Abstract
Pompe disease (PD) is a lysosomal disorder caused by acid α-glucosidase (GAA) deficiency. Progressive muscular weakness is the major symptom of PD, and enzyme replacement therapy can improve the clinical outcome. However, to achieve a better clinical outcome, alternative therapeutic strategies are being investigated, including gene therapy and pharmacological chaperones. We previously used lentiviral vector-mediated GAA gene transfer in PD patient-specific induced pluripotent stem cells. Some therapeutic efficacy was observed, although glycogen accumulation was not normalized. Transcription factor EB is a master regulator of lysosomal biogenesis and autophagy that has recently been associated with muscular pathology, and is now a potential therapeutic target in PD model mice. Here, we differentiated skeletal muscle from PD patient-specific induced pluripotent stem cells by forced MyoD expression. Lentiviral vector-mediated GAA and transcription factor EB gene transfer independently improved GAA enzyme activity and reduced glycogen content in skeletal muscle derived from PD-induced pluripotent stem cells. Interestingly, GAA and transcription factor EB cooperatively improved skeletal muscle pathology, both biochemically and morphologically. Thus, our findings show that abnormal lysosomal biogenesis is associated with the muscular pathology of PD, and transcription factor EB gene transfer is effective as an add-on strategy to GAA gene transfer.
Introduction
Pompe disease (PD) is a lysosomal storage disease caused by acid α-glucosidase (GAA) deficiency, with subsequent glycogen accumulation in skeletal muscle resulting in progressive muscular weakness.1 According to its onset and severity, PD is divided into infantile-onset and late-onset subtypes.2 Enzyme replacement therapy can improve major clinical conditions such as walking capacity and respiratory function.3,4 However, antibody production against the therapeutic enzyme, rhGAA, is frequently observed and closely associated with limited treatment efficacy.5 Gene therapy is an alternative therapeutic option for PD. Indeed, respiratory function has been shown to improve by direct adeno-associated virus gene transfer into the diaphragm of PD patients.6 Other therapeutic strategies such as pharmacological chaperones and adjunctive β2 agonists are being considered, and are close to clinical translation.7,8
Induced pluripotent stem cells (iPSCs) are a powerful tool to investigate disease mechanisms of monogenic diseases. Disease modeling of cardiac hypertrophy, which is frequently observed in infantile PD, has been performed by several groups.9,10 Accordingly, we have previously shown that late-onset PD patient-derived iPSCs are useful in disease modeling, with lentiviral GAA transfer ameliorating disease-specific changes in differentiated cardiomyocytes.11 Although higher GAA expression was observed in PD iPSCs after lentiviral-mediated GAA gene transfer, residual glycogen accumulation was still observed. A similar phenomenon has been observed with in vivo gene transfer experiments: adeno-associated virus gene transfer in PD model mice increases GAA enzyme activity in skeletal muscle, although residual glycogen accumulation remains.12
Gene therapy is a promising strategy for PD treatment because life-long therapeutic effect could be achieved. However, residual glycogen accumulation could be observed after GAA overexpression in PD. It is hypothesized that already accumulated glycogen is not cleared by GAA overexpression in PD. Therefore, we have sought an alternative approach other than GAA overexpression and focused on a cellular clearance mechanism against residual glycogen accumulation. Transcription factor EB (TFEB) regulates expression of lysosomal genes, and is a master regulator of lysosomal biogenesis and autophagy.13 TFEB overexpression promotes cellular exocytosis in several lysosomal storage diseases including PD.14,15
Here, we assessed the efficacy of TFEB gene transfer in skeletal muscle derived from human PD iPSCs. Consequently, we show that TFEB gene transfer synergistically improves muscular pathology together with GAA gene transfer. This suggests that impaired glycogen clearance can be alleviated by combined TFEB gene transfer and GAA rescue in PD iPSC-derived skeletal muscle.
