SUMMARY
Mitochondrial dysfunction is pervasive in human pathologies such as neurodegeneration, diabetes, cancer and pathogen infections as well as during normal aging. Cells sense and respond to mitochondrial dysfunction by activating a protective transcriptional program known as the mitochondrial unfolded protein response (UPRmt), which includes genes that promote mitochondrial protein homeostasis and the recovery of defective organelles [1, 2]. Work in C. elegans has shown that the UPRmt is regulated by the transcription factor ATFS-1, which is regulated by organelle partitioning. Normally, ATFS-1 accumulates within mitochondria, but during respiratory chain dysfunction, high levels of ROS or mitochondrial protein folding stress, a percentage of ATFS-1 accumulates in the cytosol and traffics to the nucleus where it activates the UPRmt [2]. While similar transcriptional responses have been described in mammals [3, 4], how the UPRmt is regulated remains unclear. Here, we describe a mammalian transcription factor, ATF5, which is regulated similarly to ATFS-1 and induces a similar transcriptional response. ATF5 expression can rescue UPRmt signaling in atfs-1-deficient worms requiring the same UPRmt promoter element identified in C. elegans. Furthermore, mammalian cells require ATF5 to maintain mitochondrial activity during mitochondrial stress and to promote organelle recovery. Combined, these data suggest that regulation of the UPRmt is conserved from worms to mammals.
RESULTS AND DISCUSSION
Mitochondrial dysfunction is associated with normal aging and contributes to a number of pathologic conditions including Parkinson’s, cancer, and infection. A number of stress responses and adaptations to mitochondrial dysfunction have been defined in mammalian systems such as the UPRmt, which includes the induction of mitochondrial chaperones and proteases [4, 5]. A genetic screen in C. elegans identified the transcription factor ATFS-1 as a key regulator of the UPRmt, which is regulated by organelle partitioning. ATFS-1 is normally imported into mitochondria [2, 6]. However, during mitochondrial dysfunction a percentage of ATFS-1 accumulates in the cytosol and subsequently traffics to the nucleus where it induces transcription of mitochondrial chaperones and proteases by binding the UPRmt promoter element (UPRmtE) [1, 2] as well as anti-bacterial innate immune genes [7]. Consistent with mediating a protective transcriptional response, worms lacking ATFS-1 incur respiratory defects during mitochondrial stress [1] and susceptibility to the pathogen Pseudomonas aeruginosa that perturbs mitochondrial function [7].
Considerable evidence suggests the UPRmt is conserved in mammals where it was originally discovered. Expression of the mitochondrial protein ornithine transcarbamylase (OTC) lacking 84 amino acids rendering it unable to fold (ΔOTC), or exposure to ethidium bromide (EtBr) which depletes mitochondrial DNA (mtDNA), induces transcription of mitochondrial chaperone and protease genes in cultured cells [4, 8]. Furthermore, perturbation of mitochondrial ribosomes activates a similar transcription response in cultured cells and mice [3, 9], strongly suggesting the existence of a homologous regulatory mechanism to that described in C.elegans. Perhaps most intriguing, an element in the promoters of those genes induced in a mouse model of mitochondrial myopathy is nearly identical to the UPRmtE to which ATFS-1 binds to induce chaperone and protease transcription in C. elegans [1, 10].
To identify potential regulators of a mammalian UPRmt, we searched for bZip proteins homologous to ATFS-1 with potential mitochondrial targeting sequences (MTS). ATF4 and ATF5 had considerable homology within the bZip domain to ATFS-1, however ATF5 also had a putative, but relatively weak, MTS as determined by Mitoprot [11] (Figures 1A and S1A). Interestingly, several studies suggest a role for ATF5 during mitochondrial dysfunction. A recent transcription profiling study from patients with autosomal dominant ataxia caused by a mutation in a gene encoding a mitochondrial protease had increased ATF5 transcripts [12], consistent with atfs-1 transcripts being induced in C. elegans when the orthologous mitochondrial AAA protease (spg-7) is impaired [2]. Furthermore, mouse models of respiratory chain dysfunction caused by impaired mtDNA replication or a defective mitochondrial aspartyl-tRNA synthetase also caused induction of ATF5 transcripts [10, 13].
