Abstract
Two-photon calcium imaging provides an optical readout of neuronal activity in populations of neurons with subcellular resolution. However, conventional two-photon imaging systems are limited in their field of view to ~1 mm2, precluding the visualization of multiple cortical areas simultaneously. Here, we demonstrate a two-photon microscope with an expanded field of view (>9.5 mm2) for rapidly reconfigurable simultaneous scanning of widely separated populations of neurons. We custom designed and assembled an optimized scan engine, objective, and two independently positionable, temporally multiplexed excitation pathways. We used this new microscope to measure activity correlations between two cortical visual areas in mice during visual processing.
Ensemble neuronal activity is of key interest in system neuroscience to understand sensory coding, motor output, and cognitive function. Measuring neuronal activity in populations of neurons in vivo is technically challenging, due to the densely packed neuropil and sensitive neuroanatomy that is best probed with minimally invasive approaches. Two-photon1 population calcium imaging in vivo2 supports dense optical sampling of neuronal activity deep in scattering tissue such as mammalian neocortex, and unambiguous identification of recorded neurons, particularly when genetically encoded indicators are employed3,4. Two-photon population calcium imaging is used to measure single cell-level stimulus selectivity in dense local ensembles of neurons5,6 and learning and task-related activity in awake mice7, including those navigating in a virtual reality environment8.
However, conventional two-photon imaging systems retain cellular resolution over only a restricted field of view (FOV) of ~1 mm2. This limitation precludes the simultaneous examination of areas large enough to encompass multiple cortical areas. Neuronal activity that supports sensory coding and motor output is distributed across multiple areas and millimeters of neocortex even in mice9,10 (Fig. 1a), thus new instrumentation is needed to obtain a cellular-resolution view into ongoing neural activity across extended cortical networks, which is essential for elucidating principles of neural coding.
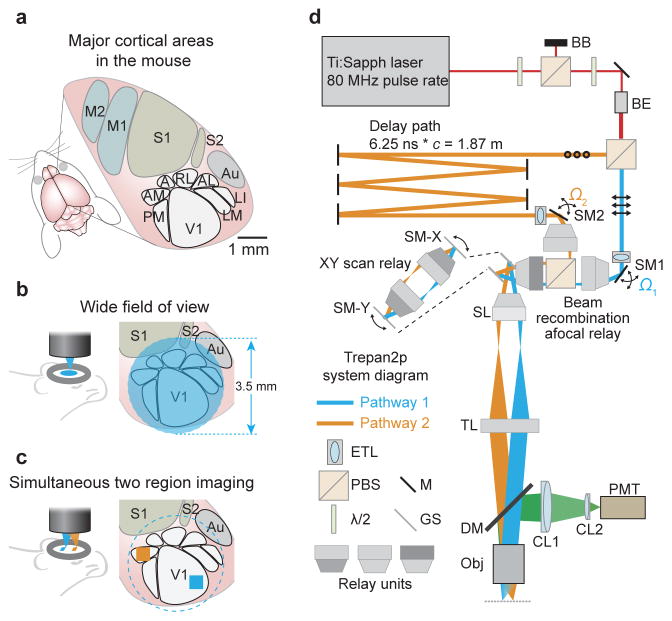
Figure 1. Treapn2p system layout.
(a) In the mouse, primary visual cortex (V1) is surrounded by higher visual areas (HVAs; PM = posteromedial; AM = anteromedial; A = anterior; RL = rostrolateral; AL = anterolateral; LM = lateromedial; LI = laterointermediate), which are distributed across several millimeters of cortex (M1,M2 = primary and secondary motor cortex; S1,S2 = primary and secondary somatosensory cortex; Au = auditory cortex). A wide field of view (FOV) is required to image neuronal activity in these distributed cortical areas simultaneously. (b) The 3.5 mm FOV can encompass V1 and HVAs. (c) The individual imaging regions can be independently positioned and repositioned anywhere within the full FOV by the steering mirrors (SM1, SM2 in d) for XY position, and the tunable lenses (ETL in d) for independent Z positioning. (d) Two imaging beams are temporally multiplexed and independently positioned in XY and Z prior to the scan mirrors (SM-X, SM-Y). First, overall power is attenuated using a half-wave plate (λ/2), a polarizing beam splitting cube (PBS) and a beam block (BB). After a second λ/2 (used to determine the power ratio sent to the two pathways) and a beam expander (BE), a second PBS divides the beam into two pathways. Pathway 1 (in blue, p-polarization, indicated by the arrows) passes directly to a motorized steering mirror (SM1) for positioning in XY. Pathway 2 (in orange, s-polarization, indicated by the circles) passes to a delay arm where it travels 1.87 meters further than pathway 1 using mirrors (M), thus delaying it by 6.25 ns before being directed to SM2. Directly prior to SM1 and SM2 are electrically tunable lenses (ETL) that can adjust the Z position (focal plane) of the pathways independently. The two pathways are recombined (beam recombination relay), and sent to X and Y galvanometer scanners (GS) that are connected by an afocal relay (expanded view inset). A scan lens (SL) and tube lens (TL) focuses the two multiplexed beams onto the back aperture of the objective (Obj). Fluorescence is directed to a photomultiplier tube (PMT) via an infrared-passing dichroic mirror (DM) and two collection lenses (CL1, CL2).
Here we present a two-photon imaging system that preserves individual neuron resolution across a wide FOV (>9.5 mm2) and can simultaneously scan neuronal populations over extended cortical networks. The system uses custom optics (objective and scan engine) and dynamically and independently repositionable temporally multiplexed imaging beams. We refer to the system as a Trepan2p (Twin Region, Panoramic 2-photon) microscope. We demonstrate the utility and flexibility of this system with ultra-wide FOV imaging, arbitrary line scan imaging, dual region imaging offset in XY and/or Z, quad area imaging, and imaging with resonant scanning. As an example application, we used this new imaging system to measure activity correlations between neurons in different cortical areas in awake mice.
In the design of the Trepan2p imaging system, two criteria needed to be met: (1) optical access to an area > 9 mm2, and (2) the ability to measure neuronal activity across cortical areas at rates relevant for neuroscience analysis (e.g., activity correlations). The large FOV of the imaging system provides optical access to neurons in multiple cortical areas (Fig. 1b) without the need to move the imaging system or the sample. However, the increase in space-bandwidth product (compared to conventional two-photon imaging systems) naturally increases the time required for raster scanning the large FOV. Temporal multiplexing is the fastest method to simultaneously acquire multiple signals, but prior implementations have been fixed and not easily repositionable during an imaging session, or did not offer completely arbitrary beam positioning11–13. Therefore we designed a new optical scheme for temporal multiplexing in which two beams are fully independently repositionable (Fig. 1c). These beams can be directed toward any region and depth within the large FOV to simultaneously scan and sample neuronal activity at time resolutions relevant for behavior and neural coding.
The imaging pathway is as follows (Fig. 1d). The light from an 80 MHz laser (Mai Tai, Coherent) is first split into two beams through polarization optics. One path (Pathway 2) is delayed 6.25 ns relative to the other path (Pathway 1). In this way the two pathways each have 80 MHz pulse trains delayed relative to one another by one half of the period (Supplementary Fig. 1a). Custom motorized steering mirrors (SM1 and SM2) impart independent solid angle deflections (Ω1 or Ω2) for the two beams and are located in conjugate planes to the scan mirrors using afocal relays. Adjustments to the focal plane for each pathway are made using an electronically tunable lens (ETL; Optotune)14. Thus, each path is independently positionable in X, Y, and Z during imaging without moving the microscope or the preparation. Single photon PMT pulses are demultiplexed using a synchronization signal from the laser module, assigning detected photons to pixels in the associated imaging pathway (1 or 2)(Supplementary Fig. 1b,c) with minimal crosstalk between pathways (Online Methods, Supplementary Fig. 2).
Commercial microscope objectives and scan engines often used in two-photon imaging were insufficient for this application because they are designed for small scan angles and beam diameters, which ultimately limit the FOV over which individual neurons can be resolved in vivo to ~1 mm2 (larger FOV two-photon imaging has been limited to lower resolution applications such as blood flow imaging15). Commercial macroscope objectives offer large FOVs for one-photon imaging, but they are not optimized for multiphoton excitation. Therefore, to provide high resolution imaging over a larger FOV, both a new scan engine and a new objective had to be designed in concert, along with optics for rapidly reconfiguring the multiplexed imaging pathways.