Results
PD-iPSCs successfully differentiate into skeletal muscle
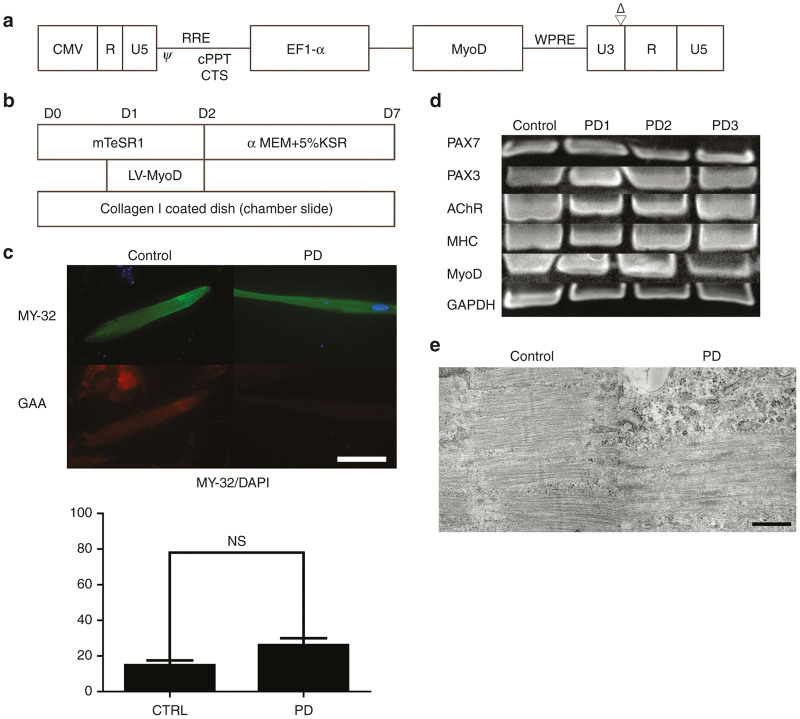
We cloned the myogenic differentiation 1 (MYOD1) gene into a third generation lentiviral vector (Figure 1a). Myogenic differentiation was performed according to the described differentiation protocol (Figure 1b). Approximately 10 days after MyoD transfer, spontaneously contracting spindle-shaped cells were observed in control and PD iPSCs. Reverse-transcriptase polymerase chain reaction (RT-PCR) supported myogenic marker expression in differentiated skeletal muscle (Figure 1c). Compared with the other clones (PD1 and PD3), PD2 showed more efficient differentiation capacity into skeletal muscle, therefore, we mainly investigated PD2 in further experiments. Skeletal muscle protein expression, including fast skeletal myosin (MY-32), was confirmed in differentiated skeletal muscle derived from both control and PD iPSCs (Figure 1d). Based on our immunofluorescence results, differentiation efficacy into fast (type 2) skeletal muscle fibers was estimated to be ~20–30%. There was no significant difference in the percentage of MY-32-positive fast skeletal muscle in control and PD iPSC-derived skeletal muscle. Transmission electron microscopy analysis also showed that functional skeletal muscle with a sarcomere structure had differentiated from control and PD iPSCs (Figure 1e). These results suggest that a MyoD-based differentiation protocol successfully differentiates fast skeletal muscle from control and PD iPSCs.
Figure 1.
Myogenic differentiation of Pompe disease iPSC. (a) Vector construct. MyoD was cloned into downstream of EF-1-α promoter. (b) Differentiation protocol. (c) RT-PCR of skeletal muscle derived from iPS cell lines (Control, PD1, PD2 and PD3). PAX7, PAX3, AChR, MHC, MyoD and GAPDH were analyzed. (d) Immunofluorescence of differentiated skeletal muscle (Control and PD2). Fast skeletal myosin (MY-32) (Alexa488) and GAA (Alexa568) were stained with DAPI. Scale bar, 100 µm. MY-32 positive cells were counted to evaluate myogenic differentiation. Data were expressed as mean ± SEM. (NS: Not significant) (n = 3) (e) Electron microscopy of differentiated skeletal muscle (Control, PD2). Sarcomeric structure was observed in differentiated skeletal muscle. Scale bar, 0.5µm. iPSCs, induced pluripotent stem cells; GAA, acid α-glucosidase; PD, Pompe disease; DAPI, 4’,6-diamidino-2-phenylindole; RT-PCR, reverse-transcriptase PCR; SEM, standard error of mean.
PD iPSC-derived skeletal muscle shows disease-specific pathology
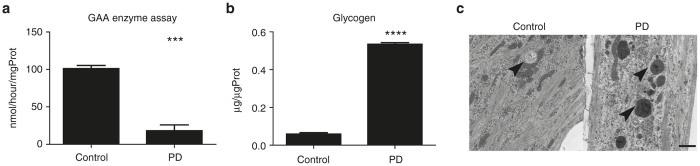
To investigate biochemical properties, we examined GAA enzyme activity and glycogen concentration in differentiated skeletal muscle. PD iPSC-derived skeletal muscle showed lower GAA enzyme activity and higher glycogen content than control iPSC-derived skeletal muscle (Figure 2a,b). These findings suggest that PD iPSC-derived skeletal muscle maintains biochemical and molecular features of PD.
Figure 2.
Disease specific changes after myogenic differentiation. (a) GAA enzyme assay of differentiated skeletal muscle (Control, PD2). Data were expressed as mean ± SEM. (***P < 0.001) (n = 3). (b) Glycogen assay of differentiated skeletal muscle (Control, PD2). Data were expressed as mean ± SEM. (****P < 0.0001) (n = 3) (c) Electron microscopy of differentiated skeletal muscle (Control, PD2). Arrow is indicating lysosome. Scale bar, 1 µm. GAA, acid α-glucosidase; PD, Pompe disease; SEM, standard error of mean.
