Abstract
The recently developed human norovirus (HuNoV) B cell culture and mouse models hold promise for drug discovery and development but their suitability for antiviral studies has not been assessed. We demonstrate the inhibitory effect of the nucleoside analogue 2′-C-methylcytidine (2CMC) on HuNoV replication in the human B cell BJAB cell line and in Balb/c Rag/gamma chain-deficient (Rag-γc−/−) mice. These data suggest the applicability of both models for future study and development of antiviral drugs for the treatment of HuNoV infections.
Keywords: human norovirus, polymerase inhibitor, positive-sense RNA virus, enteric virus, antiviral
Human noroviruses (HuNoVs) are the leading cause of viral gastroenteritis and food-borne disease worldwide constituting a global economic problem (Bartsch et al., 2016; Havelaar et al., 2015; Kirk et al., 2015; Koo et al., 2010; Patel et al., 2008; Payne et al., 2013). No vaccines or antiviral drugs are currently approved for clinical use. However, potent and safe antiviral therapy is urgently needed to reduce the burden of norovirus disease, particularly in vulnerable populations – young children, the elderly and immunocompromised – where prolonged and severe disease is linked to morbidity and mortality (Bok & Green, 2012; Hall et al., 2012; Moreno-Espinosa et al., 2004). Research into HuNoVs has been hampered by difficulties in culturing HuNoVs in cells (Duizer et al., 2004; Herbst-Kralovetz et al., 2013; Takanashi et al., 2014). The cultivable genogroup V murine norovirus (MuNoV) is therefore a widely used surrogate for HuNoV (Karst & Wobus, 2015; Karst et al., 2014; Wobus et al., 2006). However, the genetic diversity between these viruses represents one challenge for HuNoV drug development. Thus, the recently reported successes in propagation of HuNoV in immunodeficient mice (Taube et al., 2013) and in a human B cell line (Jones et al., 2015; Jones et al., 2014) provide exciting tools for the evaluation of potential antiviral strategies and vaccine candidates directed against HuNoV. A number of anti-NoV strategies, including targeting essential viral proteins, have been described in recent years (for a recent review see (Thorne et al., 2016)). In particular, the nucleoside analogue 2′-C-methylcytidine (2CMC) has been well characterized for its antiviral efficacy against MuNoV in vitro and in a mouse model as well as against the human Norwalk virus in a replicon system (Costantini et al., 2012; Rocha-Pereira et al., 2012; Rocha-Pereira et al., 2013; Rocha-Pereira et al., 2015). Therefore, the goal of these studies was to perform proof-of-concept studies to validate the recently developed HuNoV culture systems for antiviral studies using 2CMC as a tool compound.
A previous study (Jones et al., 2014) and accompanying protocols article (Jones et al., 2015) demonstrate infection of genogroup II genotype 4 (GII.4) HuNoVs in vitro in the B cell line BJAB in the presence and absence of synthetic H type histo-blood group antigen or Enterobacter cloacae. This infection can be prevented by UV treatment of the virus stock prior to infection. We established this culture system in our laboratory to test the applicability of this new HuNoV culture system for antiviral efficacy testing (Fig. 1). We first determined the nontoxic concentrations of 2CMC in BJAB cells using the WST-1 assay (Roche, San Francisco, CA, USA). BJAB cells were exposed to increasing concentrations of 2CMC for 3 days, the length of the viral infection assay (Fig. 1A, line graph). The 50% cytotoxic concentration (CC50) was 75.5 ± 0.6 μM. Next, the viral inoculum with a given genome copy number was identified that resulted in the greatest increase over input in the BJAB system over a 3 day infection period (Fig. 1B). BJAB cells were infected with unfiltered HuNoV-positive stool containing the indicated genome copy numbers of HuNoV GII.4 Sydney as described (Jones et al., 2015; Jones et al., 2014) without extra addition of synthetic H type histo-blood group antigen or Enterobacter cloacae. After removing the unbound virus inoculum, cells were resuspended in fresh media. The day 0 sample was taken immediately to quantify the amount of viral genome bound to cells. The remaining cells were incubated at 37°C for 3 days to allow the infection to proceed. The combined content of each well, including cells and supernatant, was then harvested, and RNA was extracted from a 500 μl aliquot per sample collected on days 0 and 3 postinfection (p.i.). HuNoV genomes were measured by qRT-PCR as described (Taube et al., 2013). The fold increase in genome titers over day 0 (i.e., bound genomes) was calculated and graphed. Similar to previous work (Jones et al., 2015; Jones et al., 2014), an input genome copy number of ~3500 genome copies of GII.4 Sydney resulted in the greatest fold increase over input. To determine the background signal of the assay, the virus stock was UV treated prior to infection or left untreated (Fig. 1C). Based on these data, a genome increase over input of 3 fold or greater is considered to provide an indication of viral replication in this assay.
