Abstract
While the invariant natural killer T (iNKT)-cell response to primary stimulation with the glycolipid, α-galactosylceramide (αGalCer), is robust, the secondary response to this stimulus is muted resulting in a hyporesponsive state characterized by anti-inflammatory interleukin-10 (IL-10) production and high expression of programmed cell death 1 (PD1) and neuropilin 1 (NRP1). The E protein transcription factors and their negative regulators, the Id proteins, have previously been shown to regulate iNKT cell thymic development, subset differentiation and peripheral survival. Here, we provide evidence that the expression of the transcriptional regulator Id2 is downregulated upon stimulation of iNKT cells with their cognate antigen. Moreover, loss of Id2 expression by iNKT cells resulted in a hyporesponsive state, with splenic Id2-deficient iNKT cells expressing low levels of TBET, high levels of PD1 and NRP1 and production of IL-10 upon stimulation. We propose that downregulation of Id2 expression is an essential component of induction of the anti-inflammatory, hyporesponsive state in iNKT cells.
Invariant natural killer T (iNKT) cells are innate T lymphocytes, capable of rapid response to invading pathogens and production of effector cytokines such as interferon-γ (IFNγ) and interleukin-4 (IL-4) upon stimulation.1,2 This T-cell subset develops in the thymus, undergoing rearrangement of their invariant T-cell receptor (TCR) (Vα14-Jα18 in mice) before sequential stages of development and entry into the peripheral tissue. Recent data now indicate that peripheral iNKT cells can be further divided into specific subsets: NKT1 cells, analogous to Th1 cells, express the transcription factor TBET and predominantly produce IFNγ upon stimulation, NKT2 cells express GATA3 and the signature iNKT cell protein PLZF (promyelocytic leukemia zinc-finger) and produce IL-4 and IL-13, and NKT17 cells express RORγt (retinoid-acid receptor-related orphan receptor γt) and produce IL-17.3–5 Upon activation with a strong TCR stimulus, such as the glycolipid α-galactosylceramide (αGalCer), a fourth subset of iNKT cells has been reported to differentiate. This subset, called regulatory or NKT10 cells, appears refractive to restimulation and produce anti-inflammatory cytokines such as IL-10.6,7 NKT10 cells exist under homeostatic conditions in the adipose tissue, where they help maintain an anti-inflammatory environment.8 Indeed, NKT10 cells found in the adipose tissue are necessary for the maintenance of the M2 anti-inflammatory macrophage population and for regulatory T cells, whereas their absence increases inflammation in this tissue.8 These cells can also be induced to differentiate from peripheral iNKT cells through strong TCR stimulation.7,9
E protein transcription factors and their negative regulators, the Id proteins, are essential for regulating development, differentiation, survival and proliferation of many cell types.10 Importantly, for iNKT cell biology, E protein transcription factors regulate the development of these cells in the thymus, whereas the Id proteins are required for iNKT cell subset differentiation and survival in the hepatic tissue.11–14 Here, we investigated how the protein Id2, which inhibits E protein activity, impacted differentiation of NKT10 regulatory cells. We found that Id2 is downregulated in induced NKT10 cells and that loss of Id2 increases the frequency of NKT10 regulatory cells under homeostatic conditions in the spleen. Increased understanding of how this iNKT cell subset differentiates and the factors required for this process will be essential for manipulation of these cells for therapeutic gain.