To determine the status of cellular organelles, differentiated skeletal muscle was analyzed by TEM. Compared with control iPSC-derived skeletal muscle, lysosomal enlargement, and glycogen accumulation was observed in PD iPSC-derived skeletal muscle (Figure 2c). Thus, MyoD overexpression causes differentiation of PD iPSCs into skeletal muscle with ultrastructural disease-associated features.
TFEB gene transfer improves lysosomal glycogen accumulation
We have previously shown that lentiviral-mediated GAA gene transfer ameliorates glycogen accumulation in PD iPSCs and cardiomyocytes.11 However, glycogen accumulation was not normalized, despite complete restoration of GAA enzyme activity. We hypothesized that already accumulated glycogen cannot be cleared by GAA overexpression.
To overcome this limitation, we adopted an add-on strategy using TFEB gene transfer. TFEB is a master regulator of lysosomal biogenesis and autophagy, and TFEB overexpression promotes lysosomal exocytosis and cellular clearance in multiple sulfatase deficiency and mucopolysaccharidosis type III.14 Furthermore, TFEB overexpression has already been shown to be effective in muscular glycogen clearance in PD model mouse skeletal muscle.15 Thus, we tested if this phenomenon can be translated to human.
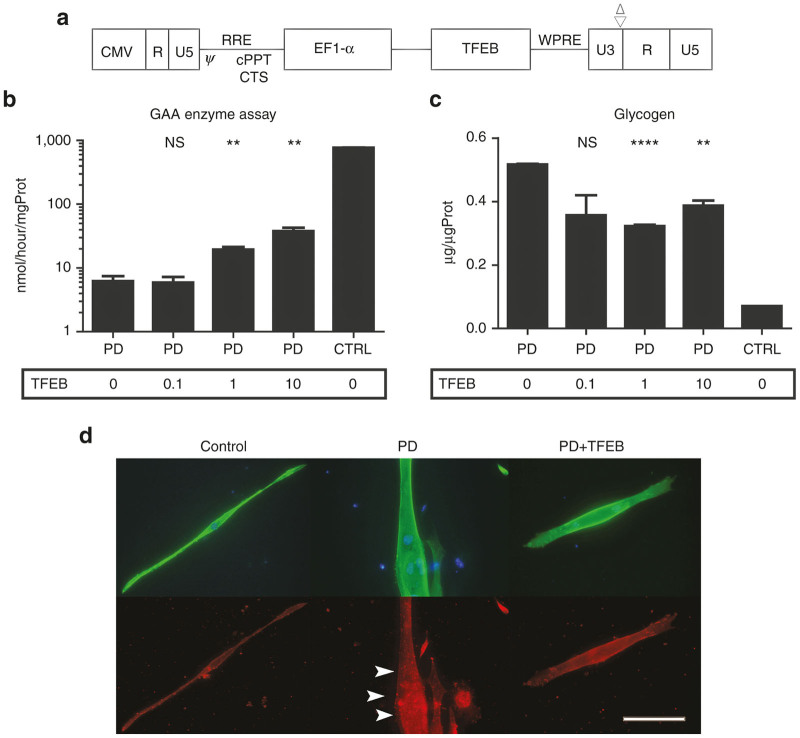
Consequently, we generated a third generation lentiviral vector expressing TFEB (Figure 3a). We transduced TFEB gene into PD iPSCs, and then induced myogenic differentiation. TFEB gene transfer increased GAA enzyme activity and reduced glycogen accumulation in iPSC-derived skeletal muscle, especially at higher multiplicity of infection (MOI) (Figure 3b,c). LC-3b expression was also alleviated by TFEB transfer (Figure 3d). It is suggested that TFEB transfer could ameliorate glycogen accumulation and autophagic build-up in PD iPSC derived skeletal muscle thorough the activation of lysosomal biogenesis and exocytosis.
Figure 3.
Myogenic differentiation after lentiviral TFEB gene transfers. (a) Vector construct. TFEB was cloned into downstream of EF-1-α promoter. (b) GAA enzyme assay of differentiated skeletal muscle after TFEB gene transfer (Control, PD2). Transduction is conducted at the MOI of 0, 0.1, 1, and 10. Data were expressed as mean ± SEM. (NS not significant, **P < 0.01) (n = 3). (c) Glycogen assay of differentiated skeletal muscle after lentiviral TFEB gene transfer (Control, PD2). Transduction is conducted at the MOI of 0, 0.1, 1, and 10. Data were expressed as mean ± SEM. (NS not significant, **P < 0.01, ****P < 0.0001) (n = 3). (d) Immunofluorescence of differentiated skeletal muscle (Control, PD2). Transduction is conducted at the MOI of 10. Fast skeletal myosin (MY-32) (Alexa488) and LC-3b (Alexa568) were stained with DAPI. Arrow is indicating autophagosome. Scale bar, 100 µm. GAA, acid α-glucosidase; PD, Pompe disease; SEM, standard error of mean; TFEB, transcription factor EB; MOI, multiplicity of infection; DAPI, 4’,6-diamidino-2-phenylindole.