Fig. 1.
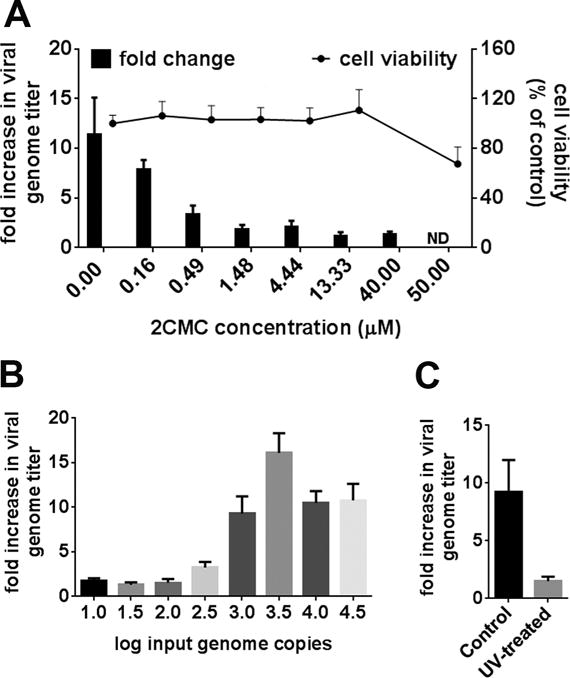
Anti HuNoV efficacy of 2CMC in vitro. A) BJAB cells were treated for 3 days with the indicated concentrations of 2CMC and cell viability was determined by WST-1 assay (Roche) (line graph). BJAB cells were infected with unfiltered HuNoV-positive stool for 3 days in the presence of the indicated concentrations of 2CMC. HuNoV genomes were measured by qRT-PCR. Genome titers at 3 days post infection were compared to inoculum titers to determine the fold increase in viral titer over inoculum (bar graph). B) BJAB cells were infected with unfiltered HuNoV-positive stool containing the indicated genome copy numbers. HuNoV genomes were measured by qRT-PCR 3 days postinfection. Genome titers were compared to inoculum titers to determine the fold increase in viral titer over inoculum. C) BJAB cells were infected with the unfiltered HuNoV-positive stool inactivated with UV light for 30 seconds or left untreated for 3 days. All data are presented as means ± SE of at least two independent experiments.
To determine the antiviral efficacy of 2CMC on GII.4 HuNoV, BJAB cells were infected with unfiltered stool containing ~3500 genome copies as described above. After 1 h of infection and removal of the unbound virus inoculum, cells were resuspended in fresh media containing a dilution series of 2CMC (0.16 – 40 μM) or PBS as vehicle control and were incubated for 3 days. In vehicle-treated cells, a maximal difference of approximately 3 PCR cycle thresholds (3 Cts) between 0 (mean Ct 23) and 3 (mean Ct 20) days p.i. was observed (not shown). This is equivalent to a log10 difference in copy number in line with a previous report (Brown et al., 2016). 2CMC significantly reduced HuNoV genome titers in BJAB cells in a concentration-dependent manner (Fig. 1A, bar graph). The half maximal 50% effective concentration (EC50) was 0.3 ± 0.02 μM and the therapeutic index (CC50/EC50) ~300. Thus, 2CMC exerted an antiviral effect at non-toxic concentrations. This antiviral potency of 2CMC against HuNoV was more pronounced than the in vitro activity reported against MuNoV (EC50 = ~2 μM) and in the replicon system (EC50 = ~18 μM) (Rocha-Pereira et al., 2012; Rocha-Pereira et al., 2013). The latter may be due to one or more differences in cell type (BJAB vs. Huh-7), virus strains (GI.1 vs. GII.4), or assay (replicon vs. infectious cycle). Importantly, these data demonstrate that the BJAB culture system is well suited to assess activity of potential inhibitors of HuNoV replication. Furthermore, this confirms that the RNA-dependent RNA polymerase (RdRp) is an excellent target to inhibit the replication of NoVs from multiple genogroups.