Multi-element optical subsystems (afocal relays, scan lens, tube lens, and objective) were designed (Supplementary Figs. 3–6, Online Methods) to minimize aberrations at the excitation wavelength (910 ± 10 nm) across scan angles up to ± 4 degrees at the objective back aperture (Supplementary Fig. 7a). In designing the custom optics, we prioritized the uniform performance (RMS wavefront error, Supplementary Fig. 7) over the entire designed scanning range. In this way we could preserve cellular resolution anywhere within the large FOV. Subsystems can be diffraction limited on their own, but demonstrate additive aberrations when used together in a full imaging system. Optimizing the system as a whole (including all relays, scan lens, tube lens, and the objective), rather than optimizing components individually, ensured we would meet the desired performance. Because the imaging system would be used in volumetric imaging applications, small variations of the focal plane across the large FOV (field curvature) were allowed (Supplementary Fig. 7b). Additionally, we relaxed requirements for F-theta distortion (Supplementary Fig. 7c). Together, these strategies facilitated the design process.
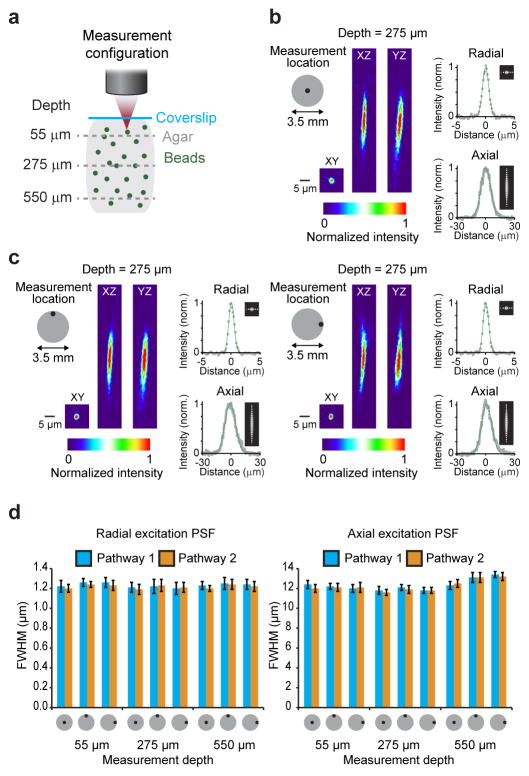
We evaluated the experimental resolution of the Trepan2p microscope by measuring the excitation point spread function (PSFex) as the full-width at half-maximum (FWHM) of the intensity profile of 0.2 μm beads (Fig. 2a–c, Online Methods). Radial FWHM was ~1.2 ± 0.1 μm (mean ± SD) both at the center and at the edges of the FOV. The axial FWHM was 12.1 ± 0.3 μm at the center, and 11.8 ± 0.4 μm at the edges of the FOV (both measurements are mean ± SD; Fig. 2d). Because the custom objective is air immersion, changes in imaging depth (from the designed imaging depth) will introduce additional spherical aberrations. However, this is largely minimized due to the moderate NA and thus the PSFex shows only minor changes as a function of imaging depth (Fig. 2d). The use of the tunable lens for focal plane alterations can affect the PSFex14, though in the range it was typically used (±50 μm) it has a small impact on the PSFex (Supplemental Table 1).
Figure 2. Focal excitation PSF profile of the Trepan2p system.
(a) 0.2 μm fluorescent beads were embedded in 0.75% agarose gel. 50 μm z-stacks were acquired, each centered at one of three depths (55 μm, 275 μm, 550 μm). This was done on axis, and at the edges of the 3.5 mm field of view. (b,c) Radial and axial excitation PSF measurements were made at the indicated locations and depths by fitting a Gaussian curve to the intensity profiles of the beads in the XY plane (measured in both the X and Y directions and averaged) and in the Z direction (measured in Z in both the XZ and YZ planes and averaged). (d) A summary of the excitation PSF measurements at three depths, for three locations, and for both of the temporally multiplexed beam pathways are shown (full width at half maximum of the Gaussian fits +/− the standard deviation for measurements from 8 different beads). The excitation PSF typically increases in axial extent with imaging depth beyond the optimized focal plane (275 μm), but the optimized aberration correction and moderate NA combine to largely mitigate that effect and preserves the excitation PSF across the full field and hundreds of microns of imaging depth.
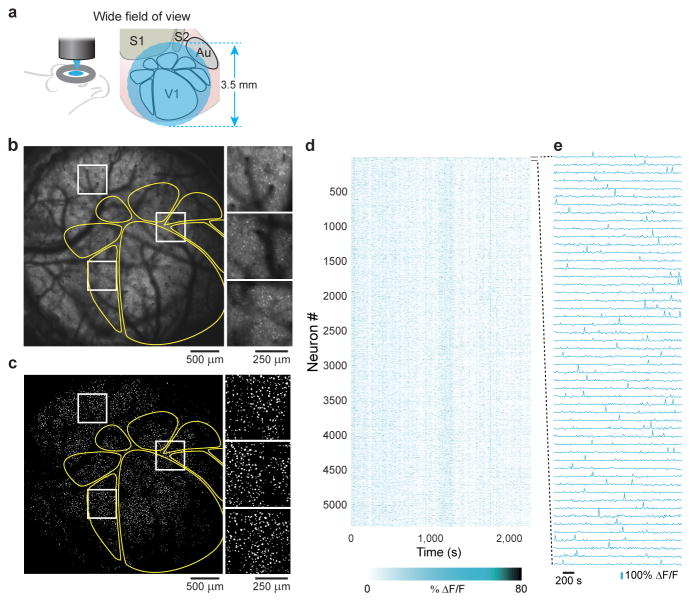
The Trepan2p system enables structural and functional imaging with individual neuron resolution in mouse neocortex (Supplementary Video 1, Supplementary Figs. 8, 9, 10)16, including structural imaging of cell bodies > 700 μm below the cortical surface in vivo in an animal sparsely expressing GFP (Thy1-GFP O-line, Supplementary Fig. 8, Supplementary Video 2). To demonstrate the functionality of the Trepan2p, we performed two sets of in vivo imaging in transgenic mice that expressed the genetically encoded calcium indicator, GCaMP6s3, in neocortical pyramidal neurons (Emx1-IRES-Cre x RCL-GCaMP6s/Ai96). In the first set of experiments, we explored the optical access to neurons across the large FOV. To locate primary visual cortex (V1) and higher visual areas (HVAs) in the mouse10, we used intrinsic signal optical imaging17 (Online Methods, Supplementary Fig. 11). We then imaged neuronal activity-related GCaMP6s signals in a large cortical region (Fig. 3a–c, Supplementary Fig. 12, Supplementary Video 3). The full large FOV was scanned with a single beam at ~0.1 frame/s (3.5 mm wide FOV; 2048 × 2048 pixels; 3 μs dwell time per pixel), and contained in it V1 and at least six HVAs. Undersampling and alternative scan strategies such as arbitrary line scanning (Supplementary Fig. 13) can increase the temporal resolution18,19, and stimulus presentation durations can facilitate the capture of stimulus-related events. Neurons were readily detected throughout the entire FOV based on spiking activity reported by GCaMP6s (5361 neurons detected, Online Methods; Fig. 3d, e). Thus the Trepan2p system provides cellular-resolution optical access across the entire FOV (9.6 mm2), which encompasses more than six different mouse cortical areas within a single FOV.
Figure 3. Two-photon imaging of neural activity across 9.6 mm2 of mouse cortex with single neuron resolution.
(a) The wide FOV provides optical access to thousands of neurons and multiple cortical areas. (b) A transgenic mouse expressing the genetically encoded fluorescent calcium indicator GCaMP6s in excitatory neurons was used to examine neuronal activity (maximum projection). Prior to 2p imaging, intrinsic signal optical imaging was used to map out the higher visual areas (yellow outlines). The expanded inlays (white) show cellular resolution is preserved across the FOV (also see Supplementary Figure 12 and Supplementary Video 3). (c) Segmenting the image sequence yields 5,361 active neurons, (d) whose visually-evoked responses were recorded, and (e) revealing clear fluorescence transients (traces for the first 50 neurons in panel d). Imaging depth was 265 μm. The animal was shown the naturalistic movie during these recordings.