GAA and TFEB cooperatively improve the muscle pathology of PD
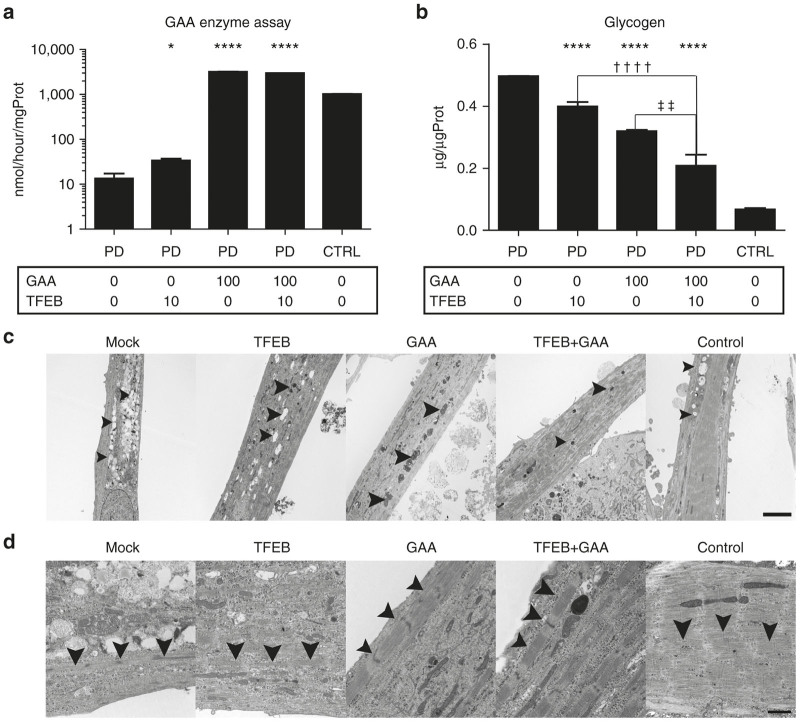
To observe the synergistic effect of TFEB and GAA, we transduced both TFEB and GAA genes into PD iPSCs, and then induced myogenic differentiation. GAA enzyme activity increased and glycogen accumulation decreased with combined TFEB and GAA gene transfer (Figure 4a,b). Higher enzyme activity due to GAA gene transfer was not affected by TFEB gene transfer. In addition, combined GAA and TFEB gene transfer effectively reduced glycogen accumulation.
Figure 4.
Myogenic differentiation after lentiviral GAA and TFEB gene transfers. (a) GAA enzyme assay of differentiated skeletal muscle after lentiviral GAA and TFEB gene transfer (Control and PD2). Transduction is conducted at the MOI of 0, 10, and 100. Data were expressed as mean ± SEM. (*P < 0.05, ****P < 0.0001) (n = 3). (b) Glycogen assay of differentiated skeletal muscle after lentiviral GAA and TFEB gene transfer (Control, PD2). Transduction is conducted at the MOI of 0, 10, and 100. Data were expressed as mean ± SEM. (****P < 0.0001, ††P < 0.01, ††††P < 0.0001) (n = 3). (c) Electron microscopy of skeletal muscle after gene transfer (Control and PD2). Arrow is indicating lysosome. Scale bar, 5 µm. (d) Electron microscopy of skeletal muscle after gene transfer (Control, PD2). Arrow is indicating sarcomeric structure. Scale bar, 1 µm. GAA, acid α-glucosidase; PD, Pompe disease; SEM, standard error of mean; TFEB, transcription factor EB; MOI, multiplicity of infection; DAPI, 4’,6-diamidino-2-phenylindole.
Furthermore, TEM findings showed morphologically improved glycogen accumulation and lysosomal enlargement with combined GAA and TFEB overexpression (Figure 4c). Similar to our biochemical results, combined GAA and TFEB overexpression significantly improved muscular pathology. Interestingly, a mature structural sarcomere was observed with combined GAA and TFEB overexpression (Figure 4d). In mock and TFEB over-expression, Z band was observed but sarcomeric structure was not as prominent as control. On the other hand, Z-line, I-band, and A-band were more clearly formed in GAA over-expression. Mature sarcomeric structure (Z-line, I-band, and A-band) was most remarkably observed in combinational GAA and TFEB over-expression and control. This indicates that profound glycogen reduction is associated with efficient myogenic differentiation.