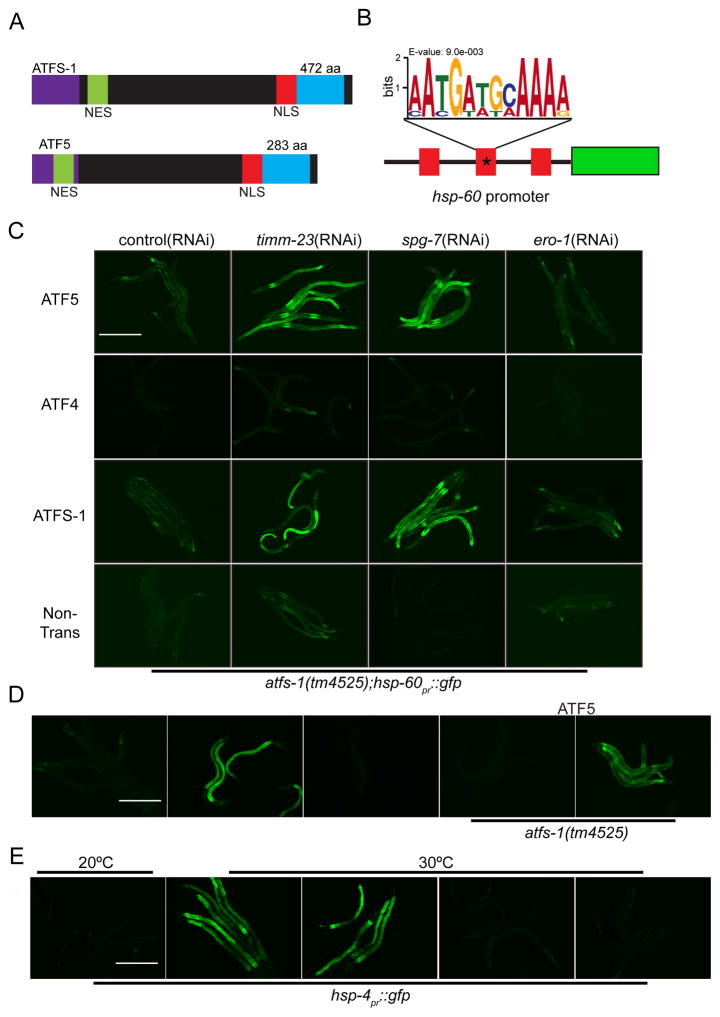
Figure 1. Expression of ATF5 rescues UPRmt activation in worms lacking ATFS-1.
(A) Schematic comparing the bZip transcription factors ATFS-1 and ATF5 including the mitochondrial targeting sequence (MTS), nuclear export sequence (NES) and the nuclear localization sequence (NLS).
(B) Schematic of the hsp-60pr::gfp reporter highlighting the three UPRmt elements in the promoter. The mutated element used in Figure 1D is marked with an asterisk (*).
(C) Photomicrographs of atfs-1(tm4525);hsp60pr::gfp worms expressing transgenic ATF5, ATF4 or ATFS-1 and raised on control, timm-23, spg-7, or ero-1(RNAi). Scale bar, 0.5 mm.
(D) Photomicrographs of wildtype and atfs-1(tm4525) worms expressing either hsp-60pr::gfp or hsp-60pr::gfp lacking a UPRmtE (*) (Figure 1B) raised on control or spg-7(RNAi). Worms in the right two panels express transgenic ATF5. Scale bar, 0.5 mm.
(E) Photomicrographs of control or transgenic ATF5 expressing hsp-4pr::gfp worms, raised on control or xbp-1(RNAi) incubated at 20°C or 30°C. Scale bar, 0.5 mm. See also Figure S1.
To determine if either ATF4 or ATF5 can regulate a UPRmt, the mammalian transcription factors were expressed in worms lacking ATFS-1. Identification of components that regulate the UPRmt has been facilitated by the use of mitochondrial chaperone transcriptional reporter C. elegans strains [14] (Figure 1B). Induction of hsp-60pr::gfp during mitochondrial stress requires both atfs-1 and the UPRmtE [1]. Interestingly, atfs-1-deletion worms expressing transgenic ATF5, but not worms expressing transgenic ATF4, were able to induce the hsp-60pr::gfp reporter during mitochondrial stress caused by depletion of a mitochondrial protein import component, timm-23, or protease, spg-7 (Figure 1C). And, UPRmt activation by ATF5 required the UPRmtE in the hsp-60 promoter (Figures 1B and D). ATF5 activation was mitochondrial stress-specific as perturbing endoplasmic reticulum (ER) protein folding with ero-1(RNAi) failed to activate hsp-60pr::gfp in transgenic ATFS-1 or ATF5 worms (Figure 1C), but did activate the ER chaperone reporter hsp-4pr::gfp (Figure S1B). Furthermore, transgenic ATF5 was unable to rescue induction of the ER UPR in worms lacking the ER stress-specific transcription factor XBP-1, further suggesting that ATF5 functions specifically during mitochondrial stress (Figure 1E).