RESULTS
Id2 expression is required for maintenance of splenic NKT1 cells
Using Id2 reporter mice in which yellow fluorescent protein (YFP) was knocked into the first exon of the Id2 gene (Id2YFP), we found a population of cells within the spleen and liver that expressed high levels of Id2. Importantly, there was no difference in cell size or granularity that could explain the higher Id2 expression (data not shown). Characterizing these cells, we identified the majority of them as TCRβ+ CD1d tetramer+ NK1.1+ iNKT cells (Figure 1a). NK1.1 is typically expressed by NKT1 cells.3,7 During thymic development, NK1.1+ NKT1 cells express higher levels of Id2 compared with NKT2 cells, which preferentially express Id3.12 To investigate the expression of Id proteins in peripheral iNKT cells, we made use of Id2YFPId3GFP double reporter mice.12 Gating on TBET and PLZF to identify NKT1 and NKT2 cells, respectively, we found that NKT1 cells in the spleen and liver had the highest expression of Id2, whereas NKT2 cells expressed higher Id3 and lower levels of Id2 (Figure 1b and Supplementary Figure S2). To assess the significance of high Id2 expression in NKT1 cells, we analyzed iNKT cell subsets in mice with conditional deletion of Id2 (CD4creId2f/f mice). Gating on splenic iNKT cells, we found a significant reduction in expression of the transcription factor TBET in the absence of Id2 (Figure 1c). Most Id2-deficient iNKT cells expressed intermediate levels of PLZF and TBET and there was a moderate increase in PLZFhi NKT2 cells in the CD4creId2f/f mice (Figure 1c). To assess if loss of Id2 would also result in reduced TBET expression in mature NKT1 cells, we crossed Id2f/f mice to granzyme Bcre mice. Granzyme B is upregulated in mature NK1.1+ NKT1 cells and TBET is required for its expression.15 Loss of Id2 in mature NKT1 cells also resulted in a clear reduction in TBET expression (Supplementary Figure S1). These findings suggested that Id2 expression in NKT1 cells is required for the expression of the lineage-defining transcription factor TBET. Consistent with previous observations, loss of Id2 did not impact the proportion of iNKT cells in the spleen (Figure 1c), but resulted in a severely reduced iNKT cell compartment in the liver (data not shown).13
Figure 1.
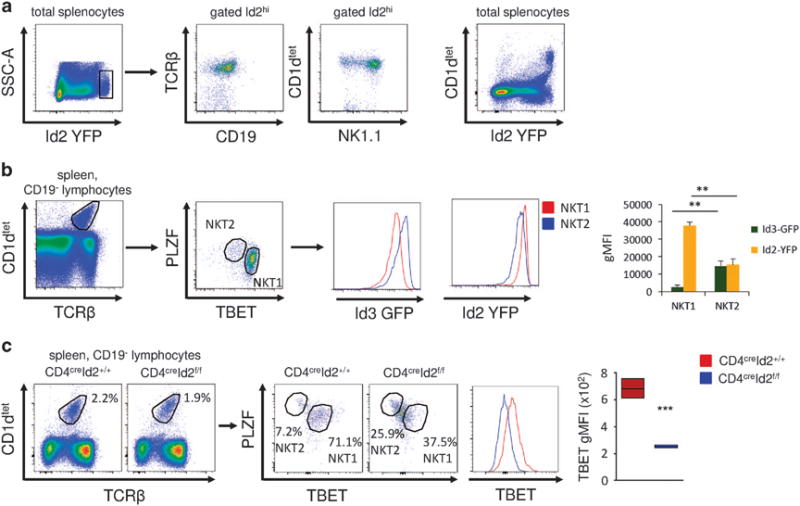
Id2 is required for NKT1 cells. (a) Representative fluorescence-activated cell sorting (FACS) plots of total splenocytes and gated Id2hi cells. (b) Representative FACS plots showing the gating strategy for NKT1 and NKT2 cells. Histograms and bar graphs show the geometric mean fluorescent intensity (gMFI) of Id2YFP and Id3GFP in NKT1 and NKT2 cells. Mean ± s.d. of three mice, representative of three independent experiments. (c) Splenic iNKT cells from untreated CD4creId2+/+ and CD4creId2f/f mice were analyzed for expression of the indicated transcription factors. Representative FACS plots and histogram of splenic iNKT cells gated as in (b). Graph of iNKT cells from three mice, representative of two experiments. Statistical significance was evaluated with unpaired Student’s t-test, where **P>0.01 and ***P>0.001.
Loss of Id2 resulted in hyporesponsive iNKT cells
In conventional T and iNKT cells, the transcription factor TBET is required for the production of IFNγ.16,17 We therefore hypothesized that the reduction in TBET expression in the absence of Id2 would result in decreased IFNγ production. We analyzed cytokine production in CD4creId2f/f and CD4creId2+/+ mice after challenge with 1 μg αGalCer in vivo. Consistent with the reduction we observed in NKT1 cells and TBET expression, Id2-deficient mice had significantly fewer IFNγ-producing iNKT cells (Figure 2a). Additionally, 475% of Id2-deficient iNKT cells produced neither IFNγ nor IL-4 (Figure 2a). These findings closely resembled the state of hyporesponsiveness or ‘anergy’ seen in iNKT cells previously exposed to αGalCer stimulation. To evaluate iNKT cell subsets and Id2 expression in mice previously exposed to αGalCer, we pretreated Id2YFP reporter mice with 1 μg of αGalCer. Twelve days after treatment, we found αGalCer-pretreated iNKT cells expressed moderate levels of PLZF and TBET, similar to our observation in Id2-deficient iNKT cells (Figures 1c and 2b). In addition, Id2 expression in αGalCer-pretreated iNKT cells was strongly reduced. Id2 downregulation occurred within 3 days of αGalCer treatment and remained low (Figure 2c). In contrast, injection with lipopolysaccharide (LPS) did not result in the downregulation of Id2 (Figure 2c). Our results show that in αGalCer-pretreated hyporesponsive iNKT cells, expression of Id2 and TBET are reduced.