For many neuroscience experiments, the activity dynamics of interest are restricted to two or more subregions of the large FOV. To capture neuronal activity correlations, these regions should be imaged simultaneously. The fastest approach to simultaneously image multiple regions is temporal multiplexing, as implemented here. To demonstrate the flexibility of this approach, we performed experiments with a variety of configurations of the twin imaging pathways (Fig. 4a, Supplementary Video 4) in animals expressing GCaMP6s in neocortical pyramidal neurons (TITL-GCaMP6s x Emx1-IRES-Cre x ROSA:LNL:tTA). While presenting visual stimuli to the mouse (a naturalistic movie), we simultaneously recorded activity in several different pairs of cortical areas, and at different depths (512 × 512 pixels, 3.8 frames/s per region, Fig. 4b). The imaging regions can be placed arbitrarily close to each other. To demonstrate this, we placed the two imaging paths at the same XY location, but offset their positions in Z (512 × 256 pixels, 9.5 frames/s per region, Fig. 4c, Supplementary Video 5). Next we explored quad region imaging. Each of the dual imaging pathways were configured to alternate (every other frame) between two different regions. Thus, we imaged four different XY regions, each at 10 frames/s per region (250 × 100 pixels, Fig. 4d; 512 × 512 pixels, 1.9 frames/s per region, Supplementary Videos 6). We also imaged four regions that differed in the XY locations and Z (250 × 100 pixels, 10 frames/s per region, Fig. 4e). Resonant scanners provide a way to increase the frame rate in raster scanning modes, and this is completely compatible with the optical system we present here. We used a resonant scanner to achieve 30 frames/s imaging in two regions simultaneously (512 × 256 pixels, 30 frames/s per region, Fig. 4f).
Figure 4. Temporally multiplexed, independently repositionable imaging pathways for simultaneous scanning two regions.
(a) The individual imaging regions can be independently positioned and repositioned anywhere within the full FOV. (b) Within the same session, without moving the mouse or the microscope, the two pathways were moved to various configurations (left) to image neuronal activity (3.8 frames/s per region) (left, segmented active ROIs within the 500 μm imaging region; right, five example traces from each region). (c) There is no lower limit to the XYZ separation between imaging pathways (from the mechanical point-of-view). In this imaging session (9.5 frames/s per region), the XY locations were identical and the pathways only differed in the Z depth (left, segmented active ROIs within the 500 μm imaging region; right, five example traces from each region; Supplementary Video 5). (d, e) By combing temporal multiplexing (Pathways 1 and 2) with serially changing the offset voltage on the galvanometer scanner, four regions can be rapidly imaged (10 frames/s per region). (d) Pathway 1 and 2 are positioned at the same XY location and offset in Z. The galvanometers serially position the imaging region (of each pathway) anywhere within the larger field of view (left, segmented active ROIs within the 400 μm imaging region; right, five example traces from each region; Supplementary Video 6) (e) Pathway 1 and 2 are positioned at different XY locations as well as offset in Z. The galvanometers serially position the imaging region (of each pathway) anywhere within the larger field of view (left, segmented active ROIs within the 400 μm imaging region; right, five example traces from each region). (f) Resonant scanning was performed for faster frame rates (30 frames/s per region) (left, segmented active ROIs within the 500 μm imaging region; right, six example traces from each region; Supplementary Video 7). (g) Neuronal activity was imaged in two regions (20 frames/s per region), V1 and in an ROI encompassing retinotopically matched regions of AM and PM, simultaneously. Visual stimuli, either drifting gratings or a naturalistic movie, were used to evoke responses (left, segmented active ROIs within the 400 μm imaging region; right, five example traces from each region and for each visual stimulus). (h) Ca2+ signals were used to infer spike times and examine correlations. Activity correlations were measured between pairs of cells, each pair consisting of a V1 neuron and a neuron in AM or PM. These correlations were higher during presentation of the naturalistic movie compared to those during the drifting gratings (cross correlation with gratings, mean ± SEM: 0.0157 ± 0.0003; with naturalistic movie: 0.0218 ± 0.0003; N = 12,160 neuron pairs; P < 10–10; rank-sum test). The neurons on both axes were ordered from low to high average correlation for presentation clarity on the left (naturalistic movie), and the same ordering is used on the right (gratings). Imaging depth for both pathways was 235 μm. For all panels the vertical scale bar is 200% ΔF/F and the horizontal scale bar is 10 s. All imaging depths are indicated in the panels. The depth offset in Pathway 2 (orange) was accomplished using the tunable lens.
As an example experiment, we imaged neuronal activity in different cortical areas simultaneously, V1 and in medial higher visual areas (HVAs) AM and PM (250 × 100 pixels, 20 frames/s, Fig. 4g) while presenting a two different visual stimulus videos (a naturalistic movie or drifting gratings). Pairwise correlations between cortical areas were calculated from spike time courses inferred20 from the GCaMP6s signals. This analysis revealed an increase in correlated activity between neurons in V1 and neurons in areas AM and PM when the naturalistic movie is presented, compared to gratings (cross-correlation with gratings (mean ± SEM): 0.0157 ± 0.0003; with naturalistic movie: 0.0218 ± 0.0003; N = 12160 neuron pairs; P < 10−10; rank-sum test; Fig. 4h). Such cross-area correlation analysis between densely sampled populations of neurons was made possible with both the large FOV and temporally multiplexed scanning. These imaging regions can be as large or small as needed for the experiment and temporal resolution (for example, a 2.5 mm FOV at two different z-depths; 2048 × 1024 pixels; 0.8 frames/s); Supplementary Fig. 14; Supplementary Video 7), and two regions can be set to scan complementary halves of the FOV (as in ref. 11). Thus, the repositionable temporally multiplexed imaging beams can be reconfigured within an experiment for a variety of measurements.
In this work we have addressed a major barrier to progress in two-photon imaging of neuronal activity: the limited FOV. Using novel optical systems, the Trepan2p has expanded optical access to neurons in an area greater than 9.5 mm2 using mostly cost-effective COTS components. This large FOV can contain, in mice, primary visual cortex and six higher visual areas. Rapidly repositionable multiplexed imaging paths make it possible to capture neural activity in two arbitrarily selected subregions simultaneously within the large FOV.
The results presented in this manuscript demonstrate the utility of the Trepan2p system. The large working distance (8 mm) of the air-immersion objective can facilitate integration with head-fixed behavior experiments. Moreover, in principle the same instrumentation can be applied to image neural activity in many animal models whose cortical organization extends beyond the spatial limits of conventional 2p microscopy, such as multiple orientation columns in primary visual cortex of cats or non-human primates. The methodological approach and technologies presented here provide a flexible platform for measuring neural activity correlations and dynamics across extended cortical circuitry with individual neuron resolution. Beyond measuring neural activity, the Trepan2p can be applied for other physiological measurements such as blood flow and structural imaging in cleared tissue.
ONLINE METHODS
Optical design and simulations
Relay, scan, tube, and objective lens systems were modeled in OpticStudio (Zemax, LLC). Three optical subsystems ([1] Relay, [2] Scan and Tube Lens, [3] Objective) were first designed, modeled, and optimized individually. After initial isolated design, the system was simulated and optimized as a whole to ensure there would be no unforeseen additive aberrations between subsystems. One of the main difficulties in designing a highly corrected optical system is dealing with chromatic aberrations. We decided to focus our design around a narrow wavelength range suitable for GCaMP excitation (910 ± 10 nm). This eased our requirements for correction of chromatic aberrations. For the relay and scan and tube systems, all commercial off-the-shelf (COTS) lenses (Thorlabs and OptoSigma) were used to minimize cost (Supplementary Figs. 3–5). These lenses were coated with an IR anti-reflective coating that had <0.5% reflectance at 910 nm.
To obtain diffraction limited performance (Supplementary Fig. 7a) in the simulated design and sufficient performance with in-house assembly, the cost-effective COTS components were supplemented with two custom cemented doublets in the objective (Supplementary Fig. 6). The custom cemented doublets were manufactured by Rainbow Research Optics, Inc. (Centennial, CO) and were coated with a broad-band anti-reflective coating (<1% 400–1100 nm). The other elements were COTS (Thorlabs and OptoSigma) and coated with a broad-band anti-reflective coating (<1% 525–925 nm; OptoSigma). We relaxed the requirement to have the focal plane be flat as slight deviations of the focal plane across the FOV would not affect our ability to capture neural dynamics. The resulting Trepan2p field curvature is <20 μm (Supplementary Fig. 7b). Additionally, the F-theta distortion is minor (<5%) (Supplementary Fig. 7c). After the individual sub-systems were designed and optimized, the system was optimized as a whole, while further refinement of spacing was made to allow for optomechanical limitations (lens housing, scanning mirrors). Complete lens data of the Trepan2p system is given in Supplementary Figures 3–6.