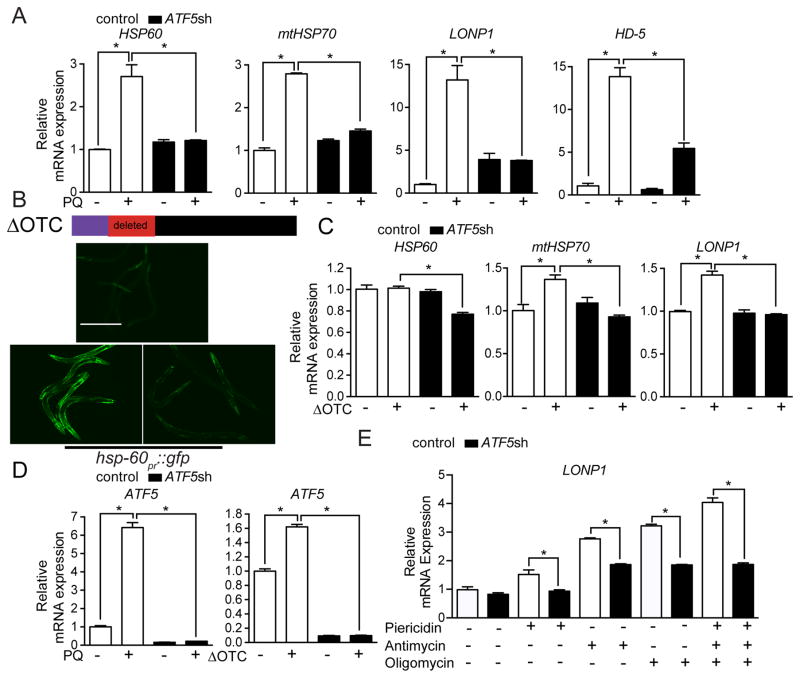
Analysis of all mammalian gene promoters [15] revealed putative UPRmtEs (Figure 1B) in the promoters of HSP60 (HSPD1), HSP10 (HSPE1), mtHSP70 (HSPA9), the mitochondrial protease LONP1, as well as ATF5 (data not shown) suggesting they may be regulated by ATF5. Exposure of HEK 293T cells to paraquat, which perturbs the respiratory chain causing toxic reactive oxygen (ROS) generation [16], resulted in increased transcription of HSP60, mtHSP70, LONP1 and HD-5, a secreted anti-microbial peptide (Figure 2A), consistent with activation of a UPRmt. Impressively, transcription of all four genes was significantly reduced when ATF5 was impaired by two different shRNAs (Figures 2A, S2A and S2B). Similarly, expression of ΔOTC, but not OTC (Figures S2C–G) caused modest induction of mtHSP70, HSP10, and LONP1, but not HSP60 (Figure 2C) consistent with ΔOTC being less toxic than paraquat. However, all four transcripts were reduced in ΔOTC expressing cells when treated with either ATF5 shRNA (Figures 2C and S2C–D), similar to C. elegans (Figure 2B). Lastly, ATF5 transcripts were also increased during mitochondrial stress (Figure 2D) consistent with what has been observed for atfs-1 in worms [1], for Atf5 in mouse models of mitochondrial disease [10, 13] and ATF5 in patient samples [12]. Of note, mRNAs encoding ER resident chaperones were not induced during mitochondrial stress in an ATF5-dependent manner (Figures S2H–I) suggesting ATF5 specifically promotes mitochondrial protein homeostasis.
Figure 2. ATF5 is required for UPRmt activation in mammalian cells.
(A) Expression levels of HSP60, mtHSP70, LONP1, and HD-5 mRNA in control or ATF5 shRNA HEK 293T cells with or without paraquat (PQ) (n=3, mean ± SEM, *p<0.05).
(B) Schematic showing ΔOTC construct. Photomicrographs of hsp-60pr::gfp worms raised on control or atfs-1(RNAi) with or without ΔOTC expression via the muscle-specific myo-3 promoter. Scale bar, 0.5 mm.