We also recently described the first mouse model of HuNoV infection, wherein Balb/c Rag-γc−/− infected intraperitoneally supported replication of a HuNoV (Taube et al., 2013). Therefore, the antiviral efficacy of 2CMC was next determined in this new animal model. Animal studies were performed according to the local and federal guidelines and protocols were approved by the University of Michigan Committee on Use and Care of Animals (UCUCA Number: PRO 00004534). Six to eight week old mice were administered 2CMC (100 mg/kg/day, divided into 2 daily treatments) or PBS vehicle control for 48 h intraperitoneally. One hour after the first drug treatment, mice were infected with HuNoV-positive stool containing 5 × 103 genome copies of GII.4 HuNoVs. Tissues were harvested 48 h p.i., and viral titers were determined by qRT-PCR as described (Taube et al., 2013). 2CMC reduced HuNoV genome loads by ~ 1.0 – 1.5 log10 in the colon, mesentery, liver and spleen, whereas no antiviral effect was observed in the jejunum, ileum, kidney and the bone marrow (Fig. 2A, data not shown). No viral genomes were detected in the stomach, duodenum, brain, heart and lung of 2CMC-treated or vehicle-treated animals (data not shown), indicating these sites were not infected. We next calculated the total viral load in 2CMC-treated or vehicle-treated animals and compared it to the inoculum titer. Analysis of the fold increase in HuNoV genome titers over input demonstrated significant decreases (~ 1 log10) in genome loads in the presence of 2CMC compared to vehicle controls (Fig. 2B). This decrease provided independent confirmation of productive HuNoV replication in this mouse model. Importantly, these data demonstrated that 2CMC significantly inhibits the replication of HuNoV infection in vivo, suggesting infection of Balb/c Rag-γc−/− can be used as an infection model for pre-clinical testing of antivirals against HuNoV.
Fig. 2.
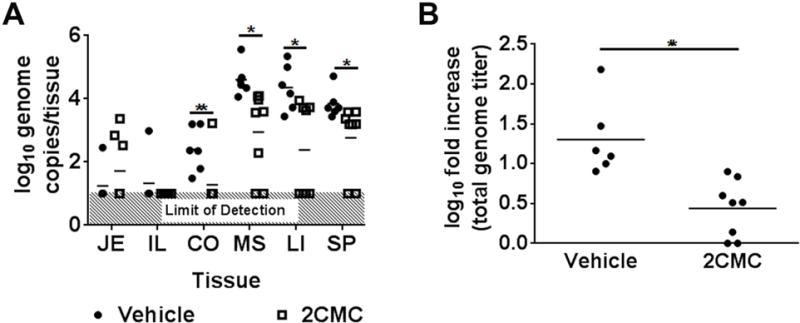
2CMC inhibits HuNoV replication in Balb/c Rag-γc−/− mice. Mice were intraperitoneally administered either 2CMC (open square) or vehicle control (closed circle) and infected with HuNoV-positive stool 1 hour later. (A) HuNoV genomes were measured by qRT-PCR in jejunum (JE), ileum (IL), colon (CO), mesentery (MS), liver (LI), and spleen (SP) from mice sacrificed 48 hours post infection (hpi). (B) Total genome titers recovered from each mouse at the time of harvest were compared to inoculum titers to determine the log fold increase in viral titer over inoculum. Data are from two independent experiments, and each symbol represents individual mouse. Data were analyzed using the Mann-Whitney U test. **, P < 0.01
Taken together, these data open up the exciting possibility of using both models for future study and development of antiviral drugs for the treatment of HuNoV infections, although both models in their current form have some caveats. For example, HuNoV replication and consequent increase in viral genome copies over input in BJAB cells and in Balb/c Rag-γc−/− mice is not as robust as what was observed using MuNoV infection of RAW 264.7 cells (~ 5 log10) and Balb/c Rag-γc−/− (~ 2 log10) (data not shown). This suggests that the number of HuNoV replication cycles is limited, for example by transformation of B cells or species-specific restrictions of HuNoV infection in mice. Thus, HuNoV adaptation and/or identification of host restriction factors may be required in both models to increase overall viral yields in both systems in the future. Current efforts to optimize both models by trying to adapt HuNoV to replicate more efficiently in BJAB cells and Balb/c Rag-γc−/− mice are ongoing in our laboratories. In addition, further improvements in the mouse model to more closely mimic aspects of HuNoV infection, especially the natural route of entry, would also help with testing orally delivered antiviral treatments. These efforts will contribute to the development of next generation propagation systems that broaden the applicability to the large number of existing HuNoV genotypes, to increase the dynamic range of the assays, and to make the cell culture system more suitable for automation. Ultimately, this will allow the testing of larger libraries of antiviral molecules and provide complementary platforms to currently available technologies (e.g., Norwalk virus replicon system (Chang et al., 2006)) in assessing the activity of clinical drug candidates.