Assembly
To assemble the lens subassemblies, we used Thorlabs slotted lens tube system (2 inch) and retaining rings. Axial separations between elements were measured using digital calipers along with a custom metrology system using a digital camera (Thorlabs, DCC1545M) and telecentric lens. All lenses were mounted and measured based on the plano or concave side to minimize tip/tilt and secured on the convex side with a retaining ring after alignment. Decenter of lens elements was done manually with aid of the camera measurement system. The smaller negative elements (surfaces 1,3,23,25,28,30,48,50,53,68; Supplementary Fig. 3–6) were mounted in the 2 inch lens tube system using a 3D printed adapter (Quickparts). Subassemblies were mounted on 60mm cage system (Thorlabs). Galvonometer and resonant scanners were mounted on a XY translator (Thorlabs, CXY2) and this was attached to a 60mm cage cube (Thorlabs, LC6W), which was in turn connected to the other optical subassemblies through the cage system. The XY translator allowed for precise positioning of the scanning mirrors and allowed for offset due to the thickness of the mirror.
Within the systems there are natural axial separations between subassemblies that can be utilized as compensators. As the system is assembled, locations of afocal space at conjugate planes (Supplemental Figs. 3–6) are checked for collimation as designed in the system (Thorlabs, SI100).
Tolerancing and sensitivity analysis
A key component in optical design and manufacturing is tolerance analysis21. A system may have high simulated performance, but if the required precision of optical alignment is too high, the system may not be easily manufactured or too costly. For rapid turnaround and to keep costs low, we sought to design and build the Trepan2p system in house, and to use primarily commercial off the shelf (COTS) components. The RMS wavefront error of the designed system shows diffraction limited performance across the field of view (Supplementary Fig. 7a). The predicted excitation PSF measurements are ~8.8 μm on axis, and ~8.9 μm at a radial distance of 1.75 mm (edge of the 3.5 mm field of view) (Supplementary Table 1). These excitation PSF measurements are the FWHM of the square of the illumination PSF (Huygens PSF) output of the Zemax simulation of the system (Online Methods: Excitation PSF and individual neuron resolution).
To ensure adequate performance of the system, we performed tolerancing analysis and designed in compensating features to adjust for residual aberrations after assembly. For the COTS elements used, we used the following tolerances: radius of curvature = 5 fringes, element thickness = 0.1 mm, tip/tilt (X and Y) = 0.05 degrees, S + A irregularity = 0.25, and index of refraction = 0.001. For the element to element tolerances we used the following: axial separation = 0.175 mm, decenter (X and Y) = 0.11 mm, and tip/tilt (X and Y) = 0.150 mm. Within the systems there are natural axial separations between subassemblies that can be utilized as compensators (red boxed spacing, Supplemental Figs. 3–6). Setting these axial separations to be compensators, Monte Carlo simulations of the system performance (tolerance analysis) was performed, utilizing a merit function that measured RMS wavefront error across the FOV and penalized the system for vignetting. The nominal merit function value for the designed system is 0.043. We performed 2500 cycles of Monte Carlo simulation for the system with only axial displacement compensators (blue distribution, Supplementary Fig. 7d), and found that the expected system performance would not be sufficient. To increase the system performance, we designed into the objective optomechanical assembly two additional compensators: the axial separation between surfaces 76 and 77 and the XY decenter of custom element 1 (Supplementary Fig. 6). This was accomplished by mounting custom element 1 in an XY translator (Thorlabs, CXY2) and by using an adjustable adapter attached to this translator (Thorlabs, SM2P). With these three additional degrees of freedom, the Monte Carlo simulated performance significantly improved (green distribution, Supplementary Fig. 7d). The measured performance (resolution) of the system as assembled (Fig. 2), best matches simulated systems with merit functions in the range of 0.14–0.15 (red bar, Supplementary Fig. 7d). The inclusions of designed compensators (especially those within the objective) were critical to the success of our in-house assembly strategy.
The inclusion of the compensators in the assembly process provide a way to approach ideal performance, yet these cannot completely compensate for all imprecisions in the optical system. Tolerances in element manufacturing and misalignments in assembly remain and will lead to deviations from simulated ideal performance. During sensitivity analysis on this system, with the inclusion of the compensators, we found that the worst offenders are the decenter of the elements within the objective assembly. Decentering of these elements leads to an increased level of coma aberration. During exploration of the Monte Carlo simulations (part of the tolerancing process), we observed the aberrations in the simulated wavefront. The most common aberrations were typically coma aberration with some level of spherical aberration. This is likely true in our assembled system leading to the differences between simulated and experimental excitation PSF.
Animals
All procedures involving living animals were carried out in accordance with the guidelines and regulations of the US Department of Health and Human Services and approved by the Institutional Animal Care and Use Committee at University of North Carolina. C57Bl/6 mice were housed under a reversed 12 hr-light/12 hr-dark light cycle with ad libitum access to food and water. Two lines of transgenic mice were used that express GCaMP6s in cortical pyramidal neurons: (1) Cre-dependent GCaMP6s mice, Ai96 (RCL-GCaMP6s), crossed with Emx1-Cre mice (Jackson Labs stock #005628; Fig. 3) and (2) Cre/Tet-dependent GCaMP6s mice, Ai94 (TITL-GCaMP6s) triple crossed with Emx1-Cre and ROSA:LNL:tTA mice (Jackson Labs stock #011008; Figs. 4, 5)22. Ai94 and Ai96 lines were kindly provided by H. Zeng and the Allen Institute. We used a transgenic mouse in which a tyrosine hydroxylase (TH) promoter directs expression of Cre recombinase (TH-Cre; (Jackson Labs stock #008601). In this mouse, the ventral tegmental area (VTA) was injected with adeno-associated virus particles (AAV5-DIO-ChR2(H134R)-EYFP; 500 nL bilateral at an infusion rate of 100 nL/min). The animal was sacrificed three months post-surgery. TH-expressing terminals arising from the VTA were imaged in the striatum (Supplementary Fig. 2). Additionally, a Thy1-GFP O-line mouse, which expresses GFP largely in Layer V, was used to generate Supplementary Video 2.
Surgery
Mice were deeply anesthetized using isoflurane (5% for induction, 1–2% for surgery) augmented with acepromazine (0 – 0.4 mg/kg body weight, i.p.). Physically-activated heat packs (SpaceGel, Braintree Scientific) were used to maintain the body temperature during surgery. The scalp overlaying right visual cortex was removed, and a custom head-fixing imaging chamber23 with 5-mm diameter opening was mounted to the skull with cyanoacrylate-based glue (Oasis Medical) and dental acrylic (Lang Dental). A 4-mm diameter craniotomy was performed over visual cortex. Carprofen (4.4mg/kg body weight, s.c.) was administered postoperatively to all mice that underwent recovery surgeries before returning to the home cage. Mice were mounted on a holder via the chamber23. For intrinsic signal optical imaging, this chamber was filled with a physiological saline containing (in mM): 150 NaCl, 2.5 KCl, 10 HEPES, 2 CaCl2 and 1 MgCl2.
Intrinsic signal optical imaging (ISOI)
Custom instrumentation was adapted from the work of Kalatsky and Stryker17. Briefly, two F-mount lenses with respective focal lengths of 135 and 50 mm (Nikon) formed a tandem lens macroscope, which was attached to Dalsa 1M30 CCD camera (Teledyne DALSA). This configuration provided a 4.7 mm × 4.7 mm field of view (21.2 mm2). Acquired images were binned 2 × 2 spatially, resulting in a final pixel size of 9.2 μm × 9.2 μm. The pial vasculature was illuminated through a green filter (550 ± 50 nm, Edmund Optics) and the vasculature map was captured through a second green filter (560 ± 5 nm). From the pial surface, the macroscope was then focused down 600 μm where intrinsic signals were illuminated with halogen light (Asahi Spectra) delivered via light guides and focusing probes (Oriel) through a red filter (700 ± 38 nm, Chroma). Reflected light was captured through a second red filter (700 ± 5 nm, Edmund Optics) at the rate of 30 frames per second with custom-made image acquisition software (adapted by J.N.S. from code kindly provided by Dr. David Ferster, Northwestern University). Mice were head-fixed 20 cm from a flat 60 cm × 34 cm (width × height) monitor which was tilted towards the mouse 17.5° from vertical with their head angled to their right to cover the visual field (110° by 75°) of the contralateral eye. A light anesthetic plane was maintained with 0.5% isoflurane during imaging, augmented with acepromazine (1.5 – 3mg/kg), and the body temperature was kept at 37°C using feedback-controlled electric heat pad systems (custom-built).
