Abstract
The activities and genome-wide specificities of CRISPR-Cas Cpf1 nucleases1 are not well defined. We show that two Cpf1 nucleases from Acidaminococcus sp. BV3L6 and Lachnospiraceae bacterium ND2006 (AsCpf1 and LbCpf1, respectively) have on-target efficiencies in human cells comparable with those of the widely used Streptococcus pyogenes Cas9 (SpCas9)2–5. We also report that four to six bases at the 3’ end of the short CRISPR RNA (crRNA) used to program Cpf1 nucleases are insensitive to single base mismatches, but that many of the other bases in this region of the crRNA are highly sensitive to single or double substitutions. Using GUIDE-seq and targeted deep sequencing analyses performed with both Cpf1 nucleases, we were unable to detect off-target cleavage for more than half of 20 different crRNAs. Our results suggest that AsCpf1 and LbCpf1 are highly specific in human cells.
Clustered, regularly interspaced, short palindromic repeat (CRISPR) systems encode RNA-guided endonucleases that are essential for bacterial adaptive immunity6. CRISPR-associated (Cas) nucleases can be readily programmed to cleave target DNA sequences for genome editing in various organisms2–5. One class of these nucleases, referred to as Cas9 proteins, complex with two short RNAs: a crRNA and a trans-activating crRNA (tracrRNA)7, 8. The most commonly used Cas9 ortholog, SpCas9, uses a crRNA that has 20 nucleotides at its 5’ end that are complementary to the “protospacer” region of the target DNA site. Efficient cleavage also requires that SpCas9 recognizes a protospacer adjacent motif (PAM). The crRNA and tracrRNA are usually combined into a single ~100 nt guide RNA (gRNA)7, 9–11 that directs the DNA cleavage activity of SpCas9. The genome-wide specificities of SpCas9 nucleases paired with different gRNAs have been characterized using multiple different approaches12–15. SpCas9 variants with substantially improved genome-wide specificities have also been engineered16, 17.
Recently, a Cas protein named Cpf1 has been identified that can also be programmed to cleave target DNA sequences1, 18–20. Unlike SpCas9, Cpf1 only requires a single 42 nt crRNA, which has 23 nucleotides at its 3’ end that are complementary to the protospacer of the target DNA sequence1. Furthermore, whereas SpCas9 recognizes an NGG PAM sequence that is 3’ of the protospacer, AsCpf1 and LbCp1 recognize TTTN PAMs that are found 5’ of the protospacer1. Early experiments with AsCpf1 and LbCpf1 showed that these nucleases can be programmed to edit target sites in human cells1 but they were only tested on a small number of sites and their genome-wide specificities were not characterized. In this report, we investigated the on-target activities and genome-wide specificities of AsCpf1 and LbCpf1 to better inform research and therapeutic applications of these nucleases.
Results
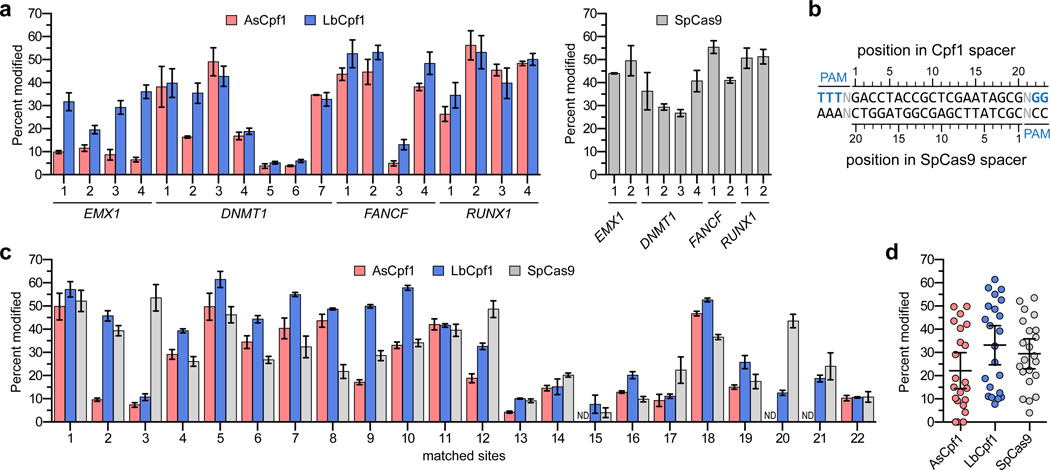
First, we assessed the robustness of AsCpf1 and LbCpf1 nucleases in U2OS human cells by testing their mutagenic editing activities on 19 target sites in four different human genes. Each Cpf1 target site is proximal to a DNA sequence that can also be efficiently mutagenized by SpCas9 (Supplementary Fig. 1). Six of the target sites (DNMT1 sites 1 – 4 and EMX1 sites 1 and 2) have previously been mutagenized with Cpf1 nucleases in HEK293 cells1. Using T7 Endonuclease I (T7E1) mismatch assays, we showed that both AsCpf1 and LbCpf1 efficiently induced insertion or deletion mutations (indels) at all 19 sites with mutagenesis percentages ranging from 3.8% to 56.2% (mean = 26.7%) for AsCpf1 and 5.2% to 53.2% (mean = 33.8%) for LbCpf1 (Fig. 1a). Using the same assay, the mutagenesis percentages for the ten proximal sites targeted by SpCas9 ranged from 26.8% to 55.4% (mean = 42.5%) (Fig. 1a).
