within nSi containing GelMA hydrogels as evident by enhanced ALP activities, upregulation of osteogenic proteins production and enhanced deposition of inorganic calcium. In addition, we have demonstrated the in vivo biocompatibility and local non-inflammatory interactions of the implanted nSi hydrogels. These findings highlight the development of composite hydrogel structures for in situ bone regeneration therapy. The biomimetic hydrogels loaded with nSi may be used to design miniaturized 3D bone-like structures, resembling architectural and compositional aspects of natural bone tissue, for bone biology, pathophysiology and drug toxicity studies.
Supplementary Material
Acknowledgments
A.K. acknowledges funding from the office of Naval Research Young National Investigator Award, the National Institutes of Health (EB012597, AR057837, DE021468, HL099073, AI105024, AR063745) and the Presidential Early Career Award for Scientists and Engineers (PECASE). A.P. would like to acknowledge the Institutional Development Award (IDeA) from the National Institute of General Medical Sciences, NIH (P20GM103638-04) and New Faculty General Research Fund (NFGRF) form University of Kansas. A.A. acknowledges postdoctoral funding from the German Heart Foundation, Frankfurt, Germany. J.A.U acknowledges postdoctoral funding from SENESCYT, Quito, Ecuador. M.A.H. thanks the National Strategic Technologies and Innovation Program of King Abdulaziz City for Science and Technology (KACST), grant number 11-NAN1544-03 for their support and funding. Authors thank Dr. Byambaa Batzaya for the valuable scientific inputs and support with graphic design artworks.
Footnotes
The authors declare no competing financial interest.
References
- 1.Amini R, Laurencin CT, Nukavarapu SP. Critical reviews in biomedical engineering. 2012;40:363–408. doi: 10.1615/critrevbiomedeng.v40.i5.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, Lim J, Teoh SH. Biotechnology advances. 2013;31:688–705. doi: 10.1016/j.biotechadv.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Seong JM, Kim BC, Park JH, Kwon IK, Mantalaris A, Hwang YS. Biomedical materials. 2010;5:062001. doi: 10.1088/1748-6041/5/6/062001. [DOI] [PubMed] [Google Scholar]
- 4.Guvendiren M, Burdick JA. Nature communications. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 5.Annabi N, Tamayol A, Uquillas JA, Akbari M, Bertassoni LE, Cha C, Camci-Unal G, Dokmeci MR, Peppas NA, Khademhosseini A. Advanced materials. 2014;26:85–123. doi: 10.1002/adma.201303233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt JA, Chen R, van Veen T, Bryan N. Journal of Materials Chemistry B. 2014;2:5319–5338. doi: 10.1039/c4tb00775a. [DOI] [PubMed] [Google Scholar]
- 7.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536–5544. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosiewicz KA, Kolb L, van der Vlies AJ, Martino MM, Lienemann PS, Hubbell JA, Ehrbar M, Lutolf MP. Nat Mater. 2013;12:1072–1078. doi: 10.1038/nmat3766. [DOI] [PubMed] [Google Scholar]
- 9.Camci-Unal G, Annabi N, Dokmeci MR, Liao R, Khademhosseini A. NPG Asia Mater. 2014;6:e99. [Google Scholar]
- 10.Shi X, Zhou J, Zhao Y, Li L, Wu H. Advanced healthcare materials. 2013;2:846–853. doi: 10.1002/adhm.201200333. [DOI] [PubMed] [Google Scholar]
- 11.Weinand C, Pomerantseva I, Neville CM, Gupta R, Weinberg E, Madisch I, Shapiro F, Abukawa H, Troulis MJ, Vacanti JP. Bone. 2006;38:555–563. doi: 10.1016/j.bone.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Haycock JW. Methods in molecular biology. 2011;695:1–15. doi: 10.1007/978-1-60761-984-0_1. [DOI] [PubMed] [Google Scholar]
- 13.Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, Burdick JA. Nat Mater. 2013;12:458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Im O, Li J, Wang M, Zhang LG, Keidar M. International Journal of Nanomedicine. 2012;7:2087–2099. doi: 10.2147/IJN.S29743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henstock JR, Rotherham M, Rashidi H, Shakesheff KM, El Haj AJ. Stem cells translational medicine. 2014;3:1363–1374. doi: 10.5966/sctm.2014-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao G, Schilling AF, Yonezawa T, Wang J, Dai G, Cui X. Biotechnology journal. 2014;9:1304–1311. doi: 10.1002/biot.201400305. [DOI] [PubMed] [Google Scholar]
- 17.Heo DN, Ko WK, Bae MS, Lee JB, Lee DW, Byun W, Lee CH, Kim EC, Jung BY, Kwon IK. Journal of Materials Chemistry B. 2014;2:1584–1593. doi: 10.1039/c3tb21246g. [DOI] [PubMed] [Google Scholar]