Visual stimuli
A drifting white bar on a black background (elevation and azimuth direction; 3° thick) was used to map retinotopy during ISOI. These were produced and presented using MATLAB and the Psychophysics Toolbox24,25. A corrective distortion was applied to compensate for the flatness of the monitor26 (code is available online, http://labrigger.com/blog/2012/03/06/mouse-visual-stim/). During calcium imaging, drifting gratings (0.04 cycles/°, 2 Hz, 8 directions, 10 s /direction) or a naturalistic movie were displayed on a small video display located 6.3 cm from the left eye. The naturalistic movie was from a helmet-mounted camera during a mountain biking run. This movie, while not something a mouse would typically encounter, provided a visual stimulus with a large amount of optic flow. Light from the display was shielded from the imaging apparatus using a shroud over the monitor.
Image analysis for mapping cortical areas
Retinotopic maps obtained from intrinsic signal optical imaging were used to locate V1 and HVAs (Supplementary Fig. 11). Borders between these areas were drawn at the meridian of elevation and azimuth retinotopy. Magnitude response maps for grating patches were consulted for cross-verification. The vasculature acquired from Trepan2p imaging was compared to that acquired during ISOI and provided landmarks for identification of the locations within V1 and the HVAs.
In vivo two photon imaging
All imaging was performed on the custom Trepan2p system. Control of the instrumentation (see Trepan2p instrumentation below) and image acquisition were controlled by custom LabVIEW software. Animals were lightly anesthetized (0.25% isoflurane) for the imaging in Figure 1; Figure 4b, c; Supplementary Figures 8, 9, 10, 12; and Supplementary Videos 1–4. Animals were awake for the imaging in Figure 4d–f; Figure 5; Supplementary Figure 13, 14; and Supplementary Videos 5. The imaging was performed with <200 mW out of the front of the objective to minimize crosstalk (see Crosstalk below; Supplementary Fig. 2). Estimating the focal volume to be an ellipsoid with semi-axes of 6 μm, 0.6 μm, and 0.6 μm (Fig. 2), this equates to a power density of ~21.5 mW/μm3. For a typical high NA objective (e.g. 16x, NA=0.8) the ellipsoid volume is ~1.0 μm3 (semi-axes of 1.5 μm, 0.4 μm, and 0.4 μm). Thus < 200 mW in the Trepan2p provides the same power density as 21.5 mW in a high NA imaging system. The larger excitation volume allows us to increase the imaging power without the damage seen in higher NA objectives27. With typical imaging parameters (512×512 at 3.8 frames/s, 0.5 mm imaging region) up to 250 mW per channel could be used with no observed damage when imaging somas ~250 microns deep. Damage was observed with these parameters if the imaging plane was moved to the surface of the dura. When imaging superficially, the power was kept below 100 mW. Assessment of damage due to laser intensity was based on visual morphological changes to the appearance of the dura mater and/or continuously bright cell bodies. As a positive control, we used high (>300 mW at ~200 μm below the pial surface) continuous laser illumination, which caused photodamage evidenced by bright cell bodies and brighter-than-normal fluorescence of the dura (mouse was not used for subsequent activity imaging). At the laser powers used for the imaging in this manuscript (<200 mW) such damage was not observed.
Image analysis for neuronal calcium signals
Ca2+ signals were analyzed using custom software in MATLAB (Mathworks). Neurons were identified and segmented using either a pixel-wise correlation map6 or a pixel-wise kurtosis map. For the kurtosis map, each imaging frame of the calcium imaging video was first Gaussian filtered in XY (sigma = 2 pixels), and then the kurtosis for the time series of each pixel location was computed using the filtered version of the video. This yields a single real-valued map with enhanced contrast for regions of interest that exhibit larger fluorescence transients. The maps were segmented into individual ROIs (neurons and processes) using a locally adaptive threshold and ΔF/F traces were calculated from the raw, unfiltered data. An exponential moving average of time width ~150 ms was applied to the high speed ΔF/F traces in Fig. 4d–f and Fig. 5a, all other data was left unfiltered. The activity transients shown in Fig. 3 are typically captured in only one frame due to acquisition frame rate. For the correlation analysis, spike inference6,20,28 was performed on the raw F/F traces prior to computing correlations. For pairwise correlations >0.5, traces were inspected for crosstalk. If the ROIs were present in the same XY positions in both imaging pathways and had high correlation values, the ROI with the weaker signal was determined to be due to crosstalk and was omitted (see Crosstalk below).
Arbitrary line scan analysis
Line scan data was arranged into a location-on-path versus time plot. The kurtosis of the data was taken along the time axis to yield a vector containing peaks (high values of kurtosis along the scan path). A small portion of the path (3–5 pixels) surrounding a peak of kurtosis was summed along the path length axis to yield a fluorescence time series for an active neuron. Each potential active region was manually inspected. This yielded 172 active cells distributed over the field of view (Supplementary Fig. 13c). An exponential moving average of time width ~150ms was applied to the traces in Supplementary Figure 13a, b.
Excitation point spread function measurements and simulations
In two-photon imaging, the optical resolution of the system is evaluated by the excitation point spread function (PSFex), which is the square of the illumination point spread function (IPSF2)(Ref.29). To evaluate the PSFex, sub-micron beads were imaged. Sub-micron fluorescent beads (0.2 μm, Invitrogen F-8811) were imbedded in a thick (~1.2 mm) 0.75% agarose gel. 50 μm z-stacks were acquired, each centered at one of three depths (55 μm, 275 μm, 550 μm). The stage was moved axially in 0.4 μm increments (Δstage). At each focal plane 5 frames were acquired and averaged to yield a high signal to noise image. Due to the difference between the refractive index of the objective immersion medium (air) and the specimen medium (water), the actual focal position within the specimen was move an amount Δfocus = 1.38 × Δstage (Ref. 30). The factor 1.38 was determined in Zemax and slightly differs from the paraxial approximation of 1.33. These images were imported into ImageJ and the μm/pixel scaling for the XY and μm/frame (step) were input. For the axial PSFex, XZ and YZ images were created at the center of a bead and a line plot was made at an angle maximizing the axial intensity spread thereby preventing underestimation of the PSFex due to tilted focal shifts. For the radial PSFex, an XY image was found at the maximum intensity position axially. A line scan in X and Y was made. Gaussian curves were fit to the individual line scans to extract FWHM measurements. The radial PSFex values are an average of the X PSFex and Y PSFex and the axial PSFex is an average of the axial PSFex found from the XZ and YZ images. Excitation PSF measurements were performed both on axis and at the edges of the field of view for both imaging pathways. For measurements using the electronically tunable lens (Supplementary Table 1), the focal plane was first set at 275 μm using the XYZ stage with a current to the tunable lens of ~75 mA for zero offset. The focal plane was then shifted to either 225 μm or 325 μm (for a ±50 um change in focal plane) by applying a previously calibrated current to the tunable lens. A z-stack was then acquired as previously described. The minimal laser power for excitation was used to minimize the effects of non-linear expansion of the apparent excitation PSF29. Data reported (Fig. 2, Supplementary Table 1) are the mean ± standard deviation of 8 beads.
For the simulated PSF measurements (Supplementary Table 1), at each of the three simulated depths (Surface 85 in Supplementary Fig. 6) the optimal focus (Surface 83 in Supplementary Fig. 6) was found (minimal RMS wavefront error). At this location and at simulated locations ±25 μm from this location in 0.4 μm steps (this represents the steps of the stage as explained previously, Δstage), the Huygens illumination PSF (IPSF) was simulated. These images were imported into ImageJ and processed as previously described (accounting for the step size change: Δfocus = 1.38 × Δstage) to generate a Gaussian curve fit. This Gaussian was then squared to yield the excitation PSF (= IPSF2). The FWHM of this squared Gaussian is reported as the simulated PSFex.
Trepan2p instrumentation
Laser pulses from a Ti:Sapphire laser (Mai-Tai; Newport) with an automated pre-chirper unit (DeepSee; Newport) were attenuated using a half wave plate followed by a polarization beam splitting cube. Similar polarization optics were used to split the beam into two paths and control the relative power between the two paths. Prior to splitting, the beam was expanded using a 3x beam expander (Thorlabs). One beam travels directly to a custom motorized steering mirror, and the other beam is first diverted to a delay arm, and subsequently to a separately controlled steering mirror (Fig. 1d). The delay arm is designed to impart a 6.25 ns temporal offset to the pulses in one beam (1.875 m additional path length). Since the laser pulses are delivered at 12.5 ns intervals (80 MHz), they are evenly spaced in time at 160 MHz after the two beams are recombined (Supplementary Fig. 1a).