Figure 1.
On-target indel mutation percentages induced by Cpf1 nucleases in human cells. (a) Endogenous gene percent modification induced by AsCpf1 and LbCpf1 at 19 endogenous human gene sites (left panel, determined by T7E1 assay). Target sites were chosen in amplicons known to be efficiently modified by SpCas9 (right panel, and see Supplementary Fig. 1). Error bars, s.e.m.; n = 3. (b) and (c) Matched target sites for AsCpf1, LbCpf1, and SpCas9 that share a common protospacer sequence (panel b) were examined for mutagenesis by AsCpf1, LbCpf1, and SpCas9 nucleases (determined by T7E1 assay; panel c). Error bars, s.e.m.; n = at least 3; ND, not detected. (d) Summary of matched site on-target activities from panel c, with means and 95% confidence intervals shown.
To enable a better comparison of the on-target mutagenic activities of AsCpf1 and LbCpf1 with that of SpCas9, we identified 22 endogenous human gene target sites (Supplementary Table 1) that contained overlapping target sites for both Cpf1 and Cas9 nucleases (Fig. 1b). These matched sites were selected to have variable numbers of predicted off-target sites in the genome, as estimated using Cas-OFFinder21 (Supplementary Table 2). Analysis of on-target activities for these 22 sites revealed variable mutagenic editing efficiencies for AsCpf1, LbCpf1, and SpCas9 (Figs. 1c and 1d). Time-course analysis revealed that near-maximum indel mutagenesis was achieved by 72 hours post-transfection with each nuclease for three different sites (Supplementary Fig. 2)..
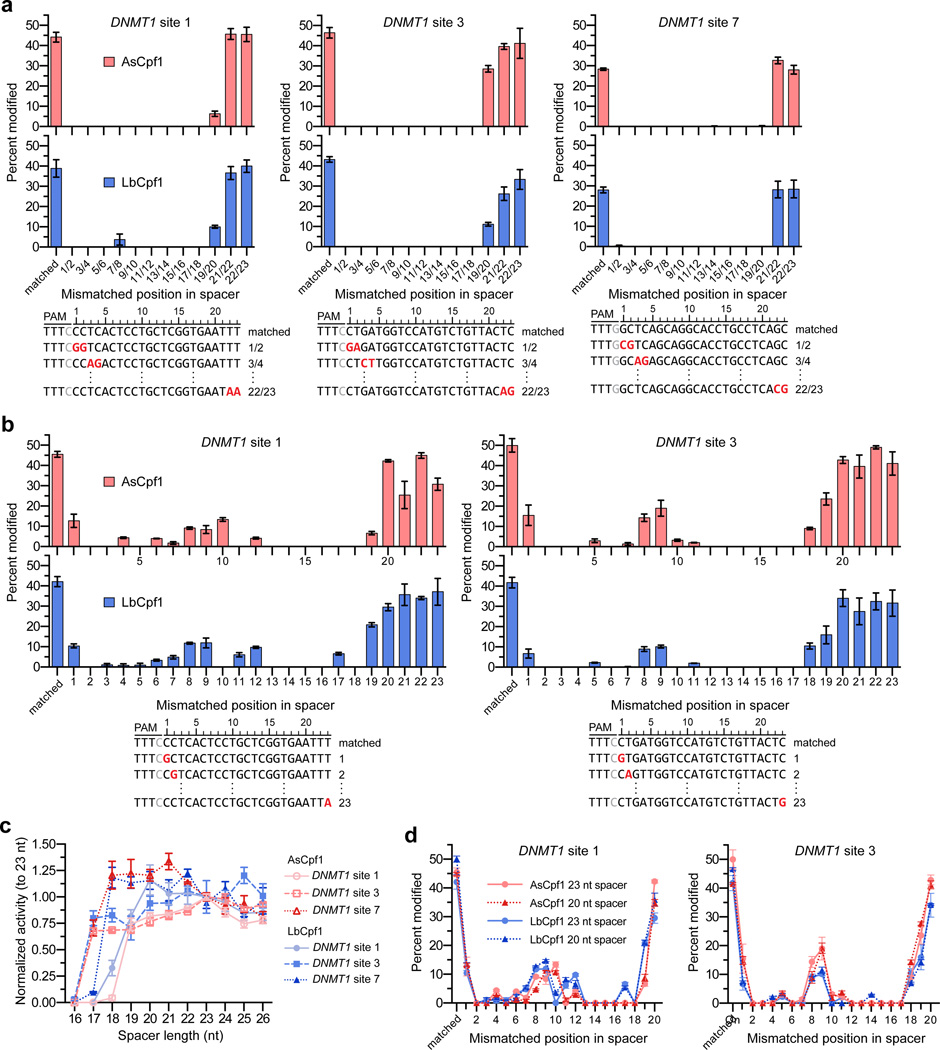
Next, to assess the specificities of AsCpf1 and LbCpf1, we examined the tolerance of these nucleases to mismatches at the crRNA-protospacer DNA interface. We mismatched adjacent pairs of bases within the complementarity regions of crRNAs targeted to three different endogenous target sites in the human DNMT1 gene (Fig. 2a). With all three target sites, we found that two adjacent mismatched bases at positions 1 through 18 (with 1 being the most PAM-proximal base) virtually eliminated detectable site modification as assessed by T7E1 assay for both AsCpf1 and LbCpf1 (Fig. 2a). By contrast, two adjacent mismatches of the PAM distal bases between positions 19 and 23 had substantially less pronounced effects on the mutagenic activities of both Cpf1 nucleases (Fig. 2a). Further testing of AsCpf1 and LbCpf1 editing at two of the three DNMT1 target sites using crRNAs with single mismatches along the length of the protospacer complementarity region revealed substantial and consistent tolerance to mismatches at positions 1, 8, 9, and 19 through 23 (Fig. 2b). Taken together, these experiments suggest that both AsCpf1 and LbCpf1 are highly sensitive to mismatched crRNA nucleotides at most positions between 1 and 18, whilst recognition of bases at the 3’ end of the crRNA may be dispensable for efficient mutagenic activity.