Discussion
PD is a monogenic disease caused by GAA deficiency, with glycogen accumulation in lysosomes causing progressive muscular weakness and hypertrophic cardiomyopathy.1 Although, glycogen accumulation is the primary cause of skeletal muscle weakness and cardiomyopathy, the precise mechanism of this phenomenon is not clearly understood. Several cellular mechanisms, such as autophagic buildup and abnormal calcium homeostasis, are associated with PD skeletal muscle pathology.16,17 To identify the cellular disease mechanism, pluripotent stem cells that can be differentiated into cardiomyocytes have been generated from both infantile and late-onset PD patients.9–11 Using these cells, dysregulation of cellular metabolism such as glycolysis and oxidative phosphorylation has previously been shown.9 To investigate skeletal muscle pathology, we differentiated skeletal muscle from late-onset PD iPSCs by MyoD overexpression, and subsequently investigated disease mechanisms using biochemical and pathological analyses.
Disease modeling by MyoD-based myogenic differentiation has already been performed for musculoskeletal and inherited metabolic disorders.18–21 In PD, differences in properties of slow (type 1) and fast (type 2) muscle fibers is important to understand skeletal muscle pathology. It is suggested that fast (type 2) muscle fibers are more closely associated with PD pathology than slow (type 1) muscle fibers.22 Therefore, MyoD-based myogenic differentiation that predominantly induces fast (type 2) skeletal muscle fibers may be optimal to replicate the muscular pathology of PD. Indeed, it has been shown that conversion of muscle fibers from fast (type 2) to slow (type 1) partially improves the muscular pathology of PD23. Here, we show that skeletal muscle differentiated from late-onset PD iPSCs exhibits relevant biochemical abnormalities such as reduced GAA enzyme activity and increased glycogen content. Additionally, TEM analysis showed lysosomal enlargement and glycogen accumulation in differentiated skeletal muscle. LC-3b was also up-regulated, reflecting the abnormal autophagic buildup observed in PD skeletal muscle. Thus, our findings show that MyoD-based myogenic differentiation replicates biochemical and molecular disease specific changes of PD patients. On the other hand, entire differentiation capacity was not high in our experiment. More efficient and robust differentiation protocol is warranted for the more precise analysis.
Gene therapy is a promising therapeutic strategy for monogenic diseases including PD. Accordingly, lentiviral ex vivo gene therapy and adeno-associated virus gene therapy may become available therapeutic options for PD patients in the future. Overexpression of GAA may ameliorate disease-associated symptoms, and we have already shown the efficacy of GAA transfer in patient-derived iPSCs and differentiated cardiomyocytes.11 However, as we found previously, glycogen accumulation is not normalized. Residual glycogen accumulation is also observed in skeletal muscle of advanced PD model mice receiving adeno-associated virus gene transfer.12 Furthermore, neonatal lentiviral-mediated GAA gene transfer also failed to normalize glycogen accumulation in cardiomyocytes and skeletal muscle.24 This indicates that it is necessary to develop an alternative therapeutic strategy for greater efficacy.
TFEB is a master regulator of lysosomal biogenesis and autophagy, and promotes lysosomal exocytosis in several lysosomal storage diseases.14 TFEB overexpression effectively reduces glycogen accumulation in skeletal muscle of PD model mice.15 Moreover, GAA enzyme activity was partially restored and glycogen accumulation improved to some extent by TFEB overexpression. Certain chemical compounds increase TFEB expression and provide therapeutic efficacy in other lysosomal storage diseases. Some of these compounds have already been used in clinical trials, for example, β-cyclodextrin and genistein activate TFEB and improve lysosomal function.25,26 We performed TFEB overexpression to determine its therapeutic efficacy in vitro, and show that TFEB improves the muscular pathology of PD iPSC-derived skeletal muscle. It has been already shown that TFEB overexpression improved several lysosomal enzyme activities by enhancing protein holding and trafficking of lysosomal proteins.27 It is hypothesized that mutant GAA was also stabilized by TFEB overexpression in our experiment.
We also found that combined GAA and TFEB gene transfer results in greater phenotypic correction in skeletal muscle differentiated from late-onset PD iPSCs. Consequently, GAA and TFEB cooperatively reduce glycogen accumulation in PD iPSC-derived skeletal muscle. It is also hypothesized that both mutant and overexpressed lysosomal enzyme might be enhanced by TFEB. Alternatively, the observed synergistic effects may be owing to amelioration of autophagic buildup, which may interfere with therapeutic enzyme distribution in PD. TFEB expression also induces exocytosis of autolysosomes, which may reduce LC-3, and reflect amelioration of autophagic buildup in PD. Overall, these findings suggested that a GAA gene replacement strategy has limited efficacy in skeletal muscle, but can be enhanced by TFEB gene transfer.