Figure 2.
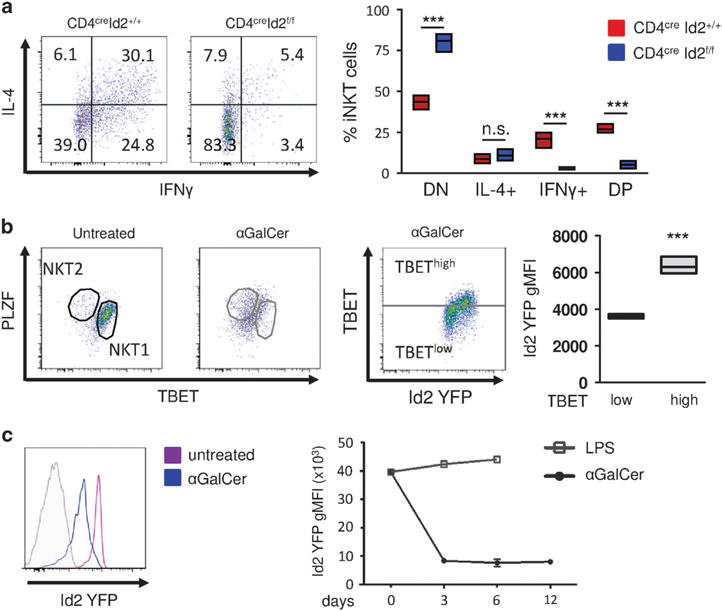
Absence of Id2 results in hyporesponsive iNKT cells. (a) CD4creID2f/f and CD4creID2+/+ mice were injected with 1 μg of αGalCer for 2 h. Total splenocytes were cultured for 4 h in the presence of monensin. iNKT cells were gated as in Figure 1b and analyzed for the expression of IL-4 and IFNγ. Representative fluorescence-activated cell sorting (FACS) plots and graph of iNKT cells from three mice. DN, double negative; DP, double positive for IFNγ and IL-4. (b) Analysis of iNKT cells from Id2YFP reporter mice 12 days after treatment with 1 μg of αGalCer or without treatment. Splenic iNKT cells were gated as in Figure 1b. Bar graphs indicate minimum to maximum and mean values of Id2 YFP geometric mean fluorescent intensity (gMFI) of three mice representative of two independent experiments. (c) Id2 YFP expression after iNKT cell activation. Mice were treated with 1 μg αGalCer or 40 μg LPS intraperitoneally. Splenic iNKT cells were gated as in Figure 1b and analyzed for Id2 YFP expression after 3 days (histogram), and 6 and 12 days (chart). Statistical significance was evaluated with unpaired Student’s t-test, where ***P>0.001.
Id2-deficient iNKT cells resemble NKT10 cells
αGalCer-pretreated iNKT have recently been described as a novel subset with regulatory potential termed induced NKT10 cells.7,9 This iNKT cell subset acquires characteristics of regulatory cells including the secretion of IL-10 rather than IFNγ upon restimulation. The cell-surface proteins programmed cell death 1 (PD1) and neuropilin 1 (NRP1) are considered typical markers of induced NKT10 cells.7 In accordance with these observations, most peripheral iNKT cells were positive for both PD1 and NRP1 expression after pretreatment with αGalCer (Figure 3a). Interestingly, induced PD1hi NRP1hi NKT10 cells had lower Id2 expression compared with iNKT cells not coexpressing PD1 and NRP1 (Figure 3a). To assess if loss of Id2 expression would result in a similar phenotype, we analyzed iNKT cells in CD4creId2f/f mice. In the absence of Id2, ~ 26% of iNKT cells coexpressed PD1 and NRP1 compared with ~ 12% of the CD4creId2+/+ wild-type controls (Figure 3b). We also observed increased expression of CXCR5, but not of Bcl6 or E4BP4 in iNKT cells of CD4creId2f/f mice (data not shown). We next examined IL-10 production by these cells. Upon stimulation with αGalCer significantly more Id2-deficient iNKT cells produced IL-10 compared with controls (Figure 3c). We confirmed this finding by crossing IL-10GFP reporter mice to CD4creId2f/f mice. Again, in the absence of Id2 significantly more iNKT cells expressed IL-10GFP after stimulation with αGalCer (Figure 3c). Thus, in the absence of Id2, the iNKT cell population in the spleen is enriched for cells phenotypically and functionally resembling induced NKT10 cells. These findings indicate that downregulation of Id2 observed in iNKT cells after pretreatment with αGalCer contributes to the induction of NKT10 cells.