The steering mirror in each pathway imparts a solid angle (Ω1 or Ω2) deflection to the beam prior to recombination (Fig. 1d). It is this angle that determines the central locations (X1, Y1; X2, Y2) of individual imaging subareas within the larger FOV. The two beams are combined and relayed to the X-axis galvanometer scanner using the third polarization cube located in the middle of an afocal pupil relay (Supplementary Fig. 3). The 2-inch polarization beam combining cube (Edmund Optics) was positioned in the afocal relay such that the focal point of the relay was outside the cube. This was to avoid high intensity damage to the cemented surface within the cube. A similar afocal pupil relay (Supplementary Fig. 4) is present between the X-axis and Y-axis galvanometers (Cambridge Technologies). In this manner the two beams are simultaneously raster scanned (field size determined by scan amplitude), but with independently controlled spatial positions (determined by Ω1, Ω2). Immediately prior to the steering mirror in Pathways 1 and 2 are electrically tunable lenses (Optotune)14 which provides a z-range of ~450 μm.
A scan lens and tube lens form a 4x telescope (Supplementary Fig. 5) and relayed the expanded beams (24 mm) to the back aperture of the custom objective (EFL = 27.5 mm; NA = 0.43; Supplementary Fig. 6). The entrance scan angles at the objective back aperture were ~±3.7 degrees yielding our 3.5 mm FOV. Overall, the system transmitted 41% of entering laser power (Supplementary Fig. 15). The main factors influencing this figure were the galvanometer or resonant scanners and the objective, which were both overfilled. The system was not power limited in this configuration. Decreasing the overfilling of these elements can increase power transmission, though with a possible decrease in excitation efficiency (effective NA). Fluorescence was collected using a dichroic mirror, two lenses31, and a GaAsP photomultiplier tube (PMT; H10769PA-40, Hamamatsu).
Photon counting electronics
Output from the photomultiplier tube (PMT; H10769PA-40, Hamamatsu) was first amplified with a high bandwidth amplifier (C5594-44, Hamamatsu) and then split into two channels (ZFSC-2-2A, Mini-Circuits) (Supplementary Fig. 1b). One channel was delayed relative to the other by 6.25 ns by using different lengths of BNC cable (~ Δ122 cm) (Supplementary Fig. 1b, c). These were connected to a NIM discriminator (4608C octal discriminator, LeCroy). The ~80 MHz synchronization output pulses from the laser was amplified (Mini-Circuits ZFL-1000) and then delivered to a second NIM discriminator (161L dual discriminator, LeCroy) (Supplementary Fig. 1b) which has a continuous potentiometer adjustment to adjust the output NIM pulse width from ~5 ns to >150 ns. This output pulse was delivered to the common veto input on the LeCroy 4608C discriminator where the PMT outputs were collected. The veto width was adjusted by the potentiometer on the LeCroy 161L discriminator and the relative phase of the veto window was adjusted by time delaying the synchronization pulses from the laser module using small lengths of cables (~0.5 ns resolution). Two photon detection schemes were used. The first directly counts output pulses32 (digital counting) and the second is an analog counting scheme33. To implement digital counting, the demultiplexed output pulses from the LeCroy 4608C were sent to a NIM-to-TTL converter (PRL-350-TTL-NIM, Pulse Research Labs) before going to the counter inputs on a fast counter (PCI-6110, National Instruments) (Supplementary Fig. 15b). Counter measurements were organized into images in the custom LabView software. Pixel dwell time was determined by setting the counting window time in software. The maximum count rate for the PCI-6110 is 20 MHz therefore it is possible some pulses are missed, lowering the overall signal amplitude. In the analog counting mode output from the LeCroy 4608C is passed through a low-pass filter (EF508, EF516; Thorlabs) before being digitized at 5 MHz (PCI-6110, National Instruments) or 25 MHz (ATS860, AlazarTech). Analog measurements were arranged into images with the indicated pixel count in the custom LabView software. In this manner we could demultiplex the single PMT output into two channels corresponding to the two excitation pathways (Supplementary Fig. 1c).
Scanning hardware and strategy
Multiple scanning strategies were employed in this work: raster scanning using galvanometer and resonant scanners, and arbitrary line scanning.
Galvanometer scanning
The galvanometer based scanning was performed using 6 mm galvanometers (6210H, Cambridge). Angular positions of the galvanometer were controlled by sending analog commands (PCI-6110, National Instruments) to the galvanometer driver board. The common reference clock on the PCI-6110 was used to synchronize the analog out commands and either the counters (digital counting) or the analog digitizer (analog counting) on the PCI-6110.
Resonant scanning
Only the analog counting scheme was used with resonant scanning. Resonant scanning was accomplished using a galvanometer scanner on the slow X axis (6210H, Cambridge) and a 4kHz resonant scanner (CRS04K-S4-S-045) on the fast Y axis. The resonant scanner was controlled using a CRS driver (Cambridge). This driver outputs a 50% duty cycle pulse with the rising edge occurring every period (~4kHz) thus outputting a rising edge every other line and a falling edge on the alternate line. This synchronization signal from the CRS driver was input to an Arduino Uno board that output a 5 μs pulse on every rising and falling edge thus converting the signal to an ~8kHz signal. This signal was input to the PCI-6110 as the clock for the analog output which drove the slow X-axis galvanometer. In this way the X and Y axes were synchronized. Additionally, the CRS diver synchronization pulse was used as a trigger for analog acquisition (ATS860, AlazarTech). After a trigger event occurs, a number of digitization events occur. This forms the pixels along a bidirectional line and the number is found by dividing the digitization frequency by the resonant scan frequency and subtracting the re-arm time. For example, in this system the resonant scanner had a measured frequency of 3941.8 Hz and for a digitization clock of 5 MHz and a re-arm time of 64 clock ticks = 5,000,000/3941.8 – 64 = 1204. This number plus 16 must be divisible by 32 as required by the digitizer and thus is rounded down to 1200. Therefore 1200 pixels make up every two lines. Interlacing is accomplished by finding the best line alignment offline and then subsequently using these settings online. Because of the small re-arm time of the digitizer, some pixels are lost at the edge of every even line (~70). These are given a value of 0 in the reconstructed image. As higher digitization frequencies are used, the percentage of these pixels becomes less. For digitization at 25 MHz, every two lines contain ~6256 pixels and the even lines have ~86 pixels given a value of 0. Additionally, for image synchronization, we use the counter on the PCI-6110 to output a pulse every # lines/2 pulses of the CRS synchronization signal. This is used as an arm-trigger input into the ATS860 board. These signals ensure synchronization between the galvanometer scanner, resonant scanner, and the digitization board. An additional analog output on the PCI-6110 is used to control the amplitude of the resonant scanner. Corrections for the sinusoidal scanning path were performed online in software34.
Arbitrary line scanning
The line scan pathway was drawn in custom LabVIEW software and converted to voltage commands for the galvanometer scanners. The imaging rate (28.7 Hz; Supplementary Fig. 13b) was determined to yield approximately equivalent pixel dwell time as the slower raster scans.
Crosstalk
Many factors affect the crosstalk between the two pathways including GCaMP6 fluorescence lifetime, veto pulse width, veto pulse phase, PMT gain, and excitation power. The veto width was set to inhibit pulses arising from the off-target pathway while maximizing the signal from the desired pathway. The working value is ~6.25 ns though small deviations from this made little difference. The phase of the veto (inhibit counting) signal relative to the PMT signal played a more significant role and was adjusted to give maximal signal in one channel arising from the non-delayed excitation path (Pathway 1) and maximal signal in the other channel arising from the 6.25 ns delayed excitation path (Pathway 2), while at the same time minimizing the signal from pathway 1 excitation when Pathway 2 was blocked and similarly for Pathway 2 into Pathway 1 (Supplementary Fig. 2a). With increases in the laser power and decreases in gain, the effective crosstalk was greatly diminished (Supplementary Fig. 2a–c) due to changes in the instrument response function (IRF). By adjusting the laser intensity and gain on the PMT, we can minimize the level of crosstalk, while still obtaining high signal-to-noise measurements of fluorescence transients. For the experiments shown in this study, the laser power was <200 mW out of the front of the objective (also see In vivo two photon imaging in Methods) yielding <5% crosstalk between pathways.