In addition to profound glycogen clearance by GAA and TFEB gene transfer, improvement of sarcomere structure was observed in our experiments. There were two possibilities in this improvement. One possibility was reduction of glycogen directly related to this structual improvement. The other possibility was that myogenic differentiation was promoted by TFEB expression. However, the later possibility was less likely because only TFEB expression did not promote muscular differentiation. Thus, high level glycogen clearance induced by coexpression GAA and TFEB could be a main cause of structural improvement. This explanation was supported by following evidence that neural differentiation capacity is reduced in GM2-gangliosidosis patient-derived iPSCs, while overexpression of the deficient enzyme (β-hexosaminidase) rescued this reduced neural differentiation capacity.28 These observations indicate that lysosomal storage (such as GM2 ganglioside accumulation in lysosomes) interferes with neural differentiation. It is suggested that lysosomal storage might be associated with low differentiation capacity in vitro and improved by overexpression of the deficient lysosomal enzyme.
Limitation of our study is the numbers of cell lines of iPSC. Three different clones from one patient were investigated but we mainly analyzed single clone (PD2) in our experiment. A comparison was only conducted with single clone (Control) from healthy control. Clonal variations should be taken into account in our study. The other limitation is transduction efficacy. We have shown that ~60% transduction efficacy was achieved in the same vector construct expressing fluorescent protein.11 Transduction efficacy was assumed to be high but not 100%. This might result in not only limited differentiation efficacy, but incomplete clearance of the glycogen.
Materials and Methods
iPSC culture
Patient specific iPSC (HSP0175, 0176, 0177; PD1, 2, and 3) were kindly provided from (RIKEN BRC, , Japan). Healthy control iPSC (HPS0223; Control) was also provided from RIKEN BRC. iPSCs were cultured on radio-inactivated Mouse embryonic fibroblast (Kitayama Labes, Japan) in Dulbecco’s modified essential medium (DMEM)/F12 medium (Sigma Aldrich, St Louis, MO) supplied with 20% KnockOut serum replacement (KSR) (Life Technologies, Carlsbad, CA), 1% Glutamax (Life Technologies), 0.01% beta-mercaptoethanol (Life Technologies), nonessential amino acids (Life Technologies), and Fibroblast growth factor 2 (Wako Pure Chemical Industries, Japan). Every 4–5 days, iPSCs were detached using trypsin and passaged onto radio-inactivated Mouse embryonic fibroblast.
Lentiviral vector construction
The third generation lentiviral vector, CSII-EF1α-MCS, was kindly provided by Dr Miyoshi (RIKEN BRC). MyoD, GAA, and TFEB were cloned into the EcoRI site of the multiple cloning sites by overnight ligation using Mighty Mix (Takara Bio, Japan). As described previously,11 large-scale culture of HEK293T cells was performed to collect viral supernatant. The virus was then concentrated by ultra-centrifugation using Centricon (EMD Millipore, Germany). Viral titers were determined using a p24 enzyme-linked immunosorbent assay kit (Cell Biolabs, San Diego, CA).
Myogenic differentiation
Myogenic differentiation was performed according to the following differentiation protocol. On day 1, iPSCs were detached using trypsin solution, and 1 × 106 cells transferred to 24-well collagen-I coated multiwell plates (AGC TECHNO GLASS, Japan). On day 2, MyoD transfer was performed in each well at a MOI of 50. On day 3, the medium was changed to α-minimum essential medium (MEM) (Thermo Scientific, Waltham, MA) containing 5% KSR, and replaced every day until days 7–10.
Lentiviral transduction
On day 1, iPSCs were detached using trypsin solution, and 1 × 106 cells transferred to 24-well collagen-I coated multiwell plates. On day 2, TFEB and/or GAA transfer was performed in each well at the calculated MOI. On day3, medium was changed in each well. On day5, MyoD transfer performed as described earlier. On day 10, cells were collected for further analysis.
Immunofluorescence
Cells were fixed using 4% paraformaldehyde (Wako Pure Chemical Industries) for 15 minutes at room temperature. Next, cells were permeabilized with 0.2% Triton-X (Sigma Aldrich) for 15 minutes at room temperature. The primary antibody was reacted overnight at 4°C, and the secondary antibody for 30 minutes at room temperature. Cells were mounted in 4’,6-diamidino-2-phenylindole-containing mounting medium (Life Technologies). Images were obtained by fluorescence microscopy. The antibodies used in this study are described in the Supplementary Materials and Methods (see Supplementary Table S1).