Figure 3.
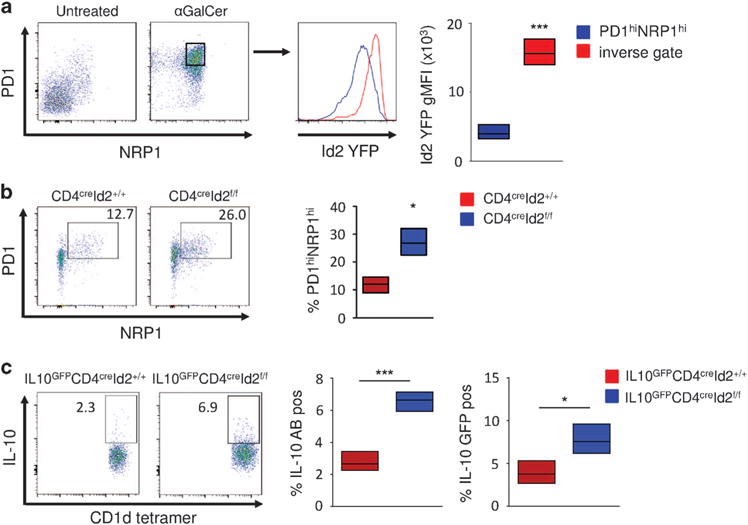
Decrease in Id2 expression induces NKT10 cells. (a) Analysis of induced NKT10 cells from Id2YFP reporter mice 12 days after no treatment or treatment with 1 μg of αGalCer. Splenic iNKT cells were gated as in Figure 1b. Bar graph indicating Id2 YFP geometric mean fluorescent intensity (gMFI) of iNKT cells from three individual mice. (b) Splenic iNKT cells from CD4creID2f/f and CD4creID2+/+ mice were analyzed for the expression of PD1 and NRP1. Fluorescence-activated cell sorting (FACS) plot gated as in Figure 1b and graphed data of three mice representative of two independent experiments. (c) IL-10GFPCD4creID2f/f and IL-10GFPCD4creID2+/+ mice were treated as in Figure 3b. Total splenocytes were cultured for 4 h in the presence of ionomycin, phorbol 12-myristate 13-acetate (PMA) and monensin. Expression of IL-10 in iNKT cells (representative FACS plots and graph) and IL-10 GFP iNKT cells (graph) are depicted. Data on graphs are percent positive iNKT cells from three mice. Bar graphs indicate minimum to maxium and mean values. Statistical significance was evaluated with unpaired Students t-test, where *P>0.05 and ***P>0.001.
Downregulation of Id2 occurs in response to strong TCR stimulus
To assess if downregulation of Id2 was a general phenomenon during iNKT cell activation, we analyzed Id2 expression in response to other known activators of iNKT cells. We found that iNKT cells down-regulate Id2 after stimulation with the CD1d-restricted antigens β-D-glucopyranosylceramide (βGlcCer), αGalCer and its truncated analog OCH in vitro (Figure 4a). In contrast, α-glucuronosylceramide (GSL1) another CD1d-restricted antigen derived from Sphingomonas species did not result in significant downregulation of Id2. LPS did not impact on Id2 expression, confirming our in vivo results (Figures 2c and 4a). LPS activates iNKT cells in a TCR-independent manner and the GSL1-CD1d complex has a low binding affinity for the invariant TCR.18,19 We therefore hypothesized that the strength of the TCR signal could be crucial for the induction of Id2 downregulation. To test our hypothesis, we exposed sorted iNKT cells to increasing concentrations of anti-CD3 antibody in the presence of costimulation. Consistent with our hypothesis, we observed a dose-dependent inverse correlation of anti-CD3 concentration with Id2 expression (Figure 4b). Strong TCR signal potently induces iNKT cell proliferation. We examined proliferation of iNKT cells in response to αGalCer in vitro. Extensive proliferation was associated with a strong reduction in Id2 expression (Figure 4c). Similarly, Ki-67+ iNKT cells proliferating 72 h after αGalCer in vivo had substantially lower Id2 expression compared with resting Ki-67− iNKT cells (Figure 4d). From these results, we conclude that increased TCR signaling strength positively correlates with the downregulation of Id2 expression by iNKT cells.