Figure 2.
Tolerance of AsCpf1 and LbCpf1 to mismatched or truncated crRNAs. (a, b) Endogenous gene modification by AsCpf1 and LbCpf1 using crRNAs that contain pairs of mismatched bases (panel a) or singly mismatched bases (panel b). Activity determined by T7E1 assay; error bars, s.e.m.; n = 3. (c) Summary of mutagenesis percentages by AsCpf1 and LbCpf1 at 3 different endogenous sites with crRNAs bearing variable length 3’ end truncations or extensions (see also Supplementary Fig. 3a), where activities are normalized to mutagenesis observed when using the canonical 23 nucleotide spacer. Error bars, s.e.m.; n = 3; nt, nucleotide. (d) Activity of AsCpf1 and LbCpf1 when programed with singly mismatched crRNAs either of 23 or 20 nucleotides in length. For clarity, data from panel b is re-presented here. Activity determined by T7E1 assay; error bars, s.e.m.; n = 3.
We further evaluated the importance of bases at the 3’ end of the crRNA for editing by testing AsCpf1 and LbCpf1 with a series of crRNAs bearing variable-length 3’ end deletions and extensions. When Cpf1 was paired with truncated crRNAs against three DNMT1 sites, T7E1 analysis revealed that mutagenesis efficiencies remained robust even when four to six bases were removed from the 3’ end of the crRNA (Fig. 2c; Supplementary Fig. 3a). Interestingly, the addition of up to three matched bases to the 3’ end of the canonical length 23 nt crRNAs did not substantially alter indel formation by either AsCpf1 or LbCpf1 (Fig. 2c and Supplementary Fig. 3a). These findings suggest that just 17 to 19 bases of crRNA protospacer complementarity are required for robust Cpf1 activity and that four to six bases at the 3’ end of a 23 nt spacer may not be necessary for target site cleavage. Previous studies with SpCas9 showed that gRNAs bearing truncated target complementarity regions are usually more sensitive to single mismatches at the gRNA/target DNA interface than their full-length counterparts22. However, we observed virtually no difference in AsCpf1 and LbCpf1 activities when using singly mismatched crRNAs bearing 20 instead of 23 nucleotide spacer sequences (Fig. 2d; Supplementary Fig. 3b).