In summary, our findings demonstrate that 2CMC inhibits infection of HuNoV in cell culture and in a mouse model. Specifically, our data demonstrate that both the new in vitro HuNoV propagation assay in BJAB cells and the HuNoV infection model in Balb/c Rag-γc−/− mice allow the evaluation of the anti-HuNoV activities of small molecule inhibitors of viral infection. Thus, these two models will be important tools in the future development of antiviral strategies for the treatment and prophylaxis of HuNoV infections.
Highlights.
the polymerase inhibitor 2′-C-methylcytidine inhibits HuNoV replication in the recently established B cell culture system
the nucleoside analogue 2′-C-methylcytidine inhibits HuNoV replication in a mouse model of HuNoV infection.
both models show promise for future study and development of antiviral drugs for the treatment of HuNoV infections
Acknowledgments
This work was supported in part by NIH grants R01 AI080611 and R21 AI110907 to CEW, by the Belgian Science Policy IUAP Interuniversity Attraction Poles (IAP) VII – P7/45 grant and KU Leuven IOF project HB/14/031 to JN. JRP is a Marie Curie COFUND William Harvey International Translational Research Academy (WHRI-ACADEMY #343) Postdoctoral Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. Global Economic Burden of Norovirus Gastroenteritis. PLoS ONE. 2016;11:e0151219. doi: 10.1371/journal.pone.0151219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok K, Green KY. Norovirus gastroenteritis in immunocompromised patients. The New England journal of medicine. 2012;367:2126–2132. doi: 10.1056/NEJMra1207742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Gilmour K, Breuer J. Norovirus infections occur in B cell deficient patients. Clin Infect Dis. 2016 doi: 10.1093/cid/ciw060. [DOI] [PubMed] [Google Scholar]
- Chang KO, Sosnovtsev SV, Belliot G, King AD, Green KY. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology. 2006;353:463–473. doi: 10.1016/j.virol.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Costantini VP, Whitaker T, Barclay L, Lee D, McBrayer TR, Schinazi RF, Vinje J. Antiviral activity of nucleoside analogues against norovirus. Antiviral therapy. 2012;17:981–991. doi: 10.3851/IMP2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MP, Estes MK. Laboratory efforts to cultivate noroviruses. The Journal of general virology. 2004;85:79–87. doi: 10.1099/vir.0.19478-0. [DOI] [PubMed] [Google Scholar]
- Hall AJ, Curns AT, McDonald LC, Parashar UD, Lopman BA. The roles of Clostridium difficile and norovirus among gastroenteritis-associated deaths in the United States, 1999–2007. Clin Infect Dis. 2012;55:216–223. doi: 10.1093/cid/cis386. [DOI] [PubMed] [Google Scholar]
- Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ, Praet N, Bellinger DC, de Silva NR, Gargouri N, Speybroeck N, Cawthorne A, Mathers C, Stein C, Angulo FJ, Devleesschauwer B, World Health Organization Foodborne Disease Burden Epidemiology Reference, G World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLoS medicine. 2015;12:e1001923. doi: 10.1371/journal.pmed.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst-Kralovetz MM, Radtke AL, Lay MK, Hjelm BE, Bolick AN, Sarker SS, Atmar RL, Kingsley DH, Arntzen CJ, Estes MK, Nickerson CA. Lack of norovirus replication and histo-blood group antigen expression in 3-dimensional intestinal epithelial cells. Emerging infectious diseases. 2013;19:431–438. doi: 10.3201/eid1903.121029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MK, Grau KR, Costantini V, Kolawole AO, de Graaf M, Freiden P, Graves CL, Koopmans M, Wallet SM, Tibbetts SA, Schultz-Cherry S, Wobus CE, Vinje J, Karst SM. Human norovirus culture in B cells. Nat Protoc. 