Figure 4.
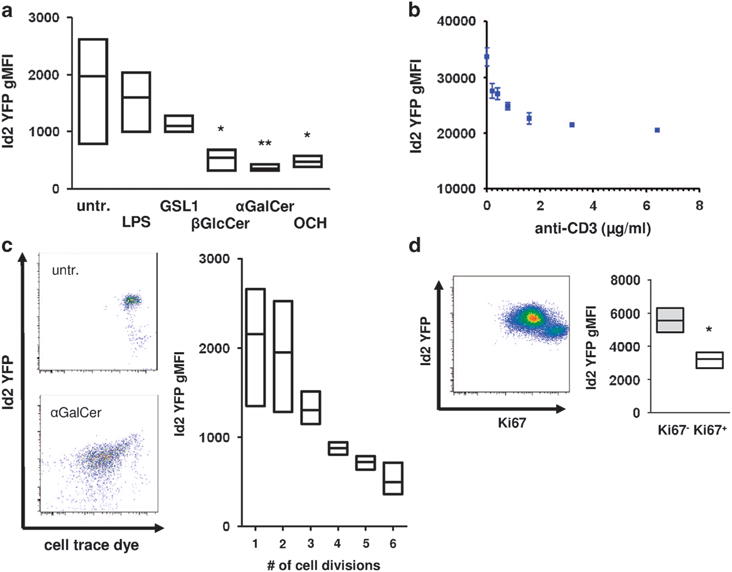
TCR signaling mediates downregulation of Id2. (a) Total splenocytes were stimulated with the indicated substances for 3 days in vitro. iNKT cells were gated as in Figure 1b and were analyzed for Id2 YFP expression. Histogram and bar graph indicating Id2 YFP geometric mean fluorescent intensity (gMFI) of iNKT cells representative of two independent experiments with three individual mice each. (b) Sorted splenic iNKT cells were stimulated with the indicated amount of plate-bound anti-CD3 and 1 μg ml−1 of anti-CD28 for 3 days in vitro. Representative data of two independent experiments with three individual mice each. (c) Total splenocytes were stimulated with 100 ng ml−1 αGalCer for 3 days in vitro. Id2 YFP expression and cell trace dye were analyzed. Representative fluorescence-activated cell sorting (FACS) plots and bar graph from two independent experiments with three individual mice each. (d) Mice were treated with 1 μg αGalCer intraperitoneally and Id2 YFP and Ki-67 expression were analyzed in splenic iNKT cells on day 3 after injection. Representative FACS plot and bar graph from three individual mice. Bar graphs indicate minimum to maximum and mean values of gMFI. Statistical significance was evaluated using unpaired Student’s t-test and one-way analysis of variance with Bonferoni post hoc test. *P>0.05 and **P>0.01.
DISCUSSION
Activation of iNKT cells with αGalCer in vivo results in the rapid secretion of multiple cytokines followed by a sustained state of hyporesponsiveness.20 This induction of hyporesponsiveness may be one reason for the modest success of αGalCer in clinical trials for the treatment of cancer patients.21 Therefore, understanding the molecular mechanisms leading to hyporesponsiveness is critical for the development of therapies aimed at activating iNKT cells for the treatment of disease.22,23 Here we report that in peripheral NKT1 cells the transcriptional regulator Id2 was required for the expression of TBET and production of IFNγ. In the absence of Id2, splenic iNKT cells exhibited phenotypic and functional features of hyporesponsiveness. Furthermore, strong TCR signaling led to sustained downregulation of Id2 accompanied by hyporesponsiveness of iNKT cells in vivo.