(C) Expression levels of HSP60, mtHSP70, and LONP1 mRNA in control or ATF5 shRNA HEK 293T cells with or without ΔOTC expression (n=3, mean ± SEM, *p<0.05)
(D) Expression levels of ATF5 mRNA in control or ATF5 shRNA HEK 293T cells with or without paraquat (PQ), or expressing ΔOTC (n=3, mean ± SEM, *p<0.05).
(E) Expression levels of LONP1 mRNA in control or ATF5 shRNA HEK 293T cells treated with oligomycin, antimycin, piericidin, or all three inhibitors (n=3, mean ± SEM, *p<0.05). See also Figure S2.
We next sought to determine the impact of OXPHOS activity and mitochondrial membrane potential (Δψ) on ATF5-dependent UPRmt activation. Interestingly, simultaneous inhibition of respiratory chain complexes I, III and V, which dissipates Δψ [17] induced LONP1 expression in an ATF5-dependent manner (Figure 2E) indicating that the UPRmt can be activated upon loss of membrane potential. However, because separate treatment with either piericidin (complex I), antimycin (complex III), or oligomycin (complex V), which does not deplete Δψ, also increased LONP1 expression (Figure 2E), depletion of Δψ is not required for UPRmt activation. Of note, unlike LONP1 mRNA (Figure 2E), we did not detect an increase in the steady state protein expression of LONP1 during oligomycin treatment (Figure SJ), consistent with multiple post-transcriptional events occurring during mitochondrial stress concomitant to the UPRmt including impaired mitochondrial protein import and subsequent degradation of those newly synthesized proteins [18, 19], the attenuation of protein synthesis [20, 21] and mitophagy [22][23].
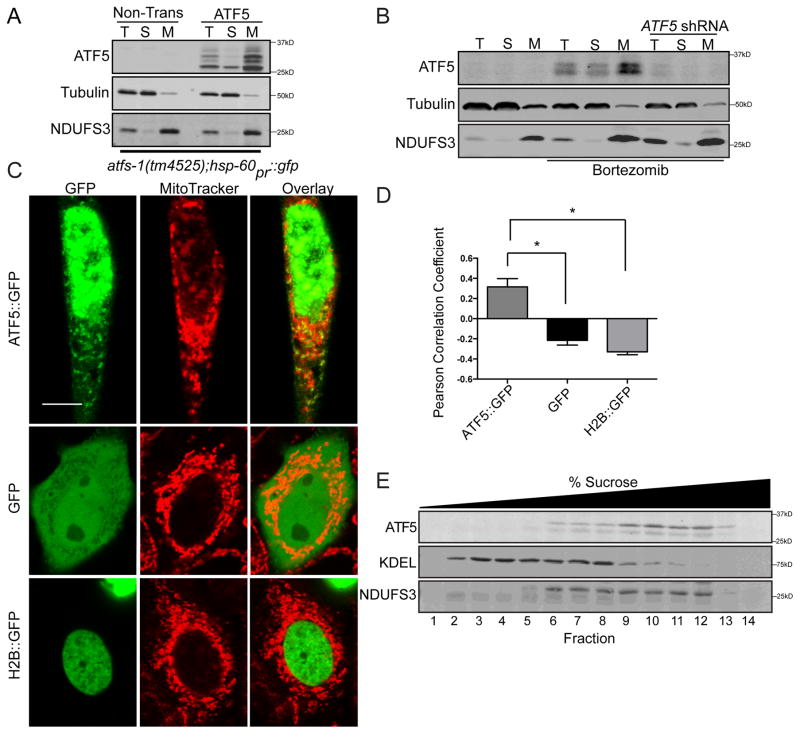
Because ATF5 regulates mitochondrial chaperone and protease transcription during mitochondrial stress similarly to ATFS-1, we hypothesized ATF5 may also be regulated via organelle partitioning. ATF5-dependent transcription has been shown to coincide with its nuclear accumulation [24, 25], consistent with the presence of a nuclear localization sequence (Figure 1A). Therefore, we sought to determine if ATF5 localizes to mitochondria in the absence of mitochondrial stress. Subcellular fractionation in C. elegans indicated that in the absence of stress, ATF5 co-fractionated with a mitochondrial protein (Figure 3A), consistent with ATF5 being inactive in the absence of stress (Figure 1C).