2015;10:1939–1947. doi: 10.1038/nprot.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinje J, Tibbetts SA, Wallet SM, Karst SM. Enteric bacteria promote human and mouse norovirus infection of B cells. Science (New York, NY. 2014;346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst SM, Wobus CE. Viruses in rodent colonies: lessons learned from murine noroviruses. Ann Rev Virol. 2015;2:5.1–5.24. doi: 10.1146/annurev-virology-100114-055204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst SM, Wobus CE, Goodfellow IG, Green KY, Virgin HW. Advances in norovirus biology. Cell host & microbe. 2014;15:668–680. doi: 10.1016/j.chom.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS medicine. 2015;12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo HL, Ajami NJ, Atmar RL, DuPont HL. Noroviruses: The Leading Cause of Gastroenteritis Worldwide. Discovery Medicine. 2010;50:61–70. [PMC free article] [PubMed] [Google Scholar]
- Moreno-Espinosa S, Farkas T, Jiang X. Human caliciviruses and pediatric gastroenteritis. Seminars in pediatric infectious diseases. 2004;15:237–245. doi: 10.1053/j.spid.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinje J, Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerging infectious diseases. 2008;14:1224–1231. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne DC, Vinje J, Szilagyi PG, Edwards KM, Staat MA, Weinberg GA, Hall CB, Chappell J, Bernstein DI, Curns AT, Wikswo M, Shirley SH, Hall AJ, Lopman B, Parashar UD. Norovirus and medically attended gastroenteritis in U.S. children. The New England journal of medicine. 2013;368:1121–1130. doi: 10.1056/NEJMsa1206589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Pereira J, Jochmans D, Dallmeier K, Leyssen P, Cunha R, Costa I, Nascimento MS, Neyts J. Inhibition of norovirus replication by the nucleoside analogue 2′-C-methylcytidine. Biochemical and biophysical research communications. 2012;427:796–800. doi: 10.1016/j.bbrc.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Rocha-Pereira J, Jochmans D, Debing Y, Verbeken E, Nascimento MS, Neyts J. The Viral Polymerase Inhibitor 2′-C-Methylcytidine Inhibits Norwalk Virus Replication and Protects against Norovirus-Induced Diarrhea and Mortality in a Mouse Model. Journal of virology. 2013;87:11798–11805. doi: 10.1128/JVI.02064-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Pereira J, Van Dycke J, Neyts J. Treatment with a nucleoside polymerase inhibitor reduces shedding of murine norovirus in stool to undetectable levels without emergence of drug-resistant variants. Antimicrobial agents and chemotherapy. 2015 doi: 10.1128/AAC.02198-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanashi S, Saif LJ, Hughes JH, Meulia T, Jung K, Scheuer KA, Wang Q. Failure of propagation of human norovirus in intestinal epithelial cells with microvilli grown in three-dimensional cultures. Archives of virology. 2014;159:257–266. doi: 10.1007/s00705-013-1806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube S, Kolawole AO, Hohne M, Wilkinson JE, Handley SA, Perry JW, Thackray LB, Akkina R, Wobus CE. A mouse model for human norovirus. MBio. 2013;4:e00450–00413. doi: 10.1128/mBio.00450-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne L, Arias A, Goodfellow I. Advances Toward a Norovirus Antiviral: From Classical Inhibitors to Lethal Mutagenesis. The Journal of infectious diseases. 2016;213(Suppl 1):S27–31. doi: 10.1093/infdis/jiv280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus CE, Thackray LB, Virgin HWt. Murine norovirus: a model system to study norovirus biology and pathogenesis. Journal of virology. 2006;80:5104–5112. doi: 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]