NKT1 cells exhibited higher levels of Id2 compared with other immune cell populations in the spleen and liver under homeostatic conditions. This is consistent with our previous finding that during iNKT cell development, Id2 was highly expressed in thymic NKT1 but not NKT2 or NKT17 cells.12 In the absence of Id2, the numbers of splenic NKT1 cells were diminished as indicated by a marked reduction of the NKT1 signature transcription factor TBET. Reduced TBET expression as a result of Id2 deficiency has also been observed in conventional CD4+ and CD8+ T cells24,25 and TBET is known to be required for IFNγ production.15 The reduced ability of Id2-deficient iNKT cells to produce IFNγ is therefore possibly a result of lowered TBET expression. In spite of reduced TBET expression, most Id2-deficient iNKT cells did not assume a PLZFhigh NKT2 phenotype but had intermediate levels of both transcription factors. Intriguingly, we found similar intermediate PLZF and TBET expression, as well as low Id2 expression, in hyporesponsive iNKT cells.
Recent data demonstrate that a substantial population of hyporesponsive iNKT cells produce IL-10 in response to restimulation and are thus termed NKT10 cells.7,9 Id2-deficient iNKT cells were enriched for surface markers of NKT10 cells including PD1 and NRP1 and readily produced IL-10. In line with our findings, increased IL-10 production has been shown in Id2-deficient conventional CD8+ T cells.26 Mechanistically, Id2 inhibits the transcriptional activity of E2A, an E protein transcription factor, known to bind to the Il10 promoter.26,27 Thus, downregulation of Id2 after stimulation with αGalCer may contribute to the induction of NKT10 cells by enabling E2A to continually positively regulate the expression of IL-10.
We found that Id2 downregulation occurred in response to moderate and strong TCR stimulation. The TCR-independent stimulus LPS did not induce Id2 downregulation consistent with its inability to induce hyporesponsiveness. Interestingly, OCH potently downregulated Id2 expression in iNKT cells in vitro but does not lead to a hyporesponsive state in vivo.9 While in vitro stimulation of iNKT cells is useful for the analysis of TCR-induced signaling, it does not induce hyporesponsiveness, which only occurs in vivo.7,28 Thus, our findings could indicate that initial downregulation of Id2 in response to OCH is not maintained in vivo or that under certain conditions downregulation of Id2 can be compensated for by other Id proteins. In addition, factors other than Id2 such as the expression of tuberous sclerosis 1 may also contribute to hyporesponsiveness in iNKT cells.29
In summary, we demonstrate that the transcriptional regulator Id2 impacts TBET expression in NKT1 cells and that the absence of Id2 results in hyporesponsiveness to restimulation and IL-10 production by iNKT cells. Our study for the first time sheds light on the transcriptional network underlying the hyporesponsive state in iNKT cells. A better understanding of this network could ultimately be used to selectively induce or prevent hyporesponsiveness of iNKT cells in human disease.
METHODS
Mice
Mice were bred and housed in specific pathogen-free conditions in accordance with the Institutional Animal Care and Use Guidelines of the University of California San Diego. Id2 YFP mice were generated as described previously.30 Id3 GFP mice were a gift from C Murre and were generated as described.31 CD4creId2+/+ and CD4creId2f/f mice were a gift from A Lasorella and were generated as described previously. IL-10GFP reporter mice were purchased from Jackson Laboratories (Sacramento, CA, USA) and crossed to CD4creId2f/f. αGalCer (1 μg; Avanti Polar Lipids, Alabaster, AL, USA) and LPS (40 μg, from Escherichia coli 0111; Sigma-Aldrich, St Louis, MO, USA) were administered intraperitoneally.
Cell culture
Total splenocytes or sorted iNKT cells (CD1d tetramer+ TCRβ+) were cultured in RPMI-1640 media containing 10% fetal calf serum, 25 mM HEPES, pH 7.2, 1% penicillin–streptomycin–glutamine and 55 μM β-mercaptoethanol. Total splenocytes were treated with αGalCer (100 ng ml−1), LPS (10 μg ml−1), GSL1 (10 μg ml−1; NIH Tetramer Core Facility, Atlanta, GA, USA), OCH (10 μg ml−1; NIH Tetramer Core Facility) and βGlcCer (10 μg ml−1; Avanti Polar Lipids). For the analysis of proliferation, cells were stained with CellTrace violet (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol.