Figure 3. ATF5 localizes to mitochondria and nuclei.
(A) Immunoblots of lysates from control or ATF5 expressing atfs-1(tm4525);hsp60pr::gfp worms following fractionation into total lysate (T), postmitochondrial supernatant (S), and mitochondrial pellet (M). NDUFS3 serves as a mitochondrial marker and tubulin as a cytosolic marker.
(B) Immunoblots of control or ATF5 shRNA HeLa cells treated with DMSO or Bortezomib and fractionated into total lysate (T), postmitochondrial supernatant (S), and mitochondrial pellet (M).
(C) Photomicrographs of HeLa cells expressing either ATF5::GFP, Histone 2B::GFP (H2B::GFP), or GFP and stained with Mitotracker. Scale bar, 0.01 mm.
(D) Pearson Correlation Coefficient [40] of co-localization of ATF5::GFP, Histone 2B::GFP (H2B::GFP), or GFP and MitoTracker (Figure 3C) (n=5, mean ± SEM, *p<0.05).
(E) Immunoblot of mouse liver fractions following centrifugation on a sucrose gradient. Endogenous KDEL serves as an ER marker and NDUFS3 as a mitochondria marker.
Endogenous ATF5 is difficult to detect in cultured mammalian cells as translation of the ATF5 transcript is impaired by the presence of upstream open reading frames (uORFs) [26], and its relatively short half-life [27]. To increase ATF5 expression, HeLa cells were cultured in the presence of a proteasome inhibitor that leads to phosphorylation of the translation initiation factor eIF2α and increases synthesis of proteins encoded by uORF containing mRNAs [28]. Bortezomib treatment resulted in increased expression of ATF5, which was impaired by ATF5 shRNA (Figure 3B). Interestingly, ATF5 was enriched in the mitochondrial fraction further supporting localization of ATF5 in mitochondria in the absence of mitochondrial stress. Of note, we were unable to detect cleavage of the MTS upon import into mitochondria (Figure 3B), suggesting the MTS remains intact not unlike several other mitochondrial proteins [29].
Because we were only able to detect endogenous ATF5 in cultured cells upon proteasome inhibition (Figure 3B), we expressed ATF5::GFP via the CMV promoter in HeLa cells. Given the strong over-expression, considerable ATF5::GFP localized to the nucleus as expected, but ATF5::GFP also co-localized with mitochondria (Figures 3C and D), unlike GFP fusion proteins directed to the cytosol or nucleus (Figures 3C and D). Unlike in cultured cells, ATF5 is highly expressed in mouse and human liver cells [30], which coincides with an isoform of ATF5 mRNA lacking uORFs [31]. Interestingly, sucrose fractionation of mouse liver homogenates demonstrated co-fractionation of ATF5 with a mitochondrial, but not an ER-resident protein (Figure 3E). Given the difficulties in detecting ATF5 perhaps due to its low expression and short half-life [26, 32], it has been challenging to observe stress dependent shifts in ATF5 localization, but because ATF5-dependent transcription is induced during mitochondrial stress and ATF5 localizes to mitochondria in the absence of stress, our data are consistent with ATF5 being regulated post-translationally similarly to ATFS-1.