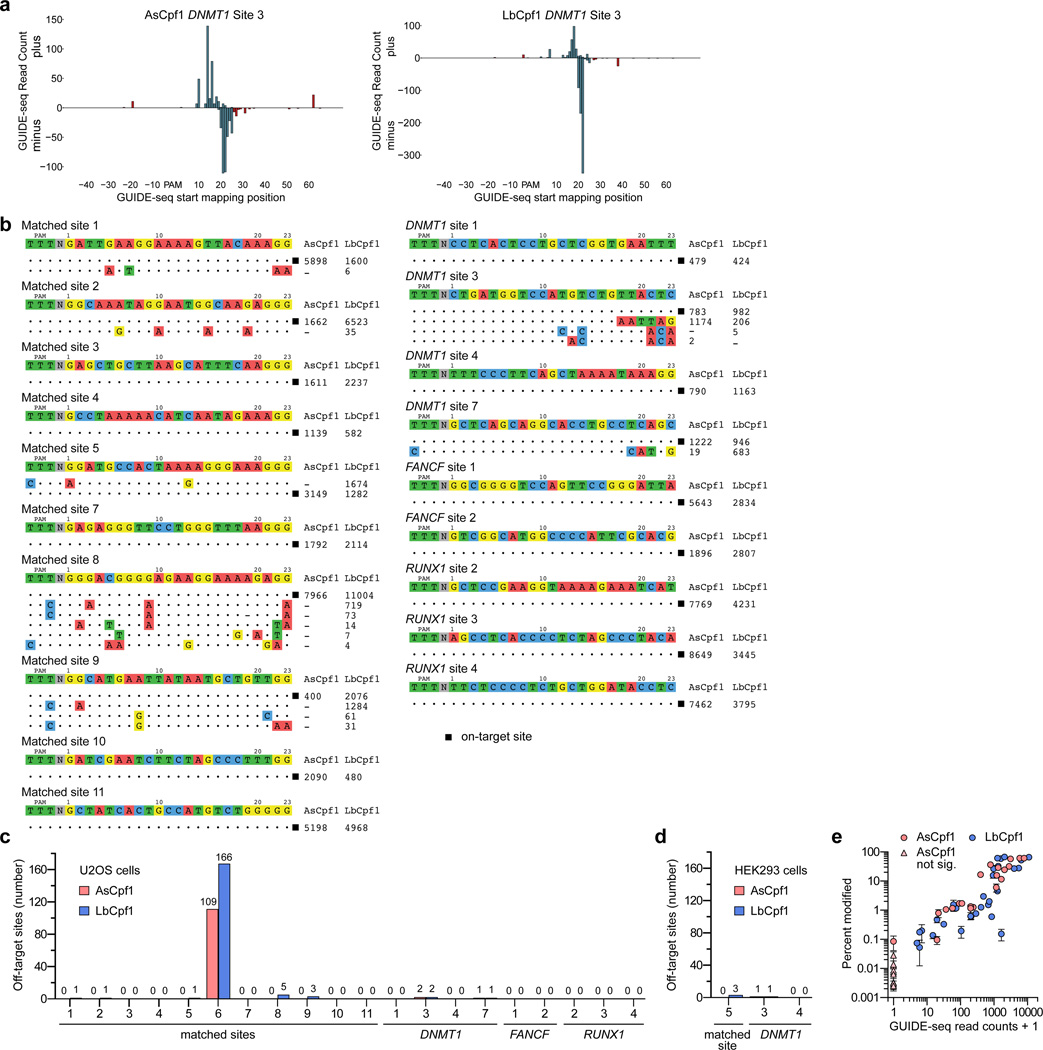
To ascertain the genome-wide specificities of Cpf1 nucleases in human cells, we used the Genome-wide Unbiased Identification of Double-stranded breaks Enabled by sequencing (GUIDE-seq) method12. GUIDE-seq has been used previously to identify off-target cleavage sites of wild-type and engineered Cas9 nucleases12, 16, 23–27. GUIDE-seq is based on the capture of an end-protected double-stranded oligodeoxynucleotide (dsODN) tag into nuclease-induced breaks in living cells; subsequent tag-specific amplification and sequencing of adjacent genomic sequences can then identify genome-wide cleavage sites. We assessed dsODN incorporation at 11 of the 22 matched sites from Fig. 1c that were mutagenized by all three nucleases (Fig. 1c) and observed efficient tag integration by AsCpf1, LbCpf1, and SpCas9 at all 11 genomic loci in human U2OS cells as measured using an RFLP assay (Supplementary Fig. 4a). The overall on-target mutagenic efficiencies observed in these experiments (the combination of dsODN integration and indels) were also assessed using the T7EI mismatch assay (Supplementary Fig. 4b). The ratios of dsODN incorporation efficiency to overall mutagenic efficiency for AsCpf1 and LbCpf1 at these 11 sites, which is a measure of the efficiency of tag integration at the on-target sites, were lower than for SpCas9 (means of 0.31, 0.28, and 0.53 for AsCpf1, LbCpf1, and SpCas9, respectively; Supplementary Figs. 4c and 4d). We also assessed dsODN tag integration (Supplementary Fig. 4e) with AsCpf1 and LbCpf1 at 9 of the 19 human gene target sites we used in the experiments shown in Fig. 1a and observed reasonably efficient tag incorporation and on-target mutagenesis in U2OS cells (Supplementary Fig. 4f). Ratios of dsODN incorporation efficiency to overall mutagenesis for AsCpf1 and LbCpf1 with these nine crRNAs (0.20 and 0.22, respectively) were comparable to the ratios observed with the 11 other crRNAs (Supplementary Fig. 4g). Collectively, these results show that dsODN tag integration occurs at Cpf1-induced breaks, albeit with lower efficiency than for SpCas9, perhaps due to differences in the nature of the DSBs induced by these nucleases (blunt ends for SpCas9 and 5’ overhangs for Cpf1; ref. 1). Variability in the location of dsODN tag integration was higher than is usually observed with SpCas9, but seemed to be constrained to 10–20 bps surrounding the expected DSB position at least for the sites assayed (Fig. 3a).
Figure 3.