Flow cytometry
Single-cell suspensions were prepared from the spleen and liver. Hepatic iNKT cells were isolated from the liver as described previously using 34% Percoll solution (Sigma, St Louis, MO, USA). The following antibodies were used: TCRβ FITC (clone H57-597), CD4 FITC (L3T4), CD1d tetramer Alexa 488 (NIH Tetramer Core Facility (mCD1d/PBS57)), CD1d tetramer PE (NIH Tetramer Core Facility (mCD1d/PBS57)), TCRβ PE (H57-597), PLZF PE (Mags.21F7), NK1.1 PerCP Cy5.5(PK136), NK1.1 APC (PK136), TCRβ APC (H57-597), TBET APC (4B10), CD1d tetramer Alexa 647 (NIH Tetramer Core Facility (mCD1d/PBS57)), CD19 PeCy7(ID3) and Ki-67 Pacific Blue (SolA15). All antibodies were purchased from eBioscience (San Diego, CA, USA), unless otherwise specified. Samples were collected on a FACS (fluorescence-activated cell sorting) LSRII, FACS Fortessa or FACS Aria (BD Biosciences, San Jose, CA, USA) fitted with custom mirrors from Omega Filters (510/21) with 502LP or 505LP for GFP, 530/30 with 525LP for YFP) and were analyzed with FlowJo software (TreeStar, Ashland, OR, USA). For iNKT cell sorting, single-cell suspensions were MACS-enriched for CD1d tetramer phycoerythrin-stained cells using magnetically labeled anti-phycoerythrin microbeads (Miltenyi Biotec, Auburn, CA, USA). CD19−CD3+CD1d tetramer+ iNKT were then sorted with an FACSAria (BD Biosciences) to at least 98% purity.
Statistical analysis
Normal distribution of the data was assessed using the Kolmogorov–Smirnov test. Data in all treatment groups were normally distributed. Two-group comparisons were assessed with an unpaired Student’s t-test, and grouped data were assessed with one-way analysis of variance followed by Bonferroni post-test (to adjust for multiple comparisons). GraphPad Prism software (La Jolla, CA, USA, version 5) was used for statistical analyses. P-values <0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank members of the Goldrath, Stradner and D’Cruz Labs for critical reading of the manuscript. This work was supported by PEW Scholars (to AWG), Leukemia and Lymphoma Society Scholar (to AWG) and Fellow (to LMD) awards and grants from the NIH (AI067545 and AI072117; to AWG), R00AI097286A (to LMD) and the Austrian Science Fund (FWF) (J3189-B11; to MHS).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
The Supplementary Information that accompanies this paper is available on the Immunology and Cell Biology website (http://www.nature.com/icb)
References
- 1.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 2.Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the ‘Swiss-Army knife’ of the immune system. Curr Opin Immunol. 2008;20:358–368. doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engel I, Kronenberg M. Transcriptional control of the development and function of Valpha14i NKT cells. Curr Top Microbiol Immunol. 2014;381:51–81. doi: 10.1007/82_2014_375. [DOI] [PubMed] [Google Scholar]
- 5.Verykokakis M, Zook EC, Kee BL. ID’ing innate and innate-like lymphoid cells. Immunol Rev. 2014;261:177–197. doi: 10.1111/imr.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wingender G, Sag D, Kronenberg M. NKT10 cells: a novel iNKT cell subset. Oncotarget. 2015;6:26552–26553. doi: 10.18632/oncotarget.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest. 2014;124:3725–3740. doi: 10.1172/JCI72308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch L, Michelet X, Zhang S, Brennan PJ, Moseman A, Lester C, et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of Treg cells and macrophages in adipose tissue. Nat Immunol. 2015;16:85–95. doi: 10.1038/ni.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wingender G, Birkholz AM, Sag D, Farber E, Chitale S, Howell AR, et al. Selective conditions are required for the induction of invariant NKT cell hyporesponsiveness by antigenic stimulation. J Immunol. 2015;195:3838–3848. doi: 10.4049/jimmunol.1500203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belle I, Zhuang Y. E proteins in lymphocyte development and lymphoid diseases. Curr Top Dev Biol. 2014;110:153–187. doi: 10.1016/B978-0-12-405943-6.00004-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol. 2010;11:240–249. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Cruz LM, Stradner MH, Yang CY, Goldrath AW. E and Id proteins influence invariant NKT cell sublineage differentiation and proliferation. J Immunol. 