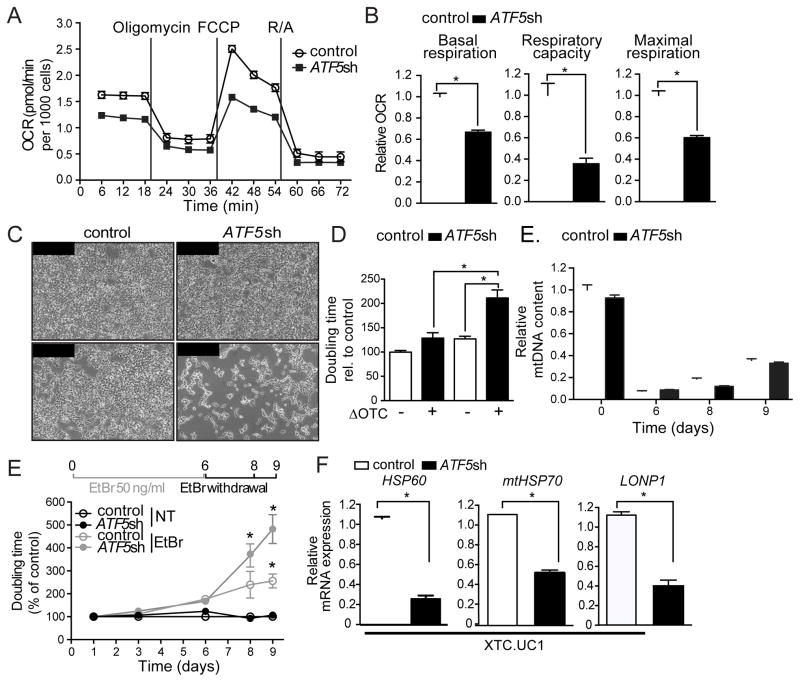
Lastly, we sought to determine the role of ATF5 in protecting or maintaining mitochondrial function. Interestingly, in HEK 293T cells, knockdown of ATF5 reduced basal respiration, overall respiratory capacity and maximal respiration (Figures 4A–B, S3A–B) suggesting a basal role for ATF5 in mitochondrial maintenance in these cells. ATF5 knockdown also impaired cell proliferation specifically in cells expressing ΔOTC (Figures 4C–D), further suggesting ATF5 promotes mitochondrial function during stress. We next examined the role of ATF5 during the recovery from mitochondrial stress caused by depletion of mitochondrial genomes (mtDNAs) via EtBr exposure. mtDNA was depleted to ~10% of normal levels in both control and ATF5 shRNA cells (Figures 4E and 4F). Upon EtBr removal, both cell types recovered mtDNA levels at similar rates (Figure 4E), suggesting ATF5 does not affect mtDNA replication. However, the ATF5 shRNA cells proliferated much more slowly (Figures 4F and S3C–D) suggesting ATF5 and the regulation of mitochondrial protein homeostasis machinery promotes the recovery from mitochondrial stress. Furthermore, ATF5 shRNA reduced the steady state expression of HSP60, mtHSP70, LONP1 and impaired cell growth (Figures 4G and S3E) in an oncocytic cell line harboring multiple mtDNA lesions that impair respiratory chain activity further supporting the role for ATF5-dependent transcription during mitochondrial dysfunction [33].
Figure 4. ATF5 promotes proliferation and recovery from mitochondrial stress.
(A–B) Oxygen consumption rates (OCR) in control or ATF5 shRNA HEK 293T cells (n=15, mean ± SEM,*p<0.05).
(C–D) Photomicrographs (C) and doubling times (D) of control or ATF5 shRNA HEK 293T cells, with or without ΔOTC expression. Scale bar, 0.1 mm (n=3-7, mean ± SEM *p<0.05).
(E) mtDNA quantification of control and ATF5 shRNA HEK 293T after 6 days of EtBr treatment, 4 days of withdrawal (n=3, mean ± SEM).
(F) Time course of doubling times of control and ATF5 shRNA HEK 293T cells following 6 days of mtDNA depletion by EtBr treatment (n=3, mean ± SEM, *p<0.05).
(G) Expression levels of HSP60, mtHSP70, or LONP1 mRNA in control or ATF5 shRNA XTC.UC1 cells (n=3, mean ± SEM, *p<0.05). See also Figure S3.
Combined, our data suggest that ATF5 regulates a UPRmt in mammalian cells that is similar to the response regulated by ATFS-1 in C. elegans. Our data support a model that when expressed, ATF5 localizes to mitochondria in the absence of stress. However, during mitochondrial stress, there is induction of mitochondrial protective transcripts, or a UPRmt, in an ATF5-dependent manner. This role is consistent with a previously established function of ATF5 as being anti-apoptotic by inducing BCL-2 transcription [34] and promoting survival of a number of cancer cells including glioblastomas [35, 36], which are known to have mitochondrial dysfunction [37]. Of note, our work also suggests an interaction between the UPRmt and the integrated stress response (ISR) because increased eIF2α-phosphorylation results in preferential ATF5 synthesis [26]. This is consistent with several studies demonstrating increased eIF2α phosphorylation during mitochondrial dysfunction mediated by the kinases GCN2 and PERK [38, 39] and suggests an additional layer of regulation in addition to organelle partitioning. Our findings indicate a protective role for ATF5 during mitochondrial dysfunction by regulating a UPRmt.
Experimental Procedures
A full description of the experimental procedures can be found in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank R. Wek, D. Chen and National BioResource Project for reagents. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SCHU 3023/1-1) to AMS, a NIH training fellowship to CJF (5T32GM008539-17) and the NIH (R01AG040061 and R01HL127891) to CMH.