Genome-wide specificities of AsCpf1 and LbCpf1. (a) Relative GUIDE-seq read start mapping position. The first 5’ base of the protospacer adjacent to the PAM is position 1. Reads that start within the protospacer region are colored in blue, reads that originate outside are colored in red. Reads mapping to the plus strand are depicted above the x-axis, reads mapping in the reverse orientation are depicted below. (b) Off-target sites for AsCpf1 and LbCpf1 with 20 different crRNAs, determined using GUIDE-seq in U2OS cells. Mismatched positions in the target sites of off-targets are highlighted in color, and GUIDE-seq read counts shown to the right of the on- and off-target sequences represent a measure of cleavage efficiency at a given site. (c) and (d) Summary of the total number of off-target sites detected by GUIDE-seq in U2OS cells with 20 crRNAs (panel c) and in HEK293 cells with 3 crRNAs (panel d). The off-target sequences and GUIDE-seq read counts for matched site #6 are shown in Supplementary Fig. 5. (e) Scatterplots of mean mutagenesis versus GUIDE-seq read counts analyzed from independent samples. Mutagenesis percentages for AsCpf1 that are not significantly different from negative controls are indicated with triangles. Error bars, s.e.m.; n = 3.
Having established the frequency of tag incorporation at Cpf1-induced DNA breaks for 20 different crRNAs, we used GUIDE-seq for these crRNAs and both Cpf1 nucleases in U2OS cells (40 GUIDE-seq experiments in total). The on-target sites for each of these 20 crRNAs tested with AsCpf1 and LbCpf1 were identified by GUIDE-seq (Fig. 3b). Strikingly, no crRNA-specific off-target sites were detected using GUIDE-seq for 17 of the 20 crRNAs with AsCpf1 and for 12 of the 20 crRNAs with LbCpf1 (Figs. 3b and 3c; Supplementary Table 4). The remaining crRNAs induced cleavage at only a very small number of off-target sites (range of 1 to 5 per crRNA), with the exception of the crRNA for matched site 6 for which 109 and 166 off-target sites were identified for AsCpf1 and LbCpf1, respectively (Figs. 3b and 3c; Supplementary Fig. 5). The larger numbers of off-target cleavage sites identified for the matched site 6 crRNA may be due to the relatively greater number of closely matched potential off-target sequences (as predicted by Cas-OFFinder21) that occur in the human genome for this crRNA compared with the others (Supplementary Table 2). Some of the off-target sites we identified have multiple mismatched bases at the PAM-distal end of the protospacer, consistent with the results of our experiments using systematicall y mismatched crRNAs. AsCpf1 generally targeted fewer numbers of off-target sites than LbCpf1 in matched comparisons and also appeared to more stringently specify the PAM, consistent with previous in vitro experiments showing that AsCpf1 is more specific for a TTTN PAM than LbCpf1 (ref. 1).