GAA enzyme assay
GAA enzyme activity was measured as described previously.11 Briefly, cells were homogenized in 200 µl distilled water and the supernatant was collected after centrifugation at 14,000×g for 10 minutes at 4°C. Protein concentration of the supernatant was measured by the bicinchoninic acid assay (BCA) protein assay. Then 80 µl homogenate containing 15 µg of protein and 120 µl 4-MU buffer containing 100 µl of 6mmol/l 4MU-α-D-glucopyranoside (Sigma Aldrich) and 20 µl of 100 µmol/l Acarbose (Sigma Aldrich) were incubated at 37°C for 30 minutes. After 3.8 ml of 0.15 mol/l glycine carbonate stop buffer (pH 10.4) was added, fluorescent intensity was measured using a spectrophotometer and enzyme activity (nmol/hr/mg protein) was calculated according to the intensity and protein concentration.
Glycogen assay
Glycogen was measured as described previously.11 Briefly, cells were homogenized in distilled water and the protein concentration measured using a BCA protein assay kit. Glycogen content was measured using a glycogen assay kit (BioVision, Milpitas, CA). Glycogen concentration was calculated based on the protein concentration obtained by BCA protein assay.
RT-PCR
Total RNA was extracted from differentiated skeletal muscle using the Maxwell 16 Simply RNA purification kit (PROMEGA, Madison, WI). RT-PCR was performed using a RT-PCR kit (Takara Bio). Primers are described in the Supplementary Materials and Methods. (see Supplementary Table S2).
Transmission electron microscopy
Cells were fixed in 2% glutaraldehyde, followed by 1% osmium tetroxide. Epoxy resin embedded sections were analyzed by transmission electron microscopy.
Acknowledgments
We thank Division of Molecular Cell Biology, Core Research Facilities for Basic Science, for providing fluorescent and transmission electron microscopy. S.Y. is supported by Grant-in-aid for JSPS research fellow (DC2).
O.T. and I.H. have active research support from Genzyme Japan and Shire Japan. These activities have been fully disclosed and are managed under a Memorandum of Understanding with the Conflict of Interest Resolution Board of The Jikei University School of Medicine.
References
- Hers, HG (1963). Alpha-glucosidase deficiency in generalized glycogenstorage disease (Pompe’s disease). Biochem J; 86: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishnani, PS and Howell, RR (2004). Pompe disease in infants and children. J Pediatr 144(5 Suppl): S35–S43. [DOI] [PubMed] [Google Scholar]
- Kishnani, PS, Corzo, D, Nicolino, M, Byrne, B, Mandel, H, Hwu, WL et al. (2007). Recombinant human acid [alpha]-glucosidase: major clinical benefits in infantile-onset Pompe disease. Neurol 68: 99–109. [DOI] [PubMed] [Google Scholar]
- van der Ploeg, AT, Clemens, PR, Corzo, D, Escolar, DM, Florence, J, Groeneveld, GJ et al. (2010). A randomized study of alglucosidase alfa in late-onset Pompe’s disease. N Engl J Med 362: 1396–1406. [DOI] [PubMed] [Google Scholar]
- Kishnani, PS, Goldenberg, PC, DeArmey, SL, Heller, J, Benjamin, D, Young, S et al. (2010). Cross-reactive immunologic material status affects treatment outcomes in Pompe disease infants. Mol Genet Metab 99: 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, BK, Collins, SW, Conlon, TJ, Mah, CS, Lawson, LA, Martin, AD et al. (2013). Phase I/II trial of adeno-associated virus-mediated alpha-glucosidase gene therapy to the diaphragm for chronic respiratory failure in Pompe disease: initial safety and ventilatory outcomes. Hum Gene Ther 24: 630–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenti, G, Zuppaldi, A, Gabriela Pittis, M, Rosaria Tuzzi, M, Annunziata, I, Meroni, G et al. (2007). Pharmacological enhancement of mutated alpha-glucosidase activity in fibroblasts from patients with Pompe disease. Mol Ther 15: 508–514. [DOI] [PubMed] [Google Scholar]
- Koeberl, DD, Austin, S, Case, LE, Smith, EC, Buckley, AF, Young, SP et al. (2014). Adjunctive albuterol enhances the response to enzyme replacement therapy in late-onset Pompe disease. FASEB J 28: 2171–2176. [DOI] [PubMed] [Google Scholar]
- Huang, HP, Chen, PH, Hwu, WL, Chuang, CY, Chien, YH, Stone, L et al. (2011). Human Pompe disease-induced pluripotent stem cells for pathogenesis modeling, drug testing and disease marker identification. Hum Mol Genet 20: 4851–4864. [DOI] [PubMed] [Google Scholar]