Footnotes
Author Contributions
CJF, AMS and CMH designed the experiments. CJF, AMS, MP, YFL, and NR performed the experiments. CJF, AMS and CMH interpreted the results and CJF and CMH wrote the manuscript. CJF performed the C. elegans, imaging and fractionation experiments and AS performed the mRNA quantification, cell proliferation and oxygen consumption experiments.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and Nuclear Accumulation of the Transcription Factor ATFS-1 Promotes OXPHOS Recovery during the UPR. Molecular cell. 2015 doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, Argmann CA, Faridi P, Wolski W, Kutalik Z, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. The EMBO journal. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pellegrino MW, Haynes CM. Mitophagy and the mitochondrial unfolded protein response in neurodegeneration and bacterial infection. BMC biology. 2015;13:22. doi: 10.1186/s12915-015-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Molecular cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, Haynes CM. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. 2014;516:414–417. doi: 10.1038/nature13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Hoj PB, Hoogenraad NJ. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. European journal of biochemistry / FEBS. 1996;240:98–103. doi: 10.1111/j.1432-1033.1996.0098h.x. [DOI] [PubMed] [Google Scholar]
- 9.Moullan N, Mouchiroud L, Wang X, Ryu D, Williams EG, Mottis A, Jovaisaite V, Frochaux MV, Quiros PM, Deplancke B, et al. Tetracyclines Disturb Mitochondrial Function across Eukaryotic Models: A Call for Caution in Biomedical Research. Cell reports. 2015 doi: 10.1016/j.celrep.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyynismaa H, Carroll CJ, Raimundo N, Ahola-Erkkila S, Wenz T, Ruhanen H, Guse K, Hemminki A, Peltola-Mjosund KE, Tulkki V, et al. Mitochondrial myopathy induces a starvation-like response. Human molecular genetics. 2010;19:3948–3958. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- 11.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. European journal of biochemistry / FEBS. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 12.Mancini C, Roncaglia P, Brussino A, Stevanin G, Lo Buono N, Krmac H, Maltecca F, Gazzano E, Bartoletti Stella A, Calvaruso MA, et al. Genome-wide expression profiling and functional characterization of SCA28 lymphoblastoid cell lines reveal impairment in cell growth and activation of apoptotic pathways. BMC medical genomics. 2013;6:22. doi: 10.1186/1755-8794-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dogan SA, Pujol C, Maiti P, Kukat A, Wang S, Hermans S, Senft K, Wibom R, Rugarli EI, Trifunovic A. Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell metabolism. 2014;19:458–469. doi: 10.1016/j.cmet.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. Journal of cell science. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 15.Bucher P, Trifonov EN. Compilation and analysis of eukaryotic POL II promoter sequences. Nucleic acids research. 1986;14:10009–10026. doi: 10.1093/nar/14.24.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castello PR, Drechsel DA, Patel M. Mitochondria are a major source of paraquat-induced reactive oxygen species production in the brain. The Journal of biological chemistry. 2007;282:14186–14193. doi: 10.1074/jbc.M700827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfanner N, Neupert W. Transport of F1-ATPase subunit beta into mitochondria depends on both a membrane potential and nucleoside triphosphates. FEBS letters. 1986;209:152–156. doi: 10.1016/0014-5793(86)81101-2. [DOI] [PubMed] [Google Scholar]
- 18.Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 2015;524:485–488. doi: 10.1038/nature14951. [DOI] [PubMed] [Google Scholar]
- 19.Wright G, Terada K, Yano M, Sergeev I, Mori M. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Experimental cell research. 2001;263:107–117. doi: 10.1006/excr.2000.5096. [DOI] [PubMed] [Google Scholar]