We also carried out GUIDE-seq experiments with AsCpf1 and LbCpf1 in human HEK293 cells using three of the 20 crRNAs we tested in U2OS cells. RFLP and T7E1 experiments showed GUIDE-seq tag incorporation and overall mutagenesis efficiencies to be slightly lower in HEK293 cells compared with U2OS cells (Supplementary Figs. 6a–6c), consistent with previous studies26. GUIDE-seq analysis with each of the three crRNAs and AsCpf1 or LbCpf1 in HEK293 cells identified a similar number of off-target sites as were observed in U2OS cells (Figs. 3c and 3d). Although a subset of off-target sites were the same in both cell types, some were also unique to one cell type and showed some differences in GUIDE-seq read count numbers (Supplementary Fig. 6d). Of note, the one crRNA that did not identify any off-target sites with both AsCpf1 and LbCpf1 in U2OS cells also failed to identify any sites in HEK293 cells (Supplementary Fig. 6d).
Given that dsODN incorporation with Cpf1 nucleases was less efficient than it was with Cas9, we assessed the sensitivity of GUIDE-seq for detecting Cpf1-induced off-target mutations. As previously described for Cas9, we used targeted amplicon sequencing to quantify mutagenesis at off-target sites (detected by GUIDE-seq) in U2OS cells transfected without the dsODN. We sequenced 30 GUIDE-seq-identified off-target sites induced by LbCpf1 with eight different crRNAs, and also sequenced these same sites for AsCpf1 even though 10 were not identified by GUIDE-seq. Mutation percentages and GUIDE-seq read counts are well correlated (R2 values between 0.64 and 1.00 for crRNAs with 4 or more sites sequenced)and revealed that GUIDE-seq can detect off-target sites mutagenized in the ~0.1% range (Fig. 3e and Supplementary Table 3). Importantly, for the AsCpf1-induced off-target sites not identified by GUIDE-seq, we observed mutation percentages that were either not statistically significant or at levels below 0.01% (Fig. 3e and Supplementary Table 3).
Because we found relatively few Cpf1-induced off-target sites using GUIDE-seq, we examined whether detectable indel mutations were occurring at closely mismatched sites in the human genome. To do this, we performed targeted amplicon sequencing of 61 of the most closely mismatched potential off-target sites for 15 of the 20 crRNAs we had assessed by GUIDE-seq. For 51 of these 61 mismatched sites, we failed to find statistically significant evidence of indels in U2OS cells transfected with Cpf1 nuclease and a crRNA but no dsODN tag (Supplementary Fig. 7). Among the ten mismatched sites at which we observed indel mutagenesis above background (0.026% to 32.0%), six had been found by our GUIDE-seq experiments and the other four were not identified by GUIDE-seq but had very low levels of mutations (0.026% to 0.206%) (Supplementary Fig. 7 and Supplementary Table 3). Based on these data, Cpf1 nucleases do not appear to efficiently mutagenize most closely related sequences. Further, GUIDE-seq can detect Cpf1 off-target sites that are mutagenized with frequencies as low as ~0.1–0.2%.
In silico analysis using Cas9-OFFinder21 revealed that Cpf1 nucleases with a TTTN PAM have at least one target site in a high percentage of first- and second-coding exons, microRNAs and promoter regions in human cells, including some regions that cannot be targeted with wild-type SpCas9 (although the expansion in targeting range diminishes with removal of the 5’ G required when using an RNA polymerase III U6 promoter to express gRNAs for SpCas9; Supplementary Fig. 8).
Discussion
In summary, we show that AsCpf1 and LbCpf1 nucleases can induce indel mutations at endogenous gene target sites in human cells with efficiencies comparable to those of the wild-type SpCas9 nuclease. GUIDE-seq analysis and targeted deep sequencing using a large number of different crRNAs with both AsCpf1 and LbCpf1 suggests that these nucleases are highly specific in human cells, with most nucleases showing no off-target effects. Furthermore, the sensitivity of Cpf1 to single base mismatches in certain positions of the protospacer might mean that these nucleases are suitable for allele-specific editing of heterozygous alleles. Although it is difficult to rigorously compare the genome-wide specificities of Cas9 and Cpf1 nucleases (owing to their different modes of target recognition and PAM requirements) our analysis suggests that the specificities of Cpf1 nucleases (and of AsCpf1 in particular) may approach that of the recently described high-fidelity SpCas9 variants16, 17. Improving the detection limit of GUIDE-seq will enable better assessment of whether lower frequency off-target effects are induced by these nucleases. In addition, it may be possible to further improve Cpf1 nuclease specificities using strategies similar to those recently applied to SpCas916, 17. Collectively, our results encourage broader use of Cpf1 for research applications and evaluation of these nucleases for the development of highly specific therapeutics.