2014;192:2227–2236. doi: 10.4049/jimmunol.1302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monticelli LA, Yang Y, Knell J, D’Cruz LM, Cannarile MA, Engel I, et al. Transcriptional regulator Id2 controls survival of hepatic NKT cells. Proc Natl Acad Sci USA. 2009;106:19461–19466. doi: 10.1073/pnas.0908249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verykokakis M, Krishnamoorthy V, Iavarone A, Lasorella A, Sigvardsson M, Kee BL. Essential functions for ID proteins at multiple checkpoints in invariant NKT cell development. J Immunol. 2013;191:5973–5983. doi: 10.4049/jimmunol.1301521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuda JL, Zhang Q, Ndonye R, Richardson SK, Howell AR, Gapin L. T-bet concomitantly controls migration, survival, and effector functions during the development of Valpha14i NKT cells. Blood. 2006;107:2797–2805. doi: 10.1182/blood-2005-08-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 17.Peng SL. The T-box transcription factor T-bet in immunity and autoimmunity. Cell Mol Immunol. 2006;3:87–95. [PubMed] [Google Scholar]
- 18.Nagarajan NA, Kronenberg M. Invariant NKT cells amplify the innate immune response to lipopolysaccharide. J Immunol. 2007;178:2706–2713. doi: 10.4049/jimmunol.178.5.2706. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Li Y, Kinjo Y, Mac TT, Gibson D, Painter GF, et al. Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proc Natl Acad Sci USA. 2010;107:1535–1540. doi: 10.1073/pnas.0909479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parekh VV, Wilson MT, Olivares-Villagomez D, Singh AK, Wu L, Wang CR, et al. Glycolipid antigen induces long-term natural killer T cell anergy in mice. J Clin Invest. 2005;115:2572–2583. doi: 10.1172/JCI24762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa A, Motohashi S, Ishikawa E, Fuchida H, Higashino K, Otsuji M, et al. A phase I study of alpha-galactosylceramide (KRN7000)-pulsed dendritic cells in patients with advanced and recurrent non-small cell lung cancer. Clin Cancer Res. 2005;11:1910–1917. doi: 10.1158/1078-0432.CCR-04-1453. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan BA, Kronenberg M. Activation or anergy: NKT cells are stunned by alpha-galactosylceramide. J Clin Invest. 2005;115:2328–2329. doi: 10.1172/JCI26297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McEwen-Smith RM, Salio M, Cerundolo V. The regulatory role of invariant NKT cells in tumor immunity. Cancer Immunol research. 2015;3:425–435. doi: 10.1158/2326-6066.CIR-15-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omilusik KD, Shaw LA, Goldrath AW. Remembering one’s ID/E-ntity: E/ID protein regulation of T cell memory. Curr Opin Immunol. 2013;25:660–666. doi: 10.1016/j.coi.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusunoki T, Sugai M, Katakai T, Omatsu Y, Iyoda T, Inaba K, et al. TH2 dominance and defective development of a CD8+ dendritic cell subset in Id2-deficient mice. J Allergy Clin Immunol. 2003;111:136–142. doi: 10.1067/mai.2003.29. [DOI] [PubMed] [Google Scholar]
- 26.Masson F, Ghisi M, Groom JR, Kallies A, Seillet C, Johnstone RW, et al. Id2 represses E2A-mediated activation of IL-10 expression in T cells. Blood. 2014;123:3420–3428. doi: 10.1182/blood-2014-03-561456. [DOI] [PubMed] [Google Scholar]
- 27.Miyazaki M, Miyazaki K, Chen S, Itoi M, Miller M, Lu LF, et al. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nat Immunol. 2014;15:767–776. doi: 10.1038/ni.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uldrich AP, Crowe NY, Kyparissoudis K, Pellicci DG, Zhan Y, Lew AM, et al. NKT cell stimulation with glycolipid antigen in vivo: costimulation-dependent expansion, Bim-dependent contraction, and hyporesponsiveness to further antigenic challenge. J Immunol. 2005;175:3092–3101. doi: 10.4049/jimmunol.175.5.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Shin J, Xie D, Wang H, Gao J, Zhong XP. Tuberous sclerosis 1 promotes invariant NKT cell anergy and inhibits invariant NKT cell-mediated antitumor immunity. J Immunol. 2014;192:2643–2650. doi: 10.4049/jimmunol.1302076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8(+) T cell subsets. Nat Immunol. 2011;12:1221–1229. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