- 20.Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet. 2012;8:e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michel S, Canonne M, Arnould T, Renard P. Inhibition of mitochondrial genome expression triggers the activation of CHOP-10 by a cell signaling dependent on the integrated stress response but not the mitochondrial unfolded protein response. Mitochondrion. 2015;21:58–68. doi: 10.1016/j.mito.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 22.Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9:1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin YF, Schulz AM, Pellegrino MW, Lu Y, Shaham S, Haynes CM. Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature. 2016;533:416–419. doi: 10.1038/nature17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155:321–332. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monaco SE, Angelastro JM, Szabolcs M, Greene LA. The transcription factor ATF5 is widely expressed in carcinomas, and interference with its function selectively kills neoplastic, but not nontransformed, breast cell lines. International journal of cancer Journal international du cancer. 2007;120:1883–1890. doi: 10.1002/ijc.22469. [DOI] [PubMed] [Google Scholar]
- 26.Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. The Journal of biological chemistry. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- 27.Uekusa H, Namimatsu M, Hiwatashi Y, Akimoto T, Nishida T, Takahashi S, Takahashi Y. Cadmium interferes with the degradation of ATF5 via a post-ubiquitination step of the proteasome degradation pathway. Biochemical and biophysical research communications. 2009;380:673–678. doi: 10.1016/j.bbrc.2009.01.158. [DOI] [PubMed] [Google Scholar]
- 28.Teske BF, Fusakio ME, Zhou D, Shan J, McClintick JN, Kilberg MS, Wek RC. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Molecular biology of the cell. 2013;24:2477–2490. doi: 10.1091/mbc.E13-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amaya Y, Arakawa H, Takiguchi M, Ebina Y, Yokota S, Mori M. A noncleavable signal for mitochondrial import of 3-oxoacyl-CoA thiolase. The Journal of biological chemistry. 1988;263:14463–14470. [PubMed] [Google Scholar]
- 30.Pascual M, Gomez-Lechon MJ, Castell JV, Jover R. ATF5 is a highly abundant liver-enriched transcription factor that cooperates with constitutive androstane receptor in the transactivation of CYP2B6: implications in hepatic stress responses. Drug metabolism and disposition: the biological fate of chemicals. 2008;36:1063–1072. doi: 10.1124/dmd.107.019380. [DOI] [PubMed] [Google Scholar]
- 31.Shimizu YI, Morita M, Ohmi A, Aoyagi S, Ebihara H, Tonaki D, Horino Y, Iijima M, Hirose H, Takahashi S, et al. Fasting induced up-regulation of activating transcription factor 5 in mouse liver. Life sciences. 2009;84:894–902. doi: 10.1016/j.lfs.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Li G, Xu Y, Guan D, Liu Z, Liu DX. HSP70 protein promotes survival of C6 and U87 glioma cells by inhibition of ATF5 degradation. The Journal of biological chemistry. 2011;286:20251–20259. doi: 10.1074/jbc.M110.211771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonora E, Porcelli AM, Gasparre G, Biondi A, Ghelli A, Carelli V, Baracca A, Tallini G, Martinuzzi A, Lenaz G, et al. Defective oxidative phosphorylation in thyroid oncocytic carcinoma is associated with pathogenic mitochondrial DNA mutations affecting complexes I and III. Cancer research. 2006;66:6087–6096. doi: 10.1158/0008-5472.CAN-06-0171. [DOI] [PubMed] [Google Scholar]
- 34.Persengiev SP, Devireddy LR, Green MR. Inhibition of apoptosis by ATFx: a novel role for a member of the ATF/CREB family of mammalian bZIP transcription factors. Genes & development. 2002;16:1806–1814. doi: 10.1101/gad.992202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angelastro JM, Canoll PD, Kuo J, Weicker M, Costa A, Bruce JN, Greene LA. Selective destruction of glioblastoma cells by interference with the activity or expression of ATF5. Oncogene. 2006;25:907–916. doi: 10.1038/sj.onc.1209116. [DOI] [PubMed] [Google Scholar]
- 36.Sheng Z, Li L, Zhu LJ, Smith TW, Demers A, Ross AH, Moser RP, Green MR. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nature medicine. 2010;16:671–677. doi: 10.1038/nm.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griguer CE, Oliva CR. Bioenergetics pathways and therapeutic resistance in gliomas: emerging role of mitochondria. Current pharmaceutical design. 2011;17:2421–2427. doi: 10.2174/138161211797249251. [DOI] [PubMed] [Google Scholar]
- 38.Baker BM, Nargund AM, Sun T, Haynes CM. Protective Coupling of Mitochondrial Function and Protein Synthesis via the eIF2? Kinase GCN-2. PLoS Genet. 2012:8. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hori O, Ichinoda F, Tamatani T, Yamaguchi A, Sato N, Ozawa K, Kitao Y, Miyazaki M, Harding HP, Ron D, et al. Transmission of cell stress from endoplasmic reticulum to mitochondria enhanced expression of Lon protease. J Cell Biol. 2002;157:1151–1160. doi: 10.1083/jcb.200108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.