ONLINE METHODS
Plasmids and oligonucleotides
A list and partial sequences of plasmids used in this study can be found in Supplementary Note 1; crRNA sequences are listed in Supplementary Table 1 and oligonucleotide sequences are found in Supplementary Table 5. AsCpf1 and LbCpf1 human expression plasmids (SQT1659 and SQT1665, respectively) were generated by inserting the open-reading frames of these nucleases from plasmids pY010 and pY016 (Addgene plasmids # 69982 and # 69988, respectively) into the NotI and AgeI sites of pCAG-CFP (Addgene # 11179). Plasmid SQT817 was used to express SpCas9 (ref 28). Oligonucleotide duplexes corresponding to spacer sequences were annealed and ligated into BsmBI-digested BPK3079, BPK3082, and BPK1520 (ref 24) for U6 promoter-driven expression of AsCpf1, LbCpf1, and SpCas9 gRNAs, respectively. New plasmids described in this study will be deposited with the non-profit plasmid repository Addgene: http://www.addgene.org/crispr-cas.
Human cell culture and transfection
U2OS cells (obtained from Toni Cathomen, Freiburg) and HEK293 cells were cultured at 37°C with 5% CO2 in Advanced DMEM supplemented with 10% heat-inactivated fetal bovine serum, 2 mM GlutaMax, and penicillin/streptomycin (all cell culture products were obtained from Life Technologies). Cell line identity was validated by STR profiling (ATCC) and mycoplasma testing was performed twice per month. 2 × 105 cells were transfected using the SE Cell Line Nucleofector Kit with 500 ng Cpf1- or SpCas9-encoding plasmid and 250 ng gRNA expression plasmid and the DN-100 program for U2OS cells, or with 300 ng Cpf1- or SpCas9-encoding plasmid and 150 ng gRNA expression plasmid using the CM-137 program for HEK293 cells, on a 4D-Nucleofector according to the manufacturer’s instructions (Lonza). For negative control transfections, Cpf1- or SpCas9-encoding plasmids were co-transfected with a U6-null plasmid that did not encode a crRNA. Genomic DNA extraction was performed approximately 72 hours following nucleofection using an Agencourt DNAdvance Genomic DNA Isolation Kit (Beckman Coulter).
T7E1 assays
T7 endonuclease I (T7E1) mutation detection assays were performed as previously described29 to determine Cpf1 or SpCas9 editing efficiencies at endogenous loci in human cells. Briefly, target loci were PCR amplified using Phusion Hot Start Flex DNA Polymerase (New England Biolabs) with 100 ng of genomic DNA as template. Primers used for PCR amplification are listed in Supplementary Table 5. Following an Agencourt AMPure XP (Beckman Coulter) purification, 200 ng of PCR product was denatured, reannealed, and digested with T7E1 (New England Biolabs). AMPure XP purified digestion reactions were analyzed using a QIAxcel capillary electrophoresis instrument (Qiagen), enabling calculation of estimated modification percentages.
GUIDE-seq
GUIDE-seq experiments were performed essentially as previously described12. Briefly, U2OS or HEK293 cells were transfected as described above with the addition of either 100 or 5 pmol, respectively, of an end-protected double-stranded oligodeoxynucleotide (dsODN) GUIDE-seq tag that encodes an internal NdeI restriction site. Restriction-fragment length polymorphism (RFLP) assays, performed as previously described24, and T7E1 assays were performed at the target loci to determine tag integration efficiencies and modification percentages, respectively. High-throughput sequencing libraries generated after tag-specific amplification were sequenced using an Illumina MiSeq sequencer as previously described. Data was analyzed using open-source guideseq software30 with a window size of 75 bp and allowing for up to 7 mismatches relative to the intended target site. The threshold of 7 mismatches was chosen as the value where less than 0.001% of alignments would be expected to occur by chance (one locus excluded by this threshold showed medium to high mapping read counts in all 40 GUIDE-seq experiments (i.e., all 20 crRNAs with AsCpf1 or LbCpf1; Supplementary Table 4). This segment mapped to the U6 promoter that is present within chromosome 15 region but more detailed sequence analysis revealed that these reads are derived from the U6 promoter present on the crRNA expression vector used in our GUIDE-seq experiments (data not shown). These reads likely result from ligation of dsODN to the presence of some linearized crRNA vector present in our plasmid preparations). High-confidence cell-type specific SNPs were called using SAMTools. Updated guideseq software to analyze Cpf1 GUIDE-seq experiments is available online at: http://www.jounglab.org/guideseq/.