- Raval, KK, Tao, R, White, BE, De Lange, WJ, Koonce, CH, Yu, J et al. (2015). Pompe disease results in a Golgi-based glycosylation deficit in human induced pluripotent stem cell-derived cardiomyocytes. J Biol Chem 290: 3121–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, Y, Kobayashi, H, Higuchi, T, Shimada, Y, Era, T, Kimura, S et al. (2015). Disease modeling and lentiviral gene transfer in patient-specific induced pluripotent stem cells from late-onset Pompe disease patient. Mol Ther Methods Clin Dev 2: 15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, B, Zhang, H, Bird, A, Li, S, Young, SP and Koeberl, DD (2009). Impaired clearance of accumulated lysosomal glycogen in advanced Pompe disease despite high-level vector-mediated transgene expression. J Gene Med 11: 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello, M, Palmieri, M, di Ronza, A, Medina, DL, Valenza, M, Gennarino, VA et al. (2009). A gene network regulating lysosomal biogenesis and function. Science 325: 473–477. [DOI] [PubMed] [Google Scholar]
- Medina, DL, Fraldi, A, Bouche, V, Annunziata, F, Mansueto, G, Spampanato, C et al. (2011). Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 21: 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampanato, C, Feeney, E, Li, L, Cardone, M, Lim, JA, Annunziata, F et al. (2013). Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med 5: 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben, N, Schreiner, C, Baum, R, Takikita, S, Xu, S, Xie, T et al. (2010). Suppression of autophagy permits successful enzyme replacement therapy in a lysosomal storage disorder–murine Pompe disease. Autophagy 6: 1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, JA, Li, L, Kakhlon, O, Myerowitz, R and Raben, N (2015). Defects in calcium homeostasis and mitochondria can be reversed in Pompe disease. Autophagy 11: 385–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudenege, S, Lebel, C, Huot, NB, Dufour, C, Fujii, I, Gekas, J et al. (2012). Myoblasts derived from normal hESCs and dystrophic hiPSCs efficiently fuse with existing muscle fibers following transplantation. Mol Ther 20: 2153–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffioletti, SM, Gerli, MF, Ragazzi, M, Dastidar, S, Benedetti, S, Loperfido, M et al. (2015). Efficient derivation and inducible differentiation of expandable skeletal myogenic cells from human ES and patient-specific iPS cells. Nat Protoc 10: 941–958. [DOI] [PubMed] [Google Scholar]
- Tanaka, A, Woltjen, K, Miyake, K, Hotta, A, Ikeya, M, Yamamoto, T et al. (2013). Efficient and reproducible myogenic differentiation from human iPS cells: prospects for modeling Miyoshi Myopathy in vitro. PLoS One 8: e61540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuno, T, Osafune, K, Sakurai, H, Asaka, I, Tanaka, A, Yamaguchi, S et al. (2014). Functional analysis of iPSC-derived myocytes from a patient with carnitine palmitoyltransferase II deficiency. Biochem Biophys Res Commun 448: 175–181. [DOI] [PubMed] [Google Scholar]
- van den Berg, LE, Drost, MR, Schaart, G, de Laat, J, van Doorn, PA, van der Ploeg, AT et al. (2013). Muscle fiber-type distribution, fiber-type-specific damage, and the Pompe disease phenotype. J Inherit Metab Dis 36: 787–794. [DOI] [PubMed] [Google Scholar]
- Takikita, S, Schreiner, C, Baum, R, Xie, T, Ralston, E, Plotz, PH et al. (2010). Fiber type conversion by PGC-1α activates lysosomal and autophagosomal biogenesis in both unaffected and Pompe skeletal muscle. PLoS One 5: e15239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyosen, SO, Iizuka, S, Kobayashi, H, Kimura, T, Fukuda, T, Shen, J et al. (2010). Neonatal gene transfer using lentiviral vector for murine Pompe disease: long-term expression and glycogen reduction. Gene Ther 17: 521–530. [DOI] [PubMed] [Google Scholar]
- Song, W, Wang, F, Lotfi, P, Sardiello, M and Segatori, L (2014). 2-Hydroxypropyl-β-cyclodextrin promotes transcription factor EB-mediated activation of autophagy: implications for therapy. J Biol Chem 289: 10211–10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskot, M, Montefusco, S, Jakóbkiewicz-Banecka, J, Mozolewski, P, Węgrzyn, A, Di Bernardo, D et al. (2014). The phytoestrogen genistein modulates lysosomal metabolism and transcription factor EB (TFEB) activation. J Biol Chem 289: 17054–17069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W, Wang, F, Savini, M, Ake, A, di Ronza, A, Sardiello, M et al. (2013). TFEB regulates lysosomal proteostasis. Hum Mol Genet 22: 1994–2009. [DOI] [PubMed] [Google Scholar]
- Ogawa, Y, Tanaka, M, Tanabe, M, Suzuki, T, Togawa, T, Fukushige, T et al. (2013). Impaired neural differentiation of induced pluripotent stem cells generated from a mouse model of Sandhoff disease. PLoS One 8: e55856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.