For crosstalk to influence the acquired Ca2+ imaging and measured correlations there must be an ROI identified in the off-target pathway that is in the same XY position as a true ROI (neuron) in the on target pathway. False ROIs can occur, and these are ROIs that arise from the off-target pathway and are solely due to crosstalk. These can be identified by correlations in neural activity Ca2+ transients that satisfy two criteria: (1) have a high degree of correlation (>0.5), and (2) occur in the same XY locations in both images (pathway 1 and pathway 2). Thus, identification of the false ROIs is computationally simple, and they can be automatically removed from further analysis. Crosstalk was not detected in the data set displayed in Figure 4. A more subtle form of crosstalk can occur when two neurons, one in each pathway, are scanned simultaneously. To determine whether this form of crosstalk is affecting cross-correlation measurements, we searched the data set for pairs of neurons, one in each pathway, that overlap in XY position, and thus are simultaneously scanned and could contaminate each other’s signals. For these pairs, we computed the correlation between amount of overlap (the intersection of XY pixel locations between the two neurons’ ROIs) and activity cross-correlation. If activity signals from one neuron are contaminating the other, there should be a trend for more overlapped neurons to also exhibit higher cross-correlations. However, this relationship was not significant (R2 = 0.0094, P = 0.55, N = 40 pairs), and thus even when cells are simultaneously scanned, crosstalk is low and high fidelity measurements of neural activity are obtained.
Excitation PSF and individual neuron resolution
To compare the effect of different axial PSFex on acquired data, we simultaneously imaged GCaMP6s activity with two different excitation PSF (Ref. 16): 14.0 ± 0.3 μm and 4.4 ± 0.2 μm (Supplementary Figure 9a). To establish the two different PSFex, we used a Nikon 16x objective (NA = 0.8) and under-filled the back aperture of the objective using Pathway 1 (14.0 ± 0.3 μm, which is slightly larger than that obtained in the Trepan2p), and over-filled the back aperture of the objective using Pathway 2 (4.4 ± 0.2 μm). The PSFex measurements were made as described above (see Methods: Excitation point spread function measurements). The XY locations of the two pathways were slightly offset in order to ensure crosstalk due to multiplexing (though minimal) did not influence our measurements and to minimize the potential photo-damage. The two pathways scanned (~3 frames/s) the same neurons in V1 (Supplementary Fig. 9b) while the mouse was shown the naturalistic video. Calcium transients were extracted from the same cells in both pathways from 3 different fields of view (Supplementary Fig. 9c, Supplementary Fig. 10). The correlation coefficients (n = 456 cells) were calculated between the two different excitation PSF pathways for the raw ΔF/F traces (Supplementary Fig. 9d).
These findings agree with a prior report16, which showed that in mammalian neocortex (imaging signals from pyramidal neurons expressing GCaMP6s), excitation PSF that are ~10 microns in the axial dimension can be sufficient to keep potential crosstalk between cells < 10%. They found that although crosstalk can occur, in practice, the signals are rarely and minimally contaminated. Nearby cell bodies could conceivably contribute to crosstalk, but typically do not. This is because in mouse cortex, < 10% of neurons are located axially < 30 microns from each other16. Thus, the Nyquist criterion for keeping maximal potential crosstalk below 10% is an axial resolution that is < 15 microns. In summary, since the vast majority of neurons are separated axially, crosstalk due to PSFex that are ~10–15 microns in the axial dimension do not preclude single neuron resolution.
In addition to the spatial distribution of neurons, these results can also be influenced by the staining pattern and the fluorescence of the indicator in the low calcium state. Thus other indicators, staining patterns, and imaged structures can impose different requirements on the imaging system.
Supplementary Material
Supplementary Video 1. High resolution imaging using the Trepan2p.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. Images were acquired at 1024 × 1024 pixels at 1.9 frames/s. Video playback is 15 frames/s (~7x real time). Imaging depth was 242 μm.
Supplementary Video 2. Z stack of GFP expressing layer 5 neurons.
A Z-stack acquired in a Thy1-GFP O-line mouse in vivo. Layer V somas are readily visible >600 μm below the pial surface in this sparsely labeled animal. The laser power was not adjusted during data acquisition. Intensity of the images was adjusted after acquisition for ease of visualization of the somas.
Supplementary Video 3. Imaging neural activity across a 3.5 mm FOV.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. Images were acquired at 2048 × 2048 pixels at ~0.1 frames/s. Video playback is 12 frames/s (~120x real time). The anatomical location imaged is shown on the right. Zooming in digitally shows individual neuron resolution is maintained across the FOV. The undersampling strategy utilized here for full FOV imaging leads to a more pixelated appearance of the individual neurons compared to Supplementary Video 1. Imaging depth was 265 μm.
Supplementary Video 4. Simultaneous dual region imaging at high resolution.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. The two imaging pathways (Pathway 1: left; Pathway 2: right) are located at separate cortical areas. Imaging depth was 278 μm for Pathway 1 and 235 μm for Pathway 2. Images were acquired at 1024 × 1024 pixels at ~0.5 frames/s. Video playback is 12 frames/s (~24x real time).
Supplementary Video 5. Simultaneous dual region imaging at the same XY position and different depths.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. The two imaging pathways (Pathway 1: left; Pathway 2: right) are located at the same XY location within V1 (Fig. 5c). Imaging depth was 230 μm for Pathway 1 and 278 μm for Pathway 2. Images were acquired at 512 × 256 pixels at ~9.5 frames/s. Video playback is 30 frames/s (~3x real time).
Supplementary Video 6. Quad region imaging.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. Four regions were imaged by utilizing the two imaging pathways (Pathway 1: top row; Pathway 2: bottom row) and serially applying an offset voltage to the galvanometer scanners (Fig. 4d). Imaging depth was 238 μm for Pathway 1 and 284 μm for Pathway 2. Each of the four regions was acquired at 512 × 512 pixels at ~1.9 frames/s. Video playback is 15 frames/s (~7x real time).
Supplementary Video 7. Imaging neural activity in two regions with a resonant scanner.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. The two imaging pathways (Pathway 1: left; Pathway 2: right) are located in different XY locations and are located at the same focal plane (Fig. 4f). Imaging depth was 256 μm for Pathway 1 and 256 μm for Pathway 2. Images were acquired at 512 × 256 pixels at 30 frames/s. Video playback is 90 frames/s (~3x real time).
Supplementary Video 8. Imaging neural activity across a 2.5 mm FOV simultaneously at two depths.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. The two imaging pathways (Pathway 1: left; Pathway 2: right) are located at the same XY location and differ by 55 μm in Z (Supplementary Fig. 14). Imaging depth was 245 μm for Pathway 1 and 190 μm for Pathway 2. Images were acquired at 2048 × 1024 pixels at 0.8 frames/s. Video playback is 12 frames/s (~15x real time).
Acknowledgments
We are grateful to Hongkui Zeng for kindly providing transgenic mice, Kei Eto for providing mice for imaging in pilot experiments, Janet Berrios for providing the YFP labeled brain section, Sally Kim and Ben Philpot for providing the Thy1-GFP O-line mouse, and Zemax LLC for providing upgraded software. This work was supported by the following: the National Institute of Child Health and Human Development (T32-HD40127) and Burroughs Wellcome Fund Career Award at the Scientific Interface (J.N.S.); a Helen Lyng White Fellowship (I.T.S.); a Career Development Award from the Human Frontier Science Program (00063/2012), and grants from the National Science Foundation (1450824), the Whitehall Foundation, the McKnight Foundation, the Kingenstein Foundation, the Simons Foundation (SCGB 325407SS), and the NIH (R01NS091335, R01EY024294) (S.L.S.).
Footnotes
AUTHOR CONTRIBUTIONS
S.L.S. conceived the Trepan2p imaging system. J.N.S. and S.L.S. designed and engineered the system. J.N.S. developed the demultiplexing electronics, designed and optimized the optical systems, wrote the software, and constructed the system. M.W.K. consulted on optical optimizations. J.N.S., I.T.S., and S.L.S. performed the animal experiments. J.N.S. and S.L.S. analyzed and interpreted the data. J.N.S. and S.L.S. wrote the manuscript with input from all authors. I.T.S. and S.L.S. supervised the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing interests.