Targeted deep sequencing experiments
Targeted deep sequencing experiments were performed as previously described16. On-target, GUIDE-seq identified off-target, and potential off-target loci predicted using Cas-OFFinder21 were PCR amplified using Phusion Hot Start Flex DNA Polymerase (New England Biolabs) with the primers listed in Supplementary Table 5. A high-throughput library preparation kit (KAPA BioSystems) was used to generate dual-indexed Tru-seq libraries that were sequenced on an Illumina MiSeq sequencer. Amplicons with less than 5000 mapping read counts were excluded from the analysis. P-values were calculated by fitting a negative binomial regression model comparing indel rates between nuclease treated and control samples. To avoid model fitting issues when all control samples report zero reads, 1 count was added to each member of the set of observed read counts. Deep sequencing data and the results of statistical tests are reported in Supplementary Table 3.
Targeting range calculations
Targetable sites for SpCas9 and Cpf1 were identified using Cas-OFFinder21 by searching for genomic sequences matching GN19-NGG/N20-NGG and TTTN-N23 motifs, respectively. Targeting range calculations are based on overlaps between predicted cut sites and features defined in the RefSeq gene table downloaded from the UCSC Genome Browser on January 23, 2016. The first (defined as the most 5’) and second exons were extracted for each unique gene name. The set of TSS upstream sequences (200bp and 1000bp) were defined using all transcripts, and only counted once in cases where two genes share the same TSS upstream sequence. The set of microRNAs was defined as those transcripts with a “MIR” prefix in their RefSeq gene names.
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Health (NIH) Director’s Pioneer Award (DP1 GM105378) and NIH R01 GM107427 to J.K.J., the Jim and Ann Orr Research Scholar Award (to J.K.J.), a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (to B.P.K.), and an MGH Tosteson Award (to S.Q.T.). New reagents described in this work have been deposited with the non-profit plasmid distribution service Addgene (http://www.addgene.org/crispr-cas).
Conflict of Interest Statement
J.K.J. is a consultant for Horizon Discovery. J.K.J. has financial interests in Editas Medicine, Hera Testing Laboratories, Poseida Therapeutics, and Transposagen Biopharmaceuticals. JKJ’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. S.Q.T., M.J.A., and J.K.J. are co-founders of Beacon Genomics, a company that is commercializing methods for determining nuclease specificity.
Footnotes
Accession Codes
SRA: [currently obtaining]
Author Contributions
B.P.K., S.Q.T., and J.K.J. conceived of and designed experiments. B.P.K., S.Q.T., M.S.P., N.T.N., and M.M.W. performed all human cell experiments and analyzed data. S.Q.T., J.M.L., Z.M., and M.J.A. analyzed the GUIDE-seq and targeted deep-sequencing data. B.P.K., S.Q.T., and J.K.J. wrote the manuscript with input from all authors.
References
- 1.Zetsche B, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 5.Maeder ML, Gersbach CA. Genome-editing Technologies for Gene and Cell Therapy. Mol Ther. 2016 doi: 10.1038/mt.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright AV, Nunez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature's Toolbox for Genome Engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 7.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015;33:175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- 15.Kim D, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. 231. doi: 10.1038/nmeth.3284. p following 243. [DOI] [PubMed] [Google Scholar]
- 16.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schunder E, Rydzewski K, Grunow R, Heuner K. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int J Med Microbiol. 2013;303:51–60. doi: 10.1016/j.ijmm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Makarova KS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fagerlund RD, Staals RH, Fineran PC. The Cpf1 CRISPR-Cas protein expands genome-editing tools. Genome Biol. 2015;16:251. doi: 10.1186/s13059-015-0824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinstiver BP, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015 doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin H, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016 doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolukbasi MF, et al. DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat Methods. 2015 doi: 10.1038/nmeth.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedland AE, et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai SQ, Topkar VV, Joung JK, Aryee MJ. Open-source guideseq software for analysis of GUIDE-seq data. Nat Biotechnol. 2016;34:483. doi: 10.1038/nbt.3534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.