- 18.Bose S, Fielding G, Tarafder S, Bandyopadhyay A. Trends in biotechnology. 2013;31:594–605. doi: 10.1016/j.tibtech.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaharwar K, Peppas NA, Khademhosseini A. Biotechnology and bioengineering. 2014;111:441–453. doi: 10.1002/bit.25160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xavier JR, Thakur T, Desai P, Jaiswal MK, Sears N, Cosgriff-Hernandez E, Kaunas R, Gaharwar AK. ACS nano. 2015;9:3109–3118. doi: 10.1021/nn507488s. [DOI] [PubMed] [Google Scholar]
- 21.Gaharwar K, Mihaila SM, Swami A, Patel A, Sant S, Reis RL, Marques AP, Gomes ME, Khademhosseini A. Advanced materials. 2013;25:3329–3336. doi: 10.1002/adma.201300584. [DOI] [PubMed] [Google Scholar]
- 22.Shin SR, Jung SM, Zalabany M, Kim K, Zorlutuna P, Kim SB, Nikkhah M, Khabiry M, Azize M, Kong J, Wan KT, Palacios T, Dokmeci MR, Bae H, Tang XS, Khademhosseini A. ACS nano. 2013;7:2369–2380. doi: 10.1021/nn305559j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikkhah M, Eshak N, Zorlutuna P, Annabi N, Castello M, Kim K, Dolatshahi-Pirouz A, Edalat F, Bae H, Yang Y, Khademhosseini A. Biomaterials. 2012;33:9009–9018. doi: 10.1016/j.biomaterials.2012.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul A, Shao W, Abbasi S, Shum-Tim D, Prakash S. Molecular pharmaceutics. 2012;9:2479–2488. doi: 10.1021/mp3000502. [DOI] [PubMed] [Google Scholar]
- 25.Gaharwar K, Avery RK, Assmann A, Paul A, McKinley GH, Khademhosseini A, Olsen BD. ACS nano. 2014;8:9833–9842. doi: 10.1021/nn503719n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang NS, Varghese S, Li H, Elisseeff J. Cell and tissue research. 2011;344:499–509. doi: 10.1007/s00441-011-1153-2. [DOI] [PubMed] [Google Scholar]
- 27.Bhat A, Hoch AI, Decaris ML, Leach JK. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:4844–4852. doi: 10.1096/fj.12-213611. [DOI] [PubMed] [Google Scholar]
- 28.Assmann A, Zwirnmann K, Heidelberg F, Schiffer F, Horstkotter K, Munakata H, Gremse F, Barth M, Lichtenberg A, Akhyari P. Biomaterials. 2014;35:7416–7428. doi: 10.1016/j.biomaterials.2014.05.034. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Hou R, Cheng Y, Fu J. Soft Matter. 2012;8:6048–6056. [Google Scholar]
- 30.Shin SR, Bae H, Cha JM, Mun JY, Chen YC, Tekin H, Shin H, Farshchi S, Dokmeci MR, Tang S, Khademhosseini A. ACS nano. 2012;6:362–372. doi: 10.1021/nn203711s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin SR, Aghaei-Ghareh-Bolagh B, Dang TT, Topkaya SN, Gao X, Yang SY, Jung SM, Oh JH, Dokmeci MR, Tang XS, Khademhosseini A. Advanced materials. 2013;25:6385–6391. doi: 10.1002/adma.201301082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaharwar K, Rivera C, Wu CJ, Chan BK, Schmidt G. Materials science & engineering C, Materials for biological applications. 2013;33:1800–1807. doi: 10.1016/j.msec.2012.12.099. [DOI] [PubMed] [Google Scholar]
- 33.Desimone MF, Helary C, Rietveld IB, Bataille I, Mosser G, Giraud-Guille MM, Livage J, Coradin T. Acta biomaterialia. 2010;6:3998–4004. doi: 10.1016/j.actbio.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Desimone MF, Helary C, Quignard S, Rietveld IB, Bataille I, Copello GJ, Mosser G, Giraud-Guille MM, Livage J, Meddahi-Pelle A, Coradin T. ACS applied materials & interfaces. 2011;3:3831–3838. doi: 10.1021/am2009844. [DOI] [PubMed] [Google Scholar]
- 35.El-Fiqi A, Lee JH, Lee EJ, Kim HW. Acta biomaterialia. 2013;9:9508–9521. doi: 10.1016/j.actbio.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi DT. World journal of stem cells. 2014;6:94–110. doi: 10.4252/wjsc.v6.i2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holloway JL, Ma H, Rai R, Burdick JA. Journal of controlled release : official journal of the Controlled Release Society. 2014;191:63–70. doi: 10.1016/j.jconrel.2014.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khetani SR, Bhatia SN. Nature biotechnology. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 39.March S, Ng S, Velmurugan S, Galstian A, Shan J, Logan DJ, Carpenter AE, Thomas D, Sim BK, Mota MM, Hoffman SL, Bhatia SN. Cell host & microbe. 2013;14:104–115. doi: 10.1016/j.chom.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neffe T, Pierce BF, Tronci G, Ma N, Pittermann E, Gebauer T, Frank O, Schossig M, Xu X, Willie BM, Forner M, Ellinghaus A, Lienau J, Duda GN, Lendlein A. Advanced materials. 2015 doi: 10.1002/adma.201404787. [DOI] [PubMed] [Google Scholar]
- 41.Paul A. Nanomedicine (Lond) 2015;10:1371–1374. doi: 10.2217/nnm.15.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pacelli S, Manoharan V, Desalvo A, Lomis N, Prakash S, Paul A. J Mater Chem B. 2015 doi: 10.1039/C5TB01686J. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