References
- 1.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–6. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 2.Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci U S A. 2003;100:7319–24. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Theer P, Denk W. On the fundamental imaging-depth limit in two-photon microscopy. J Opt Soc Am A Opt Image Sci Vis. 2006;23:3139–49. doi: 10.1364/josaa.23.003139. [DOI] [PubMed] [Google Scholar]
- 5.Ohki K, Chung S, Ch’ng YH, Kara P, Reid RC. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature. 2005;433:597–603. doi: 10.1038/nature03274. [DOI] [PubMed] [Google Scholar]
- 6.Smith SL, Hausser M. Parallel processing of visual space by neighboring neurons in mouse visual cortex. Nat Neurosci. 2010;13:1144–9. doi: 10.1038/nn.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komiyama T, et al. Learning-related fine-scale specificity imaged in motor cortex circuits of behaving mice. Nature. 2010;464:1182–6. doi: 10.1038/nature08897. [DOI] [PubMed] [Google Scholar]
- 8.Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron. 2007;56:43–57. doi: 10.1016/j.neuron.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuster JM. The cognit: a network model of cortical representation. Int J Psychophysiol. 2006;60:125–32. doi: 10.1016/j.ijpsycho.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q, Burkhalter A. Area map of mouse visual cortex. J Comp Neurol. 2007;502:339–57. doi: 10.1002/cne.21286. [DOI] [PubMed] [Google Scholar]
- 11.Cheng A, Goncalves JT, Golshani P, Arisaka K, Portera-Cailliau C. Simultaneous two-photon calcium imaging at different depths with spatiotemporal multiplexing. Nat Methods. 2011;8:139–42. doi: 10.1038/nmeth.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amir W, et al. Simultaneous imaging of multiple focal planes using a two-photon scanning microscope. Opt Lett. 2007;32:1731–3. doi: 10.1364/ol.32.001731. [DOI] [PubMed] [Google Scholar]
- 13.Voigt FF, Chen JL, Krueppel R, Helmchen F. A modular two-photon microscope for simultaneous imaging of distant cortical areas in vivo. Proc SPIE. 2015;9329 [Google Scholar]
- 14.Grewe BF, Voigt FF, van ‘t Hoff M, Helmchen F. Fast two-layer two-photon imaging of neuronal cell populations using an electrically tunable lens. Biomed Opt Express. 2011;2:2035–46. doi: 10.1364/BOE.2.002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai PS, et al. Ultra-large field-of-view two-photon microscopy. Opt Express. 2015;23:13833–47. doi: 10.1364/OE.23.013833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lecoq J, et al. Visualizing mammalian brain area interactions by dual-axis two-photon calcium imaging. Nat Neurosci. 2014;17:1825–9. doi: 10.1038/nn.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalatsky VA, Stryker MP. New paradigm for optical imaging: temporally encoded maps of intrinsic signal. Neuron. 2003;38:529–45. doi: 10.1016/s0896-6273(03)00286-1. [DOI] [PubMed] [Google Scholar]
- 18.Gobel W, Kampa BM, Helmchen F. Imaging cellular network dynamics in three dimensions using fast 3D laser scanning. Nat Methods. 2007;4:73–9. doi: 10.1038/nmeth989. [DOI] [PubMed] [Google Scholar]
- 19.Cotton RJ, Froudarakis E, Storer P, Saggau P, Tolias AS. Three-dimensional mapping of microcircuit correlation structure. Front Neural Circuits. 2013;7:151. doi: 10.3389/fncir.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogelstein JT, et al. Fast nonnegative deconvolution for spike train inference from population calcium imaging. J Neurophysiol. 2010;104:3691–704. doi: 10.1152/jn.01073.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson JD, Youngworth RN, Aikens DM. The cost of tolerancing. Proc SPIE 7433, Optical System Alignment, Tolerancing, and Verification III. 2009;7433 [Google Scholar]
- 22.Madisen L, et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–58. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Judkewitz B, Rizzi M, Kitamura K, Hausser M. Targeted single-cell electroporation of mammalian neurons in vivo. Nat Protoc. 2009;4:862–9. doi: 10.1038/nprot.2009.56. [DOI] [PubMed] [Google Scholar]
- 24.Brainard DH. The Psychophysics Toolbox. Spat Vis. 1997;10:433–6. [PubMed] [Google Scholar]
- 25.Pelli DG. The VideoToolbox software for visual psychophysics: transforming numbers into movies. Spat Vis. 1997;10:437–42. [PubMed] [Google Scholar]
- 26.Marshel JH, Garrett ME, Nauhaus I, Callaway EM. Functional specialization of seven mouse visual cortical areas. Neuron. 2011;72:1040–54. doi: 10.1016/j.neuron.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hopt A, Neher E. Highly nonlinear photodamage in two-photon fluorescence microscopy. Biophys J. 2001;80:2029–36. doi: 10.1016/S0006-3495(01)76173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogelstein JT, et al. Spike inference from calcium imaging using sequential Monte Carlo methods. Biophys J. 2009;97:636–55. doi: 10.1016/j.bpj.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotechnol. 2003;21:1369–77. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 30.Visser TD, Oud JL. Volume measurements in three-dimensional microscopy. Scanning. 1994;16:198–200. [Google Scholar]
- 31.Zinter JP, Levene MJ. Maximizing fluorescence collection efficiency in multiphoton microscopy. Opt Express. 2011;19:15348–62. doi: 10.1364/OE.19.015348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Driscoll JD, et al. Photon counting, censor corrections, and lifetime imaging for improved detection in two-photon microscopy. J Neurophysiol. 2011;105:3106–13. doi: 10.1152/jn.00649.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moon S, Kim DY. Analog single-photon counter for high-speed scanning microscopy. Opt Express. 2008;16:13990–4003. doi: 10.1364/oe.16.013990. [DOI] [PubMed] [Google Scholar]
- 34.Tian X, Xu L, Li X, Shang G, Yao J. IEEE International Conference on Imaging Systems and Techniques. IEEE, Thessaloniki; 2010. Geometric distortion correction for sinusoidally scanned atomic force microscopic images; pp. 398–402. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1. High resolution imaging using the Trepan2p.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. Images were acquired at 1024 × 1024 pixels at 1.9 frames/s. Video playback is 15 frames/s (~7x real time). Imaging depth was 242 μm.
Supplementary Video 2. Z stack of GFP expressing layer 5 neurons.
A Z-stack acquired in a Thy1-GFP O-line mouse in vivo. Layer V somas are readily visible >600 μm below the pial surface in this sparsely labeled animal. The laser power was not adjusted during data acquisition. Intensity of the images was adjusted after acquisition for ease of visualization of the somas.
Supplementary Video 3. Imaging neural activity across a 3.5 mm FOV.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. Images were acquired at 2048 × 2048 pixels at ~0.1 frames/s. Video playback is 12 frames/s (~120x real time). The anatomical location imaged is shown on the right. Zooming in digitally shows individual neuron resolution is maintained across the FOV. The undersampling strategy utilized here for full FOV imaging leads to a more pixelated appearance of the individual neurons compared to Supplementary Video 1. Imaging depth was 265 μm.
Supplementary Video 4. Simultaneous dual region imaging at high resolution.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. The two imaging pathways (Pathway 1: left; Pathway 2: right) are located at separate cortical areas. Imaging depth was 278 μm for Pathway 1 and 235 μm for Pathway 2. Images were acquired at 1024 × 1024 pixels at ~0.5 frames/s. Video playback is 12 frames/s (~24x real time).
Supplementary Video 5. Simultaneous dual region imaging at the same XY position and different depths.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. The two imaging pathways (Pathway 1: left; Pathway 2: right) are located at the same XY location within V1 (Fig. 5c). Imaging depth was 230 μm for Pathway 1 and 278 μm for Pathway 2. Images were acquired at 512 × 256 pixels at ~9.5 frames/s. Video playback is 30 frames/s (~3x real time).
Supplementary Video 6. Quad region imaging.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. Four regions were imaged by utilizing the two imaging pathways (Pathway 1: top row; Pathway 2: bottom row) and serially applying an offset voltage to the galvanometer scanners (Fig. 4d). Imaging depth was 238 μm for Pathway 1 and 284 μm for Pathway 2. Each of the four regions was acquired at 512 × 512 pixels at ~1.9 frames/s. Video playback is 15 frames/s (~7x real time).
Supplementary Video 7. Imaging neural activity in two regions with a resonant scanner.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. The two imaging pathways (Pathway 1: left; Pathway 2: right) are located in different XY locations and are located at the same focal plane (Fig. 4f). Imaging depth was 256 μm for Pathway 1 and 256 μm for Pathway 2. Images were acquired at 512 × 256 pixels at 30 frames/s. Video playback is 90 frames/s (~3x real time).
Supplementary Video 8. Imaging neural activity across a 2.5 mm FOV simultaneously at two depths.
Neuronal activity in a mouse expressing GCaMP6s in neocortical pyramidal neurons was imaged through a cranial window while a naturalistic visual stimulus was presented. The two imaging pathways (Pathway 1: left; Pathway 2: right) are located at the same XY location and differ by 55 μm in Z (Supplementary Fig. 14). Imaging depth was 245 μm for Pathway 1 and 190 μm for Pathway 2. Images were acquired at 2048 × 1024 pixels at 0.8 frames/s. Video playback is 12 frames/s (~15x real time).